Abstract
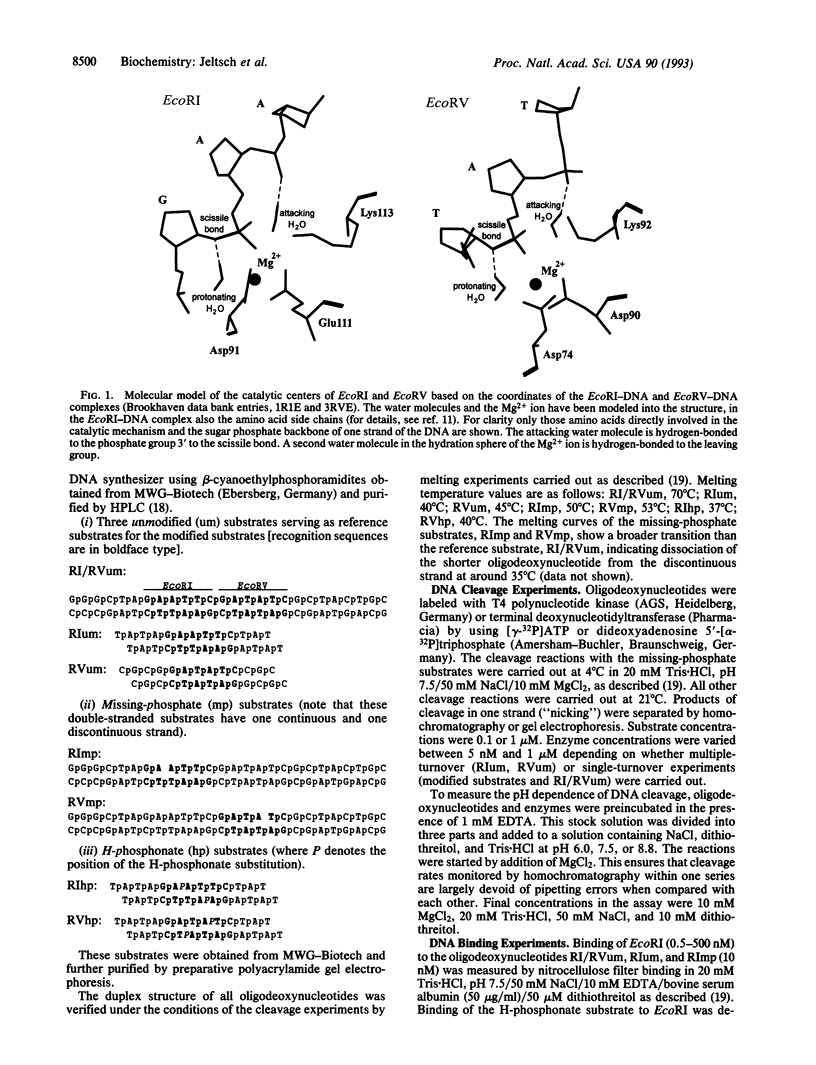
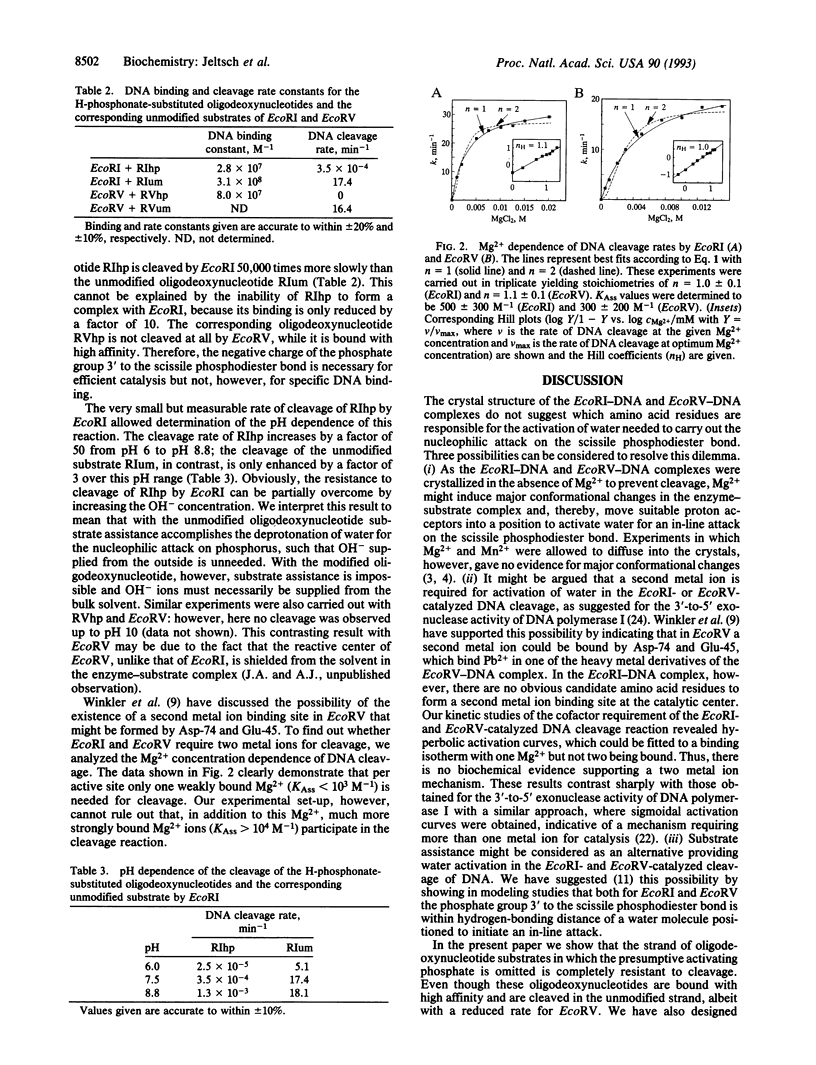
The crystal structure analyses of the EcoRI-DNA and EcoRV-DNA complexes do not provide clear suggestions as to which amino acid residues are responsible for the activation of water to carry out the DNA cleavage. Based on molecular modeling, we have proposed recently that the attacking water molecule is activated by the negatively charged pro-Rp phosphoryl oxygen of the phosphate group 3' to the scissile phosphodiester bond. We now present experimental evidence to support this proposal. (i) Oligodeoxynucleotide substrates lacking this phosphate group in one strand are cleaved only in the other strand. (ii) Oligodeoxynucleotide substrates carrying an H-phosphonate substitution at this position in both strands and, therefore, lacking a negatively charged oxygen at this position are cleaved at least four orders of magnitude more slowly than the unmodified substrate. These results are supported by other modification studies: oligodeoxynucleotide substrates with a phosphorothioate substitution at this position in both strands are cleaved only if the negatively charged sulfur is in the RP configuration as shown for EcoRI [Koziolkiewicz, M. & Stec, W.J. (1992) Biochemistry 31, 9460-9466] and EcoRV (B. A. Connolly, personal communication). As the phosphate residue 3' to the scissile phosphodiester bond is not needed for strong DNA binding by both enzymes, these findings strongly suggest that this phosphate group plays an active role during catalysis. This proposal, furthermore, gives a straightforward explanation of why in the EcoRI-DNA and EcoRV-DNA complexes the DNA is distorted differently, but in each case the 3' phosphate group closely approaches the phosphate group that is attacked. Finally, an alternative mechanism for DNA cleavage involving two metal ions is unlikely in the light of our finding that both EcoRI and EcoRV need only one Mg2+ per active site for cleavage.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiken C. R., Gumport R. I. Base analogs in study of restriction enzyme-DNA interactions. Methods Enzymol. 1991;208:433–457. doi: 10.1016/0076-6879(91)08023-b. [DOI] [PubMed] [Google Scholar]
- Alves J., Urbanke C., Fliess A., Maass G., Pingoud A. Fluorescence stopped-flow kinetics of the cleavage of synthetic oligodeoxynucleotides by the EcoRI restriction endonuclease. Biochemistry. 1989 Sep 19;28(19):7879–7888. doi: 10.1021/bi00445a050. [DOI] [PubMed] [Google Scholar]
- Beese L. S., Steitz T. A. Structural basis for the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991 Jan;10(1):25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett S. P., Halford S. E. Recognition of DNA by type II restriction enzymes. Curr Top Cell Regul. 1989;30:57–104. doi: 10.1016/b978-0-12-152830-0.50005-0. [DOI] [PubMed] [Google Scholar]
- Brunelle A., Schleif R. F. Missing contact probing of DNA-protein interactions. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6673–6676. doi: 10.1073/pnas.84.19.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter P., Wells J. A. Engineering enzyme specificity by "substrate-assisted catalysis". Science. 1987 Jul 24;237(4813):394–399. doi: 10.1126/science.3299704. [DOI] [PubMed] [Google Scholar]
- Connolly B. A., Eckstein F., Pingoud A. The stereochemical course of the restriction endonuclease EcoRI-catalyzed reaction. J Biol Chem. 1984 Sep 10;259(17):10760–10763. [PubMed] [Google Scholar]
- Fliess A., Wolfes H., Seela F., Pingoud A. Analysis of the recognition mechanism involved in the EcoRV catalyzed cleavage of DNA using modified oligodeoxynucleotides. Nucleic Acids Res. 1988 Dec 23;16(24):11781–11793. doi: 10.1093/nar/16.24.11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey P. A., Sammons R. D. Bond order and charge localization in nucleoside phosphorothioates. Science. 1985 May 3;228(4699):541–545. doi: 10.1126/science.2984773. [DOI] [PubMed] [Google Scholar]
- Geiger R., Rüter T., Alves J., Fliess A., Wolfes H., Pingoud V., Urbanke C., Maass G., Pingoud A., Düsterhöft A. Genetic engineering of EcoRI mutants with altered amino acid residues in the DNA binding site: physicochemical investigations give evidence for an altered monomer/dimer equilibrium for the Gln144Lys145 and Gln144Lys145Lys200 mutants. Biochemistry. 1989 Mar 21;28(6):2667–2677. doi: 10.1021/bi00432a046. [DOI] [PubMed] [Google Scholar]
- Grasby J. A., Connolly B. A. Stereochemical outcome of the hydrolysis reaction catalyzed by the EcoRV restriction endonuclease. Biochemistry. 1992 Sep 1;31(34):7855–7861. doi: 10.1021/bi00149a016. [DOI] [PubMed] [Google Scholar]
- Han H., Rifkind J. M., Mildvan A. S. Role of divalent cations in the 3',5'-exonuclease reaction of DNA polymerase I. Biochemistry. 1991 Nov 19;30(46):11104–11108. doi: 10.1021/bi00110a012. [DOI] [PubMed] [Google Scholar]
- Hayes J. J., Tullius T. D. The missing nucleoside experiment: a new technique to study recognition of DNA by protein. Biochemistry. 1989 Nov 28;28(24):9521–9527. doi: 10.1021/bi00450a041. [DOI] [PubMed] [Google Scholar]
- Heitman J. How the EcoRI endonuclease recognizes and cleaves DNA. Bioessays. 1992 Jul;14(7):445–454. doi: 10.1002/bies.950140704. [DOI] [PubMed] [Google Scholar]
- Jacob-Dubuisson F., Lamotte-Brasseur J., Dideberg O., Joris B., Frère J. M. Arginine 220 is a critical residue for the catalytic mechanism of the Streptomyces albus G beta-lactamase. Protein Eng. 1991 Oct;4(7):811–819. doi: 10.1093/protein/4.7.811. [DOI] [PubMed] [Google Scholar]
- Jeltsch A., Alves J., Maass G., Pingoud A. On the catalytic mechanism of EcoRI and EcoRV. A detailed proposal based on biochemical results, structural data and molecular modelling. FEBS Lett. 1992 Jun 8;304(1):4–8. doi: 10.1016/0014-5793(92)80576-3. [DOI] [PubMed] [Google Scholar]
- Kim R., Modrich P., Kim S. H. 'Interactive' recognition in EcoRI restriction enzyme-DNA complex. Nucleic Acids Res. 1984 Oct 11;12(19):7285–7292. doi: 10.1093/nar/12.19.7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. C., Grable J. C., Love R., Greene P. J., Rosenberg J. M. Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing. Science. 1990 Sep 14;249(4974):1307–1309. doi: 10.1126/science.2399465. [DOI] [PubMed] [Google Scholar]
- Koziolkiewicz M., Stec W. J. Application of phosphate-backbone-modified oligonucleotides in the studies on EcoRI endonuclease mechanism of action. Biochemistry. 1992 Oct 6;31(39):9460–9466. doi: 10.1021/bi00154a019. [DOI] [PubMed] [Google Scholar]
- Krueger J. K., Stock J., Schutt C. E. Evidence that the methylesterase of bacterial chemotaxis may be a serine hydrolase. Biochim Biophys Acta. 1992 Mar 12;1119(3):322–326. doi: 10.1016/0167-4838(92)90220-8. [DOI] [PubMed] [Google Scholar]
- Lesser D. R., Grajkowski A., Kurpiewski M. R., Koziolkiewicz M., Stec W. J., Jen-Jacobson L. Stereoselective interaction with chiral phosphorothioates at the central DNA kink of the EcoRI endonuclease-GAATTC complex. J Biol Chem. 1992 Dec 5;267(34):24810–24818. [PubMed] [Google Scholar]
- Lesser D. R., Kurpiewski M. R., Jen-Jacobson L. The energetic basis of specificity in the Eco RI endonuclease--DNA interaction. Science. 1990 Nov 9;250(4982):776–786. doi: 10.1126/science.2237428. [DOI] [PubMed] [Google Scholar]
- Lu A. L., Jack W. E., Modrich P. DNA determinants important in sequence recognition by Eco RI endonuclease. J Biol Chem. 1981 Dec 25;256(24):13200–13206. [PubMed] [Google Scholar]
- McSwiggen J. A., Cech T. R. Stereochemistry of RNA cleavage by the Tetrahymena ribozyme and evidence that the chemical step is not rate-limiting. Science. 1989 May 12;244(4905):679–683. doi: 10.1126/science.2470150. [DOI] [PubMed] [Google Scholar]
- Ruffner D. E., Uhlenbeck O. C. Thiophosphate interference experiments locate phosphates important for the hammerhead RNA self-cleavage reaction. Nucleic Acids Res. 1990 Oct 25;18(20):6025–6029. doi: 10.1093/nar/18.20.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selent U., Rüter T., Köhler E., Liedtke M., Thielking V., Alves J., Oelgeschläger T., Wolfes H., Peters F., Pingoud A. A site-directed mutagenesis study to identify amino acid residues involved in the catalytic function of the restriction endonuclease EcoRV. Biochemistry. 1992 May 26;31(20):4808–4815. doi: 10.1021/bi00135a010. [DOI] [PubMed] [Google Scholar]
- Stöver T., Köhler E., Fagin U., Wende W., Wolfes H., Pingoud A. Determination of the DNA bend angle induced by the restriction endonuclease EcoRV in the presence of Mg2+. J Biol Chem. 1993 Apr 25;268(12):8645–8650. [PubMed] [Google Scholar]
- Taylor J. D., Badcoe I. G., Clarke A. R., Halford S. E. EcoRV restriction endonuclease binds all DNA sequences with equal affinity. Biochemistry. 1991 Sep 10;30(36):8743–8753. doi: 10.1021/bi00100a005. [DOI] [PubMed] [Google Scholar]
- Thielking V., Selent U., Köhler E., Landgraf A., Wolfes H., Alves J., Pingoud A. Mg2+ confers DNA binding specificity to the EcoRV restriction endonuclease. Biochemistry. 1992 Apr 21;31(15):3727–3732. doi: 10.1021/bi00130a001. [DOI] [PubMed] [Google Scholar]
- Thielking V., Selent U., Köhler E., Wolfes H., Pieper U., Geiger R., Urbanke C., Winkler F. K., Pingoud A. Site-directed mutagenesis studies with EcoRV restriction endonuclease to identify regions involved in recognition and catalysis. Biochemistry. 1991 Jul 2;30(26):6416–6422. doi: 10.1021/bi00240a011. [DOI] [PubMed] [Google Scholar]
- Waring R. B. Identification of phosphate groups important to self-splicing of the Tetrahymena rRNA intron as determined by phosphorothioate substitution. Nucleic Acids Res. 1989 Dec 25;17(24):10281–10293. doi: 10.1093/nar/17.24.10281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters T. R., Connolly B. A. Continuous spectrophotometric assay for restriction endonucleases using synthetic oligodeoxynucleotides and based on the hyperchromic effect. Anal Biochem. 1992 Jul;204(1):204–209. doi: 10.1016/0003-2697(92)90162-z. [DOI] [PubMed] [Google Scholar]
- Winkler F. K., Banner D. W., Oefner C., Tsernoglou D., Brown R. S., Heathman S. P., Bryan R. K., Martin P. D., Petratos K., Wilson K. S. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993 May;12(5):1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]


