Figure 1.
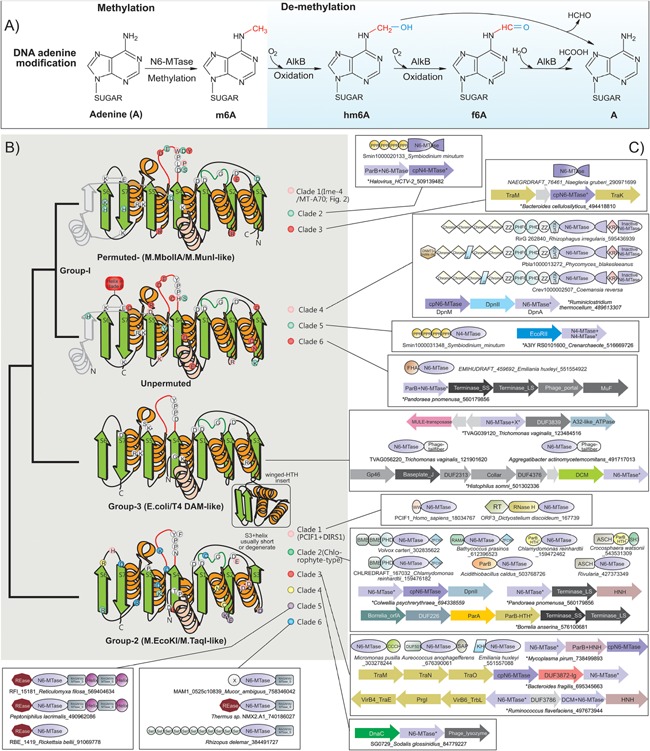
m6A methylation and demethylation reactions, topology, and conserved features of eukaryotic N6A‐MTases. A: Groups modifying the nucleotide are colored red and blue. B: Topology and anatomy of MTase domains. Cartoon representations of principle groups of eukaryotic MTases grouped according to their higher order relationships (shown to the left). Conserved strands are colored green and helices orange. Additionally, lineage‐specific structural elements are shown in gray. Ancestrally conserved residues are shown in gray circles at their structural position, whereas clade specific residues are shown in their respective colors. C: Representative domain architectures and gene neighborhoods for different clades within the three groups are illustrated. Genes in operons are shown with the arrow head pointing to the 3′ direction of the coding strand. Proteins are denoted by their gene name if present, species name, and Genbank identifier (GI) separated by underscores. Proteins from species not available in Genbank are given a temporary id, separated by the species name. The full sequence can be accessed in the Supporting Information. Standard abbreviations are used for domain names. Additional non‐standard names include: X, domains of uncharacterized function; cpN6‐MTase, circularly permuted Group I‐like MTase; RAGNYA, RAGNYA fold domain found in the methylase‐specificity subunit; Helix, α‐helical element that forms coiled coils; ZFCW, PHDX/ZFCW domain; DUF3872‐Ig, an all‐β Ig fold domain.
