Abstract
Aldehyde oxidase (AO) is a complex molybdo-flavoprotein that belongs to the xanthine oxidase family. AO is active as a homodimer, and each 150-kDa monomer binds two distinct [2Fe2S] clusters, FAD, and the molybdenum cofactor. AO has an important role in the metabolism of drugs based on its broad substrate specificity oxidizing aromatic aza-heterocycles, for example, N1-methylnicotinamide and N-methylphthalazinium, or aldehydes, such as benzaldehyde, retinal, and vanillin. Sequencing the 35 coding exons of the human AOX1 gene in a sample of 180 Italian individuals led to the identification of relatively frequent, synonymous, missense and nonsense single-nucleotide polymorphisms (SNPs). Human aldehyde oxidase (hAOX1) was purified after heterologous expression in Escherichia coli. The recombinant protein was obtained with a purity of 95% and a yield of 50 μg/l E. coli culture. Site-directed mutagenesis of the hAOX1 cDNA allowed the purification of protein variants bearing the amino acid changes R802C, R921H, N1135S, and H1297R, which correspond to some of the identified SNPs. The hAOX1 variants were purified and compared with the wild-type protein relative to activity, oligomerization state, and metal content. Our data show that the mutation of each amino acid residue has a variable impact on the ability of hAOX1 to metabolize selected substrates. Thus, the human population is characterized by the presence of functionally inactive hAOX1 allelic variants as well as variants encoding enzymes with different catalytic activities. Our results indicate that the presence of these allelic variants should be considered for the design of future drugs.
Introduction
Aldehyde oxidase (AO) (EC1.2.3.1) is a molybdo-flavoenzyme present in the cytosolic compartment of many tissues in various animal species, including humans (Garattini et al., 2008, 2009). AO is a member of the xanthine oxidase (XO) family, which consists of complex metalloflavoproteins containing two [2Fe2S] clusters, FAD, and the molybdenum cofactor (Moco) as the catalytically active units (Hille, 1996). The AO holoenzyme is a homodimer, and each 150-kDa monomer is characterized by three separate domains: the 20-kDa N-terminal domain binds the two distinct [2Fe2S] clusters, FeSI and FeSII, the 40-kDa central domain binds FAD, and the 80-kDa C-terminal domain binds Moco (Garattini et al., 2003). Members of the XO family of molybdoenzymes are characterized by an equatorial sulfur ligand at the Moco that is essential for enzyme activity (Edmondson et al., 1972; Wahl and Rajagopalan, 1982).
AO is found in most animal species, including fish and insects (Garattini et al., 2008). In humans, AO is encoded by a single functional gene, hAOX1. Although hAOX1 orthologs are found in almost all mammalian organisms, the number of functional AOX genes varies according to the species considered. Rodents contain the largest number of AOX functional genes: Aox1, Aox3, Aox4, and Aox3l1 (Garattini et al., 2009). These genes arose from a series of gene duplication events from a common ancestor and are clustered on a short region of mouse chromosome 1 and rat chromosome 9. All of the products of the mammalian AOX genes have high amino acid sequence similarity and are expressed in a tissue-specific manner in different organisms (Garattini et al., 2003; Terao et al., 2006). It is believed that the various AOX isoforms recognize distinct substrates and carry out different physiological tasks. The tissue distribution of mouse AOX3 (mAOX3) is superimposable with that of mAOX1, and the two enzymes are synthesized predominantly in liver, lung, and testis (Vila et al., 2004). The expression of mAOX4 is limited to the Harderian gland, esophagus and skin, whereas mAOX3L1 expression is restricted to the nasal mucosa (Terao et al., 2000). Except for mAOX4, which metabolizes retinaldehyde into retinoic acid and plays a role in skin homeostasis (Terao et al., 2009), very little is known about specific substrates and the physiological role of mAOX1 or any of the other AOX homologs.
In spite of this lack of information on the physiological significance of the AO enzymes, hAOX1 long has been recognized as a prominent drug-metabolizing enzyme (Obach et al., 2004; Pryde et al., 2010; Garattini and Terao, 2011). AOX1 is characterized by broad substrate specificity, catalyzing the oxidation of a wide range of endogenous and exogenous aldehydes as well as N-heterocyclic aromatic compounds (Kitamura et al., 2006). In addition, AOX1 catalyzes the reduction of a variety of functional groups, including sulfoxides, N-oxides, azo dyes, and N-hydroxycarbamoyl substituents in the presence of an appropriate donor. N-Heterocyclic drugs, such as methotrexate, 6-mercaptopurine, cinchona alkaloids, and famciclovir, also are oxidized by this enzyme (Obach et al., 2004; Kitamura et al., 2006). Finally, AOX1 is involved in the oxidation of intermediary drug metabolites, such as the conversion of cyclic iminium ions arising from cytochrome P450-catalyzed oxidation of pyrrolidines and piperidines into lactams or the oxidation of aldehydes derived from alcohol-containing drugs (Beedham, 1997).
Marked species differences have been well documented for the aldehyde-catalyzed metabolism of drugs, including methotrexate and famciclovir (Rashidi et al., 1997; Jordan et al., 1999; Kitamura et al., 1999a,b). Interindividual variability in hAOX1 in vitro activity has been reported in humans (Kitamura et al., 1999b; Al-Salmy, 2001), although the underlying determinants of these variations have never been investigated (Beedham et al., 2003). Gender may be one such determinant. In fact, male mice exhibit a 2- to 4-fold higher AO activity than female mice (Beedham, 1985; Kurosaki et al., 1999; Al-Salmy, 2001). It also has been suggested that factors such as age, cigarette smoking, drug usage, and disease states, such as cancer, also may account for the interindividual variability of hAOX1 activity (Pryde et al., 2010). One final and prominent source of interindividual variation is represented by missense single-nucleotide polymorphisms (SNPs) affecting the catalytic activity of the hAOX1 enzyme. Numerous SNPs of the hAOX1 gene are available in the National Center for Biotechnology Information dbSNP database, although for the majority of the data on frequency in the human population are not available. In addition, no single human SNP has been characterized for its effect on the catalytic activity of the purified enzyme. The only available functional studies on AOX1 SNPs were reported in Donryu rats (Adachi et al., 2007; Itoh et al., 2007a,b,c). Functional characterization of the identified SNPs permitted the classification of these rats into ultrarapid metabolizers, extensive metabolizers, and poor metabolizers according to the AOX1 mutation considered.
Interindividual differences in hAOX1 activity are of primary importance for the clinical use of drugs known to be metabolized by the enzyme. They also are very important factors to be considered in the development of new drugs. For these reasons, we decided to focus on this aspect of hAOX1 biology. In this article, we report on the identification of relatively frequent hAOX1 SNPs and define their frequency in the Italian population. The most frequent missense SNPs were selected to produce the corresponding recombinant hAOX1 variant proteins, using an efficient Escherichia coli expression system. The enzymatic and kinetic characteristics of the canonical and variant hAOX1 proteins were compared. Our data provide evidence for the existence of frequent hAOX1 allelic variants defining fast and poor metabolizers in the human population.
Materials and Methods
Identification of SNPs
Blood samples (3 ml) were collected from 180 (67 males and 113 females) volunteers after signing written consent forms. Thirty of these individuals were healthy volunteers, whereas the remaining 150 were individuals recruited for an unrelated epidemiological study. All of the samples were treated anonymously according to the guidelines of the internal Ethical Committee of the Istituto Mario Negri. The genomic DNA was extracted from blood samples by a semiautomated vacuum-based nucleic acids extractor (AB6100; Applied Biosystems, Foster City, CA). Oligonucleotides were synthesized by Invitrogen (Carlsbad, CA). The genomic DNA fragments containing each of the exon sequences were amplified using the oligonucleotides listed in Supplemental Table 1, using the Taq DNA polymerase kit (Applied Biosystems). The amplified polymerase chain reaction (PCR) products (30–60 ng) were sequenced in 96-well plates by Primm, S.r.l. (Milan, Italy).
Cloning, Expression, and Purification of hAOX1 and Its Variants
Cloning of hAOX1 cDNA into pQE-30 Xa vector and PCR mutagenesis of the variants R921H and N1135S were done after Alfaro et al. (2009). For site-directed mutagenesis of hAOX1 R802C and H1297R variants, the expression vector pQE-30 Xa including hAOX1 cDNA was used as a template, resulting in plasmids pTHAO1 and pTHAO2. All of the constructs used express hAOX1 as an N-terminal fusion protein with a His6 tag.
For expression in E. coli, the constructs were transformed into TP1000 (ΔmobAB) cells (Palmer et al., 1996). E. coli were grown at 30°C in Luria-Bertani medium supplemented with 150 μg/ml ampicillin, 1 mM molybdate, and 20 μM isopropyl β-d-thiogalactoside. Cells were harvested by centrifugation after 24 h of growth and resuspended in 50 mM sodium phosphate buffer (pH 8.0) containing 300 mM NaCl and frozen at −20°C until purification.
Cells were lysed twice in a cell disruptor system at 1.35 kbar (Constant Systems, Northhampton, UK). Cell fragments were removed by centrifugation, and the supernatant was mixed with 4 ml of nickel-nitrilotriacetic acid resin (QIAGEN GmbH, Hilden, Germany) per 14 liters of cell growth and incubated for 20 min at low stirring speed. The mixture was transferred to a column and washed with 10 column volumes of both 50 mM sodium phosphate and 300 mM NaCl (pH 8.0) containing 10 mM imidazole and the same buffer containing 20 mM imidazole. Proteins were eluted from the resin with 50 mM sodium phosphate and 300 mM NaCl (pH 8.0) containing 250 mM imidazole. The buffer of the eluted proteins was changed to 50 mM Tris (pH 7.5) by using PD-10 columns (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK). For the stabilization of the protein, 2.5 mM dithiothreitol was added to the buffer. For further purification, hAOX1 was loaded on a Mono Q 5/50 GL column (GE Healthcare) equilibrated with 50 mM Tris (pH 7.5) containing 1 mM EDTA and eluted with a linear gradient of the same buffer containing 1 M NaCl. Fractions were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE), and the ones containing hAOX1 were combined. hAOX1 was purified further on a Superdex 200 column (GE Healthcare), equilibrated in 50 mM Tris-HCl (pH 7.5) containing 200 mM NaCl and 1 mM EDTA. Only the fractions containing the dimeric form of hAOX1 were combined and used for the kinetic studies.
SDS-PAGE
SDS-PAGE was performed using 12% polyacrylamide gels described by Laemmli (1970). Staining of the proteins was done using Coomassie Blue R.
Enzyme Assays
Steady-state enzyme kinetics were performed with purified hAOX1 in 50 mM Tris buffer (pH 7.5) containing 200 mM NaCl and 1 mM EDTA at 25°C in a final volume of 500 μl. The substrates benzaldehyde, phthalazine, and chloroquinazolinone were used in a range from 0.1 to 100 μM using dichlorophenolindophenol (100 μM) as an electron acceptor. For phenanthridine, molecular oxygen was used as an electron acceptor, and the product phenanthridone was detected at 450 nm. Total enzyme concentration varied between 200 and 600 nM. Reactions were monitored over a range of 60 s. Activities were calculated using the extinction coefficients of 16,100 M−1 cm−1 at 600 nm for dichlorophenolindophenol and 4775 M−1 cm−1 at 450 nm for phenanthridone. Data obtained from three individual measurements were fitted nonlinearly using the Michaelis-Menten equation to obtain the kinetic constants KM and turnover numbers.
Metal and Molybdopterin Analysis
The molybdenum and iron content of purified hAOX1 was determined by inductively coupled plasma optical emission spectroscopy on an Optima 2100 DV (PerkinElmer Life and Analytical Sciences, Waltham, MA). Five-hundred-microliter protein samples with a final concentration of 5 to 10 μM were wet-ashed in the same volume of 65% nitric acid by incubation at 100°C for 24 h. The samples were diluted with 4 ml of water. The multielement standard solution XVI (Merck, Darmstadt, Germany) was used as a reference. The resulting mass concentrations were calculated as a percentage of protein saturation of both molybdenum and two [2Fe2S] in relation to a theoretical 100% saturation. Molybdopterin (MPT) was converted to its fluorescent derivative Form A by adjusting the pH of the supernatant to 2.5 with HCl and heating at 95°C for 30 min in the presence of iodine (Johnson et al., 1984). Excess iodine was removed by the addition of 55 μl of 1% w/v ascorbic acid, and the sample was adjusted with 1 M Tris to pH 8.3. Form A was obtained from phospho-Form A by the addition of 40 mM MgCl2 and 1 unit of calf intestine alkaline phosphatase. Form A was isolated with 10 mM acetic acid on a QAE ion exchange column (Sigma-Aldrich, St. Louis, MO), which was equilibrated in H2O. Form A was identified and quantified by high-performance liquid chromatography analysis with a C18 reversed-phase high-performance liquid chromatography column [4.6 × 250 mm; Hypersil ODS (Thermo Fisher Scientific, Waltham, MA); particle size 5 μm] with 5 mM ammonium acetate and 15% v/v methanol at an isocratic flow rate of 1 ml/min. In-line fluorescence was monitored by an 1100 series detector (Agilent Technologies, Santa Clara, CA) with excitation at 383 nm and emission at 450 nm.
EPR Spectroscopy
Continuous-wave EPR spectra at 9.4 GHz X-band were recorded on a home-built spectrometer (microwave bridge, ER041MR, Bruker, Newark, DE; lock-in amplifier, SR810, Stanford Research, Stanford, CA; microwave counter, 53181A, Agilent Technologies) equipped with a Bruker SHQ resonator. An ESR 910 helium flow cryostat with an ITC503 temperature controller (Oxford, Oxfordshire, UK) were used for temperature control. The magnetic field was calibrated using a Li/LiF standard with a known g value of 2.002293 ± 0.000002. hAOX1 samples with concentrations of 50 μM were reduced with a 20-fold excess of dithionite anaerobically on an argon flow prepared in quartz tubes with a 4-mm outer diameter. Reduction was achieved by adding a small volume of an anaerobic dithionite solution to generate reduced FeII/FeIII in the FeS clusters. Sample tubes were frozen rapidly in deep-cold ethanol after a color change confirmed successful reduction.
Results
SNPs of the hAOX1 Gene
The hAOX1 gene is characterized by a very complex structure consisting of 35 coding exons (Garattini et al., 2008). To identify relatively frequent SNPs in the coding region of the gene, we amplified by PCR and sequenced each hAOX1 exon from genomic DNA belonging to a cohort of 180 volunteers representative of the Italian population. The couple of exon-specific oligonucleotides used for the amplification and corresponding to the sequences upstream and downstream of each exon in intronic regions are shown in Supplemental Table 1. The presence of bona fide SNPs was verified by sequencing in both directions. In this population, we identified one nonsense mutation, five nonsynonymous SNPs, and one synonymous SNP (Table 1). The nonsense mutation was located in exon 5 and is predicted to result in a very short, nonfunctional protein of 126 amino acids. This mutation was relatively frequent and caused haploinsufficiency in eight individuals. The most frequent missense mutation was an A/G transition resulting in the substitution H/R at position 1297. The SNP was located in exon 34, and it was observed in 13 individuals, six of which were homozygous for the trait. Exon 34 appeared to be a mutational hotspot, because another relatively frequent SNP (C/T) resulting in the substitution S/L at position 1271 and a synonymous SNP were located in this exon. All of the individuals carrying the S1271L SNP were heterozygous for the trait. A relatively frequent missense SNP corresponding to an A/G transition, causing the substitution N/S at position 1135, was identified in exon 30. Two further and rare nonsynonymous SNPs were identified in exons 22 and 25. They were the result of C/T (R802C) and G/A (R921H) transitions, respectively.
TABLE 1.
Nonsense, missense, and synonymous polymorphisms of the hAOX1 gene in the Italian population
The SNP data were deposited in the National Center for Biotechnology Information database under the accession numbers indicated in the last column.
| Exon | Nucleotide | Amino Acid | Allelic Frequency | Heterozygotes | Homozygotes | RefSNP(rs) |
|---|---|---|---|---|---|---|
| 5 | C/G | Y126stop | 0.026 | 8/153 | 0/153 | ss469333481 |
| 22 | C/T | R802C | 0.006 | 2/180 | 0/180 | rs41309768 |
| 25 | G/A | R921H | 0.003 | 1/180 | 0/180 | rs56199635 |
| 30 | A/G | N1135S | 0.029 | 1/154 | 4/154 | rs55754655 |
| 34 | C/T | S1271L | 0.039 | 14/178 | 0/178 | rs141786030 |
| 34 | A/G | H1297R | 0.053 | 7/178 | 6/178 | rs3731722 |
| 34 | C/T | L1268silent | 0.022 | 8/178 | 0/178 | ss469333482 |
Expression and Purification of hAOX1
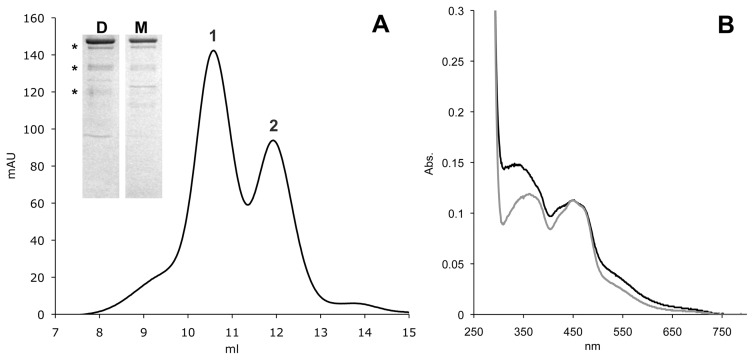
A prerequisite for the functional characterization of the identified SNPs was the optimization of the system used for the purification of a catalytically active hAOX1 protein (Alfaro et al., 2009). For this purpose, recombinant hAOX1 was expressed as an N-terminal His6 tag fusion protein in E. coli TP1000 using the protocol described for mAOX1 (Schumann et al., 2009) with slight modifications. The protein was purified using nickel-nitrilotriacetic acid, anion exchange, and size-exclusion chromatography. We isolated 95% pure protein (Fig. 1) with a yield of 50 μg/l cell growth. After size-exclusion chromatography, two major peaks were detected corresponding to calculated molecular masses of 300 and 150 kDa, which represented the dimeric and monomeric forms of hAOX1 (Fig. 1). The ratio between dimeric and monomeric forms was calculated to be 1.5:1. The formation of protein aggregates also was observed; however, aggregation was prevented by the addition of 2.5 mM dithiothreitol during the purification. Analysis of the purified hAOX1 protein by 12% SDS-PAGE revealed a dominant protein band corresponding to the calculated molecular mass of 150 kDa for hAOX1 (Fig. 1). Three other less intense bands of lower molecular masses were visible by SDS-PAGE. The three bands were analyzed by matrix-assisted laser desorption ionization peptide mapping and identified to be the degradation products of hAOX1. Similar bands also were observed in purified mAOX3 (Mahro et al., 2011) and AOX1 protein preparations of mouse and rat origin (Kundu et al., 2007; Schumann et al., 2009). These products were never detectable upon native PAGE (Kundu et al., 2007; Mahro et al., 2011) and fast protein liquid chromatography. All of this suggested that the observed degradation products are generated by the reductive conditions intrinsic to SDS-PAGE.
Fig. 1.
Characterization of hAOX1. Shown are the size-exclusion chromatogram of Superdex 200-purified hAOX1 and its UV/Vis absorption spectrum. A, elution profile of a size-exclusion chromatography (Superdex 200) shows two peaks of wild-type hAOX1 protein corresponding to the dimeric (1) and monomeric (2) forms in solution. The purity of the protein was determined by SDS-PAGE (D, dimer; M, monomer). Stars indicate the degradation products of hAOX1 as determined by mass spectroscopy. B, UV/Vis spectrum of air-oxidized hAOX1 in 50 mM Tris (pH 7.5) at 25°C showing characteristic absorptions of the protein-bound FAD at 450 nm and a shoulder for the iron-sulfur clusters at 550 nm. The spectrum of the protein dimer is shown in black, and the spectrum of the monomeric portion is shown in gray.
The UV-visible (UV/Vis) spectra of hAOX1 in its oxidized form displayed the typical features of molybdo-flavoenzymes (Fig. 1). The 280:450 nm absorbance ratio of 5.0 for the recombinant protein indicated the high purity of the enzyme. The 450:550 nm absorbance ratio in the UV/Vis spectrum was calculated to be 3.0 for the recombinant enzyme, demonstrating full saturation with FAD. In addition, the FAD content of hAOX1 was determined to be 100% by quantification of the AMP content of the protein.
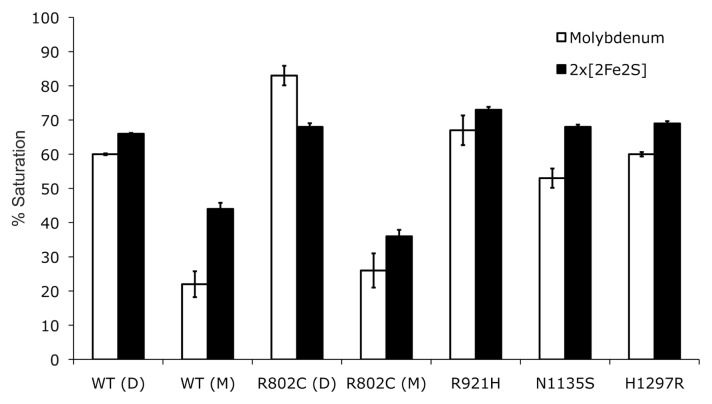
Protein expression over an extended time at low temperature allows E. coli cells to produce sufficient amounts of Mo-MPT for almost complete saturation of the enzyme with this cofactor (Leimkühler et al., 2004). The main problem of the recombinant system is poor insertion of the terminal sulfido ligand that is essential for enzyme activity. We determined 60% saturation of recombinant hAOX1 with molybdenum and 70% saturation of the 2×[2Fe2S] clusters with iron (Fig. 2). The amount of active enzyme containing the Mo=S ligand was calculated from the reduction spectra. For this purpose, we compared the ratio of oxidized to benzaldehyde-reduced hAOX1 with the amount of the enzyme reduced in relation to the fully reduced enzyme with dithionite under anaerobic conditions. This method showed a 30% saturation ratio of sulfurated Mo-MPT in hAOX1. Thus, only 50% of Mo-MPT contained the terminal sulfur ligand. To increase the catalytically active fraction of hAOX1, we coexpressed hAOX1 and human Moco sulfurase (hMCSF) cDNAs (Ichida et al., 2001). However, an analysis of purified hAOX1 showed no increase in the levels of sulfurated Mo-MPT. A band corresponding to hMCSF was not detected in cell lysates after separation by SDS-PAGE, suggesting that the majority of hMCSF was expressed in inclusion bodies in an inactive form. Thus, in all of the subsequent purification experiments, we decided to express only hAOX1 cDNA in TP1000 cells.
Fig. 2.
Saturation of hAOX1 with molybdenum and iron. Determination of the cofactor saturation of hAOX1 by inductively coupled plasma optical emission spectroscopy. The iron content corresponds to the saturation of both FeSI and FeSII clusters. The wild-type protein and all of its variants show a similar saturation of 2×[2Fe2S] but vary in their molybdenum content saturations between 60 and 80% (D, dimer; M, monomer). The percentage values are related to the theoretical full complement of Moco and the 2×[2Fe2S] clusters.
EPR Spectroscopy of the hAOX1 [2Fe2S] Clusters
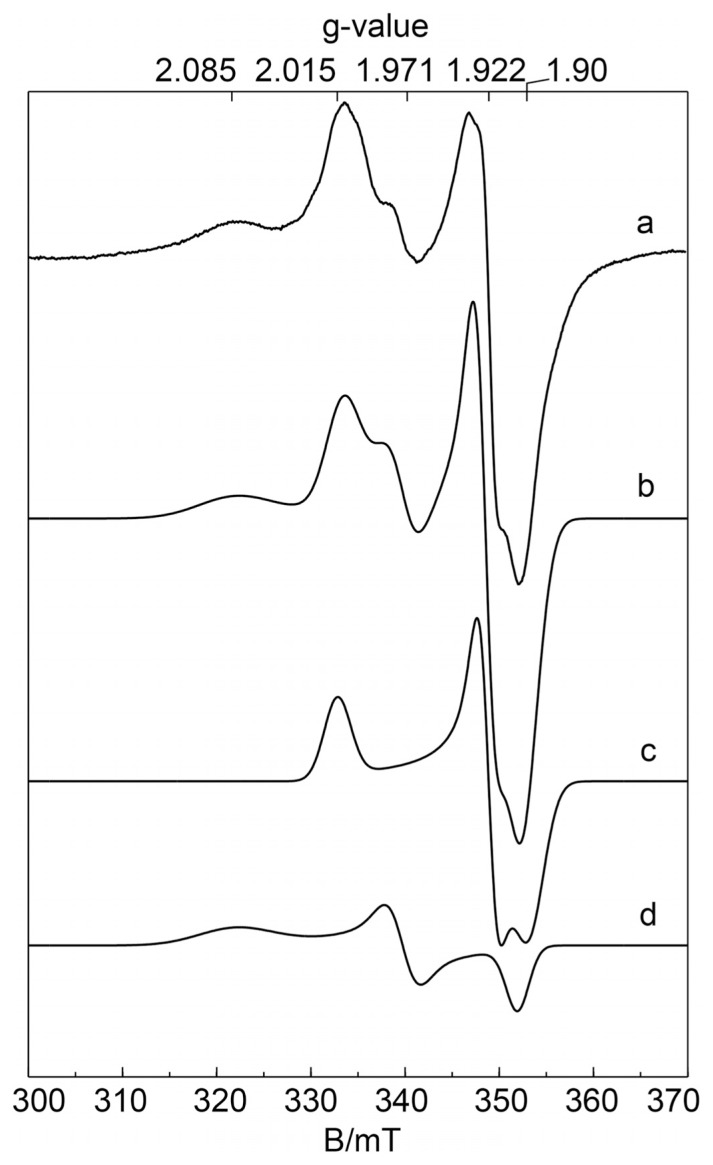
The two [2Fe2S] clusters were characterized by EPR spectroscopy. Figure 3 shows the EPR spectra of recombinant and dithionite-reduced wild-type hAOX1 along with the corresponding simulations. The spectra display signals from several superimposed paramagnetic species, which are observed usually in enzymes belonging to the XO family. However, the most prominent signals are the characteristic EPR signals assigned to the two iron-sulfur centers FeSI and FeSII, which are similar for all of the members of the XO family that have been described to date (Hille, 1996; Parschat et al., 2001). FeSI has EPR properties showing a slightly rhombic g tensor, similar to those of many other [2Fe2S] proteins, being fully developed at relatively high temperatures (up to T = 80 K), whereas FeSII has unusual EPR properties for [2Fe2S] species with a more rhombic g tensor, showing lines with varying line widths that can only be observed clearly at much lower temperatures (T <30 K). The g values and line widths were evaluated by simulating the spectra using the EasySpin toolbox for MATLAB (Stoll and Schweiger, 2006). In both proteins, FeSI has similar EPR properties, including the typical, rhombic g tensor as observed for other [2Fe2S] clusters of the ferredoxin type (Table 2). Under the experimental conditions used to reduce the proteins, no clear signals of the reduced flavin semiquinone (FAD) or Moco (MoV) were observed. The line widths and positions of the FeS signals at lower temperatures were affected by magnetic interactions. The g tensors obtained and line widths for FeSI and FeSII of hAOX1 are almost identical to the values published previously for mAOX1 (Schumann et al., 2009) or mAOX3, indicating high structural similarity between the proteins as expected from amino acid sequence identities of 83% and 61%, respectively.
Fig. 3.
EPR spectra of wild-type hAOX1. Experimental continuous-wave EPR spectra of dithionite-reduced wild-type hAOX1 samples at pH 7.0 (trace a) together with the corresponding simulation (trace b). For simulation parameters, see Table 2. The flavin semiquinone and MoV were not detected under these experimental conditions and therefore neglected in all of the simulations: a, wild-type hAOX1; b, simulation of complete spectrum; c, simulation of FeSI; d, simulation of FeSII. Experimental conditions: T = 20 K; microwave power, 1 mW; microwave frequency, 9.385 GHz; modulation amplitude, 0.5 mT; modulation frequency, 100 kHz.
TABLE 2.
EPR line widths and g values of FeSI and FeSII from hAOX1
| g Values | Line Widths (mT)a | ||||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Protein | Cluster | gx | gy | gz | ΔBx | ΔBy | ΔBz |
| hAOX1 | FeSI | 2.0115 | 1.924 | 1.900 | 4.1 | 2.4 | 3.7 |
| FeSII | 2.085 | 1.975 | 1.906 | 9.2 | 3.3 | 2.8 | |
| mAOX1† | FeSI | 2.019 | 1.927 | 1.912 | 2.6 | 2.6 | 3.2 |
| FeSII | 2.085 | 1.971 | 1.90 | 7.4 | 4.0 | 4.0 | |
The variable line width was included as g strain in the simulations.
Values for mAOX1 are taken from Schumann et al. (2009).
Purification and Characterization of hAOX1 Polymorphic Variants
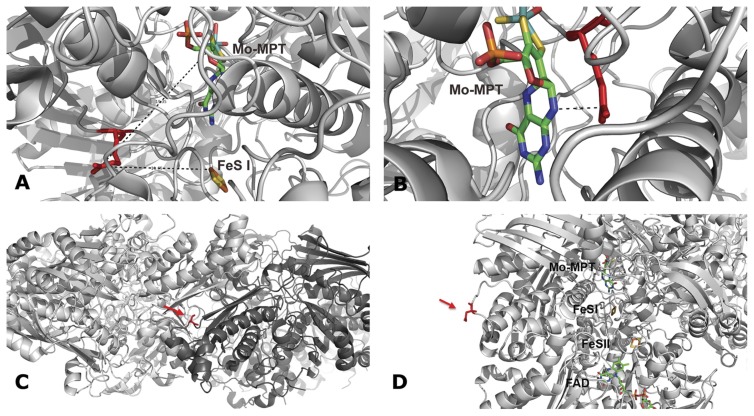
We studied the four most frequent hAOX1 SNPs, R802C, R921H, N1135S, and H1297R, as to their influences on enzyme activity, protein dimerization, and cofactor insertion. AOs belong to the XO family of molybdoenzymes and are characterized by high amino acid identity to the XO and xanthine dehydrogenase (XDH) forms of xanthine oxidoreductase (Garattini et al., 2008). Given the 50% identity between the two proteins, we modeled the structure of hAOX1 against the crystal structure of XDH from Bos taurus (Protein Data Bank code 1FO4) to define the spatial localization of the amino acid residues modified by the SNPs (Fig. 4). The hAOX1 model indicates that Arg802 and Arg921 are in proximity to the Moco and FeSI sites, Asn1135 is located close to the dimerization domain, and His1297 maps to the surface of the protein.
Fig. 4.
Model of the locations of the amino acid exchanges as a result of SNPs identified in the hAOX1 gene. Model with program MacPyMOL using bovine XDH (Protein Data Bank code 1FO4): A, Arg793 (Arg802 in human AOX1); B, Arg912 (Arg921 in human AOX1); C, Ser1126 (Asn1135 in human AOX1); D, Asn1288 (His1297 in human AOX1). The amino acid that is affected by the SNPs is marked in red. The cofactors (Moco, FeS clusters, and FAD) are shown in stick representations. The overall (α)2 structure is labeled in light gray for one subunit and in dark gray for the other subunit of the homodimer.
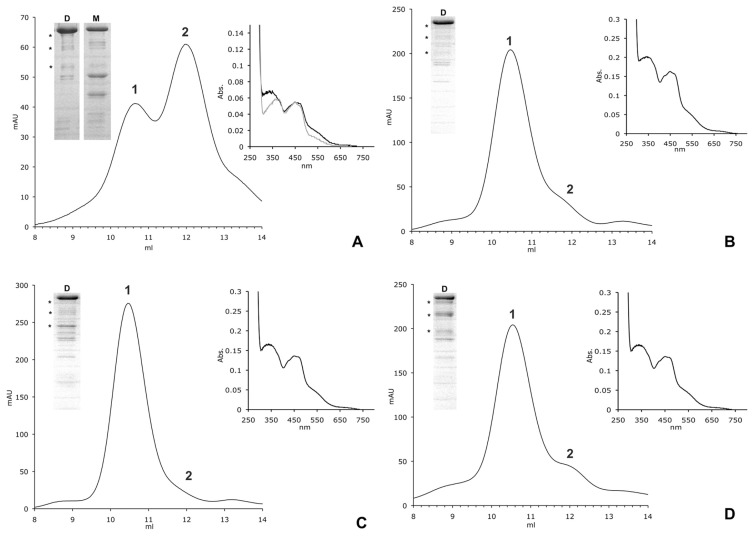
All of the hAOX1 variants were expressed and purified under similar conditions as the wild-type protein. Differences in the oligomerization states of the variants and wild-type hAOX1 were identified by size-exclusion chromatography. All of the variants showed an altered dimer/monomer ratio. In the case of the R802C variant, only 40% of the protein was in its dimeric form (Fig. 5). In contrast, all of the other hAOX1 variants showed a higher dimer/monomer ratio, indicating a higher proportion of the active protein. Mutation of each of the four amino acid residues did not affect the insertion of the [2Fe2S] clusters, because the iron content was similar to that of wild-type hAOX1. In fact, similar to wild-type hAOX1, all of the variants showed 70% saturation with iron (Fig. 2). In the R802C variant, molybdenum saturation was increased to 80%, which is slightly higher than the 70% saturation observed in the wild-type protein and all of the other variants (Fig. 2). The variant hAOX1 proteins were purified and showed purities comparable with that of the wild-type protein. The UV/Vis absorption spectra of the purified variants showed the same characteristic features of all of the molybdoflavoenzymes (Fig. 5).
Fig. 5.
Size-exclusion chromatography profiles and UV/Vis spectra of hAOX1 variants identified in SNPs. Elution profiles of hAOX1 variants using size-exclusion chromatography on a Superdex 200 column and UV/Vis spectra of purified proteins are shown (A, R802C; B, R921H; C, N1135S; D, H1297R). In comparison with the wild-type protein, hAOX1 variants show different dimer/monomer ratios in solution. R802C is mainly purified in its monomeric form (A), whereas all of the other studied variants almost completely exist in their dimeric forms (B–D). For all of the variants, similar degradation products were visualized by SDS-PAGE as in the wild-type (D, Dimer; M, Monomer; stars, degradation products of hAOX1; 1, peak corresponding to the AOX1 dimer; 2, peak corresponding to the AOX1 monomer).
Steady-State Kinetics of Wild-Type hAOX1 and Its Variants
To determine the impact of SNPs on enzymatic activity, steady-state kinetics were performed on the variants hAOX1-R802C, hAOX1-R921H, hAOX1-N1135S, and hAOX1-H1297R. All of the variants generated were tested for their abilities to catalyze the conversion of benzaldehyde, phthalazine, phenanthridine, and chloroquinazolinone as substrates (Table 3). The wild-type hAOX1 showed activities with all of the substrates tested with kcat values in the range from 5 to 12 min−1 and KM values between 1 and 7 μM. The best substrate was phenanthridine with a kcat value of 12.2 min−1.
TABLE 3.
Steady-state kinetics of hAOX1 and its variants corresponding to SNPs
Benzaldehyde, phthalazine, phenanthridine, and chloroquinazolinone were used as substrates to cover a range from aldehydes to N-heterocyclic compounds. Assays were performed photometrically using 2,6-dichlorphenolindophenol as a final electron acceptor. With phenanthridine, molecular oxygen was used as the electron acceptor. Data are mean values from three independent measurements (±S.D.).
| Substrate | Protein Portion | Kinetic Parameters | WT | R802C | R921H | N1135S | H1297R |
|---|---|---|---|---|---|---|---|
| Benzaldehyde | KM, μM | 7.1 ± 0.6 | 7.6 ± 1.9 | 6.3 ± 1.2 | 6.7 ± 2.8 | 5.2 ± 1.8 | |
| Active portion | kcat, min−1 | 6.4 ± 0.1 | 5.3 ± 0.3 | 1.7 ± 0.1 | 6.2 ± 0.3 | 6.4 ± 0.3 | |
| kcat/KM, 1/min · μM | 0.91 ± 0.17 | 0.70 ± 0.16 | 0.27 ± 0.08 | 0.92 ± 0.11 | 1.23 ± 0.17 | ||
| Total protein | kcat, min−1 | 2.7 ± 0.1 | 1.8 ± 0.3 | 1.1 ± 0.1 | 3.8 ± 0.3 | 2.6 ± 0.3 | |
| kcat/KM, 1/min · μM | 0.38 ± 0.17 | 0.24 ± 0.16 | 0.17 ± 0.08 | 0.57 ± 0.11 | 0.50 ± 0.17 | ||
| Phthalazine | KM, μM | 1.3 ± 0.3 | 0.9 ± 0.3 | 1.6 ± 0.3 | 1.2 ± 0.1 | 1.3 ± 0.2 | |
| Active portion | kcat, min−1 | 5.6 ± 0.2 | 5.2 ± 0.3 | 2.4 ± 0.1 | 7.2 ± 0.1 | 5.4 ± 0.1 | |
| kcat/KM, 1/min · μM | 4.31 ± 0.67 | 5.78 ± 1.00 | 1.50 ± 0.33 | 6.00 ± 1.00 | 4.15 ± 0.50 | ||
| Total protein | kcat, min−1 | 2.3 ± 0.2 | 1.8 ± 0.3 | 1.5 ± 0.1 | 4.4 ± 0.1 | 2.9 ± 0.1 | |
| kcat/KM, 1/min · μM | 1.77 ± 0.67 | 2.00 ± 1.00 | 0.94 ± 0.33 | 3.67 ± 1.00 | 2.23 ± 0.50 | ||
| Phenanthridine | KM, μM | 3.9 ± 0.8 | 4.4 ± 0.4 | N.D. | 6.1 ± 1.0 | 4.1 ± 0.7 | |
| Active portion | kcat, min−1 | 12.2 ± 0.5 | 10.2 ± 0.2 | N.D. | 32.6 ± 1.1 | 31.5 ± 1.0 | |
| kcat/KM, 1/min · μM | 3.13 ± 0.63 | 2.32 ± 0.50 | 5.34 ± 1.10 | 7.68 ± 1.43 | |||
| Total protein | kcat, min−1 | 5.3 ± 0.5 | 3.4 ± 0.2 | N.D. | 19.9 ± 1.1 | 16.9 ± 1.0 | |
| kcat/KM, 1/min · μM | 1.36 ± 0.63 | 0.77 ± 0.50 | 3.26 ± 1.10 | 4.12 ± 1.43 | |||
| Chloroquinazolinone | KM, μM | 5.2 ± 0.7 | 4.7 ± 0.7 | 4.5 ± 0.8 | 4.1 ± 0.5 | 5.8 ± 0.5 | |
| Active portion | kcat, min−1 | 5.6 ± 0.1 | 5.4 ± 0.2 | 3.6 ± 0.1 | 6.5 ± 0.1 | 6.7 ± 0.2 | |
| kcat/KM, 1/min · μM | 1.08 ± 0.14 | 1.15 ± 0.29 | 0.80 ± 0.13 | 1.59 ± 0.20 | 1.16 ± 0.4 | ||
| Total protein | kcat, min−1 | 2.3 ± 0.1 | 1.8 ± 0.2 | 2.2 ± 0.1 | 4.0 ± 0.1 | 3.6 ± 0.2 | |
| kcat/KM, 1/min · μM | 0.44 ± 0.14 | 0.38 ± 0.29 | 0.49 ± 0.13 | 0.98 ± 0.20 | 0.62 ± 0.4 | ||
| Oligomerization in solution | Dimer | Percentage | 58 | 39 | 83 | 90 | 78 |
| Monomer | Percentage | 38 | 58 | 13 | 7 | 17 | |
| Multimer | Percentage | 4 | 3 | 4 | 3 | 5 |
N.D., no activity was detectable; WT, wild type.
For a comparison, the dimeric portion of the hAOX1 variants also was subjected to steady-state kinetic analysis. The R802C variant showed kinetic data most comparable with those of the wild-type protein. For hAOX1-N1135S and hAOX1-H1297R, the turnover numbers were increased 2.5-fold with phenanthridine as the substrate. With the other substrates, the kcat and KM values for the two variants remained in the same range as those of the wild-type protein. hAOX1-R921H showed a 3.7- to 1.5-fold decrease in kcat values with most of the substrates in comparison with those of the wild-type protein, whereas the KM values remained comparable. Only with phenanthridine as a substrate was the protein completely inactive. The results show that amino acid substitution has different influences on the kinetic constants depending on the substrate. The inactivity or reduced activity of the R921H variant might be explained by the fact that arginine is highly conserved in all of the members of the XO family, and with its close proximity to the pterin molecule of the Mo-MPT cofactor, it might influence the geometry of the bound Mo-MPT molecule, thus affecting catalytic turnover but not affecting the binding of the substrate. However, because the different mutations affected the dimer/monomer ratio of the protein variants, we also considered the total amount of hAOX1 expressed and calculated the turnover number, taking into account the inactive portion of the monomeric protein. The normalized values are shown in Table 3. In general, the results show the same trends as those with the active portion of the protein.
Discussion
In this article, we analyzed the impact of SNPs identified in hAOX1 in an Italian population. From a cohort of 180 healthy volunteers, representative of the Italian population, seven different SNPs were identified that occurred with different frequencies. In total, eight individuals were heterozygous for the exchange of Tyr126 to a stop codon. Another eight individuals were heterozygous for a silent mutation in Leu1268. The most frequent mutations resulted in the amino acid exchange S1271L, which was identified in 14 individuals heterozygous for this mutation, and the amino acid exchange H1297R, for which seven individuals were heterozygous and six individuals were homozygous. Another mutation resulted in amino acid exchange N1135S, which was not so frequent and was identified in one individual heterozygous for this mutation and four individuals homozygous for this mutation. Two other rather rare mutations resulted in the amino acid exchanges R802C and R921H, for which two and one individual, respectively, were heterozygous. We decided to characterize the mutations resulting in the AOX1 variants H1297R, N1135S, R802C, and R921H further, because they seemed to be the most interesting SNPs, because either they were rather frequent and might have an impact on human metabolism or additional homozygous individuals were identified. However, for the characterization of the hAOX1 variants, a detailed characterization of the wild-type protein first was required. A heterologous expression system for hAOX1 had been described previously (Alfaro et al., 2009); however, here the protein was only partially purified and characterized only in terms of its activity with 6-substituted quinazolinones. In this report, we optimized the expression and purification of hAOX1 from E. coli. The protein was purified to almost homogeneity, with minor degradation products, which became visible after SDS-PAGE. Characterization of the active portion of the protein showed that it was purified in a form with a molybdenum saturation of 70%, of which 50% contained the terminal sulfido ligand that is essential for activity. Thus, hAOX1 was purified in a form in which 30% was active. This is consistent with other reports of the purification of AOX1 and AOX3 from mouse (Schumann et al., 2009; Mahro et al., 2011) or of XDH purified from a baculovirus insect cell system (Nishino et al., 2002). Thus, apparently when heterologous expression systems are used, the terminal sulfido ligand seems to be the limiting step to obtain a fully active mammalian enzyme. The iron saturation was determined to be 70%. The EPR spectra of hAOX1 were found to be very similar to those of mAOX1, showing a slightly rhombic signal for FeSI, whereas FeSII has unusual EPR properties for [2Fe2S] species, with a pronounced rhombic g tensor, showing broad lines and being observed only at much lower temperatures (20 K). Overall, the close similarity of the EPR parameters indicated the presence of the same ligands and similar geometries of the two redox centers in comparison with those of mAOX1, which was expected from the high amino acid sequence identity of the two enzymes of 83%. Although the expression of AOs from mouse shows moderate protein yields (Schumann et al., 2009; Mahro et al., 2011), only a low yield was achieved for hAOX1, suggesting major influence due to the difference in codon usage between Homo sapiens and E. coli. A codon-optimized construct for expression in E. coli will be tested in future studies to increase the protein yield.
Our characterization of wild-type hAOX1 allowed us to have a good basis for the characterization of the SNPs. First, modeling the amino acid exchanges in bovine XDH gave us an idea of their locations in the protein. The crystal structure of eukaryotic AO is not yet available; however, attempts to solve the structure of the human and mouse proteins are in progress. The amino acid exchanges resulting from the SNPs were introduced into the protein, and the respective variants were stable and showed overall molybdenum and iron contents similar to those of the wild-type protein, with the exception of the hAOX1-R802C variant, which showed a slightly higher saturation of Moco. The UV/Vis spectra also were comparable with those of wild-type hAOX1, suggesting complete saturation of the FAD cofactor. However, in contrast to the overall cofactor composition, crucial changes were observed in protein quaternary structures. Although the wild-type enzyme is stable in its monomeric and dimeric forms in a ratio of 1:1.5, hAOX1-R921H, hAOX1-N1125S, and hAOX1-H1297R were purified mainly as stable dimers. In contrast, hAOX1-R802C showed much higher levels of monomer in solution in a ratio of 1.5:1, resulting in a higher proportion of inactive protein. A change in the monomer/dimer ratio has been reported previously in AO and XDH enzymes with similar variants in proximity to the FeS clusters. In SNPs identified in Donryu rats, the amino acid exchange G101S in proximity to FeSII also resulted in the production of the monomeric form of AOX1 (Itoh et al., 2007c). In addition, in a human patient suffering from xanthinuria I, a mutation resulting in the amino acid exchange R149C was identified in the XDH gene (Sakamoto et al., 2001). The arginine is located close to FeSI, and when this mutation was introduced in the Rhodobacter capsulatus xdhB gene and the corresponding RcXDH-R135C was characterized, the purified protein also existed in two forms, a monomeric inactive form and a dimeric active form (Leimkühler et al., 2003). Further analyses of the monomer/dimer behavior of R. capsulatus XDH resulted in a model in which it was proposed that dimerization requires that the two FeS clusters are assembled correctly before Moco can be inserted and the protein can dimerize via the Moco domain (Schumann et al., 2008). Thus, amino acid exchanges in proximity to the FeS clusters influence the structure of the protein in a manner that dimerization is no longer effective.
We studied the activities of wild-type hAOX1 in comparison with those of selected variants based on the SNPs with four selected substrates, benzaldehyde, phthalazine, phenanthridine, and chloroquinazolinone. Our results show that the SNPs can be classified into three groups in general: fast metabolizers (FMs), poor metabolizers (PMs), and no affect on catalytic efficiency. Taking into account only the active portion of the protein, hAOX1-R802C and, even more pronounced, hAOX1-R921H are PMs, because both variants had a 2.4- to 1.5-fold reduced activity with most of the substrates tested. In addition, the R921H variant was identified to be inactive with phenanthridine; thus, the influence of the amino acid substitution on enzyme activity varies depending on the substrate. hAOX1-N1135S and hAOX1-H1297R can be classified as FMs, because an increased catalytic efficiency of 2- to 4-fold was observed depending on the substrate tested (taking into account only the active portion of the protein). In general, for the catalytic efficiencies, we calculated the overall activity of the purified enzyme, taking into account the active and inactive portions of the protein. In general, the same trends for the kinetic values were obtained for the variants in comparison with those of the wild-type protein. hAOX1-R921H showed that this residue is important for maintaining the catalytic activity of hAOX1, in particular with phenanthridine as the substrate. The residue Arg921 lies close to Moco and affected mainly the monomer/dimer ratio but also the overall activity of the protein. This might suggest that not only substrate turnover and binding were impaired but also intramolecular electron transfer. Thus, the positive charge of the arginine might affect substrate binding and intramolecular electron transfer, which can only poorly be substituted by His. In contrast, in hAOX1-R802C, the catalytic efficiency of the protein was not affected, and we obtained similar values in comparison with those of wild-type hAOX1. Both hAOX1-N1135S and hAOX1-H1297R are considered to be FM variants of hAOX1. Especially with phthalazine and phenanthridine, a 2- to 4-fold higher catalytic efficiency was obtained. Both amino acids are located at the surface of the protein and seem to influence the stability of hAOX1. Thus, the more polar and positive charged residues seem to affect the surface charge of the protein, which results in higher stability of hAOX1. This also might influence its interaction with other proteins and/or posttranslational modifications of the protein, as suggested by Itoh et al. (2007c) in a report on the characterization of SNPs in Donryu rats.
Our studies reveal the importance of considering individual differences based on SNPs for drug design. We analyzed 180 healthy individuals representative of the Italian population and identified the total occurrence of 51 SNPs in hAOX1, and a total of 10 were homozygous for the SNP. Two of the SNPs resulted in FM and one in PM individuals. hAOX1 is an important enzyme responsible for the metabolism of a number of drugs containing aldehydes and the more prevalent nitrogen heterocycles (Kitamura et al., 2006; Torres et al., 2007). The fraction of drugs metabolized by hAOX1 is likely to increase over the next decade. Thus, it should be carefully considered which dose of the drug is administered to an individual, because our results indicate that there might be a difference in hAOX1 activity in different individuals containing SNPs that additionally varies depending on the substrate used; thus, in FMs the drug might be cleared too fast and have no effect, whereas in PMs the drug might reach a toxic dose that would cause more severe side effects. In addition, some individuals were identified with SNPs that resulted in nonsense mutations, which might affect even the overall hAOX1 content and thus the overall activity more drastically when the active protein amount is expressed only from one allele. In the future, hAOX1 activities should be measured in individuals with known SNPs, or a pharmacokinetic study with a hAOX1-cleared drug in genotyped individuals should be conducted to confirm our results on the purified enzyme variants.
Supplementary Material
Acknowledgments
We thank Manfred Nimtz (Helmholtz Center for Infection Research, Braunschweig, Germany) for matrix-assisted laser desorption ionization peptide mapping and T. Nishino and T. Matsumura (Nippon Medical School, Tokyo, Japan) for providing hMCSF cDNA.
ABBREVIATIONS
- AO
aldehyde oxidase
- AOX1
aldehyde oxidase 1
- FM
fast metabolizer
- MCSF
Moco sulfurase
- Moco
molybdenum cofactor
- PAGE
polyacrylamide gel electrophoresis
- PCR
polymerase chain reaction
- PM
poor metabolizer
- SNP
single nucleotide polymorphism
- XDH
xanthine dehydrogenase
- XO
xanthine oxidase
- Vis
visible
- MPT
molybdopterin
Footnotes
This work was supported by the Cluster of Excellence “Unifying Concepts in Catalysis” (to C.T. and S.L.) coordinated by the Technische Universität Berlin and funded by the Deutsche Forschungsgemeinschaft and by grants from the Associazione Italiana per la Ricerca contro il Cancro and the Fondazione Italo Monzino (to E.G.).
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
Authorship Contributions
Participated in research design: Hartmann, Terao, Garattini, Teutloff, Jones, and Leimkühler.
Conducted experiments: Hartmann, Terao, Teutloff, and Alfaro.
Performed data analysis: Hartmann, Terao, Garattini, Teutloff, and Leimkühler.
Wrote or contributed to the writing of the manuscript: Hartmann, Garattini, Jones, and Leimkühler.
References
- Adachi M, Itoh K, Masubuchi A, Watanabe N, Tanaka Y. Construction and expression of mutant cDNAs responsible for genetic polymorphism in aldehyde oxidase in Donryu strain rats. J Biochem Mol Biol. 2007;40:1021–1027. doi: 10.5483/bmbrep.2007.40.6.1021. [DOI] [PubMed] [Google Scholar]
- Al-Salmy HS. Individual variation in hepatic aldehyde oxidase activity. IUBMB Life. 2001;51:249–253. doi: 10.1080/152165401753311799. [DOI] [PubMed] [Google Scholar]
- Alfaro JF, Joswig-Jones CA, Ouyang W, Nichols J, Crouch GJ, Jones JP. Purification and mechanism of human aldehyde oxidase expressed in Escherichia coli. Drug Metab Dispos. 2009;37:2393–2398. doi: 10.1124/dmd.109.029520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beedham C. Molybdenum hydroxylases as drug-metabolizing enzymes. Drug Metab Rev. 1985;16:119–156. doi: 10.3109/03602538508991432. [DOI] [PubMed] [Google Scholar]
- Beedham C. The role of non-P450 enzymes in drug oxidation. Pharm World Sci. 1997;19:255–263. doi: 10.1023/a:1008668913093. [DOI] [PubMed] [Google Scholar]
- Beedham C, Miceli JJ, Obach RS. Ziprasidone metabolism, aldehyde oxidase, and clinical implications. J Clin Psychopharmacol. 2003;23:229–232. doi: 10.1097/01.jcp.0000084028.22282.f2. [DOI] [PubMed] [Google Scholar]
- Edmondson D, Massey V, Palmer G, Beacham LM, 3rd, Elion GB. The resolution of active and inactive xanthine oxidase by affinity chromatography. J Biol Chem. 1972;247:1597–1604. [PubMed] [Google Scholar]
- Garattini E, Terao M. Increasing recognition of the importance of aldehyde oxidase in drug development and discovery. Drug Metab Rev. 2011;43:374–386. doi: 10.3109/03602532.2011.560606. [DOI] [PubMed] [Google Scholar]
- Garattini E, Fratelli M, Terao M. Mammalian aldehyde oxidases: genetics, evolution and biochemistry. Cell Mol Life Sci. 2008;65:1019–1048. doi: 10.1007/s00018-007-7398-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garattini E, Fratelli M, Terao M. The mammalian aldehyde oxidase gene family. Hum Genomics. 2009;4:119–130. doi: 10.1186/1479-7364-4-2-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garattini E, Mendel R, Romão MJ, Wright R, Terao M. Mammalian molybdo-flavoenzymes, an expanding family of proteins: structure, genetics, regulation, function and pathophysiology. Biochem J. 2003;372:15–32. doi: 10.1042/BJ20030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille R. The Mononuclear Molybdenum Enzymes. Chem Rev. 1996;96:2757–2816. doi: 10.1021/cr950061t. [DOI] [PubMed] [Google Scholar]
- Ichida K, Matsumura T, Sakuma R, Hosoya T, Nishino T. Mutation of human molybdenum cofactor sulfurase gene is responsible for classical xanthinuria type II. Biochem Biophys Res Commun. 2001;282:1194–1200. doi: 10.1006/bbrc.2001.4719. [DOI] [PubMed] [Google Scholar]
- Itoh K, Maruyama H, Adachi M, Hoshino K, Watanabe N, Tanaka Y. Lack of dimer formation ability in rat strains with low aldehyde oxidase activity. Xenobiotica. 2007a;37:709–716. doi: 10.1080/00498250701397713. [DOI] [PubMed] [Google Scholar]
- Itoh K, Maruyama H, Adachi M, Hoshino K, Watanabe N, Tanaka Y. Lack of formation of aldehyde oxidase dimer possibly due to 377G>A nucleotide substitution. Drug Metab Dispos. 2007b;35:1860–1864. doi: 10.1124/dmd.107.015503. [DOI] [PubMed] [Google Scholar]
- Itoh K, Masubuchi A, Sasaki T, Adachi M, Watanabe N, Nagata K, Yamazoe Y, Hiratsuka M, Mizugaki M, Tanaka Y. Genetic polymorphism of aldehyde oxidase in Donryu rats. Drug Metab Dispos. 2007c;35:734–739. doi: 10.1124/dmd.106.011502. [DOI] [PubMed] [Google Scholar]
- Johnson JL, Hainline BE, Rajagopalan KV, Arison BH. The pterin component of the molybdenum cofactor. Structural characterization of two fluorescent derivatives. J Biol Chem. 1984;259:5414–5422. [PubMed] [Google Scholar]
- Jordan CG, Rashidi MR, Laljee H, Clarke SE, Brown JE, Beedham C. Aldehyde oxidase-catalysed oxidation of methotrexate in the liver of guinea-pig, rabbit and man. J Pharm Pharmacol. 1999;51:411–418. doi: 10.1211/0022357991772619. [DOI] [PubMed] [Google Scholar]
- Kitamura S, Nakatani K, Sugihara K, Ohta S. Strain differences of the ability to hydroxylate methotrexate in rats. Comp Biochem Physiol C Pharmacol Toxicol Endocrinol. 1999a;122:331–336. doi: 10.1016/s0742-8413(98)10134-2. [DOI] [PubMed] [Google Scholar]
- Kitamura S, Sugihara K, Ohta S. Drug-metabolizing ability of molybdenum hydroxylases. Drug Metab Pharmacokinet. 2006;21:83–98. doi: 10.2133/dmpk.21.83. [DOI] [PubMed] [Google Scholar]
- Kitamura S, Sugihara K, Nakatani K, Ohta S, Ohhara T, Ninomiya S, Green CE, Tyson CA. Variation of hepatic methotrexate 7-hydroxylase activity in animals and humans. IUBMB Life. 1999b;48:607–611. doi: 10.1080/713803569. [DOI] [PubMed] [Google Scholar]
- Kundu TK, Hille R, Velayutham M, Zweier JL. Characterization of superoxide production from aldehyde oxidase: an important source of oxidants in biological tissues. Arch Biochem Biophys. 2007;460:113–121. doi: 10.1016/j.abb.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki M, Demontis S, Barzago MM, Garattini E, Terao M. Molecular cloning of the cDNA coding for mouse aldehyde oxidase: tissue distribution and regulation in vivo by testosterone. Biochem J. 1999;341:71–80. [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leimkühler S, Hodson R, George GN, Rajagopalan KV. Recombinant Rhodobacter capsulatus xanthine dehydrogenase, a useful model system for the characterization of protein variants leading to xanthinuria I in humans. J Biol Chem. 2003;278:20802–20811. doi: 10.1074/jbc.M303091200. [DOI] [PubMed] [Google Scholar]
- Leimkühler S, Stockert AL, Igarashi K, Nishino T, Hille R. The role of active site glutamate residues in catalysis of Rhodobacter capsulatus xanthine dehydrogenase. J Biol Chem. 2004;279:40437–40444. doi: 10.1074/jbc.M405778200. [DOI] [PubMed] [Google Scholar]
- Mahro M, Coelho C, Trincão J, Rodrigues D, Terao M, Garattini E, Saggu M, Lendzian F, Hildebrandt P, Romão MJ, et al. Characterization and crystallization of mouse aldehyde oxidase 3: from mouse liver to Escherichia coli heterologous protein expression. Drug Metab Dispos. 2011;39:1939–1945. doi: 10.1124/dmd.111.040873. [DOI] [PubMed] [Google Scholar]
- Nishino T, Amaya Y, Kawamoto S, Kashima Y, Okamoto K, Nishino T. Purification and characterization of multiple forms of rat liver xanthine oxidoreductase expressed in baculovirus-insect cell system. J Biochem. 2002;132:597–606. doi: 10.1093/oxfordjournals.jbchem.a003262. [DOI] [PubMed] [Google Scholar]
- Obach RS, Huynh P, Allen MC, Beedham C. Human liver aldehyde oxidase: inhibition by 239 drugs. J Clin Pharmacol. 2004;44:7–19. doi: 10.1177/0091270003260336. [DOI] [PubMed] [Google Scholar]
- Palmer T, Santini CL, Iobbi-Nivol C, Eaves DJ, Boxer DH, Giordano G. Involvement of the narJ and mob gene products in distinct steps in the biosynthesis of the molybdoenzyme nitrate reductase in Escherichia coli. Mol Microbiol. 1996;20:875–884. doi: 10.1111/j.1365-2958.1996.tb02525.x. [DOI] [PubMed] [Google Scholar]
- Parschat K, Canne C, Hüttermann J, Kappl R, Fetzner S. Xanthine dehydrogenase from Pseudomonas putida 86: specificity, oxidation-reduction potentials of its redox-active centers, and first EPR characterization. Biochim Biophys Acta. 2001;1544:151–165. doi: 10.1016/s0167-4838(00)00214-4. [DOI] [PubMed] [Google Scholar]
- Pryde DC, Dalvie D, Hu Q, Jones P, Obach RS, Tran TD. Aldehyde oxidase: an enzyme of emerging importance in drug discovery. J Med Chem. 2010;53:8441–8460. doi: 10.1021/jm100888d. [DOI] [PubMed] [Google Scholar]
- Rashidi MR, Smith JA, Clarke SE, Beedham C. In vitro oxidation of famciclovir and 6-deoxypenciclovir by aldehyde oxidase from human, guinea pig, rabbit, and rat liver. Drug Metab Dispos. 1997;25:805–813. [PubMed] [Google Scholar]
- Sakamoto N, Yamamoto T, Moriwaki Y, Teranishi T, Toyoda M, Onishi Y, Kuroda S, Sakaguchi K, Fujisawa T, Maeda M, et al. Identification of a new point mutation in the human xanthine dehydrogenase gene responsible for a case of classical type I xanthinuria. Hum Genet. 2001;108:279–283. doi: 10.1007/s004390100477. [DOI] [PubMed] [Google Scholar]
- Schumann S, Saggu M, Möller N, Anker SD, Lendzian F, Hildebrandt P, Leimkühler S. The mechanism of assembly and cofactor insertion into Rhodobacter capsulatus xanthine dehydrogenase. J Biol Chem. 2008;283:16602–16611. doi: 10.1074/jbc.M709894200. [DOI] [PubMed] [Google Scholar]
- Schumann S, Terao M, Garattini E, Saggu M, Lendzian F, Hildebrandt P, Leimkühler S. Site directed mutagenesis of amino acid residues at the active site of mouse aldehyde oxidase AOX1. PLoS One. 2009;4:e5348. doi: 10.1371/journal.pone.0005348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll S, Schweiger A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J Magn Reson. 2006;178:42–55. doi: 10.1016/j.jmr.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Terao M, Kurosaki M, Barzago MM, Fratelli M, Bagnati R, Bastone A, Giudice C, Scanziani E, Mancuso A, Tiveron C, et al. Role of the molybdoflavoenzyme aldehyde oxidase homolog 2 in the biosynthesis of retinoic acid: generation and characterization of a knockout mouse. Mol Cell Biol. 2009;29:357–377. doi: 10.1128/MCB.01385-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terao M, Kurosaki M, Barzago MM, Varasano E, Boldetti A, Bastone A, Fratelli M, Garattini E. Avian and canine aldehyde oxidases. Novel insights into the biology and evolution of molybdo-flavoenzymes. J Biol Chem. 2006;281:19748–19761. doi: 10.1074/jbc.M600850200. [DOI] [PubMed] [Google Scholar]
- Terao M, Kurosaki M, Saltini G, Demontis S, Marini M, Salmona M, Garattini E. Cloning of the cDNAs coding for two novel molybdo-flavoproteins showing high similarity with aldehyde oxidase and xanthine oxidoreductase. J Biol Chem. 2000;275:30690–30700. doi: 10.1074/jbc.M005355200. [DOI] [PubMed] [Google Scholar]
- Torres RA, Korzekwa KR, McMasters DR, Fandozzi CM, Jones JP. Use of density functional calculations to predict the regioselectivity of drugs and molecules metabolized by aldehyde oxidase. J Med Chem. 2007;50:4642–4647. doi: 10.1021/jm0703690. [DOI] [PubMed] [Google Scholar]
- Vila R, Kurosaki M, Barzago MM, Kolek M, Bastone A, Colombo L, Salmona M, Terao M, Garattini E. Regulation and biochemistry of mouse molybdo-flavoenzymes. The DBA/2 mouse is selectively deficient in the expression of aldehyde oxidase homologues 1 and 2 and represents a unique source for the purification and characterization of aldehyde oxidase. J Biol Chem. 2004;279:8668–8683. doi: 10.1074/jbc.M308137200. [DOI] [PubMed] [Google Scholar]
- Wahl RC, Rajagopalan KV. Evidence for the inorganic nature of the cyanolyzable sulfur of molybdenum hydroxylases. J Biol Chem. 1982;257:1354–1359. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.