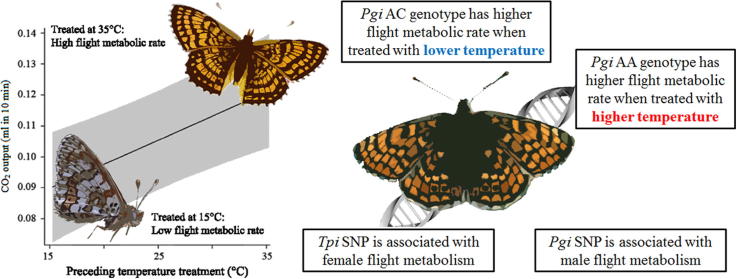
Graphical abstract
Keywords: Flight metabolic rate, Temperature treatment, Pgi, Tpi, Temperature–genotype interaction, Flight muscle maturation
Highlights
-
•
Thermal conditions following eclosion influence flight muscle maturation.
-
•
Two days of low temperature after eclosion reduced flight metabolic rate by 17%.
-
•
SNPs from two metabolic genes are associated with butterfly flight metabolic rate.
-
•
SNPs in metabolic genes interact with temperature to influence flight metabolism.
-
•
Male butterflies are more sensitive to temperature treatments than females.
Abstract
Flight is essential for foraging, mate searching and dispersal in many insects, but flight metabolism in ectotherms is strongly constrained by temperature. Thermal conditions vary greatly in natural populations and may hence restrict fitness-related activities. Working on the Glanville fritillary butterfly (Melitaea cinxia), we studied the effects of temperature experienced during the first 2 days of adult life on flight metabolism, genetic associations between flight metabolic rate and variation in candidate metabolic genes, and genotype–temperature interactions. The maximal flight performance was reduced by 17% by 2 days of low ambient temperature (15 °C) prior to the flight trial, mimicking conditions that butterflies commonly encounter in nature. A SNP in phosphoglucose isomerase (Pgi) had a significant association on flight metabolic rate in males and a SNP in triosephosphate isomerase (Tpi) was significantly associated with flight metabolic rate in females. In the Pgi SNP, AC heterozygotes had higher flight metabolic rate than AA homozygotes following low preceding temperature, but the trend was reversed following high preceding temperature, consistent with previous results on genotype–temperature interaction for this SNP. We suggest that these results on 2-day old butterflies reflect thermal effect on the maturation of flight muscles. These results highlight the consequences of variation in thermal conditions on the time scale of days, and they contribute to a better understanding of the complex dynamics of flight metabolism and flight-related activities under conditions that are relevant for natural populations living under variable thermal conditions.
1. Introduction
Temperature influences all biological processes. Ectothermic animals in particular are greatly affected by thermal conditions, as both high and low temperatures constrain activity and reduce fitness. At geographical scale, adaptation to the thermal environment is exemplified by Drosophila melanogaster in North America and Australia, where there are distinct temperature-related latitudinal clines in several life history and morphological traits as well as in molecular variation in central metabolic genes (Hoffmann and Weeks, 2007, Sezgin et al., 2004, Reinhardt et al., 2014, Cogni et al., 2015). At a smaller spatial scale, the willow beetle Chrysomela aeneicollis that inhabits the Sierra Nevada Mountains in California shows temperature-associated geographic variation in the glycolytic allozyme phosphoglucose isomerase (PGI) (Dahlhoff and Rank, 2000). In this case, variation in allele frequencies reflects genotypic differences in thermal stability at the enzyme level and heat and cold tolerance at the individual level (Dahlhoff and Rank, 2007). Moreover, the thermally sensitive genotype shows higher heat shock protein Hsp70 expression levels (Neargarder et al., 2003). In North American Colias butterflies, allozyme variation in PGI is associated with enzyme kinetics and thermal stability, which in turn has been shown to correlate with flight activity, mating success and reproductive performance (Watt, 1977, Watt et al., 2003). In butterflies in general, flight performance is critical for fitness in the wild, and flight activity is highly dependent on temperature.
The flight muscles of temperate butterflies operate best within a narrow range of temperatures, which is well above the typical ambient air temperatures (Kohane and Watt, 1999, Kingsolver, 1983). Work on the Eurasian Glanville fritillary butterfly (Melitaea cinxia) has shown that within the favorable range of temperatures, molecular variation in the Pgi locus is associated with variation in flight metabolic rate (FMR) and flight performance in the field (Niitepõld, 2010, Orsini et al., 2009). A single nucleotide polymorphism (SNP) in the Pgi locus interacts with ambient temperature in affecting both FMR and mobility: one genotype shows higher activity in lower temperatures, while the other one performs better in higher temperatures (Niitepõld et al., 2009).
Flight metabolic rate is a fundamentally important process for butterflies and many other insects, but it remains an understudied trait and we lack fundamental knowledge of its dynamics and the environmental conditions affecting it. For instance, though the effects of ambient temperature on butterfly activity and performance have been well documented, we know much less about the effects of thermal conditions experienced by individuals prior to particular flight activity. Temperature is never constant on the scale of days, and many insects are short-lived, which means that thermal conditions experienced during early life can potentially have a large effect on fitness. A previous study on the Glanville fritillary indicated that the isoform composition of troponin t, an important muscle gene associated with performance, changes significantly in early life, presumably reflecting gradual maturation of flight muscles following adult eclosion (Marden et al., 2008). Thermal conditions may influence the rate of maturation, and so may genetic variation in metabolic genes, which suggests that flight metabolism of young butterflies may be sensitive to the thermal conditions they have experienced earlier in their life, with possible interactions with genetic variation.
In the present study, we investigated how the thermal conditions during a few days following adult emergence affect flight performance in the Glanville fritillary butterfly, a species that inhabits fragmented habitats and whose ecological dynamics are therefore highly dependent on flight performance (Hanski, 1999, Hanski, 2011). We focus on three questions. First, we tested whether thermal conditions in the first two days after emergence, when the butterfly is fully formed but still undergoes physiological maturation, have an effect on flight metabolic rate (FMR), a quantitative measure of flight performance. Second, we conducted an association study on SNPs in candidate metabolic genes using mixed model regression. Third, we examined whether these candidate SNPs interact with previously experienced temperatures versus ambient (current) measurement temperature on affecting FMR. The results demonstrate significant effects of both current and past temperatures on FMR. Among the genetic associations, we found a significant association of the SNP in Pgi but also a novel association between FMR and a SNP in the gene triosephosphate isomerase (Tpi), which is located on the sex chromosome. To test the generality of this result, the SNP was genotyped in two additional data sets from previous experiments, and was found to have a significant result in one of them.
2. Materials and methods
2.1. Study materials
The primary dataset, hereafter referred to as DS1, consists of the offspring of butterflies studied in an outdoor population cage experiment at the Lammi biological station in Finland in 2008 (Klemme and Hanski, 2009). The parents of these offspring had been sampled as 5th instar larvae from the field, across the Åland Islands (50 by 70 km) in SW Finland, in the autumn of 2007. For the present experiment, we sampled the offspring from 15 mating. The offspring were reared to the 5th instar in the laboratory and maintained in diapause at 3 °C over winter. In spring 2009, post-diapause larvae were reared into adult butterflies in controlled conditions (12 h/12 h L/D, 28 °C/8 °C), fed with the leaves of the host plant Plantago lanceolata.
Two additional datasets, called DS2 and DS3, were used to validate the novel discovery from DS1, and these data were originated from the study of Kvist et al. (2015) and Mattila (2015). Both DS2 and DS3 consist of apparently unrelated individuals originating from 31 and 87 larval family groups (full sibs) sampled across the Åland Islands in 2011 and 2010, respectively. Flight metabolic rate was measured in the same way as in DS1 (below), but there was no temperature treatment. The samples and rearing conditions for DS1, DS2 and DS3 are described in Supplementary Table S1 in the Supplementary Information.
2.2. Temperature treatments and the measurement of metabolic rate
Adult butterflies from DS1 were marked individually after eclosion and assigned into three thermal treatments: 15, 24 and 35 °C. The treatment temperatures were chosen based on the activity level of butterflies at different temperatures. At 15 °C, butterflies are inactive, they do not move nor feed and often rest with their wings closed. At 35 °C, butterflies are very active and constantly fly around in the cage (diameter 15 cm, height 30 cm) in the laboratory. The intermediate temperature, 24 °C, is favorable for maintaining butterflies in the laboratory. Each group was kept in their designated temperature for 12 h from 0800 until 2000 during two days, and they all spent the night (2000–0800) at 8 °C. Butterflies were provided with water in a sponge but no sugar solution.
The flight metabolic rate of 51 females and 89 males was measured as CO2 emission rate during flight on the third day following eclosion (see Niitepõld et al. (2009) for details on the method). Half of the individuals were measured at 30 °C and the other half at 35 °C. These two temperatures were selected as we expected heterozygous Pgi AC individuals to show higher performance in lower temperatures and homozygous AA individuals to perform better in higher temperatures (Niitepõld et al., 2009). Butterflies were encouraged to fly for 10 min as continuously as possible by gently shaking and tapping the one-liter respirometry chamber whenever the individual landed. The chamber was kept under a light source emitting both visible and UV light. We extracted two variables from the data to describe metabolic performance during flight: the highest rate of CO2 production (FMRpeak), which typically occurs within a few min from the beginning of the experiment, and the total (integrated) volume of CO2 (FMRint) produced during the 10-min experiment. FMRpeak reflects maximal metabolic performance whereas FMRint reflects a combination of endurance and behavior. The experimental protocols for DS2 and DS3 were similar to that for DS1 except for the duration of the experiment and lack of the temperature treatment prior to the measurement of metabolic rate. In DS2, metabolic rate was measured for 16 females and 11 males for 15 min, while in DS3 it was measured for 36 females and 51 males for 7 min. Measurement temperature was 30 °C in both DS2 and DS3. Further details on the three datasets are given in the Supplementary Table S1.
2.3. Selection of candidate genes and genotyping
In DS1, 18 SNPs from 12 Expressed Sequence Tag (EST) sequences and 3 SNPs from the phosphoglucose isomerase (Pgi) partial messenger RNA (mRNA) sequence were selected for genotyping (Supplementary Table S2). These sequences were extracted from transcriptome data (Ahola et al., 2015) where complementary DNA (cDNA) from individuals originating from the Åland Islands in Finland, China and France were sequenced with 454 pyrosequencing (Roche Diagnostics, US; GS FLX). The resulting sequences were annotated with PANNZER (v 1.0) annotation tool (Radivojac et al., 2013). Gene set enrichment analyses (GSEA) were performed for pairs of populations to explore the enrichment of gene ontologies (GO). Significant results from GSEA were used to compile the initial set of genes. As our aim was to determine genetic associations with flight metabolic rate, we selected mostly genes of central metabolic pathways as candidate genes, such as genes from the glycolysis pathway (phosphoglucose isomerase, triosephosphate isomerase), the pentose phosphate pathway (gluocose-6-phosphate dehydrogenase), and the oxygen binding pathway (cytochrome P450 337). Stress response genes (Heatshock protein 70, c-Jun N-terminal protein kinase 1) were also included as candidate genes based on previous studies (Czaja, 2010, Karl et al., 2008). Finally, several other genes were selected based on significant results in previous gene expression studies on life-history traits in the Glanville fritillary (succinate dehydrogenase complex subunit D, flightin, peripheral-type benzodiazepine receptor, troponin t) (Klepsatel and Flatt, 2011, Marden et al., 2008, Wheat et al., 2011) (Supplementary Table S2). Candidate SNPs in these genes with their flanking sequences were extracted from the corresponding EST for genotyping, which was performed with Sequenom iPLEX Gold (Sequenom Inc. CA. USA) chemistry in a single multiplex well at the Institute for Molecular Medicine Finland (FIMM, Helsinki). Primers and probes for genotyping were designed using the MassARRAY Assay Designer Program (2006).
A SNP in the gene triosephosphate isomerase (Tpi) was significantly associated with female FMR in DS1. As this association has not been reported previously, we genotyped females from two other experiments on FMR to validate this finding. In DS2, the SNP was identified from RNA-seq data in Kvist et al. (2015). RNA was extracted from thorax and sequenced using next generation RNA sequencing technology implemented in Illumina Hiseq2000 and HiScanSQ (Illumina Inc., San Diego, CA, USA). SNP and genotype calling were performed using SAMtools ver 0.1.18 (Li et al., 2009) in two steps. The first step involved piling up the relevant input file in the .bam format in relation to the reference genome (Ahola et al., 2014). The output of this “pileup” step was piped to bcftools to create a single bcf file. In the second step, variant calling was performed by first viewing the bcf file via bcftools, and the output was piped to vcfutils for variant calling (Full command in the Supplementary material). In DS3, we used Sanger sequencing to determine the genotypes for the SNP, which is located in the 3′ UTR region of Tpi.
2.4. Statistical analyses
Mixed model multiple regression was used to test for the effects of body mass (pupal weight), preceding temperature and measurement temperature on flight metabolic rates (FMRint and FMRpeak), with brood as a random factor. As sex-dependent genetic effects occur in insects (Le Goff et al., 2006), the data were analyzed separately for males and females to facilitate the interpretation of the results. The statistical analyses were performed using R (x64 ver 3.1.0) (R Core Team, 2014).
For dataset DS1, association studies were implemented with mixed model regression using the “nlme” package in R (Pinheiro et al., 2014). To control for the relatedness of individuals, brood was used as a random variable in the model. Body mass was treated as a covariate, while preceding and measurement temperatures were treated as factors. To determine the significance of each explanatory variable in models for FMR, Anova function in the R package “car” (Fox and Weisberg, 2011) was used to obtain P values from the Wald Chi-square test. The P values were adjusted for multiple testing via false discovery rate (FDR) (Benjamini and Hochberg, 1995). We used the Lund test (Lund, 1975) for outlier detection to exclude significant outliers before the regression model.
In the validation datasets (DS2 and DS3), apparently unrelated samples were collected from the wild, but additional measures were taken to account for possible population stratification. In DS2, genome-wide autosomal SNPs were extracted from RNA-seq data to construct a matrix of relationships of the individuals. SNPs were filtered by minor allele frequency (MAF > 0.2), call rate (CR > 0.9) and linkage disequilibrium (LD) scores (R2 < 0.5, higher R2 indicates that SNPs are in LD), yielding 105 SNPs for the estimation of the relationship matrix to control for population stratification. Dataset DS2 was analyzed using mixed model regression implemented in Tassel software package ver 5.1.0 (Bradbury et al., 2007, Zhang et al., 2010). In DS3, individuals were sampled from different habitat patches across the large 4000 patch network in the Åland Islands (Hanski, 2011). As there was no genome-wide SNP data available in DS3 to construct the relationship matrix, we used sub-networks into which the 4000 patch network has been subdivided as a parameter to control population stratification in a linear model implemented in PLINK (Purcell et al., 2007). The properties of each dataset and the methods of their analysis are summarized in Supplementary Table S1. We report in the Supplementary material power analyses for the statistical tests.
3. Results
We excluded 3 of the 21 SNPs as they showed no variation (Supplementary Table S2). Two pairs of SNPs were completely linked and hence only one SNP from each pair was retained, resulting in 16 SNPs from 11 genes (detailed characterization of SNPs was depicted in Supplementary Tables S2 and S4). Below, we present the results for a series of increasingly complex models. We first analyze the effects of preceding temperature treatments and measurement temperatures on flight metabolic rates (FMRint and FMRpeak), then test for the associations of the SNPs, and finally analyze possible interactions between the SNPs and the temperature before and during the measurement of FMR.
3.1. Effects of preceding temperature and measurement temperature on FMR
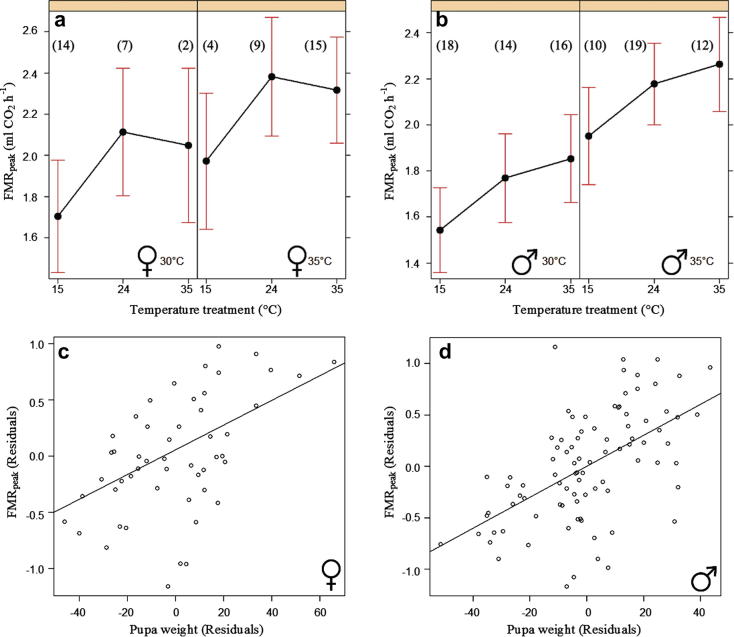
In males, body mass, the 2-day temperature treatment and the measurement temperature all had significant effects on FMRpeak (Table 1). In the case of FMRint, the result was qualitatively the same, but the effect of body mass was not quite significant. The effect size of the preceding temperature treatment was large, with 17% greater values of FMRpeak at 35 °C than at 15 °C. In females, body mass and preceding temperature treatment had significant effects on FMRint, while the effect of preceding temperature was nearly significant on FMRpeak (Table 1). According to the model, changing the preceding temperature from 15 to 35 °C increased FMRpeak by 16% in females. The effects of the explanatory variables on FMRpeak are illustrated in Fig. 1.
Table 1.
The effects of body mass (pupal weight), 2-day temperature treatment preceding the measurement, and the measurement temperature on the integrated and peak flight metabolic rates in males and females. Significant results are highlighted in bold.
| Sex | Trait | Explanatory variable | χ2 | df | Pr(>χ2) |
|---|---|---|---|---|---|
| Male | FMRint | Pupal weight | 3.273 | 1 | 0.070 |
| Preceding temp | 5.654 | 1 | 0.017 | ||
| Measurement temp | 7.606 | 1 | 0.006 | ||
| FMRpeak | Pupal weight | 38.670 | 1 | 5.02E−10 | |
| Preceding temp | 6.161 | 1 | 0.013 | ||
| Measurement temp | 19.313 | 1 | 1.11E−05 | ||
| Female | FMRint | Pupal weight | 6.125 | 1 | 0.013 |
| Preceding temp | 8.670 | 1 | 0.003 | ||
| Measurement temp | 0.014 | 1 | 0.906 | ||
| FMRpeak | Pupal weight | 21.247 | 1 | 4.04E−06 | |
| Preceding temp | 2.784 | 1 | 0.095 | ||
| Measurement temp | 2.990 | 1 | 0.084 | ||
Fig. 1.
The effect plots with 95% confidence intervals for three 2-day temperature treatments prior to the measurement and two measurement temperatures on FMRpeak in (a) females and (b) males. Partial regression plots of FMRpeak against body mass are shown for (c) females and (d) males, controlling for the preceding temperature treatment and measurement temperature. Sample size is shown in parentheses close to the data point.
3.2. Association between SNPs and FMR
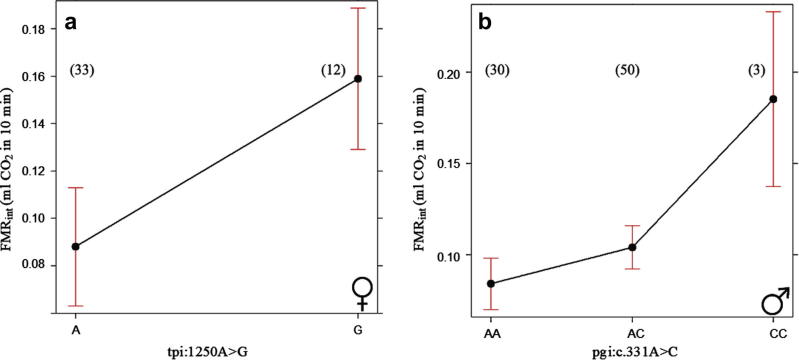
Table 2 gives the results for the SNPs with significant association with metabolic rates (false discovery rate < 0.05). These associations were detected in SNPs from two metabolic genes, phosphoglucose isomerase (Pgi) and triosephosphate isomerase (Tpi). In the case of the Pgi SNP, pgi:c.331A>C, the association with integrated flight metabolic rate (FMRint) was significant in males, while the SNP from Tpi, tpi:1250A>G, was significantly associated with both FMRpeak and FMRint in females. Fig. 2 shows the results for FMRint and pgi:c.331A>C in males and for tpi:1250A>G in females. The full association results for all the SNPs are presented in Supplementary Table S3.
Table 2.
Associations of SNPs in two metabolic genes with integrated and peak flight metabolic rates. Significant associations are highlighted in bold.
| Sex | Trait | SNP | Accession | Allele | Effect | P | FDR |
|---|---|---|---|---|---|---|---|
| Male | FMRint | pgi:c.331A>C | EU888473.1 | C | 0.028 | 0.0017 | 0.0265 |
| tpi:1250A>G | KJ803028 | G | 0.004 | 0.7257 | 0.9213 | ||
| FMRpeak | pgi:c.331A>C | EU888473.1 | C | 0.179 | 0.0559 | 0.1774 | |
| tpi:1250A>G | KJ803028 | G | −0.032 | 0.6783 | 0.6783 | ||
| Female | FMRint | pgi:c.331A>C | EU888473.1 | C | −0.006 | 0.6236 | 0.9070 |
| tpi:1250A>G | KJ803028 | G | 0.070 | 0.0008 | 0.0120 | ||
| FMRpeak | pgi:c.331A>C | EU888473.1 | C | −0.137 | 0.2259 | 0.6824 | |
| tpi:1250A>G | KJ803028 | G | 0.490 | 0.0024 | 0.0381 | ||
Fig. 2.
The genotypic plot with 95% confidence intervals of SNP (a) tpi:1250A>G in females and (b) pgi.c:331A>C in males on FMRint. SNP tpi:1250A>G is Z chromosome-linked, hence there is only one copy of the allele in females. Sample size is shown in parentheses close to the data point.
3.3. Interaction between SNPs and temperature on FMR
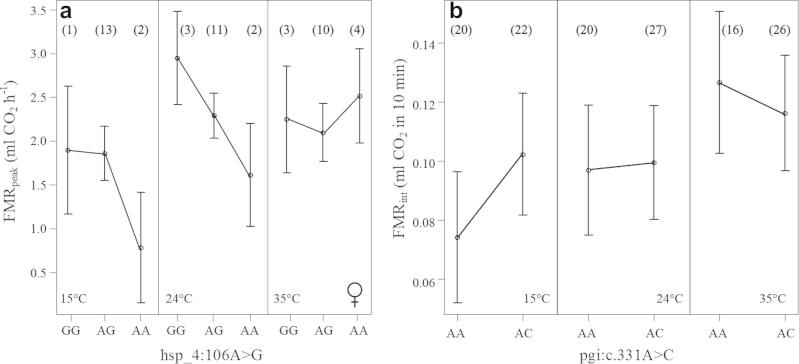
We next analyzed possible interactions between SNPs and the preceding temperature treatment and the measurement temperature. For the measurement temperature, none of the interactions were significant, but there was one significant interaction between preceding temperature and a SNP from Heatshock protein 70, hsp_4:106A>G, in females (FMRint P = 0.0026, FDR = 0.042; FMRpeak P = 0.0009, FDR = 0.014). One additional copy of the A-allele increased FMRpeak at the highest temperature treatment, in clear contrast to the trend in the lower temperature treatments (Fig.3a).
Fig. 3.
(a) The genotypic plot with 95% confidence intervals at three different preceding temperature treatments for the SNP hsp_4:106A>G on FMRpeak in females, and (b) for SNP pgi.c:331A>C in pooled data for males and females. Sample size is shown in parentheses close to the data point.
Table 3 shows trait value differences of pgi:c.331A>C genotypes on FMR in the three preceding temperature treatments. The same pattern is seen in both sexes and in both FMRpeak and FMRint: the AC heterozygotes had higher FMR than the AA homozygotes in the lowest preceding temperature treatment, but vice versa at higher preceding temperatures (Table 3). As the pattern was the same in the two sexes, we analyzed the pooled data for FMRpeak, including temperature treatment prior to flight, measurement temperature, sex and the SNP pgi:c.331A>C as well as their interactions as explanatory factors. The best model based on Akaike Information Criterion (AIC) included all the main factors as well as interactions between the SNP and sex and the SNP and the preceding temperature (Table 4). The latter interaction gives statistical support to the pattern in Table 3.
Table 3.
Genotypic differences of pgi:c.331A>C between the AC and AA individuals in the different preceding temperature treatments. Positive values indicate higher values of AC than AA individuals. The differences are significant for FMRint (P = 0.017) and FMRpeak (P = 0.016) in males at 15 °C, which are highlighted in bold. For further analysis see the text and Table 4.
| Males Effect (%) |
Females Effect (%) |
|||||
|---|---|---|---|---|---|---|
| Preceding temp (°C) | 15 | 24 | 35 | 15 | 24 | 35 |
| N (AC, AA) | 14, 13 | 19, 12 | 17, 10 | 8, 7 | 8, 8 | 9, 6 |
| FMRint | 56.3 | 19.2 | 14.8 | 7.5 | 2.7 | −7.1 |
| FMRpeak | 22.5 | 16.6 | −4.7 | 13.3 | −17.1 | −9.2 |
Table 4.
Models for the peak and integrated flight metabolic rate involving the SNP pgi.c:331A>C using pooled data for males and females. Significant results are highlighted in bold.
| Explanatory variable | FMRint |
FMRpeak |
||||
|---|---|---|---|---|---|---|
| χ2 | df | Pr(>χ2) | χ2 | df | Pr(>χ2) | |
| Pupal weight | 8.1018 | 1 | 0.00442 | 55.2452 | 1 | 1.06E−13 |
| Preceding temp | 16.1818 | 1 | 5.75E−05 | 12.115 | 1 | 0.0005 |
| Measurement temp | 8.1117 | 1 | 0.0044 | 25.6542 | 1 | 4.08E−07 |
| Sex | 0.596 | 1 | 0.44012 | 0.3529 | 1 | 0.5525 |
| pgi.c:331A>C | 11.9132 | 1 | 0.00056 | 9.9502 | 1 | 0.00161 |
| Sex: pgi.c:331A>C | 5.2068 | 1 | 0.0225 | 4.3131 | 1 | 0.03782 |
| Preceding temp: pgi.c:331A>C | 5.4732 | 1 | 0.01931 | 5.315 | 1 | 0.02114 |
3.4. Validation of the association between tpi:1250A>G and FMR
The association between the SNP tpi:1250A>G and FMR in females has not been reported previously in the literature, and hence we validated this result with material from two additional datasets, DS2 and DS3. In DS2, genotypes were available for 16 females. One significant outlier was removed from the dataset before the analysis. The SNP tpi:1250A>G was significantly associated with FMRint (P = 0.028) but not with FMRpeak (Table 5). In dataset DS3, there was no significant association with either measure of flight metabolic rate (Table 5).
Table 5.
Association of the SNP tpi:1250A>G with integrated and peak flight metabolic rates in females in datasets DS2 and DS3. Significant result is highlighted in bold.
| Trait | Dataset DS2 |
Dataset DS3 |
||
|---|---|---|---|---|
| F | P | T | P | |
| FMRint | 6.258 | 0.028 | −1.378 | 0.179 |
| FMRpeak | 1.641 | 0.224 | −1.958 | 0.060 |
4. Discussion
4.1. Effects of body mass and preceding temperature on FMR
Previous studies on the Glanville fritillary (Niitepõld et al., 2009) and other insects (Harrison and Fewell, 2002, Harrison et al., 1996) have demonstrated an effect of ambient temperature on flight metabolic rate when a wide range of temperatures has been studied. We measured flight metabolic rate at two temperatures, 30 and 35 °C, which are both favorable for flight, as butterfly flight muscles perform best when their temperature is between 30 and 40 °C (Kohane and Watt, 1999, Watt, 1968). For these two temperatures, we found a significant effect of measurement temperature on flight metabolism in males, especially on FMRpeak, but no significant effect in females. In the experiment, butterflies were moved into the respirometry chamber from the temperature of 22 °C ca. 25 min before the measurement. As female butterflies have larger body mass than males (19% difference in pupal weight in the present material), males can be expected to be more sensitive to temperature changes (Gilchrist, 1990). In the field, male butterflies spend shorter periods of time basking before flight (Gilchrist, 1990), as they can increase body temperature faster than females. In an experiment on the Glanville fritillary, thoracic temperature during take-off was positively correlated with ambient temperature in males but not in females (Mattila, 2015). These results suggest that body temperature responds more slowly to short-term changes in ambient temperature in females than in males, which may explain the sex difference in the effect of measurement temperature in our results.
Thermal conditions experienced during the first two days following adult eclosion had a significant effect on flight metabolic rate. In females, FMRint was 53% greater in individuals that had spent two days at 35 °C compared to 15 °C, and in males the difference was 28%. The corresponding figures for FMRpeak were 16% in females and 17% in males. The temperatures used in our treatments correspond to realistic thermal conditions experienced by butterflies in temperate climates. Butterflies can elevate their body temperature well above the ambient air temperature by basking in the sun, but under overcast conditions the body temperatures follows closely the ambient air temperature (Kingsolver, 1983).
We suggest that the mechanism explaining our results could be related to slower flight muscle maturation in lower temperatures. The maturation process of insect flight muscles involves changes in the abundance of alternatively spliced isoforms of the regulatory protein troponin t, as demonstrated for dragonflies (Fitzhugh and Marden, 1997, Marden et al., 1999), honeybees (Schippers et al., 2006), and Lepidoptera (Marden et al., 2008). The activity of several metabolic enzymes increases during the first days of life in honeybees, and the flight metabolic rate of honeybees increases in early life, and also later in life, when workers undergo a behavioral change from staying in the hive to foraging in the field (Schippers et al., 2010). In butterflies, flight metabolic rate has been shown to be low on the first day after emergence, after which it increases. Mated females of Speyeria mormonia reached their highest flight metabolic rate four days after emergence (Niitepõld et al., 2014). In unmated male Glanville fritillaries, flight metabolic rate increased from the age of one day to the age of three days, after which it stayed relatively stable (Niitepõld and Hanski, 2013). Physical activity may affect muscle maturation, as artificially increased wing loading and therefore increased need for muscle output changed the relative abundances of different troponin t isoforms in the flight muscles of the fall armyworm moth (Spodoptera frugiperda), and in the Glanville fritillary there was a correlation between voluntary activity and the troponin t isoform composition (Marden et al., 2008). Similarly, bumblebees initiate foraging flights while they still experience maturation at the enzyme activity level (Skandalis et al., 2011), supporting the notion that activity is an important driving force in the maturation process. In summary, these results suggest that conditions experienced during the first days after adult emergence may interact with the process of maturation. To test this hypothesis, further experiments on troponin t isoform relative abundances and gene expression under different thermal conditions and levels of physical activity are needed.
Long-term thermal conditions during the larval stage may have different consequences than thermal conditions experienced during adult maturation. Mattila and Hanski (2014) found that larval development in high temperature (daytime temperature 35 °C) resulted in significantly reduced flight metabolic rate in adults in comparison with larval development in moderate temperature (daytime temperature 28 °C). High temperature during larval development may lead to a stress response with consequences for adult flight performance. Moreover, Mattila and Hanski (2014) found that FMRint was significantly heritable in moderate temperature conditions (28 °C) but not under thermally stressful conditions (35 °C), highlighting the role of thermal plasticity in FMR. In the present study, variation among individuals in FMR was greater in females than in males (based on the variance of the random variable in the analysis). Heritability estimates for DS1 based on variance component linkage analysis (Amos, 1994, Sham et al., 2002) indicated high heritability of integrated flight metabolic rate (male h2 = 0.44, female h2 = 0.65), similar to the values reported by Mattila and Hanski (2014), but not of peak flight metabolic rate (male h2 = 0, female h2 = 0.15). Thus the more plastic of the two traits, FMRpeak, with a significant negative effect of preceding temperature, showed no significant heritability.
4.2. Association of metabolic SNPs
Previous studies on the Glanville fritillary have reported significant associations between the SNP pgi:c.331A>C in the glycolytic gene phosphoglucose isomerase and several life history traits, such as life span (Klemme and Hanski, 2009, Saastamoinen et al., 2009, Niitepõld and Hanski, 2013), clutch size (Saastamoinen, 2007, Saastamoinen and Hanski, 2008) and flight metabolic rate (FMRpeak) (Niitepõld and Hanski, 2013, Niitepõld et al., 2009, Niitepõld, 2010). Here, we found a significant association between pgi:c.331A>C and FMRint in males but not in females, in agreement with the result of Mattila (2015). Pooled results for both sexes showed a significant interaction between the SNP and the preceding temperature treatment: AC heterozygotes had higher flight metabolic rate than AA homozygotes following two days at low temperature, but vice versa in the high preceding temperature treatment. This interaction is qualitatively similar to the one between the Pgi genotype and measurement temperature when a wider range of measurement temperatures were used (Niitepõld et al., 2009). Moreover, the same interaction between pgi:c.331A>C and ambient temperature has been observed in the distance moved in a given time by free-flying female butterflies, tracked by harmonic radar: AC females flew longer distances than AA females in low to moderate ambient temperatures but not in high temperatures (Niitepõld et al., 2009). The present results suggest that the association is present also in post-emergence maturation, which has significant consequences for the performance and fitness of butterflies in the field. As discussed above, thermal conditions following adult emergence may influence flight muscle maturation, and as AC heterozygotes function better in cool conditions, they may mature at a faster rate than AA homozygotes in low temperatures. In this scenario, cool periods can affect butterfly mobility in the field through a direct effect on body temperature and through an indirect effect on maturation, affecting flight capacity later in life.
Triosephosphate isomerase (Tpi) is located in the same metabolic pathway as Pgi (glycolysis). It encodes the enzyme catalyzing the reversible conversion between dihydroxyacetone phosphate and d-glyceraldehyde 3-phosphate. Several insect studies have shown significant associations of genetic variation in Tpi on individual performance. For example, in a study on hypoxia tolerance in D. melanogaster, Zhou et al. (2008) discovered seven polymorphic sites in Tpi with distinct allele frequency differences between control and hypoxia tolerant individuals, including one SNP at the 3′UTR region. Another study on D. melanogaster showed a latitudinal cline in allele frequency in Tpi (Oakeshott et al., 1984).
Here, we detected a novel association between tpi:1250A>G and FMR in females. As Tpi is located in the sex chromosome, there is only one copy in females, while two copies in males, and hence there are no heterozygotes in females. In datasets DS1 and DS2, female butterflies with the G-allele exhibited higher flight metabolic rate than females with the A-allele. There was no significant association in DS3, possibly due to differences in the butterfly material and the experimental conditions (Supplementary Table S1), or due to insufficient power to detect true effects (sample size = 35). In any case, it is encouraging that two of the three independent datasets showed the same significant association between FMRint and tpi:1250A>G. Tpi is thus another polymorphic metabolic gene that deserves attention in studies of metabolic rate and e.g. dispersal rate in insects. The dissimilar associations in the two sexes with the two metabolic genes, Pgi and Tpi, in the same pathway, deserve further attention. The gender difference may be related to the fact that Tpi is located in the sex chromosome, but it may also be affected by differences in flight behavior and hence differences in metabolic demands in the two sexes (Niitepõld et al., 2011).
Finally, SNP hsp_4:106A>G from one of the Heat shock protein 70 (Hsp70) genes interacted significantly with the preceding temperature treatment in influencing FMRint and FMRpeak in females. An additional copy of the A-allele increased FMRpeak when the butterfly had spent 2 days at the highest temperature of 35 °C, while it had an opposite effect when the butterfly had experienced the temperatures of 15 and 24 °C (Fig.3a). Heat shock proteins act as molecular chaperones protecting against cellular damage, and their expression is induced by heat and other stressors (Sørensen et al., 2003). The significant role of Hsp expression in thermal stress resistance is well established (Feder and Hoffmann, 1999), and it has been studied in many insects. For instance, in the willow beetle C. aeneicollis, thermally less tolerant Pgi genotypes up-regulate Hsp70 to a greater extent than the other genotypes (Dahlhoff and Rank, 2000, Rank et al., 2007). Such buffering of individual differences in thermal tolerance by Hsp expression has been suggested also by other studies (Rutherford, 2003). Another study on insects shows that Hsp expression may be involved in the regulation of thermal plasticity (Hu et al., 2014). On the other hand, over-expression of Hsp is expected to have deleterious fitness consequences (Krebs and Feder, 1997, Sørensen et al., 2003). In a previous study on the Glanville fritillary, Luo et al. (2014) found significant differences in Hsp70 expression between populations originating from dissimilar thermal environments. Our results suggest that different Hsp70 genotypes perform dissimilarly in relation to the thermal conditions experienced in early life. The differences may be due to e.g. differential expression of the Hsp70 gene or its isoforms in different genotypes, which could affect stress response and regulation of flight metabolism. The genotypic differences may be reversed or only become visible during thermal stress when the expression of Hsp70 is induced. However, due to small sample size in each interaction category in the present study (Fig.3a), we consider that these results are not conclusive, in spite of statistical significance. Nonetheless, the present results suggest an interesting mechanism involving Hsp70 that has not been reported before.
5. Conclusion
The present results demonstrate that thermal conditions experienced during 2 days following adult emergence have significant effects on subsequent flight metabolic rate in the Glanville fritillary butterfly. We found that the maximal flight performance was strongly reduced, by 16% to 17%, by 2 days of low temperature (15 °C) prior to the measurement of the flight metabolic rate. This response may represent slow flight muscle maturation in low temperatures where activity is limited. Such effects of post-emergence thermal conditions highlight the complex dynamics of flight metabolism, dispersal and insect behavior under conditions that are relevant for natural populations, and they introduce an additional source of variation in flight metabolism that is easily missed in typical laboratory experiments. The SNP in phosphoglucose isomerase interacted significantly with previously experienced thermal conditions in affecting flight metabolism, which may contribute to the maintenance of polymorphism in these loci in natural populations. These results highlight significant consequences of variation in thermal conditions on the time scale of days on butterfly flight metabolism and dispersal. The present results also point to new questions concerning plasticity of insect flight muscle maturation and the consequences for performance and fitness in natural populations.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
We thank Annukka Ruokolainen for DNA extraction and genotyping assay preparation; Institute for Molecular Medicine Finland (FIMM) for Sequenom genotyping; Minna Taipale, Karolinska Institutet and FIMM for RNA-seq sequencing; Panu Somervuo for suggestions and help with RNA-seq processing pipelines; and Virpi Ahola for EST data processing. We thank two anonymous referees for helpful comments. This study was supported by a grant from the Integrative Life Science (ILS) doctoral program to WSC, and grants from the European Research Council (AdG Grant Number 232826) and the Academy of Finland (Finnish CoE Programme, Grant Numbers 256453 and 250444) to IH.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jinsphys.2015.11.015.
A. Supplementary data
Supplementary Tables S1–S5.
References
- 2006. Massarray® Assay Design 3.1 Software User’s Guide.
- Ahola V., Koskinen P., Wong S.C., Kvist J., Paulin L., Auvinen P., Saastamoinen M., Frilander M.J., Lehtonen R., Hanski I. Temperature and sex related effects of serine protease alleles on larval development in the Glanville fritillary butterfly. J. Evol. Biol. 2015 doi: 10.1111/jeb.12745. [DOI] [PubMed] [Google Scholar]
- Ahola V., Lehtonen R., Somervuo P., Salmela L., Koskinen P., Rastas P., Valimaki N., Paulin L., Kvist J., Wahlberg N., Tanskanen J., Hornett E.A., Ferguson L.C., Luo S., Cao Z., de Jong M.A., Duplouy A., Smolander O.P., Vogel H., McCoy R.C., Qian K., Chong W.S., Zhang Q., Ahmad F., Haukka J.K., Joshi A., Salojarvi J., Wheat C.W., Grosse-Wilde E., Hughes D., Katainen R., Pitkanen E., Ylinen J., Waterhouse R.M., Turunen M., Vaharautio A., Ojanen S.P., Schulman A.H., Taipale M., Lawson D., Ukkonen E., Makinen V., Goldsmith M.R., Holm L., Auvinen P., Frilander M.J., Hanski I. The Glanville fritillary genome retains an ancient karyotype and reveals selective chromosomal fusions in lepidoptera. Nat. Commun. 2014;5:4737. doi: 10.1038/ncomms5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos C.I. Robust variance-components approach for assessing genetic linkage in pedigrees. Am. J. Hum. Genet. 1994;54:535–543. [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995;57:12. [Google Scholar]
- Bradbury P.J., Zhang Z., Kroon D.E., Casstevens T.M., Ramdoss Y., Buckler E.S. Tassel: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Cogni R., Kuczynski K., Lavington E., Koury S., Behrman E.L., O’Brien K.R., Schmidt P.S., Eanes W.F. Variation in Drosophila melanogaster central metabolic genes appears driven by natural selection both within and between populations. Proc. R. Soc. London, Ser. B. 2015;282 doi: 10.1098/rspb.2014.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaja M.J. Jnk regulation of hepatic manifestations of the metabolic syndrome. Trends Endocrinol. Metab. TEM. 2010;21:707–713. doi: 10.1016/j.tem.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlhoff E.P., Rank N.E. Functional and physiological consequences of genetic variation at phosphoglucose isomerase: heat shock protein expression is related to enzyme genotype in a montane beetle. Proc. Natl. Acad. Sci. U.S.A. 2000;97:10056–10061. doi: 10.1073/pnas.160277697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlhoff E.P., Rank N.E. The role of stress proteins in responses of a montane willow leaf beetle to environmental temperature variation. J. Biosci. 2007;32:477–488. doi: 10.1007/s12038-007-0047-7. [DOI] [PubMed] [Google Scholar]
- Feder M.E., Hoffmann G.E. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu. Rev. Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Fitzhugh G.H., Marden J.H. Maturational changes in troponin t expression ca2+-sensitivity and twitch contraction kinetics in dragonfly flight muscle. J. Exp. Biol. 1997;200:1473–1482. doi: 10.1242/jeb.200.10.1473. [DOI] [PubMed] [Google Scholar]
- Fox J., Weisberg S. Sage; 2011. An R Companion to Applied Regression. [Google Scholar]
- Gilchrist G.W. The consequences of sexual dimorphism in body size for butterfly flight and thermoregulation. Funct. Ecol. 1990;4:475–487. [Google Scholar]
- Hanski I. Habitat connectivity, habitat continuity, and metapopulations in dynamic landscapes. Oikos. 1999;87:209–219. [Google Scholar]
- Hanski I.A. Eco-evolutionary spatial dynamics in the Glanville fritillary butterfly. Proc. Natl. Acad. Sci. U.S.A. 2011;108:14397–14404. doi: 10.1073/pnas.1110020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J.F., Fewell J.H. Environmental and genetic influences on flight metabolic rate in the honey bee, Apis mellifera. Comp. Biochem. Physiol. A: Mol. Integr. Physiol. 2002;133:323–333. doi: 10.1016/s1095-6433(02)00163-0. [DOI] [PubMed] [Google Scholar]
- Harrison J.F., Nielsen D.I., Page R.E. Malate dehydrogenase phenotype, temperature and colony effects on flight metabolic rate in the honey-bee, Apis mellifera. Funct. Ecol. 1996;10:81–88. [Google Scholar]
- Hoffmann A.A., Weeks A.R. Climatic selection on genes and traits after a 100 year-old invasion: a critical look at the temperate-tropical clines in Drosophila melanogaster from Eastern Australia. Genetica. 2007;129:133–147. doi: 10.1007/s10709-006-9010-z. [DOI] [PubMed] [Google Scholar]
- Hu J.T., Chen B., Li Z.H. Thermal plasticity is related to the hardening response of heat shock protein expression in two bactrocera fruit flies. J. Insect Physiol. 2014;67:105–113. doi: 10.1016/j.jinsphys.2014.06.009. [DOI] [PubMed] [Google Scholar]
- Karl I., Schmitt T., Fischer K. Phosphoglucose isomerase genotype affects life-history traits and cold stress resistance in a copper butterfly. Funct. Ecol. 2008;22:887–894. [Google Scholar]
- Kingsolver J.G. Thermoregulation and flight in Colias butterflies – elevational patterns and mechanistic limitations. Ecology. 1983;64:534–545. [Google Scholar]
- Klemme I., Hanski I. Heritability of and strong single gene (pgi) effects on life-history traits in the Glanville fritillary butterfly. J. Evol. Biol. 2009;22:1944–1953. doi: 10.1111/j.1420-9101.2009.01807.x. [DOI] [PubMed] [Google Scholar]
- Klepsatel P., Flatt T. The genomic and physiological basis of life history variation in a butterfly metapopulation. Mol. Ecol. 2011;20:1795–1798. doi: 10.1111/j.1365-294X.2011.05078.x. [DOI] [PubMed] [Google Scholar]
- Kohane M.J., Watt W.B. Flight-muscle adenylate pool responses to flight demands and thermal constraints in individual Colias eurytheme (lepidoptera, pieridae) J. Exp. Biol. 1999;202:3145–3154. doi: 10.1242/jeb.202.22.3145. [DOI] [PubMed] [Google Scholar]
- Krebs R.A., Feder M.E. Deleterious consequences of Hsp70 overexpression in Drosophila melanogaster larvae. Cell Stress Chaperones. 1997;2:60–71. doi: 10.1379/1466-1268(1997)002<0060:dcohoi>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvist J., Mattila A.L., Somervuo P., Ahola V., Koskinen P., Paulin L., Salmela L., Fountain T., Rastas P., Ruokolainen A., Taipale M., Holm L., Auvinen P., Lehtonen R., Frilander M.J., Hanski I. Flight-induced changes in gene expression in the Glanville fritillary butterfly. Mol. Ecol. 2015;24:4886–4900. doi: 10.1111/mec.13359. [DOI] [PubMed] [Google Scholar]
- Le Goff G., Hilliou F., Siegfried B.D., Boundy S., Wajnberg E., Sofer L., Audant P., ffrench-Constant R.H., Feyereisen R. Xenobiotic response in Drosophila melanogaster: sex dependence of P450 and GST gene induction. Insect Biochem. Mol. Biol. 2006;36:674–682. doi: 10.1016/j.ibmb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing S The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund R.E. Tables for an approximate test for outliers in linear models. Technometrics. 1975;17:473–476. [Google Scholar]
- Luo S., Chong Wong S., Xu C., Hanski I., Wang R., Lehtonen R. Phenotypic plasticity in thermal tolerance in the Glanville fritillary butterfly. J. Therm. Biol. 2014;42:33–39. doi: 10.1016/j.jtherbio.2014.02.018. [DOI] [PubMed] [Google Scholar]
- Marden J.H., Fescemyer H.W., Saastamoinen M., MacFarland S.P., Vera J.C., Frilander M.J., Hanski I. Weight and nutrition affect pre-mrna splicing of a muscle gene associated with performance, energetics and life history. J. Exp. Biol. 2008;211:3653–3660. doi: 10.1242/jeb.023903. [DOI] [PubMed] [Google Scholar]
- Marden J.H., Fitzhugh G.H., Wolf M.R., Arnold K.D., Rowan B. Alternative splicing, muscle calcium sensitivity, and the modulation of dragonfly flight performance. Proc. Natl. Acad. Sci. U.S.A. 1999;96:15304–15309. doi: 10.1073/pnas.96.26.15304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila A.L., Hanski I. Heritability of flight and resting metabolic rates in the Glanville fritillary butterfly. J. Evol. Biol. 2014;27:1733–1743. doi: 10.1111/jeb.12426. [DOI] [PubMed] [Google Scholar]
- Mattila A.L.K. Thermal biology of flight in a butterfly: genotype, flight metabolism, and environmental conditions. Ecol. Evol. 2015 doi: 10.1002/ece3.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neargarder G., Dahlhoff E.P., Rank N.E. Variation in thermal tolerance is linked to phosphoglucose isomerase genotype in a montane leaf beetle. Funct. Ecol. 2003;17:213–221. [Google Scholar]
- Niitepõld K. Genotype by temperature interactions in the metabolic rate of the Glanville fritillary butterfly. J. Exp. Biol. 2010;213:1042–1048. doi: 10.1242/jeb.034132. [DOI] [PubMed] [Google Scholar]
- Niitepõld K., Hanski I. A long life in the fast lane: positive association between peak metabolic rate and lifespan in a butterfly. J. Exp. Biol. 2013;216:1388–1397. doi: 10.1242/jeb.080739. [DOI] [PubMed] [Google Scholar]
- Niitepõld K., Mattila A.K., Harrison P., Hanski I. Flight metabolic rate has contrasting effects on dispersal in the two sexes of the Glanville fritillary butterfly. Oecologia. 2011;165:847–854. doi: 10.1007/s00442-010-1886-8. [DOI] [PubMed] [Google Scholar]
- Niitepõld K., Perez A., Boggs C.L. Aging, life span, and energetics under adult dietary restriction in lepidoptera. Physiol. Biochem. Zool. PBZ. 2014;87:684–694. doi: 10.1086/677570. [DOI] [PubMed] [Google Scholar]
- Niitepõld K., Smith A.D., Osborne J.L., Reynolds D.R., Carreck N.L., Martin A.P., Marden J.H., Ovaskainen O., Hanski I. Flight metabolic rate and pgi genotype influence butterfly dispersal rate in the field. Ecology. 2009;90:2223–2232. doi: 10.1890/08-1498.1. [DOI] [PubMed] [Google Scholar]
- Oakeshott J.G., McKechnie S.W., Chambers G.K. Population genetics of the metabolically related adh, gpdh and tpi polymorphisms in Drosophila melanogaster. I. Geographic variation in gpdh and tpi allele frequencies in different continents. Genetica. 1984;63:21–29. [Google Scholar]
- Orsini L., Wheat C.W., Haag C.R., Kvist J., Frilander M.J., Hanski I. Fitness differences associated with pgi snp genotypes in the Glanville fritillary butterfly (Melitaea cinxia) J. Evol. Biol. 2009;22:367–375. doi: 10.1111/j.1420-9101.2008.01653.x. [DOI] [PubMed] [Google Scholar]
- Pinheiro, J., Bates, D., DebRoy, S., Sarkar, D., R Core Team, 2014. Nlme: Linear and nonlinear mixed effects models. Version 3.1-117.
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. Plink: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team, 2014. R: A Language and Environment for Statistical Computing.
- Radivojac P., Clark W.T., Oron T.R., Schnoes A.M., Wittkop T., Sokolov A., Graim K., Funk C., Verspoor K., Ben-Hur A., Pandey G., Yunes J.M., Talwalkar A.S., Repo S., Souza M.L., Piovesan D., Casadio R., Wang Z., Cheng J., Fang H., Gough J., Koskinen P., Toronen P., Nokso-Koivisto J., Holm L., Cozzetto D., Buchan D.W., Bryson K., Jones D.T., Limaye B., Inamdar H., Datta A., Manjari S.K., Joshi R., Chitale M., Kihara D., Lisewski A.M., Erdin S., Venner E., Lichtarge O., Rentzsch R., Yang H., Romero A.E., Bhat P., Paccanaro A., Hamp T., Kassner R., Seemayer S., Vicedo E., Schaefer C., Achten D., Auer F., Boehm A., Braun T., Hecht M., Heron M., Honigschmid P., Hopf T.A., Kaufmann S., Kiening M., Krompass D., Landerer C., Mahlich Y., Roos M., Bjorne J., Salakoski T., Wong A., Shatkay H., Gatzmann F., Sommer I., Wass M.N., Sternberg M.J., Skunca N., Supek F., Bosnjak M., Panov P., Dzeroski S., Smuc T., Kourmpetis Y.A., van Dijk A.D., ter Braak C.J., Zhou Y., Gong Q., Dong X., Tian W., Falda M., Fontana P., Lavezzo E., Di Camillo B., Toppo S., Lan L., Djuric N., Guo Y., Vucetic S., Bairoch A., Linial M., Babbitt P.C., Brenner S.E., Orengo C., Rost B., Mooney S.D., Friedberg I. A large-scale evaluation of computational protein function prediction. Nat. Methods. 2013;10:221–227. doi: 10.1038/nmeth.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rank N.E., Bruce D.A., McMillan D.M., Barclay C., Dahlhoff E.P. Phosphoglucose isomerase genotype affects running speed and heat shock protein expression after exposure to extreme temperatures in a montane willow beetle. J. Exp. Biol. 2007;210:750–764. doi: 10.1242/jeb.02695. [DOI] [PubMed] [Google Scholar]
- Reinhardt J.A., Kolaczkowski B., Jones C.D., Begun D.J., Kern A.D. Parallel geographic variation in Drosophila melanogaster. Genetics. 2014;197:361–373. doi: 10.1534/genetics.114.161463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S.L. Between genotype and phenotype: protein chaperones and evolvability. Nat. Rev. Genet. 2003;4:263–274. doi: 10.1038/nrg1041. [DOI] [PubMed] [Google Scholar]
- Saastamoinen M. Life-history, genotypic, and environmental correlates of clutch size in the Glanville fritillary butterfly. Ecol. Entomol. 2007;32:235–242. [Google Scholar]
- Saastamoinen M., Hanski I. Genotypic and environmental effects on flight activity and oviposition in the Glanville fritillary butterfly. Am. Nat. 2008;171:E701–E712. doi: 10.1086/587531. [DOI] [PubMed] [Google Scholar]
- Saastamoinen M., Ikonen S., Hanski I. Significant effects of pgi genotype and body reserves on lifespan in the Glanville fritillary butterfly. Proc. R. Soc. London, Ser. B. 2009;276:1313–1322. doi: 10.1098/rspb.2008.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schippers M.P., Dukas R., McClelland G.B. Lifetime- and caste-specific changes in flight metabolic rate and muscle biochemistry of honeybees, Apis mellifera. J. Comp. Physiol. B: Biochem. Syst. Environ. Physiol. 2010;180:45–55. doi: 10.1007/s00360-009-0386-9. [DOI] [PubMed] [Google Scholar]
- Schippers M.P., Dukas R., Smith R.W., Wang J., Smolen K., McClelland G.B. Lifetime performance in foraging honeybees: behaviour and physiology. J. Exp. Biol. 2006;209:3828–3836. doi: 10.1242/jeb.02450. [DOI] [PubMed] [Google Scholar]
- Sezgin E., Duvernell D.D., Matzkin L.M., Duan Y.H., Zhu C.T., Verrelli B.C., Eanes W.F. Single-locus latitudinal clines and their relationship to temperate adaptation in metabolic genes and derived alleles in Drosophila melanogaster. Genetics. 2004;168:923–931. doi: 10.1534/genetics.104.027649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham P.C., Purcell S., Cherny S.S., Abecasis G.R. Powerful regression-based quantitative-trait linkage analysis of general pedigrees. Am. J. Hum. Genet. 2002;71:238–253. doi: 10.1086/341560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skandalis D.A., Roy C., Darveau C.A. Behavioural, morphological, and metabolic maturation of newly emerged adult workers of the bumblebee, Bombus impatiens. J. Insect Physiol. 2011;57:704–711. doi: 10.1016/j.jinsphys.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Sørensen J.G., Kristensen T.N., Loeschcke V. The evolutionary and ecological role of heat shock proteins. Ecol. Lett. 2003;6:1025–1037. [Google Scholar]
- Watt W.B. Adaptive significance of pigment polymorphisms in Colias butterflies. I. Variation of melanin pigment in relation to thermoregulation. Evolution. 1968;22:437–458. doi: 10.1111/j.1558-5646.1968.tb03985.x. [DOI] [PubMed] [Google Scholar]
- Watt W.B. Adaptation at specific loci. I. Natural selection on phosphoglucose isomerase of Colias butterflies – biochemical and population aspects. Genetics. 1977;87:177–194. doi: 10.1093/genetics/87.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt W.B., Wheat C.W., Meyer E.H., Martin J.F. Adaptation at specific loci. VII. Natural selection, dispersal and the diversity of molecular-functional variation patterns among butterfly species complexes (Colias: Lepidoptera, pieridae) Mol. Ecol. 2003;12:1265–1275. doi: 10.1046/j.1365-294x.2003.01804.x. [DOI] [PubMed] [Google Scholar]
- Wheat C.W., Fescemyer H.W., Kvist J., Tas E., Vera J.C., Frilander M.J., Hanski I., Marden J.H. Functional genomics of life history variation in a butterfly metapopulation. Mol. Ecol. 2011;20:1813–1828. doi: 10.1111/j.1365-294X.2011.05062.x. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Ersoz E., Lai C.Q., Todhunter R.J., Tiwari H.K., Gore M.A., Bradbury P.J., Yu J., Arnett D.K., Ordovas J.M., Buckler E.S. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010;42:355–360. doi: 10.1038/ng.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Xue J., Lai J.C., Schork N.J., White K.P., Haddad G.G. Mechanisms underlying hypoxia tolerance in Drosophila melanogaster: hairy as a metabolic switch. PLoS Genet. 2008;4:e1000221. doi: 10.1371/journal.pgen.1000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables S1–S5.