Abstract
Class I phosphoinositide 3-kinases (PI3Ks) are important regulators of neutrophil migration in response to a range of chemoattractants. Their primary lipid products PtdIns(3,4,5)P3 and PtdIns(3,4)P2 preferentially accumulate near to the leading edge of migrating cells and are thought to act as an important cue organizing molecular and morphological polarization. We have investigated the distribution and accumulation of these lipids independently in mouse neutrophils using eGFP-PH reportersand electron microscopy (EM).
We found that authentic mouse neutrophils rapidly polarized their Class I PI3K signalling, as read-out by eGFP-PH reporters, both at the up-gradient leading edge in response to local stimulation with fMLP as well as spontaneously and randomly in response to uniform stimulation. EM studies revealed these events occurred at the plasma membrane, were dominated by accumulation of PtdIns(3,4,5)P3, but not PtdIns(3,4)P2, and were dependent on PI3Kγ and its upstream activation by both Ras and Gβγs.
Keywords: PI3K, Neutrophil, Polarization
Abbreviations: PI3K, Phosphoinositide 3-kinase; eGFP, enhanced Green Fluorescent Protein; PH, Pleckstrin homology; EM, Electron Microscopy; fMLP, formylated-Met-Leu-Phe; GPCR, G-Protein Coupled Receptor; PKB, Protein Kinase B (also called Akt); TAPP-1, TAndem PH domain containing Protein-1
1. Introduction
Neutrophils are key players in the innate anti-bacterial and fungal defence mechanisms of mammals. They are capable of exiting the circulation in the proximity of local infections or tissue damage and chemotaxing towards the epicentre of the inflammatory response. These processes ready the neutrophils to phagocytose and destroy pathogens or dead cells and hence contribute to the resolution of the problem. Conversely, a number of inflammatory diseases are, in part, driven by excessive, inappropriate neutrophil infiltration (Cowburn et al., 2008, Lee et al., 2003 {Cowburn et al., 2008 #776).
Many further cell types can chemotax and the process is of very wide biological importance. Neutrophil chemotaxis towards inflammatory ligands for GPCRs, such as fMLP, can be studied in vitro and is amongst the most efficient in terms of cell speed and directionality and as a result has been subject to intense investigation aimed at defining the underlying molecular mechanisms and principles. This work has identified a large number of intracellular signals and/or proteins upon which neutrophil chemotaxis can depend. Amongst these phosphoinositide 3-kinases (PI3Ks) (Hirsch et al., 2000, Li et al., 2000, Sasaki et al., 2000) and Rac-family GTPases (Roberts et al., 1999, Servant et al., 2000, Weiner et al., 2002) have been implicated repeatedly but other key regulators include Ca2+ (Evans and Falke, 2007) and p38 MAPKs (Heit et al., 2002). Although PtdIns5P has been implicated in cell migration (Viaud et al., 2014 #3612) there is no evidence establishing that it has a role in neutrophil chemokinesis (Bulley et al., 2015).
Many reports have contributed to a picture of chemotaxis in which Class I PI3K signalling takes a central role (Meili et al., 1999, Parent et al., 1998, Servant et al., 2000). Class I PI3Ks, activated in response to fMLP and drive accumulation of PtdIns(3,4,5)P3 and PtdIns(3,4)P2 at the cell surface (Stephens et al., 1991, Traynor-Kaplan et al., 1988). This has fundamental event has been described using a variety of approaches to measure phosphoinositides, including relatively recently evolved mass-spec-based methods that can resolve different molecular species of PtdIns(3,4,5)P3 (Kielkowska et al., 2014). The increase in PtdIns(3,4,5)P3 and PtdIns(3,4)P2 in the peripheral membrane drives translocation and activation of PI3K effectors including PKB (AKT) and important modulators of the cytoskeleton, such as ARAP3 and DOCK2 (Krugmann et al., 2002, Kunisaki et al., 2006). Crucially, this process can polarise spontaneously, in the presence of uniform fMLP, to generate a leading-edge enriched in PtdIns(3,4)P2/(3,4,5)P3 (Servant et al., 2000) and a range of other signalling molecules (some targeted by PI3K activity directly) and of characteristic, dynamic morphology. This polarization is important for the neutrophils to move efficiently and hence for chemotaxis, and it is substantially, but not entirely, dependent on Class I PI3K signalling (Ferguson et al., 2007, Niggli and Keller, 1997, Nishio et al., 2007). Remarkably, by-passing receptor activation by direct, uniform shuttling of heterologous PtdIns(3,4,5)P3 (Derman et al., 1997, Niggli, 2000, Weiner et al., 2002) or homogenous “chemical” activation of a Class I PI3K construct in neutrophil-like cells (Inoue and Meyer, 2008) leads to spontaneous polarization of both their morphology and endogenous PtdIns(3,4)P2/(3,4,5)P3, suggesting that PI3K signalling is a core player in an auto-catalytic symmetry-breaking circuit. It has been argued this is a keystone, ubiquitous principle in Eukaryotic chemotaxis. Several issues remain unclear. Firstly, what happens to PtdIns(3,4)P2 dynamics? Does PtdIns(3,4)P2 simply “follow” PtdIns(3,4,5)P3 signals in living neutrophils or does it show evidence of independent regulation and hence function. Secondly, what is the precise cellular location of the pools of PtdIns(3,4,5)P3 and/or PtdIns(3,4)P2? Are these lipids in the plasma membrane or peripheral endo-membranes? Given the morphological characteristics of neutrophils, with very thin layers of cytoplasm unevenly spread around large lobed nuclei, this has remained unclear.
In this work we have addressed the identity and EM-level localization of the lipid products of PI3K activity accumulated in migrating mouse neutrophils.
2. Methods and materials
2.1. Materials
All materials used were of the lowest endotoxin level available and were purchased from Sigma-Aldrich unless stated otherwise. PI3K inhibitors have been previously described (Condliffe et al., 2005) (Condliffe et al., 2005).
2.2. Mouse strains
The eGFP-PH-PKB mouse strain has been previously described (Nishio et al., 2007) and was bred against p110γ−/− (Hirsch et al., 2000); p101−/− (Suire et al., 2006) and p110γDASAA/DASAA (Suire et al., 2006) mice. Animals were housed in high health status isolators.
2.3. Mouse neutrophil isolation
Was essentially as described (Ferguson et al., 2007). Neutrophil purity was assessed by cytospin.
3. Chemotaxis assays
3.1. Micropipette chemotaxis assay
Were conducted as described (Ferguson et al., 2007), micropipettes contained 10 μM fMLP. Wortmannin (250 nM) was applied for 10 min while neutrophils settled on the coverslip, prior to stimulation.
3.2. Bath application fMLP- chemokinesis assay
Neutrophils (1.5 ml, 1.25 × 105/ml) were settled onto a glass coverslip (22 mm, thickness no 1.5, VWR) held in an imaging ring at 37 °C for 5 min fMLP or peroxy-vanadate (final concentrations 1 μM or 12 mM) were added directly onto cells.
3.3. Assessing eGFP-PH-PKB reporter distribution
Polar plots were derived from movies of eGFP-PH-PKB neutrophil chemotaxis (Anagraph, Simon Andrews). QuimP software ((Dormann et al., 2002), Garching Innovation) was used to determine the ratio of membrane intensity to mean cytoplasmic intensity in every frame of the movie at 100 sites around the periphery of the neutrophil. The ratios from individual cells were translated onto a common pseudo-coloured scale where red indicates a high ratio and hence reporter concentration. Each individual cells intensity measurements for one frame were mapped onto a circle and sequential frames were then combined concentrically, with the first frame at the centre. Plots were aligned so that the fMLP containing micropipette (originally 20 μm from the cell) was positioned vertically below.
Production of Retrovirus, harvesting of foetal liver Cells, transduction of foetal liver cells and the injection of foetal liver cells into recipient mice.
3.3.1. Harvesting foetal liver cells
Following timed matings pregnant mice were sacrificed at day 14.5 and foetal liver cells were harvested in Stempro-34 serum-free medium (Invitrogen) (supplemented with provided StemPro-nutrient supplement and 1% penicillin/streptomycin/amphotericin B). Cells were collected, resuspended at 1.7 × 107/ml in FBS +10% DMSO and slowly frozen to −80 °C.
3.3.2. Transfection of Plat-E cells to produce retrovirus
Plat-E cells were cultured in DMEM high glucose with l-glutamine and pyruvate (Gibco) (supplemented with 10% FBS and 1% penicillin/streptomycin) at 37 °C with 5% CO2. Plat-E cells were transfected using Lipofectamine 2000 (Invitrogen, as per manufacturer's instructions). Briefly, retroviral transfer DNA (16.8 μg) and Lipofectamine 2000 (50.4 μl) were incubated in 4.2 ml of Optimem-I with Glutamax (Invitrogen) for 30 min at RT. After incubation Plat-E cells were resuspended at 1.19 × 106 in 14 ml of DMEM high glucose with l-glutamine and pyruvate (supplemented with 10% FBS) and mixed with the Lipofectamine 2000/DNA mix. Cells were plated into a 14 cm tissue culture dish and incubated at 37 °C with 5% CO2. After 24 h media was replaced with 9 ml of complete Stempro-34 media and incubated for a further 24 h. Viral supernatant was harvested, centrifuged (3000× g, 5 min) and 0.45 μm filtered (Millipore). Supernatant was then slowly frozen on dry ice before being stored at −80 °C.
3.3.3. Reconstitution using foetal liver
Foetal liver cells were rapidly thawed and resuspended at 5 × 105/ml in complete Stempro-34 serum-free medium (Invitrogen) containing a cocktail of cytokines (mSCF final concentration 100 ng/ml; TPO 100 ng/ml; hIL-6 1 ng/ml; mIL3 6 μg/ml; and Flt3L 20 ng/ml) (Peprotech). Cells were cultured for 24 h at 37 °C with 6% CO2 in untreated tissue culture plates before being harvested, resuspended at 5 × 105/ml in viral supernatant supplemented with cytokines (see above) and incubated (3 ml/well) in Retronectin (Takara Bio Inc) coated plates (12.5 μg/ml overnight at 4 °C, unbound sites were blocked with 2% BSA for 30 min at RT) for 17 h at 37 °C with 6% CO2.
Cells were harvested (adherent cells were harvested using 0.25 ml of PBS-based cell dissociation buffer, Gibco, Invitrogen), pooled, washed in PBS and resuspended at 5 × 106/ml in PBS with 10% FBS. The cell suspension (300 μl) was then injected into the tail vein of a Bl-6 mouse 24 h post-irradiation (2 × 500 rad (5Gy) separated over 3 h). To minimize infection mice were housed under IVC conditions and kept on 80 mg/ml neomycin for 4 weeks post irradiation. Four weeks after injection blood was collected via tail bleeds and reconstitution was assessed using FACS.
3.3.4. eGFP-PH-PKB neutrophil sample preparation for electron microscopy (EM)
Cells were fixed with 8% paraformaldehyde/0.2% gluteraldehyde in 0.2 M PIPES pH 7.2, incubated and pelleted.
Neutrophils were pre-incubated with wortmannin (100 nM) or vehicle control (DMSO, 0.1%) for 5 min at 37 °C then stimulated with fMLP (10 μM). Cells were then gently vortexed and fixed with an equal volume of 8% paraformaldehyde/0.2% gluteraldehyde in 0.2 M PIPES pH 7.2. Cells were incubated at RT for 5 min, micro-centrifuged at 15,000× g for 10 min at 4 °C and then incubated at RT for 45 min, fixative was aspirated and cell pellets washed twice with PBS and stored at 4 °C.
Neutrophil samples were embedded in pig skin gelatine (PSG) by aspirating PBS from the pellets, adding 500 μl PSG, incubating for 1 h at 37 °C and then cooling on ice for 20 min, all following steps were carried out at 4 °C. Pellets were removed from the Eppendorf tube, cut into smaller pieces and transferred to an Eppendorf containing 2.1 M sucrose/PBS and incubated overnight. The following day cell blocks were prepared by cutting pellets into smaller fragments and mounting onto iron panel pins (stored in acetone). Excess sucrose was removed and pyramid shaped blocks were formed with filter paper before they were frozen and stored in N2 (l). Ultrathin cryosections (80 nm) were cut from frozen blocks on a Leica EM FCS ultracut UCT microtome at −110 °C using a diamond knife. Cryosections were collected from the microtome on a frozen droplet of 2.3 M sucrose/2% methylcellulose (1:1) on a wire loop and allowed to thaw before being transferred onto Pioloform (polyvinyl butyral)/carbon-coated 100 mesh hexagonal support grids. The grids were transferred to 0.1 M NH4Cl (in PBS) for 10 min before washing with PBS and blocking in 0.5% fish skin gelatine (FSG) in PBS for 10 min. After incubation with anti-GFP for 1 h the cryosections were washed with PBS. Bound probe was detected by incubation with 10 μl of 10 nm-particle-diameter Protein A-Gold (1:50 in FSG) for 20 min. Grids were washed in PBS and distilled H2O before being coated with 2% methyl cellulose/3% uranyly acetate. Grids were dried overnight and sections viewed on a JEOL 1200EX electron microscope at 80 kV.
4. Results
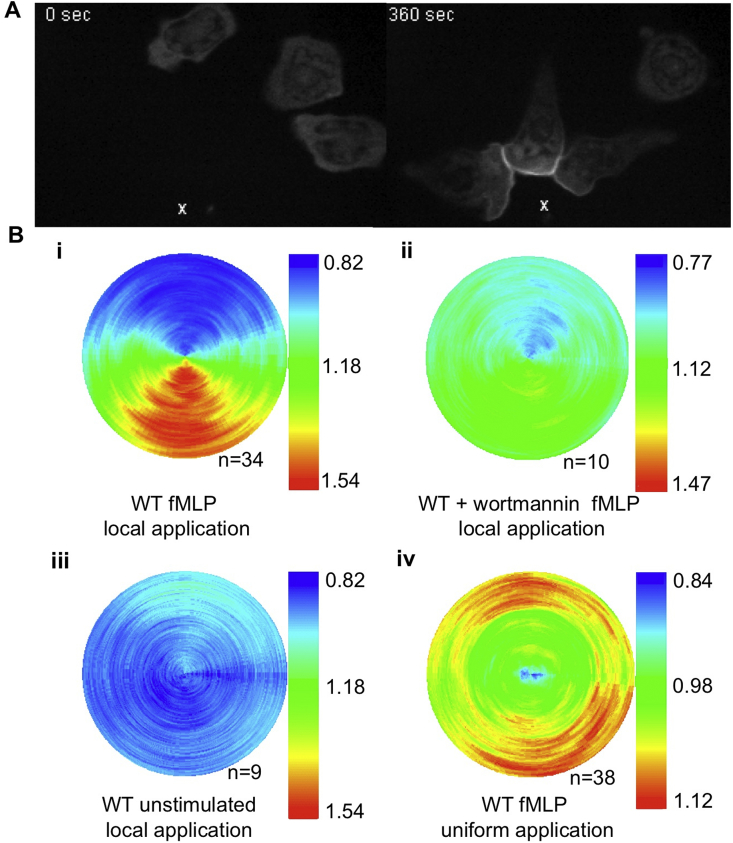
We, and others, have reported that a transgenically-expressed eGFP-PH-PKB reporter preferentially accumulated at the leading edge of mouse neutrophils migrating towards formylated bacterial peptides released from a micropipette (Ferguson et al., 2007, Nishio et al., 2007). We used cell-tracking and software designed to estimate the extent and vector of translocation of the reporter to the periphery, averaged for many cells, in comparison to DiIC16 membrane probes in the same cells. It was clear that the PH domain reporter, and hence PtdIns(3,4)P2/(3,4,5)P3, was concentrated in membrane at the leading edge, in a manner dependent on PI3Kγ. In Fig. 1 we demonstrate that the sustained, gradient-biased accumulation of the reporter at the leading edge is dependent on gradient-based stimulation and blocked by the PI3K-inhibitor wortmannin.
Fig. 1.
Localising Class I PI3K activity in neutrophils with a PH-domain reporter. (A) An fMLP gradient was generated with a micropipette positioned about 20 μm from eGFP-PH-PKB-expressing neutrophils (all of the figures are orientated so that this was vertically below the cell or polar plot), confocal imaging revealed, as described previously, accumulation of the reporter at the up-gradient leading edge. (B) Polar plots showed that, averaged across many movies (n = number of independent movies) and all of their component frames, the reporter accumulated at the leading, up-gradient edge when the micropipette contained fMLP (i), but not when the cells were pre-treated with wortmannin (ii, 250 nM), or the micropipette contained vehicle alone (iii). Uniform stimulation with fMLP resulted in spatially random translocation of the reporter (iv).
Expression of membrane-targeted-FP (fluorescent protein) constructs has indicated that membrane accumulation near to the leading edge can contribute to the apparent polarization of PH domain reporters (Dewitt et al., 2009, Onsum et al., 2006). We tested to what extent phenomenon impacted our observations by co-expressing of mCherry-CAAX with eGFP-PH-PKB in mouse neutrophils and then analysing their distribution during chemotaxis towards formylated peptides (Fig. 2A). Consistent with others work, the PH domain reporter was very significantly more polarized. Comparison of the distributions of the DiIC16 and mCherry-CAAX distributions indicated that the FP construct became more polarized. This could have been due to DiIC16 being excluded from, and/or mCherry-CAAX being concentrated at, the leading edge. As mCherry-CAAX is based upon a “biologically meaningful” targeting motif we assumed it represented an upper limit for potential morphological artifacts.
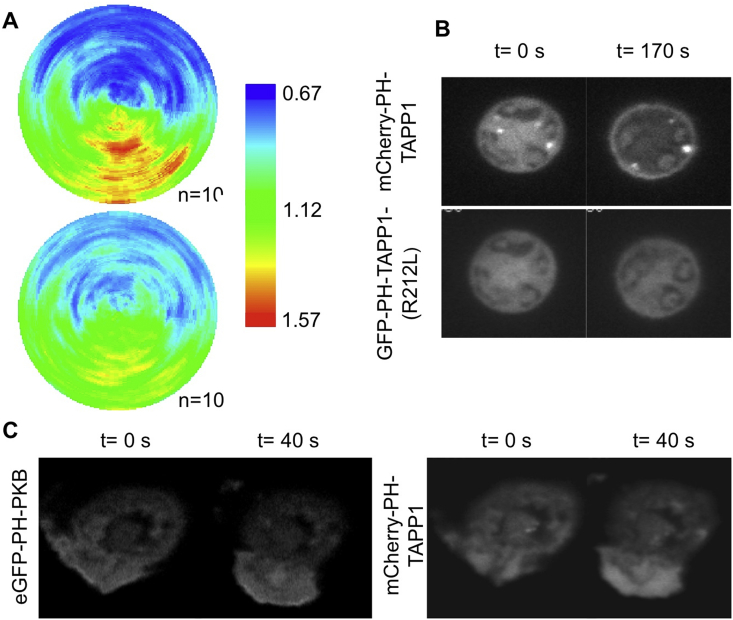
Fig. 2.
The lipid signals and morphological factors underlying polarised accumulation of PH-PKB reporters. (A) An fMLP-containing micropipette was used to stimulate neutrophils co-expressing eGFP-PH-PKB and mCherry-CAAX, polar plots were created for each reporter (eGFP-PH – upper). (B) Validation of a PtdIns(3,4)P2 reporter in neutrophils. Confocal images of neutrophils co-expressing mCherry-PH-TAPP-1 (upper) and eGFP-(R212L)PH-TAPP-1 (lower) stimulated with peroxy-vanadate for 0 or 170 s (representative of 4, see Fig. 3 for more frames from this movie and further controls). (C) The eGFP-PH-PKB construct reported changes in PtdIns(3,4,5)P3 during neutrophil chemotaxis. An fMLP-containing micropipette (position out of figure directly down) was used to stimulate neutrophils co-expressing eGFP-PH-PKB (left) and mCherry-PH-TAPP-1 (right) (representative of 3).
We attempted to identify the phosphoinositides that had been detected by the eGFP-PH-PKB reporter. The PH domain of PKB can bind with similar affinity to PtdIns(3,4)P2 and PtdIns(3,4,5)P3 and could have been responding to either lipid. To resolve this problem we made use of a PH-TAPP-1 construct demonstrated to be highly selective for PtdIns(3,4)P2 (Dowler et al., 2000, Watt et al., 2004). Co-expression of mCherry-PH-TAPP-1 with eGFP-PH-R212L-TAPP-1 (a lipid-binding mutant) in peroxy-vanadate-stimulated mouse neutrophils (this treatment drives substantial accumulation of PtdIns(3,4)P2 but not PtdIns(3,4,5)P3 (Van der Kaay et al., 1999)) showed the wild-type reporter specifically detected PtdIns(3,4)P2 accumulation (Fig. 2B). Co-expression of eGFP-PH-PKB and mCherry-PH-TAPP-1 in peroxy-vanadate-stimulated mouse neutrophils showed that both reporters responded similarly, both in terms of kinetics and the extent of translocation to the periphery; indicating that the dynamic sensitivity of the mCherry-PH-TAPP-1 reporter for PtdIns(3,4)P2 is similar to that of eGFP-PH-PKB for PtdIns(3,4,5)P3 (Fig. 3C). We found that fMLP-stimulation of cells co-expressing eGFP-PH-PKB and mCherry-PH-TAPP-1 repeatedly resulted in clear translocation of the former, but not the latter, reporter to the leading edge (Fig. 2C). We concluded that, despite the known role of SHIPI in mouse neutrophil chemotaxis (Nishio et al., 2007), the levels of PtdIns(3,4)P2 remain an order of magnitude lower than PtdIns(3,4,5)P3 at the leading edge and that our, and many other workers, results with a eGFP-PH-PKB reporter reflect changes in PtdIns(3,4,5)P3 density at the cell surface. This is consistent with measurements of these lipids by metabolic labelling with [32P]-Pi of fMLP-stimulated, non-adherent mouse neutrophils (Condliffe et al., 2005).
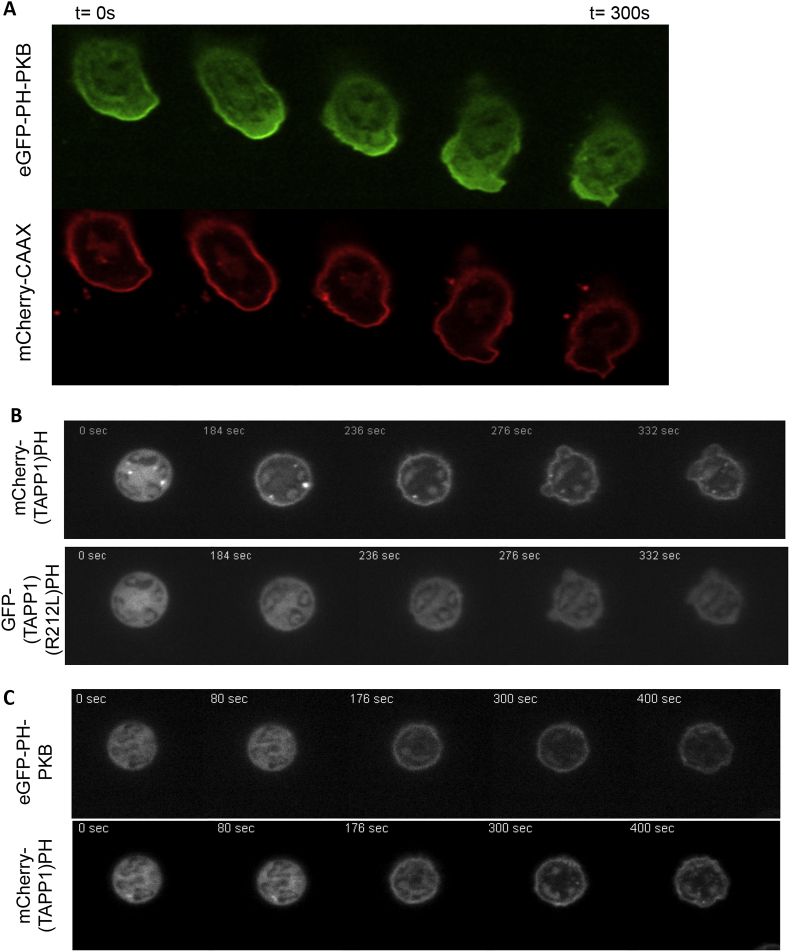
Fig. 3.
Factors underlying polarised accumulation of PH-PKB reporters. (A) An fMLP-containing micropipette was used to stimulate neutrophils co-expressing eGFP-PH-PKB (upper) and mCherry-CAAX. A representative series of frames is shown from movies analysed to produce polar plots shown in Fig. 2A and 3B. (B) Validation of a PtdIns(3,4)P2 reporter in neutrophils. Confocal images of neutrophils co-expressing mCherry-PH-TAPP-1 (upper panels) and eGFP-(R212L)PH-TAPP-1 (lower panels), a mutant unable to bind to PtdIns(3,4)P2, stimulated with peroxy-vanadate (representative of 4). (frames 1 and 2 of the series are shown in Fig. 2B) (C) Comparison of the dynamic sensitivities of the TAPP-1 and PKB-derived reporters. Neutrophils co-expressing eGFP-PH-PKB (upper) and mCherry-PH-PKB (lower) (representative of 3) were stimulated with peroxy-vanadate.
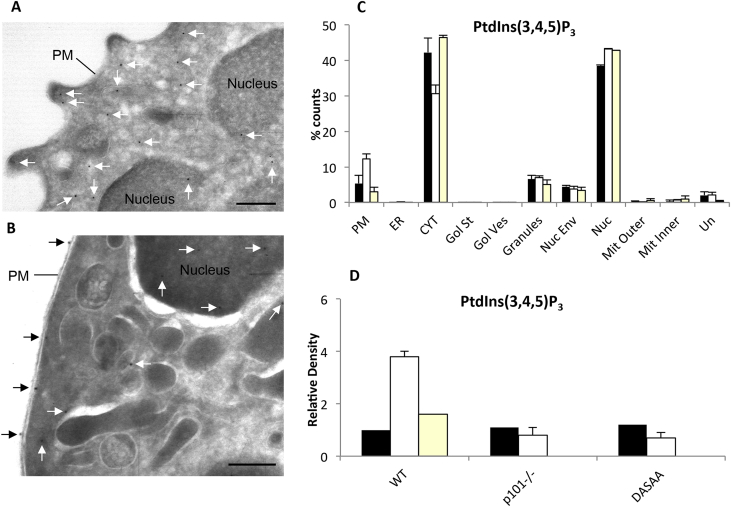
The precision with which we could locate the eGFP-PH-PKB reporter, and hence the endogenous PtdIns(3,4,5)P3, was limited by the resolving power of light microscopy and could only be stated to be peripheral. To advance this problem we used EM techniques. To achieve this we rapidly fixed, froze and cryo-sectioned eGFP-probe-expressing neutrophils before applying an anti-GFP antibody and gold-labelled secondary antibodies. The approach avoided the challenges associated with detection of endogenous phosphoinositides by application of lipid-binding probes after fixation (Lindsay et al., 2006, Watt et al., 2004, Watt et al., 2002). We found that upon fMLP-stimulation there was a reduction in the intensity of staining of the cytoplasmic compartment (Fig. 4A and C) and a large increase in the density of decoration of the plasma membrane (Fig. 4B and C), but not other membrane fractions (Fig. 4C), that was inhibited by wortmannin, genetic loss of p101, the major Gβγ-sensitive p110γ adaptor in neutrophils, or knock-in of a Ras-insensitive version of p110γ, p110γDASAA (by creating the lines eGFP-PH-PKB x p101−/− and eGFP-PH-PKB x p110γDASAA/DASAA; Fig. 4C and D). This is entirely consistent with the wortmannin-sensitivity of PI3Kγ and known roles of Gβγs and Ras in its regulation and further validates the veracity of this assay.
Fig. 4.
EM analysis of the localisation of eGFP-PH-PKB reporters in neutrophils. (A) Non-adherent eGFP-PH-PKB mouse neutrophils were stimulated with vehicle alone (or in panel (B) with fMLP (10 μM)) for 1 min, fixed, labelled with anti-GFP and protein-A-colloidal gold before imaging by electron microscopy. Arrows indicate the presence of gold particles and scale bars represents 200 nm (C) Non-adherent neutrophils were pretreated with wortmannin (yellow bars, 250 nM, 10 min) or vehicle (black and white bars) then stimulated with fMLP (white or yellow bars, 10 μM, 1 min) or vehicle alone (black bars), fixed, labelled with anti-GFP and protein-A-colloidal gold and visualised by EM. The number of gold particles on sampling lines were counted and binned (as % of total) based on cell morphology/organelle (PM = Plasma Membrane; ER = Endoplasmic Reticulum; CYT = Cytoplasm; Gol St = Golgi Structures; Gol Ves = Golgi Vesicles; Nuc Env = Nuclear Envelope; Nuc = Nucleus; Mit Outer = Outer Mitochondrial membrane; Mit Inner = Inner Mitochondrial membrane; Un = unassigned). Means (±range) from 2 experiments based on counting greater than 200 grids are presented. (D) Genetic loss of either Gβγ-sensitivity of PI3Kγ (p101−/−) or the sensitivity of p110γ to Ras (DASAA-mutant knock-in p110γDASAA/DASAA) blocks the fMLP-induced rise in eGFP-PH-PKB at the plasma membrane, assay and replication as in (C). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
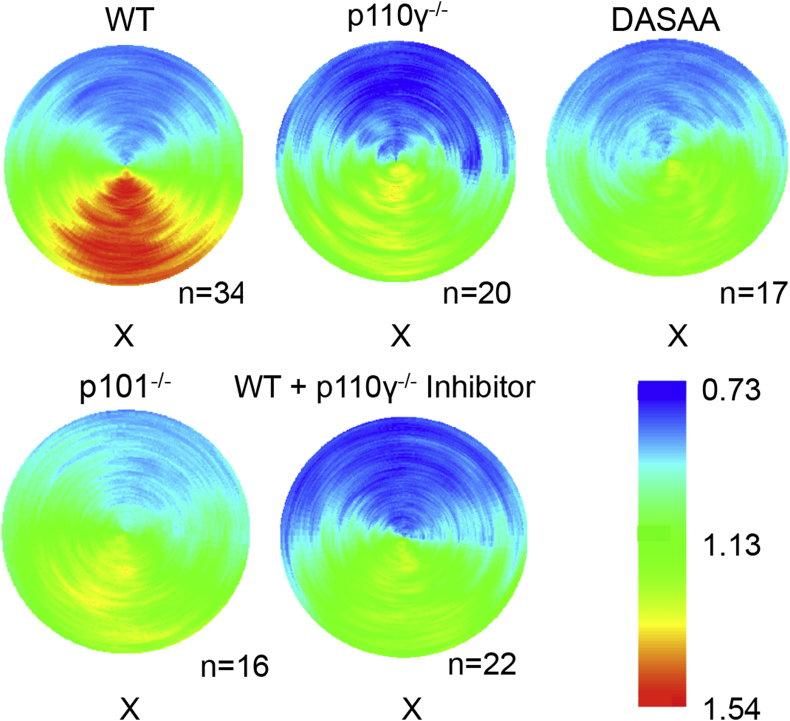
PI3Kγ can be regulated by both Gβγs and GTP-Ras in mouse neutrophils (Suire et al., 2006). We sought to test the idea that these inputs might contribute differentially to driving PtdIns(3,4,5)P3 accumulation at the leading edge using the mouse strains described above; eGFP-PH-PKB x p101−/− and eGFP-PH-PKB x p110γDASAA/DASAA. The distribution eGFP reporter was analysed in live cells chemotaxing towards fMLP using a spinning disc confocal microscope. We found that loss of p101, genetic blockade of Ras-regulation of PI3Kγ and transient chemical inhibition of PI3Kγ similarly and substantially inhibited accumulation of PtdIns(3,4,5)P3 at the leading edge (Fig. 5). This suggests both Gβγs and GTP-Ras are driving PI3Kγ in its roles in the leading edge of migrating neutrophils.
Fig. 5.
Both Gβγ and Ras regulation of PI3Kγ are crucial for accumulation of PtdIns(3,4,5)P3 at the leading, up-gradient edge. Polar plots of neutrophils, responding to an fMLP-containing micropipette, which expressed eGFP-PH-PKB in different genetic backgrounds (x wild type (WT); x p110γ−/−; x p110γDASAA/DASAA; x p101−/−) or pre-treated with a p110γ inhibitor (AS252424; 10 μM, using x WT cells).
5. Discussion
Our results are broadly in keeping with the literature. We have used EM techniques to localize an endogenously-expressed eGFP-PH-PKB reporter. Our data indicated that in controls cells the reporter was in the cytoplasmic compartment. Following stimulation with fMLP the reporter localized to the plasma membrane, and not other membranes. We found no evidence for increased accumulation of the reporter in the nucleus following fMLP-stimulation of mouse neutrophils. Our results with reporters capable of sensing PtdIns(3,4)P2 and previous work measuring PtdIns(3,4)P2 accumulation both suggest that the eGFP-PH-PKB construct is being localized by interactions with PtdIns(3,4,5)P3. These data indicate PtdIns(3,4,5)P3 accumulates in the plasma membrane; they do not, however, demonstrate that PtdIns(3,4,5)P3 only rises in the plasma membrane. There is evidence that PH domains contain motifs that bind to specific types of cell membrane and that these interactions, in addition to interactions between phosphoinositides and the PH domain, are required to allow membrane recruitment (Hammond and Balla, 2015). As a result PtdIns(3,4,5)P3 could accumulate in membranes other than the plasma membrane but would not be sensed by PtdIns(3,4,5)P3-binding PH domains. It is unclear if domains that bind PtdIns(3,4,5)P3 that are not PH domains have the same properties.
The TAPP1 construct we used to sense PtdIns(3,4)P2 was capable of reporting an increase in PtdIns(3,4)P2 in peroxy-vanadate-stimulated neutrophils. We could not find any evidence of fMLP-induced localization of the reporter to the leading edge. These results do not allow us to conclude there is no PtdIns(3,4)P2 accumulation at the periphery of fMLP-stimulated neutrophils, indeed much evidence shows PtdIns(3,4)P2 does accumulate in stimulated neutrophils (Stephens et al., 1991, Traynor-Kaplan et al., 1989), but rather the concentrations achieved are insufficient to relocate a significant proportion of the reporter. As a result it is unclear if the distribution of that PtdIns(3,4)P2 had merely followed PtdIns(3,4,5)P3 or not.
There is evidence that p84-PI3Kγ drives accumulation of PtdIns(3,4,5)P3 and/or PtdIns(3,4)P2 in different near-plasma membrane locations to p101-PI3Kγ in mast cells (Bohnacker et al., 2009). We attempted to address this question in neutrophils. Neutrophils contain significant amounts of both p84 and p101-PI3Kγ complexes. In the experiments reported here we could find no evidence of PtdIns(3,4,5)P3 accumulation in the plasma membrane as detected by EM-analysis when neutrophils only contain p84-PI3Kγ (ie in p101−/− neutrophils) or in cells in which PI3Kγ could not be activated by Ras but could be activated via p101 Gβγs (p110γDASAA/DASAA). These results precluded an analysis of the above problem and are surprising as experiments measuring the amount of PtdIns(3,4,5)P3 in neutrophils of these genetic backgrounds indicated the responses to fMLP were only partially reduced and not abolished. Perhaps the EM-based methodology leads to a highly non-linear relationship between staining and PtdIns(3,4,5)P3 content or, alternatively, that a significant amount of the PtdIns(3,4,5)P3 we have measured is localized in a non-plasma membrane location that we cannot observe/recover. We also compared the localization of an eGFP-PH-PKB reporter in living p101−/− and p110γDASAA/DASAA-expressing neutrophils. We found no evidence that the reporter localized differently in these different genetic backgrounds although our result suggested both had reduced accumulation of PtdIns(3,4,5)P3 at the leading edge compared to wild-type neutrophils. These experiments, however, relied upon either light microscopy and hence had limited spatial resolution. Hence it remains possible that there are spatiotemporal differences in the PtdIns(3,4,5)P3 accumulated following Gβγ v Ras-activation of PI3Kγ but that we failed to detect them through limitations in our methodologies.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
BI funding; LN, BBSRC BB/B511900/1 (PhD studentship with Novartis); AD, BBSRC BB/F01582/1; TC, BBSRC BB/B5119001/1 (PhD studentship with UCB); GJF, BBSRC 8443; HG, BBSRC BB/D013593/1, BB/I008209/1.
LN, AD, TC, HG, YL, JLJ, performed experiments; LN, JL, SA, ASP, GJF, PTH, LS, analysed data; CD, TB, PF, TS, OR, SW, SS supplied important resources; PTH, LN, LS wrote the manuscript; PTH, LS designed research.
References
- Bohnacker T., Marone R., Collmann E., Calvez R., Hirsch E., Wymann M.P. PI3Kgamma adaptor subunits define coupling to degranulation and cell motility by distinct PtdIns(3,4,5)P3 pools in mast cells. Sci. Signal. 2009;2 doi: 10.1126/scisignal.2000259. ra27. [DOI] [PubMed] [Google Scholar]
- Bulley S.J., Clarke J.H., Droubi A., Giudici M.L., Irvine R.F. Exploring phosphatidylinositol 5-phosphate 4-kinase function. Adv. Biol. Regul. 2015;57:193–202. doi: 10.1016/j.jbior.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condliffe A.M., Davidson K., Anderson K.E., Ellson C.D., Crabbe T., Okkenhaug K. Sequential activation of class IB and class IA PI3K is important for the primed respiratory burst of human but not murine neutrophils. Blood. 2005;106:1432–1440. doi: 10.1182/blood-2005-03-0944. [DOI] [PubMed] [Google Scholar]
- Cowburn A.S., Condliffe A.M., Farahi N., Summers C., Chilvers E.R. Advances in neutrophil biology: clinical implications. Chest. 2008;134:606–612. doi: 10.1378/chest.08-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derman M.P., Toker A., Hartwig J.H., Spokes K., Falck J.R., Chen C.S. The lipid products of phosphoinositide 3-kinase increase cell motility through protein kinase C. J. Biol. Chem. 1997;272:6465–6470. doi: 10.1074/jbc.272.10.6465. [DOI] [PubMed] [Google Scholar]
- Dewitt S., Darley R.L., Hallett M.B. Translocation or just location? Pseudopodia affect fluorescent signals. J. Cell Biol. 2009;184:197–203. doi: 10.1083/jcb.200806047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D.L., Thorsten, Weijer Cornelis J., Bretschneider Till. Simultaneous quantitation of cell motility ad protein membrane association using active contours. Cell Motil. Cytoskelet. 2002;52:221–230. doi: 10.1002/cm.10048. [DOI] [PubMed] [Google Scholar]
- Dowler S., Currie R.A., Campbell D.G., Deak M., Kular G., Downes C.P. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem. J. 2000;351:19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J.H., Falke J.J. Ca2+ influx is an essential component of the positive-feedback loop that maintains leading-edge structure and activity in macrophages. Proc. Natl. Acad. Sci. U. S. A. 2007;104:16176–16181. doi: 10.1073/pnas.0707719104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson G.J., Milne L., Kulkarni S., Sasaki T., Walker S., Andrews S. PI(3)Kgamma has an important context-dependent role in neutrophil chemokinesis. Nat. Cell Biol. 2007;9:86–91. doi: 10.1038/ncb1517. [DOI] [PubMed] [Google Scholar]
- Hammond G.R., Balla T. Polyphosphoinositide binding domains: key to inositol lipid biology. Biochim. Biophys. Acta. 2015;1851:746–758. doi: 10.1016/j.bbalip.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heit B., Tavener S., Raharjo E., Kubes P. An intracellular signaling hierarchy determines direction of migration in opposing chemotactic gradients. J. Cell Biol. 2002;159:91–102. doi: 10.1083/jcb.200202114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch E., Katanaev V.L., Garlanda C., Azzolino O., Pirola L., Silengo L. Central role for G protein-coupled phosphoinositide 3-kinase gamma in inflammation. Science. 2000;287:1049–1053. doi: 10.1126/science.287.5455.1049. [DOI] [PubMed] [Google Scholar]
- Inoue T., Meyer T. Synthetic activation of endogenous PI3K and Rac identifies an AND-gate switch for cell polarization and migration. PLoS One. 2008;3:e3068. doi: 10.1371/journal.pone.0003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielkowska A., Niewczas I., Anderson K.E., Durrant T.N., Clark J., Stephens L.R. A new approach to measuring phosphoinositides in cells by mass spectrometry. Adv. Biol. Regul. 2014;54C:131–141. doi: 10.1016/j.jbior.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Krugmann S., Anderson K.E., Ridley S.H., Risso N., McGregor A., Coadwell J. Identification of ARAP3, a novel PI3K effector regulating both Arf and Rho GTPases, by selective capture on phosphoinositide affinity matrices. Mol. Cell. 2002;9:95–108. doi: 10.1016/s1097-2765(02)00434-3. [DOI] [PubMed] [Google Scholar]
- Kunisaki Y., Nishikimi A., Tanaka Y., Takii R., Noda M., Inayoshi A. DOCK2 is a Rac activator that regulates motility and polarity during neutrophil chemotaxis. J. Cell Biol. 2006;174:647–652. doi: 10.1083/jcb.200602142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.L., Harrison R.E., Grinstein S. Phagocytosis by neutrophils. Microbes Infect. 2003;5:1299–1306. doi: 10.1016/j.micinf.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Li Z., Jiang H., Xie W., Zhang Z., Smrcka A.V., Wu D. Roles of PLC-beta2 and -beta3 and PI3Kgamma in chemoattractant-mediated signal transduction. Science. 2000;287:1046–1049. doi: 10.1126/science.287.5455.1046. [DOI] [PubMed] [Google Scholar]
- Lindsay Y., McCoull D., Davidson L., Leslie N.R., Fairservice A., Gray A. Localization of agonist-sensitive PtdIns(3,4,5)P3 reveals a nuclear pool that is insensitive to PTEN expression. J. Cell Sci. 2006;119:5160–5168. doi: 10.1242/jcs.000133. [DOI] [PubMed] [Google Scholar]
- Meili R., Ellsworth C., Lee S., Reddy T.B., Ma H., Firtel R.A. Chemoattractant-mediated transient activation and membrane localization of Akt/PKB is required for efficient chemotaxis to cAMP in Dictyostelium. Embo J. 1999;18:2092–2105. doi: 10.1093/emboj/18.8.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niggli V. A membrane-permeant ester of phosphatidylinositol 3,4, 5-trisphosphate (PIP(3)) is an activator of human neutrophil migration. FEBS Lett. 2000;473:217–221. doi: 10.1016/s0014-5793(00)01534-9. [DOI] [PubMed] [Google Scholar]
- Niggli V., Keller H. The phosphatidylinositol 3-kinase inhibitor wortmannin markedly reduces chemotactic peptide-induced locomotion and increases in cytoskeletal actin in human neutrophils. Eur. J. Pharmacol. 1997;335:43–52. doi: 10.1016/s0014-2999(97)01169-2. [DOI] [PubMed] [Google Scholar]
- Nishio M., Watanabe K., Sasaki J., Taya C., Takasuga S., Iizuka R. Control of cell polarity and motility by the PtdIns(3,4,5)P3 phosphatase SHIP1. Nat. Cell Biol. 2007;9:36–44. doi: 10.1038/ncb1515. [DOI] [PubMed] [Google Scholar]
- Onsum M.D., Wong K., Herzmark P., Bourne H.R., Arkin A.P. Morphology matters in immune cell chemotaxis: membrane asymmetry affects amplification. Phys. Biol. 2006;3:190–199. doi: 10.1088/1478-3975/3/3/004. [DOI] [PubMed] [Google Scholar]
- Parent C.A., Blacklock B.J., Froehlich W.M., Murphy D.B., Devreotes P.N. G protein signaling events are activated at the leading edge of chemotactic cells. Cell. 1998;95:81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- Roberts A.W., Kim C., Zhen L., Lowe J.B., Kapur R., Petryniak B. Deficiency of the hematopoietic cell-specific Rho family GTPase Rac2 is characterized by abnormalities in neutrophil function and host defense. Immunity. 1999;10:183–196. doi: 10.1016/s1074-7613(00)80019-9. [DOI] [PubMed] [Google Scholar]
- Sasaki T., Irie-Sasaki J., Jones R.G., Oliveira-dos-Santos A.J., Stanford W.L., Bolon B. Function of PI3Kgamma in thymocyte development, T cell activation, and neutrophil migration. Science. 2000;287:1040–1046. doi: 10.1126/science.287.5455.1040. [DOI] [PubMed] [Google Scholar]
- Servant G., Weiner O.D., Herzmark P., Balla T., Sedat J.W., Bourne H.R. Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science. 2000;287:1037–1040. doi: 10.1126/science.287.5455.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens L.R., Hughes K.T., Irvine R.F. Pathway of phosphatidylinositol(3,4,5)-trisphosphate synthesis in activated neutrophils. Nature. 1991;351:33–39. doi: 10.1038/351033a0. [DOI] [PubMed] [Google Scholar]
- Suire S., Condliffe A.M., Ferguson G.J., Ellson C.D., Guillou H., Davidson K. Gbetagammas and the Ras binding domain of p110gamma are both important regulators of PI(3)Kgamma signalling in neutrophils. Nat. Cell Biol. 2006;8:1303–1309. doi: 10.1038/ncb1494. [DOI] [PubMed] [Google Scholar]
- Traynor-Kaplan A.E., Harris A.L., Thompson B.L., Taylor P., Sklar L.A. An inositol tetrakisphosphate-containing phospholipid in activated neutrophils. Nature. 1988;334:353–356. doi: 10.1038/334353a0. [DOI] [PubMed] [Google Scholar]
- Traynor-Kaplan A.E., Thompson B.L., Harris A.L., Taylor P., Omann G.M., Sklar L.A. Transient increase in phosphatidylinositol 3,4-bisphosphate and phosphatidylinositol trisphosphate during activation of human neutrophils. J. Biol. Chem. 1989;264:15668–15673. [PubMed] [Google Scholar]
- Van der Kaay J., Beck M., Gray A., Downes C.P. Distinct phosphatidylinositol 3-kinase lipid products accumulate upon oxidative and osmotic stress and lead to different cellular responses. J. Biol. Chem. 1999;274:35963–35968. doi: 10.1074/jbc.274.50.35963. [DOI] [PubMed] [Google Scholar]
- Viaud J., Lagarrigue F., Ramel D., Allart S., Chicanne G., Ceccato L., Courilleau D., Xuereb J.M., Pertz O., Payrastre B., Gaits-Iacovoni F. Phosphatidylinositol 5-phosphate regulates invasion through binding andactivation of Tiam1. Nat. Commun. 2014 Jun;5:4080. doi: 10.1038/ncomms5080. [DOI] [PubMed] [Google Scholar]
- Watt S.A., Kimber W.A., Fleming I.N., Leslie N.R., Downes C.P., Lucocq J.M. Detection of novel intracellular agonist responsive pools of phosphatidylinositol 3,4-bisphosphate using the TAPP1 pleckstrin homology domain in immunoelectron microscopy. Biochem. J. 2004;377:653–663. doi: 10.1042/BJ20031397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt S.A., Kular G., Fleming I.N., Downes C.P., Lucocq J.M. Subcellular localization of phosphatidylinositol 4,5-bisphosphate using the pleckstrin homology domain of phospholipase C delta1. Biochem. J. 2002;363:657–666. doi: 10.1042/0264-6021:3630657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner O.D., Neilsen P.O., Prestwich G.D., Kirschner M.W., Cantley L.C., Bourne H.R. A PtdInsP(3)- and Rho GTPase-mediated positive feedback loop regulates neutrophil polarity. Nat. Cell Biol. 2002;4:509–513. doi: 10.1038/ncb811. [DOI] [PMC free article] [PubMed] [Google Scholar]