Abstract
Objective
Microbiota is potentially linked to the development of cancer. However, the features of microbiota in gastric cancer remain unclear. The aim of this study was to characterize the gastric microbiota in cancer.
Methods
A total of 315 patients, including 212 patients with chronic gastritis and 103 patients with gastric cancer, were enrolled in the study. The bacterial load of gastric mucosa was determined using quantitative PCR. To analyze the biodiversity, structure, and composition of microbiota, amplicons of the 16S rRNA gene from 12 patients were pyrosequenced. The sequences were processed and subsequently analyzed.
Results
The amount of bacteria in gastric mucosa was estimated to be 6.9×108 per gram tissue on average. It was higher in Helicobacter pylori-infected patients (7.80±0.71) compared with those uninfected (7.59±0.57, P=0.005). An increased bacterial load up to 7.85±0.70 was detected in gastric cancer compared with chronic gastritis (P=0.001). The unweighted principal coordinate analysis showed that the structure of microbiota in gastric cancer was more diversified. Five genera of bacteria with potential cancer-promoting activities were enriched in gastric cancer. The weighted principal coordinate analysis showed that the presence of Helicobacter pylori markedly altered the structure of microbiota, but had little influence on the relative proportions of the other members in the microbiota.
Conclusion
Findings from this study indicated an altered microbiota in gastric cancer with increased quantity of bacteria, diversified microbial communities, and enrichment of bacteria with potential cancer-promoting activities. These alterations could contribute toward the gastric carcinogenesis.
Keywords: chronic gastritis, gastric cancer, Helicobacter pylori, microbiota
Introduction
The human gut microbiota consists of a huge amount of bacteria 1. Under physiological conditions, microbiota is vital to human health. It participates in energy metabolism, absorption of nutrients, maturation of the intestinal immune system, and protection from infection of pathogens 2,3. Alterations in microbiota are potentially linked to cancer. Bacteria with potential cancer-inducing activities, including Fusobacteria and Escherichia coli, have been found to be increased in colorectal cancer 4,5.
The human stomach harbors a large number of bacteria in addition to Helicobacter pylori 6. Proteobacteria, Firmicutes, Bacteroidetes, Fusobacteria, and Actinobacteria are predominant in gastric microbiota, although there is considerable variation in the most abundant bacteria between individuals 7,8. Gastric microbiota are potentially linked to the development of gastric cancer. Germ-free transgenic mice had a delayed onset of H. pylori-induced gastric cancer compared with specific pathogen-free mice 9. Intervention with antimicrobial therapies delayed the onset of gastric cancer in transgenic mice irrespective of H. pylori infection 10. Feeding of germ-free transgenic mice with an artificial intestinal microbiota accelerated the occurrence of cancer 9.
Gastric cancer is one of leading causes of cancer-related death. It develops through a multifactorial, multistep process 11. H. pylori, a major carcinogenic pathogen of the stomach 12, initiate the mucosal inflammation, leading to mucosal atrophy and finally cancer. Using culture-dependent approaches, bacterial overgrowth in the stomach has been found with a lowered acid output 13,14. These bacteria potentially promote the production of nitrite, leading to accumulation of carcinogenic N-nitroso compounds 15,16. Thus, it has been supposed that overgrowth of bacteria contributes toward the development of gastric cancer 17. However, microbiota in gastric cancer has not been well studied. The microbial diversity, structure, and composition in gastric cancer remain poorly understood. The aim of the present study was to characterize the microbial community in gastric cancer and explore its potential associations with the carcinogenesis.
Methods
Patients and sample collection
A total of 315 patients, including 212 patients with chronic gastritis and 103 patients with gastric cancer, were enrolled in the study. Of these, 190 were men. The average age of the patients was 55.8±13.5 years. All participants were selected from among those who underwent endoscopy in our hospital from March 2012 to August 2014. A written informed consent was obtained from all the participants and the study protocol was approved by the Medical Research Ethical Committee of Qingdao Municipal Hospital. To minimize the potential influence on the microbiota, all patients enrolled had not received antibiotics or proton pump inhibitors treatments 4 weeks before sample collection. For patients with chronic gastritis, those enrolled in the study had an endoscopic finding of superficial gastritis only. Those with endoscopic findings of peptic ulcer, polyps, or any other local lesions were excluded. Patients who showed histological evidence of atrophy or intestinal metaplasia were further excluded from this study. These inclusion/exclusion criteria minimized the potential compounding factors for the purpose of the study. For the enrollment of patients with gastric cancer, only those with an endoscopic finding of noncardia cancer were included. Histologically, these 103 gastric cancer cases consisted of 87 intestinal-type and 16 diffuse-type cancer. Two antral biopsies were taken from patients who underwent the endoscopy examination. For gastric cancer, biopsies were obtained from the antrum if possible or 5 cm away from the cancerous lesions. One biopsy was used for routine histological examination, whereas the other biopsy was stored at −80°C for DNA extraction. The status of H. pylori was determined using a modified Giemsa staining 18.
DNA extraction
To extract genomic DNA, biopsies were ground and then treated with 1 U of DNase I to eliminate any potentially foreign bacterial DNA. To increase the yield of bacterial DNA, samples were treated with lysozyme at a final concentration of 50 mg/ml. Genomic DNA was then extracted using a Qiagen DNeasy blood and tissue kit (Qiagen, Hilden, Germany).
Quantitative PCR
To determine the bacterial load in gastric mucosa, real-time quantitative PCR (qPCR) was performed to amplify the bacterial 16S rRNA gene according to the report by Harms et al. 19. The following primers or probes were used in the amplification: Forward primer, 1055F (5′-ATGGCTGTCGTCAGCT-3′), the reverse primer 1329R (5′-ACGGGCGGTGTGTAC-3′), probe 16STaq1115 [5′-(6-FAM)-CAACGAGCGCAACCC-(TAMRA)-3′] 19. The PCR reaction consisted of a total volume of 25 µl containing 1×Premix Ex Taq (Takara, Dalian, China), 0.2 µmol/l each of primers, 0.2 µmol/l probe, and 20 ng DNA template. Cycling conditions included an initial denaturation at 95°C for 30 s, followed by 50 cycles of 95°C for 5 s and 60°C for 30 s. Standard curves were constructed with a serial dilution of a plasmid containing the full length of the 16S rRNA gene. The bacterial load was calculated as copy numbers of the 16S rRNA gene per microgram DNA. To estimate the bacterial amount in the stomach, the biopsies were weighted. The amount was calculated as the bacterial number per gram of tissue, given that the average copy number of the 16S rRNA gene in each bacterium was 3.6 20.
To quantify H. pylori in the gastric mucosa, qPCR was performed essentially as described previously 21. The sequence of primers used to amplify ureB was 5′-CAAAATCGCTGGCATTGGT-3′ and 5′-CTTCACCGGCTAAGGCTTCA-3′, respectively. The probe sequence was 5′-(6-FAM)-AACAAAGACATGCAAGATGGCGTTAAAAACA-(TAMRA)-3′. Standard curves were constructed with a serial dilution of a plasmid containing the full length of ureB from H. pylori. The amount of the bacterium was calculated as copy numbers of 16S rRNA gene per microgram of DNA.
Pyrosequencing and data analysis
To analyze the microbial communities of gastric mucosa, the variable V1-V3 region of the 16S rRNA gene was PCR amplified with primers 8F/533R, which had adapters and barcode. Amplification was carried out with 25 PCR cycles using Q5 high-fidelity DNA polymerase. Subsequently, amplicons were sequenced on a 454 GS-FLX system (Roche, Mannheim, Germany). These sequences were trimmed of sequencing primers, barcode, and adapters and filtered using the following criteria: length>200 nt,<9 homopolymers,<0 ambiguous bases, and Qavg<25. Thus, a total of 147 001 reads were produced for these 12 samples. These reads were aligned. UCHIME was used to detect and remove chimeras. Sequences with an identity more than or equal to 97% were defined as an operational taxonomic unit. They were classified using the Ribosomal Database Project Naïve Bayes Classifier 22. Rarefaction curves, alpha diversity, and beta diversity were analyzed using QIIME (University of Colorado, Boulder, Colorado, USA) 23. The richness of gastric microbiota was evaluated with the Chao1 index, which reflects the theoretical number of species in a microbiota. The Shannon index, which took into account the number of species and the abundance of a species as well, was calculated to estimate the biodiversity of gastric microbiota 23. Using Fast UniFrac analysis, both weighted and unweighted principal coordinate analysis (PCoA) were carried out to determine the similarity among the microbial communities 24. This analysis was used to measure the phylogenetic distance between sets of taxa in a phylogenetic tree. The short read sequences are available at the website of the National Center for Biotechnology Information (study accession number: SRP060550).
Statistical analyses
SPSS and Prism (GraphPad Software Inc., La Jolla, California, USA) were used for statistical analyses and graph production. Student’s t-test or χ2-test was used for statistical analyses where appropriate. A P-value less than 0.05 was considered to be significant.
Results
Increased bacterial load in gastric cancer
The averaged log value of bacterial load in the gastric mucosa was 7.69±0.64 copies per microgram of DNA. To measure the total bacterial number in gastric mucosa, all biopsies were weighted. Given that the average copy number of 16S rRNA in a bacterial cell was 3.6 20, the amount of bacteria in gastric mucosa was determined to be 6.9×108 per gram of tissue. For those 212 cases of chronic gastritis, the bacterial load in mild, moderate, and severe gastritis was 7.53±0.57, 7.61±0.41, and 7.69±0.81, respectively. Student’s t-test showed no significant difference (P>0.05), suggesting that there was no association of the amount of bacteria with the severity of the inflammation. For gastric cancer, there was no significant difference in the bacterial load between intestinal type (7.73±0.46) and diffuse type (7.87±0.73) of cancer (P>0.05).
Multivariable linear regression analysis showed that the presence of H. pylori infection had a significant impact on the bacterial load (P<0.05), but age or sex had no influence (both P>0.05). In these patients, the prevalence of H. pylori was 46.7% (147/315). The bacterial load in H. pylori-positive patients was 7.80±0.71, which was significantly increased compared with H. pylori-negative patients (7.59±0.57, P=0.005). Moreover, the bacterial load of gastric mucosa was correlated positively with the amount of H. pylori (R=0.38, P<0.001) (Fig. 1). This suggested that the infection of H. pylori was a determinant of the bacterial amount of gastric microbiota.
Fig. 1.
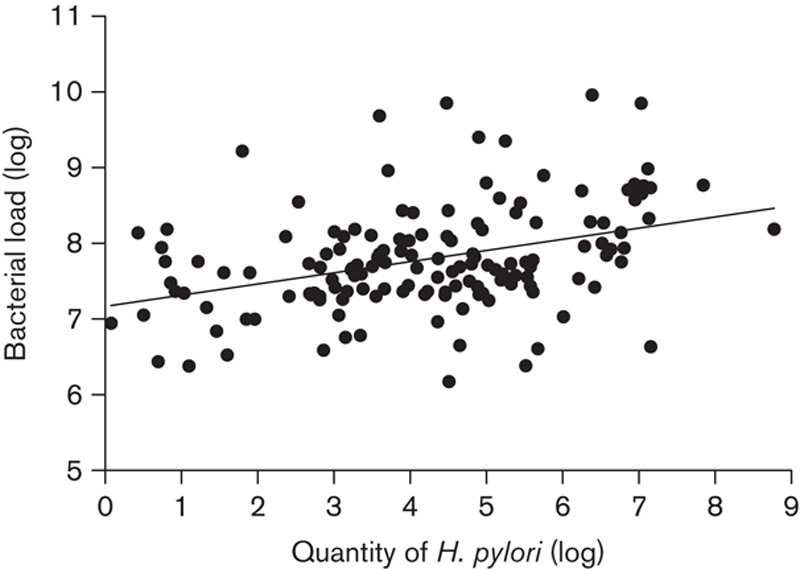
Correlation of the bacterial load in gastric mucosa with H. pylori. Bacterial load and quantity of H. pylori were determined using quantitative PCR. The amount was calculated as copy numbers of the 16S rRNA gene (or ureB for H. pylori) per microgram DNA. Linear regression analysis found that the bacterial load was positively, although weakly, correlated with the quantity of H. pylori in gastric mucosa (R=0.38, P<0.001). H. pylori, Helicobacter pylori.
The prevalence of H. pylori in chronic gastritis was 45.3% (96/212), which did not differ from that in gastric cancer (49.5%, 51/103) (P>0.05). Compared with chronic gastritis, the bacterial load in gastric cancer was significantly increased (P=0.001) (Fig. 2). These results showed that overgrowth of bacteria occurred in gastric cancer.
Fig. 2.
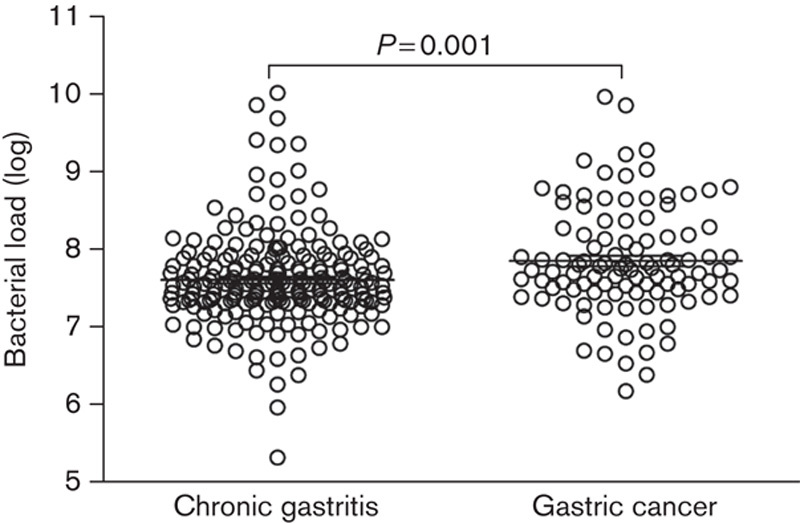
The bacterial load in gastric cancer. The bacterial load of eubacteria in gastric cancer and chronic gastritis was determined using quantitative PCR. The amount was calculated as copy numbers of 16S rRNA gene per microgram of DNA.
Alterations of microbiota in gastric cancer
The microbial communities of gastric mucosa from 12 patients were analyzed using high-throughput sequencing of 16S rRNA amplicons. These included six patients with chronic gastritis and six patients with gastric cancer. The Chao 1-estimated richness in gastric cancer (985.3±242.6) was slightly higher than that in chronic gastritis (920.5±198.7). However, the difference was not statistically significant (P>0.05). Shannon’s diversity index in gastric cancer (1.93±0.52) was similar to that in chronic gastritis (2.07±0.78) (P>0.05). The structure of microbiota was explored by an unweighted unifraction analysis. The results showed that the microbiota from chronic gastritis tended to cluster together, whereas samples from gastric cancer were scattered in the plot (Fig. 3a). This suggested that the structure of microbial communities was phylogenetically diversified in gastric cancer. Compositional analyses showed that Proteobacteria, Firmicutes, Bacteroidetes, Fusobacteria, and Actinobacteria were dominant in microbiota (Fig. 4), although the most predominant phylum varied between individuals. At the genus level, however, five genera of bacteria were enriched in gastric cancer. These included Lactobacillus, Escherichia–Shigella, Nitrospirae, Burkholderia fungorum, and Lachnospiraceae uncultured. Of particular interest, Nitrospirae was present in all patients with gastric cancer, but absent in patients with chronic gastritis.
Fig. 3.
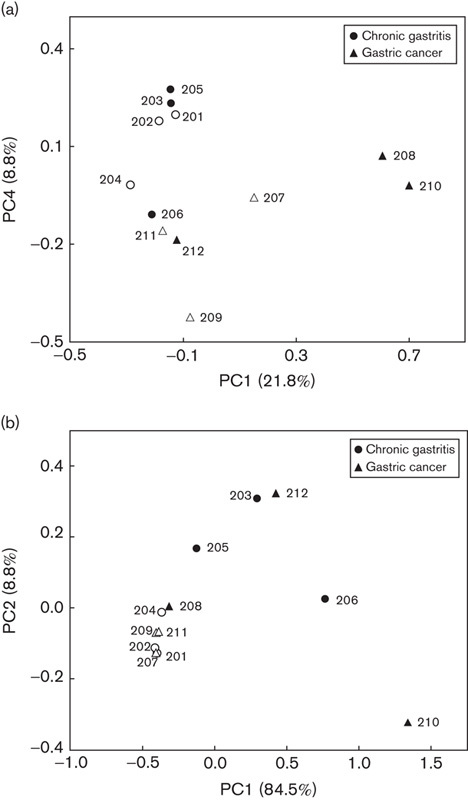
The unweighted (a) and weighted (b) principal coordinate (PC) analysis of microbiota from gastric cancer and chronic gastritis using Fast UniFrac analysis. H. pylori-positive individuals are indicated by filled circles or triangles. H. pylori, Helicobacter pylori.
Fig. 4.
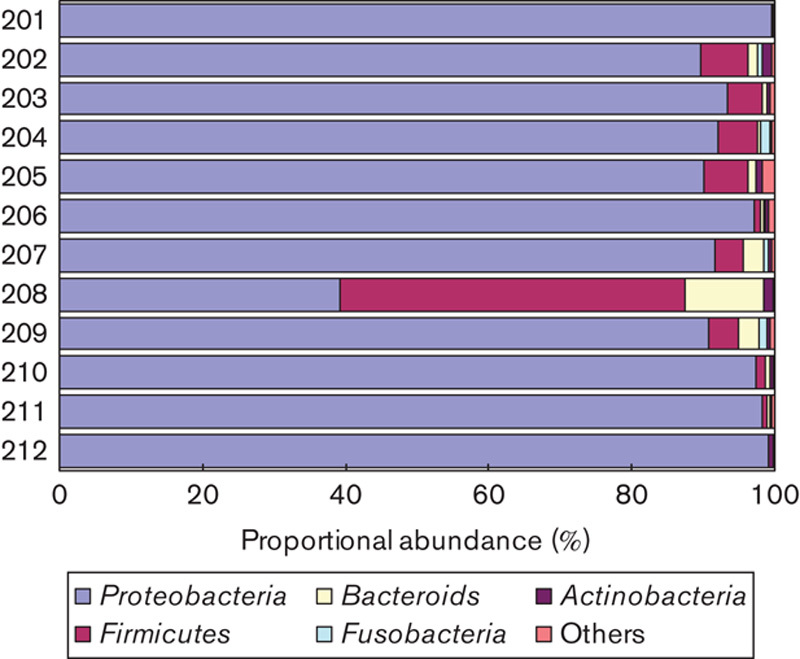
Compositions of gastric microbiota at the phylum level. High-throughput sequencing of amplicons of the 16S rRNA gene was performed on 12 samples from patients with chronic gastritis (201–206) and gastric cancer (207–212).
Influence of H. pylori on microbial communities
Of these 12 patients, three with chronic gastritis and three with gastric cancer were determined to be H. pylori positive. In those H. pylori-negative patients, however, five of six had a low level of the bacterium detected (from 0.04 to 0.67%). The Chao1-estimated richness of microbiota from H. pylori-positive patients (999.5±262.7) was not significantly different from that from H. pylori-negative patients (906.3±163.2) (P>0.05). Shannon’s diversity index was markedly increased in H. pylori-positive patients (2.42±0.58) compared with H. pylori-negative patients (1.56±0.39) (P<0.05), suggesting that the quantity of H. pylori could markedly influence the diversity of gastric microbiota. Weighted PCoA analysis found that H. pylori-negative patients tended to cluster together, whereas those infected by the bacterium were scattered (Fig. 3b). As both analyses of Shannon’s diversity index and weighted PCoA analysis take into account the abundance, these results indicated that the quantity of H. pylori could alter the abundance of other bacteria in the microbiota. This would lead finally to an alteration of the structure of microbiota. Compositional analysis of microbiota showed no significant difference between H. pylori-positive and H. pylori-negative patients.
Discussion
In this study, we found that the bacterial load in the gastric mucosa was determined to be 6.9×108 per gram of tissue. It is much lower than the abundance of bacteria present in the intestine 25, indicating that the human stomach is relatively hostile to the bacterial colonization 26. Findings from the present study, however, indicated a markedly increased bacterial load in gastric cancer. Bacterial overgrowth in the stomach has been found in various precancerous conditions 13,27, including hypochlorhydria and mucosal atrophy. It has been suggested that microbes in the stomach are involved in the production of carcinogens and promotion of inflammatory injuries 15,28 Thus, bacterial overgrowth is a potential cancer-promoting factor 17. Nonetheless, further studies are indicated to clarify whether the bacterial overgrowth is a consequence of cancerous mucosa that generates environments favoring bacteria proliferation. Both Chao1-estimated richness and Shannon’s diversity index reflect the number of species in a microbial community 29. Our results found that they were similar between gastric cancer and chronic gastritis, indicating that there is no alteration in the number of bacterial species in the microbiota from gastric cancer. The PCoA analysis takes into account the bacteria phylogeny 29. In contrast to chronic gastritis, our results showed a scattered pattern in gastric cancer. This indicated that members of microbiota in gastric cancer were more distantly related, suggesting a diversified microbiota harbored in gastric cancer. Taken together, these results indicate bacterial overgrowth of diversified microbiota in gastric cancer. The contribution of such alterations toward the development of cancer requires further investigations.
The results from this study found that at the phylum level, the composition of microbiota in gastric cancer did not differ significantly from chronic gastritis. Nonetheless, enrichment of five bacterial genera was found in gastric cancer. In agreement with recent studies 30,31, Lactobacillus and Lachnospiraceae uncultured were found to be more abundant in gastric cancer. This possibly reflects the reduced bactericidal capacities resulting from the lowered acid production in the stomach 32. A number of species from Lactobacillus have been used as probiotics functioning in the prevention of infection by pathogens 33, alleviation of inflammation, and modulating the microbiota 34,35. However, Lactobacillus is also capable of inducing inflammatory injuries of epithelial cells 36. Thus, it requires further clarification with respect to the relationship between increased abundance of Lactobacillus and gastric cancer. Burkholderia colonizes the stomach and other organs 37. It is reportedly associated with induction of inflammation 38,39. Our results also found an increased abundance of Escherichia–Shigella in gastric cancer. A similar finding has been reported in colorectal cancer 40. E. coli produces a genotoxic toxin, which promotes the development of colon cancer in mice 41. Therefore, E. coli could be involved in the development of gastric cancer.
Nitrate/nitrite and their metabolites are associated with a variety of functions. Acidified nitrite is capable of killing bacteria 32. Nitrate could shape the intestinal microbiota when acting as a source of energy 42. Nitric oxide, a final product of nitrite reduction, is intensively involved in the protection of mucosal integrity 43. Importantly, N-nitroso compounds derived from metabolisms of nitrate/nitrite are potent carcinogens 15,16. E. coli, Lactobacillus, and Nitrospirae are all known to play a role in the metabolisms of nitrate/nitrite 42,44,45. As the level of nitrate/nitrite increases in the gastric cancer and its precancerous conditions 46, it could be expected that the production of N-nitroso compounds is possibly enhanced by these bacteria. Thus, these enriched bacteria could participate in the carcinogenesis.
H. pylori is a major risk factor for gastric cancer. The influence of H. pylori on gastric microbiota has not been fully understood. Our findings showed that H. pylori infection was associated with an increased amount of mucosa-associated bacteria. This is possibly caused by changes in the gastric niche induced by H. pylori. Otherwise, it is plausible that H. pylori-infected individuals have a gastric niche favoring bacterial colonization. It has been found that H. pylori had a major impact on the structure of gastric microbiota 31. This is most likely caused simply by a takeover of other bacteria by H. pylori. When eliminating H. pylori from the analysis of microbiota, the abundance of other bacteria in H. pylori-positive patients does not alter compared with H. pylori-negative individuals 7,8. In agreement with this, our study found that the diversity and structure of microbiota altered in H. pylori-infected stomach only when analyses took into account bacterial abundance. These results showed that the major influence of H. pylori on microbiota is the increased amount of bacteria in the stomach.
Gastric microbiota in cancer has been analyzed in two recent studies 30,31. Results from this study have confirmed the findings from those studies that some bacteria were enriched in gastric cancer. Nonetheless, our study quantified the bacterial amount in gastric cancer and found bacterial overgrowth in cancer. In this study as well as in two other recent studies 30,31, microbiota has been characterized only in a small number of cases. To elucidate the nature of microbiota in gastric cancer, studies on a large cohort are indicated. In addition, findings from this study require further confirmation considering that only one biopsy was taken from each patient. This could underestimate the presence of focal atrophy, intestinal metaplasia, and H. pylori in gastric mucosa. Individual microbiota is potentially influenced by host BMI, smoking, or different strains of H. pylori 47,48. It would be interesting to take these factors into account in future studies.
Conclusion
In summary, findings from this study showed that the microbiota in gastric cancer had an increased number of diverse bacteria. It appears that the altered microbiota potentially have cancer-promoting activities. The mechanisms and pathways by which these alterations are generated remain to be investigated in the future.
Acknowledgements
Conflicts of interest
There are no conflicts of interest.
References
- 1.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol 2014; 12:661–672. [DOI] [PubMed] [Google Scholar]
- 2.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science 2012; 336:1262–1267. [DOI] [PubMed] [Google Scholar]
- 3.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature 2012; 489:220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnet M, Buc E, Sauvanet P, Darcha C, Dubois D, Pereira B, et al. Colonization of the human gut by E. coli and colorectal cancer risk. Clin Cancer Res 2014; 20:859–867. [DOI] [PubMed] [Google Scholar]
- 5.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 2012; 22:299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang LL, Yu XJ, Zhan SH, Jia SJ, Tian ZB, Dong QJ. Participation of microbiota in the development of gastric cancer. World J Gastroenterol 2014; 20:4948–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci USA 2006; 103:732–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin ME, Bhatnagar S, George MD, Paster BJ, Canfield DR, Eisen JA, et al. The impact of Helicobacter pylori infection on the gastric microbiota of the rhesus macaque. PLoS One 2013; 8:e76375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lertpiriyapong K, Whary MT, Muthupalani S, Lofgren JL, Gamazon ER, Feng Y, et al. Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the Helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut 2014; 63:54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee CW, Rickman B, Rogers AB, Muthupalani S, Takaishi S, Yang P, et al. Combination of sulindac and antimicrobial eradication of Helicobacter pylori prevents progression of gastric cancer in hypergastrinemic INS-GAS mice. Cancer Res 2009; 69:8166–8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Correa P. Human gastric carcinogenesis: a multistep and multifactorial process – First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res 1992; 52:6735–6740. [PubMed] [Google Scholar]
- 12.Correa P, Houghton J. Carcinogenesis of Helicobacter pylori. Gastroenterology 2007; 133:659–672. [DOI] [PubMed] [Google Scholar]
- 13.Forsythe SJ, Dolby JM, Webster AD, Cole JA. Nitrate- and nitrite-reducing bacteria in the achlorhydric stomach. J Med Microbiol 1988; 25:253–259. [DOI] [PubMed] [Google Scholar]
- 14.Sanduleanu S, Jonkers D, De Bruine A, Hameeteman W, Stockbrügger RW. Non-Helicobacter pylori bacterial flora during acid-suppressive therapy: differential findings in gastric juice and gastric mucosa. Aliment Pharmacol Ther 2001; 15:379–388. [DOI] [PubMed] [Google Scholar]
- 15.Forsythe SJ, Cole JA. Nitrite accumulation during anaerobic nitrate reduction by binary suspensions of bacteria isolated from the achlorhydric stomach. J Gen Microbiol 1987; 133:1845–1849. [DOI] [PubMed] [Google Scholar]
- 16.Stockbrugger RW, Cotton PB, Eugenides N, Bartholomew BA, Hill MJ, Walters CL. Intragastric nitrites, nitrosamines, and bacterial overgrowth during cimetidine treatment. Gut 1982; 23:1048–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Correa P. A human model of gastric carcinogenesis. Cancer Res 1988; 48:3554–3560. [PubMed] [Google Scholar]
- 18.Loffeld RJ, Stobberingh E, Flendrig JA, Arends JW. Helicobacter pylori in gastric biopsy specimens. Comparison of culture, modified Giemsa stain, and immunohistochemistry. A retrospective study. J Pathol 1991; 165:69–73. [DOI] [PubMed] [Google Scholar]
- 19.Harms G, Layton AC, Dionisi HM, Gregory IR, Garrett VM, Hawkins SA, et al. Real-time PCR quantification of nitrifying bacteria in a municipal wastewater treatment plant. Environ Sci Technol 2003; 37:343–351. [DOI] [PubMed] [Google Scholar]
- 20.Klappenbach JA, Saxman PR, Cole JR, Schmidt TM. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res 2001; 29:181–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whary MT, Ge Z, Fox JG. Verifying and quantifying Helicobacter pylori infection status of research mice. Methods Mol Biol 2012; 921:143–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007; 73:5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010; 7:335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 2010; 4:17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmed S, Macfarlane GT, Fite A, McBain AJ, Gilbert P, Macfarlane S. Mucosa-associated bacterial diversity in relation to human terminal ileum and colonic biopsy samples. Appl Environ Microbiol 2007; 73:7435–7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang I, Nell S, Suerbaum S. Survival in hostile territory: the microbiota of the stomach. FEMS Microbiol Rev 2013; 37:736–761. [DOI] [PubMed] [Google Scholar]
- 27.Delgado S, Cabrera-Rubio R, Mira A, Suárez A, Mayo B. Microbiological survey of the human gastric ecosystem using culturing and pyrosequencing methods. Microb Ecol 2013; 65:763–772. [DOI] [PubMed] [Google Scholar]
- 28.Sharma BK, Santana IA, Wood EC, Walt RP, Pereira M, Noone P. Intragastric bacterial activity and nitrosation before, during, and after treatment with omeprazole. Br Med J 1984; 289:717–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lozupone CA, Knight R. Species divergence and the measurement of microbial diversity. FEMS Microbiol Rev 2008; 32:557–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eun CS, Kim BK, Han DS, Kim SY, Kim KM, Choi BY. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter 2014; 19:407–416. [DOI] [PubMed] [Google Scholar]
- 31.Aviles-Jimenez F, Vazquez-Jimenez F, Medrano-Guzman R, Mantilla A, Torres J. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci Rep 2014; 4:4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rao A, Jump RL, Pultz NJ, Pultz MJ, Donskey CJ. In vitro killing of nosocomial pathogens by acid and acidified nitrite. Antimicrob Agents Chemother 2006; 50:3901–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gotteland M, Brunser O, Cruchet S. Systematic review: are probiotics useful in controlling gastric colonization by Helicobacter pylori? Aliment Pharmacol Ther 2006; 23:1077–1086. [DOI] [PubMed] [Google Scholar]
- 34.Čitar M, Hacin B, Tompa G, Štempelj M, Rogelj I, Dolinšek J, et al. Human intestinal mucosa-associated Lactobacillus and Bifidobacterium strains with probiotic properties modulate IL-10, IL-6 and IL-12 gene expression in THP-1 cells. Benef Microbes 2015; 6:325–336. [DOI] [PubMed] [Google Scholar]
- 35.Zhang H, Wang H, Shepherd M, Wen K, Li G, Yang X, et al. Probiotics and virulent human rotavirus modulate the transplanted human gut microbiota in gnotobiotic pigs. Gut Pathog 2014; 6:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lukic J, Strahinic I, Milenkovic M, Golic N, Kojic M, Topisirovic L, et al. Interaction of Lactobacillus fermentum BGHI14 with rat colonic mucosa: implications for colitis induction. Appl Environ Microbiol 2013; 79:5735–5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodyear A, Bielefeldt-Ohmann H, Schweizer H, Dow S. Persistent gastric colonization with Burkholderia pseudomallei and dissemination from the gastrointestinal tract following mucosal inoculation of mice. PLoS One 2012; 7:e37324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drevinek P, Mahenthiralingam E. Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clin Microbiol Infect 2010; 16:821–830. [DOI] [PubMed] [Google Scholar]
- 39.Alam S, Amemiya K, Bernhards RC, Ulrich RG, Waag DM, Saikh KU. Characterization of cellular immune response and innate immune signaling in human and nonhuman primate primary mononuclear cells exposed to Burkholderia mallei. Microb Pathog 2015; 78:20–28. [DOI] [PubMed] [Google Scholar]
- 40.Leung A, Tsoi H, Yu J. Fusobacterium and Escherichia: models of colorectal cancer driven by microbiota and the utility of microbiota in colorectal cancer screening. Expert Rev Gastroenterol Hepatol 2015; 9:651–657. [DOI] [PubMed] [Google Scholar]
- 41.Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan T, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012; 338:120–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 2013; 339:708–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lundberg JO, Weitzberg E. Biology of nitrogen oxides in the gastrointestinal tract. Gut 2013; 62:616–629. [DOI] [PubMed] [Google Scholar]
- 44.Fei YT, Liu DM, Luo TH, Chen G, Wu H, Li L, et al. Molecular characterization of Lactobacillus plantarum DMDL 9010, a strain with efficient nitrite degradation capacity. PLoS One 2014; 9:e113792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ushiki N, Fujitani H, Aoi Y, Tsuneda S. Isolation of Nitrospira belonging to sublineage II from a wastewater treatment plant. Microbes Environ 2013; 28:346–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kodama K, Sumii K, Kawano M, Kido T, Nojima K, Sumii M, et al. Gastric juice nitrite and vitamin C in patients with gastric cancer and atrophic gastritis: is low acidity solely responsible for cancer risk? Eur J Gastroenterol Hepatol 2003; 15:987–993. [DOI] [PubMed] [Google Scholar]
- 47.Tims S, Derom C, Jonkers DM, Vlietinck R, Saris WH, Kleerebezem M, et al. Microbiota conservation and BMI signatures in adult monozygotic twins. ISME J 2013; 7:707–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garmendia J, Morey P, Bengoechea JA. Impact of cigarette smoke exposure on host-bacterial pathogen interactions. Eur Respir J 2012; 39:467–477. [DOI] [PubMed] [Google Scholar]


