Abstract
Malaria is an infectious disease caused by parasites of the genus Plasmodium that inflicts approximately one million deaths per annum worldwide. Chemical validation of new antimalarial targets is urgently required in view of rising resistance to current drugs. One such putative target is the enzyme N-myristoyltransferase (NMT), which catalyzes N-myristoylation of protein substrates. Here we report an integrated chemical biology approach to explore protein myristoylation in the major human parasite P. falciparum, combining chemical proteomic tools for identification of the myristoylated and glycosylphosphatidylinositol-anchored proteome with selective small molecule NMT inhibitors. We demonstrate that NMT is an essential and chemically tractable target in malaria parasites both in vitro and in vivo, and show that selective inhibition of N-myristoylation leads to catastrophic and irreversible failure to assemble the inner membrane complex, a critical subcellular organelle in the parasite life cycle. Our studies provide the basis for development of new antimalarials targeting NMT.
Malaria is an infectious disease caused by parasites of the genus Plasmodium, and is among the most serious in the developing world, resulting in an estimated 200 million cases and 1.2 million deaths in 20101. The majority of cases and mortality are due to P. falciparum in sub-Saharan Africa, whilst P. vivax is widespread and causes persistent infections2. Resistance has been recorded against all frontline therapies, and the need for new drugs is widely recognized3. N-Myristoyltransferase (NMT, EC 2.3.1.97) catalyzes transfer of the lipid myristate (C14:0) from myristoyl coenzyme A (Myr-CoA) to specific substrate proteins in eukaryotes4, and has been explored pre-clinically as a target in fungal5 and trypanosome infections6, and suggested as a potential target in malaria and leishmaniasis7. Protein N-myristoylation occurs predominantly co-translationally at an N-terminal glycine following removal of initiator methionine (Fig. 1a); although an N-terminal glycine is essential, there is no pan-species consensus sequence for myristoylation4. In silico analyses suggest that up to 2% of proteins in P. falciparum may be substrates for NMT (Supplementary Table S1), although lack of suitable training sets for Plasmodium myristoylation results in inconsistent predictions8. To date, only two proteins have been shown to be N-myristoylated in their native context in P. falciparum – calcium-dependent protein kinase-1 (CDPK1)9 and glideosome-associated protein 45kDa (GAP45)10 – and there is indirect evidence for a small number of other substrates10, 11, 12, 13. N-Myristoylation regulates membrane localization and protein complex assembly in eukaryotes14, and selective inhibition of NMT is hypothesized to lead to pleiotropic effects on the parasite through multiple downstream substrates. Plasmodium species possess a single NMT gene15 and partial knockdown of NMT expression in the rodent malaria parasite P. berghei was found to result in rapid depletion of parasitemia in vivo, demonstrating that NMT is essential in this murine model of malaria16. However, target validation in human malarial parasites is particularly challenging due to their complex life cycle, the limited range of genetic tools available, and the very low efficiency of genetic modification17. Despite intensive research efforts, conditional genetic knockout of an essential gene in P. falciparum has not been demonstrated to date, and thus chemical genetic knockdown approaches based on small molecule inhibitors are a particularly powerful and versatile strategy for target validation.
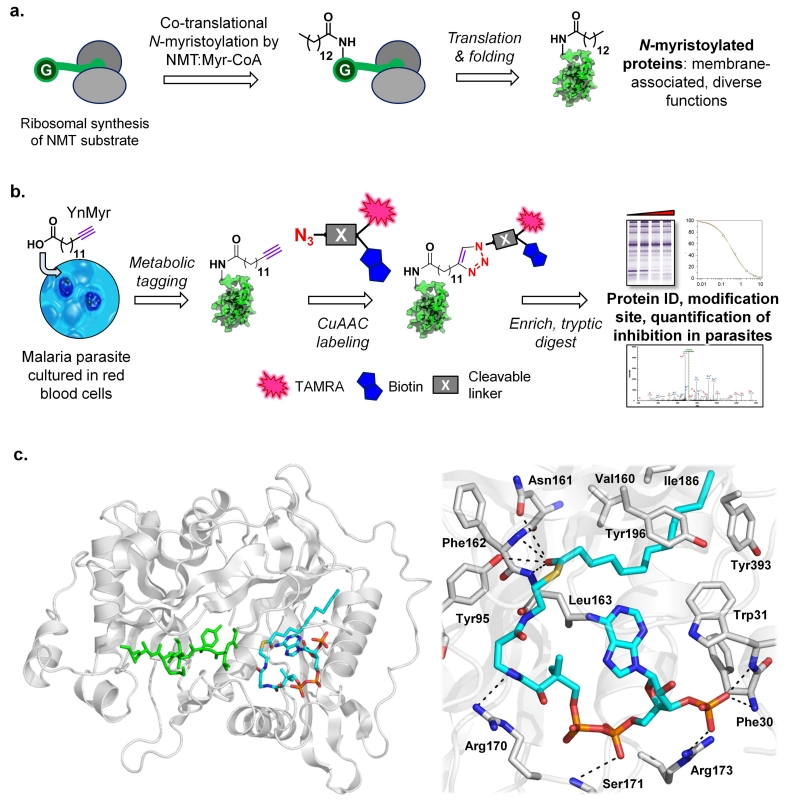
Figure 1. YnMyr-CoA is an effective substrate mimic for P. falciparum and P. vivax N-myristoyltransferases.
(a) Co-translational N-myristoylation by NMT and Myr-CoA at an N-terminal glycine. (b) YnMyr tag & label strategy: YnMyr is fed to parasites in culture and transferred to substrate proteins by metabolic incorporation. Proteins are isolated and ligated by copper (I) catalyzed azide-alkyne cycloaddition (CuAAC) to a multifunctional capture reagent, followed by enrichment and downstream analysis. The cleavable linker is an optional module in the capture reagent that enables direct identification of the modification site by MS/MS. (c) The structure of YnMyr-CoA in PvNMT was determined by X-ray crystallography to 1.75 Å resolution (PDB 2YNC). At left: overall structure of PvNMT showing YnMyr-CoA (cyan), with a peptide substrate (PDB 1IID) modeled in (green) following least-squares superposition of the respective protein backbone atoms in PyMOL (DeLano Scientific). At right: key interactions made by YnMyr-CoA in the PvNMT acyl-CoA binding site, showing accommodation of the YnMyr chain in the hydrophobic acyl binding pocket and interactions of the thioester carbonyl.
Here we present an integrated chemical biology approach to understand inhibition of N-myristoylation in P. falciparum parasites that enables both the study of protein myristoylation and the chemical dissection of NMT as a drug target in this genetically intractable organism. These studies greatly expand knowledge of both N-myristoylated and glycosylphosphatidylinositol (GPI) anchored proteins in Plasmodium, and provide the first compelling evidence that NMT is a druggable target in the symptomatic blood stage of the most important human malaria parasite. Using these tools in combination with quantitative whole proteome analyses and cellular studies, we demonstrate that NMT inhibition results in a catastrophic and irreversible failure to form critical parasite subcellular structures, followed by rapid parasite cell death. Furthermore, we define potent starting points for the development of NMT inhibitors as potential antimalarials that are active against human malaria parasites in vitro, and in vivo in an animal model of malaria.
Results
Myristoyl-CoA analogue YnMyr-CoA is a biomimetic substrate for Plasmodium NMT
Lipid probes bearing a bioorthogonal tag such as a terminal alkyne are powerful tools for analysis of protein lipidation in living systems18, 19. The myristate surrogate tetradec-13-ynoic acid (YnMyr) can be transferred from YnMyr-CoA thioester to target proteins by recombinant NMT20, in bacterial co-expression systems21 or in mammalian cells22. Subsequent chemoselective ligation through copper(I)-catalyzed [3+2] azide-alkyne cycloaddition (CuAAC) imparts a range of useful functionality to tagged proteins, including a fluorophore and/or affinity label (Fig. 1b)20, 22, 23, 24. We determined the structure of NMT from P. vivax (PvNMT, which shares >80% identity with P. falciparum NMT (PfNMT))25 in complex with YnMyr-CoA (PDB 2YNC; Supplementary Fig. S1 and Table S2), revealing that YnMyr-CoA occupies an extended groove that runs across one face of the enzyme, with the YnMyr chain occupying the fatty acyl binding pocket (Fig. 1c). The terminal sp hybridized carbon atoms are accommodated without steric clashes and the hydrophobic contacts and native interactions of the thioester carbonyl with backbone amide and polar side chain moieties are preserved (Fig. 1c)26. This structure is the first reported example of a tagged analogue of a post-translational modification precursor in complex with its cognate transferase, and together with kinetic data (Supplementary Fig. S2) indicates that a terminal alkyne moiety is fully compatible with enzymatic transfer of YnMyr to substrate proteins.
YnMyr tags N-myristoylated and GPI-anchored proteins in P. falciparum blood stage parasites
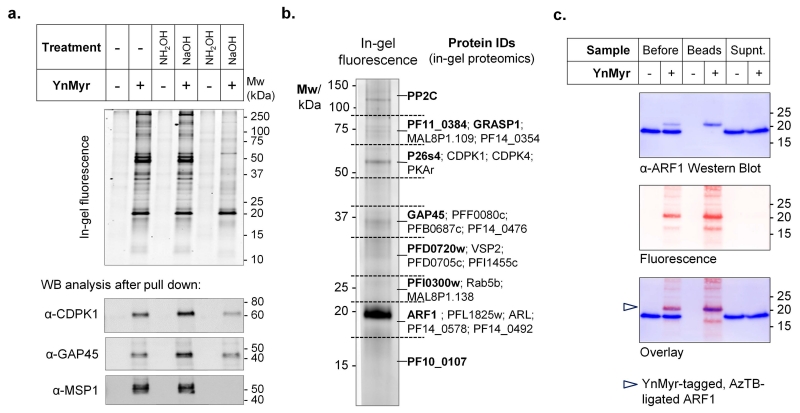
We next investigated the capacity of YnMyr to tag proteins in the asexual stage of the parasite, which invades and replicates within red blood cells (RBCs). Intracellular P. falciparum 3D7 parasites (schizonts) were treated with YnMyr at concentrations up to 50 μM and allowed to develop over 5 h; no changes were observed in morphology or life cycle progression in YnMyr-treated parasites. Parasite proteins were isolated and CuAAC ligation to tri-functional capture reagent AzTB (azido-TAMRA-PEG-Biotin; Supplementary Fig. S3)27 enabled direct in-gel fluorescence detection of protein tagging following SDS PAGE (Fig. 2a). YnMyr tagging was concentration-dependent with no detectable background, readily out-competed by excess myristate, and inhibited by the protein synthesis inhibitor cycloheximide (CHX, Supplementary Fig. S4), consistent with biomimetic co-translational modification of target proteins. The biotin label in AzTB further permitted quantitative immobilization of fluorescently labeled proteins on streptavidin-coated beads (Supplementary Fig. S5). The known N-myristoylated proteins CDPK1 and GAP4510, 28 were selectively pulled down from parasites cultured in the presence of YnMyr, and resistance of labeling to base and hydroxylamine indicated the presence of N-acyl (amide linked) YnMyr (Fig. 2a and Supplementary Fig. S6). Control experiments cross-comparing metabolic incorporation of the longer chain palmitate probe YnPal (heptadec-14-ynoic acid) confirmed that YnMyr was not significantly incorporated into sites of S-palmitoylation, which occurs through a thioester linked to cysteine side chains (Supplementary Fig. S7). However, a substantial fraction of the fluorescently labeled bands were base sensitive suggesting the presence of ester-linked YnMyr (Fig. 2a). Ester-linked incorporation of myristate at the GPI anchor inositol is characteristic of Plasmodium species29, and we found that the abundant GPI-anchored surface protein MSP1 was indeed tagged by YnMyr in a base-sensitive manner (Fig. 2a). Taken together, these data suggest that YnMyr is a high-fidelity myristate mimetic in live parasites, and can be used as a probe for both N-myristoylated and GPI-anchored proteins under standard culture conditions.
Figure 2. YnMyr tags N-myristoylated and GPI-anchored proteins in blood stage P. falciparum.
(a) YnMyr was fed to parasites in culture, ligated by CuAAC to AzTB and a portion subjected to base (0.2 M NaOH) or hydroxylamine (1 M NH2OH) treatment to differentiate amide and ester-linked YnMyr. Samples were analyzed by in-gel fluorescence (top) or enriched by pull-down on streptavidin beads and analyzed by Western blot (WB; bottom). MSP1 is detected as the processed ~45 kDa GPI-anchored C-terminal fragment. (b) Summary of N-terminal glycine-containing proteins found by gel-based proteomics following base treatment and pull-down on streptavidin beads. Proteins are grouped by the gel slice in which they were predominantly found and dominant proteins in each slice are highlighted in bold. (c) Confirmation of the major fluorescent band as ARF1 by Western blot against a novel anti-ARF1 antibody, analyzed before and after pull-down on streptavidin beads; a shift to higher molecular weight following addition of AzTB is observed only for tagged ARF1 (marked by arrow), which precisely overlays with the fluorescent band both before and after pull-down.
Chemical proteomic profiling of N-myristoylated and GPI-anchored proteins in P. falciparum
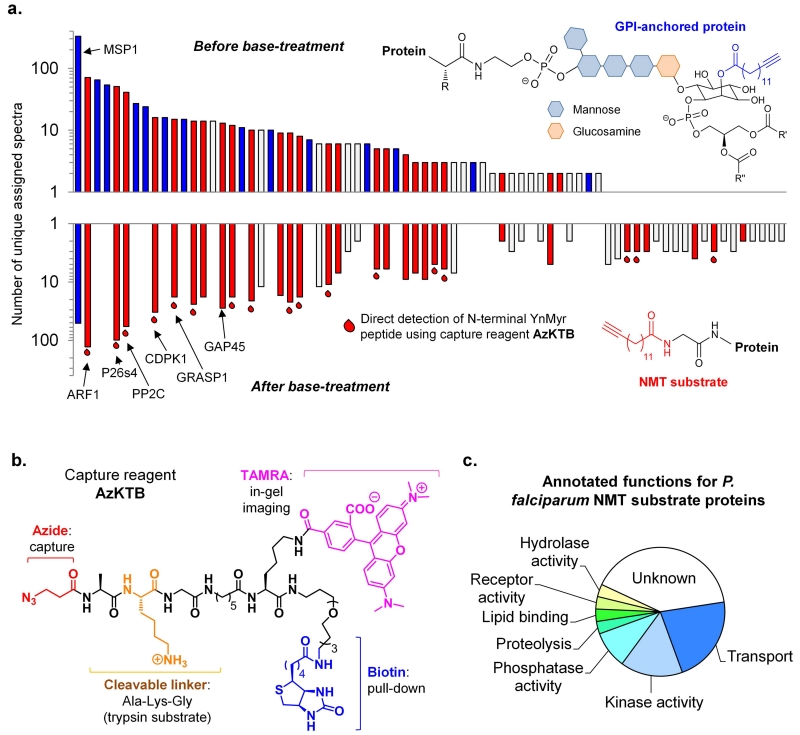
We next performed de novo identification of YnMyr targets in P. falciparum by LC-MS/MS based chemical proteomics after AzTB ligation, pull-down and protein digest, using both gel-based and gel-free approaches. Tryptic peptides were analyzed by a standard proteomics workflow, and proteins found with very high (>99 %) confidence and >4-fold enrichment compared to controls were considered for further analysis; proteomic methods are described in detail in Supplementary Methods. N-Myristoylated target proteins were mapped to specific regions of the gel following gel-based proteomics (Fig. 2b; Supplementary Fig. S8 and Table S6), identifying ADP-ribosylation factor 1 (ARF1) as the major component of the strongest fluorescent band. A novel antibody directed against PfARF1 further confirmed this assignment (Fig. 2c). Base-treatment prior to pull-down was used to dissect the contributions of GPI-anchored and N-myristoylated proteins; identification of MSP1 and predicted GPI-anchored proteins was dramatically reduced or eliminated by base treatment, whilst putative N-myristoylated proteins were unaffected (Fig. 3a; Supplementary Tables S3, S4 and S5)30, 31. YnMyr-tagged peptides were not detected in these analyses, due to the presence of the biotin label added during CuAAC ligation that anchors the peptide to the resin during tryptic digest. Indeed, direct determination of the site of modification has been recognized as a significant unsolved challenge in metabolic tagging of lipidated proteins32 due to poor release of the biotinylated peptide from the streptavidin resin and the large size of the resultant label. We therefore designed a new tetra-functional capture reagent (AzKTB, Fig. 3b) which bears a short trypsin-cleavable peptide sequence between the azide module and the TAMRA/PEG-biotin labels. Tryptic on-bead digest releases the YnMyr-modified peptide bearing a hydrophilic zwitterionic moiety, permitting the site of YnMyr modification to be determined unambiguously by tandem MS (Supplementary Fig. S9). Using this new approach, 17 N-terminal YnMyr-Gly motifs were detected (Fig. 3a and Supplementary Table S7), further confirming the fidelity of the YnMyr probe for NMT substrates in live parasites.
Figure 3. Chemical proteomic discovery and direct detection of modification site for N-myristoylated and GPI-anchored proteins in blood stage P. falciparum.
(a) Assigned spectra profile (vertical log scale) of proteins identified from a representative gel-free analysis +/− base-treatment of samples, showing those proteins predicted or known to be GPI-anchored (blue bars), those containing an N-terminal glycine indicative of N-myristoylation (red bars), and those for which N-terminal YnMyr was detected directly by MS/MS (following ligation to AzKTB); other proteins are shown in white. (b) Structure of capture reagent AzKTB. (c) Functional annotation of 31 potential NMT substrates: all substrate proteins carry a base-insensitive modification, an N-terminal glycine, and evidence of N-terminal YnMyr and/or bioinformatic prediction of N-myristoylation.
Taken together, these data provide direct experimental evidence for more than 30 NMT substrates in wild type parasites, encompassing a diverse range of functions including motility, protein transport, parasite development and phosphorylation pathways (Fig. 3c). As expected, many substrates have predicted or known localization at membranes or in membrane-bound compartments (Supplementary Fig. S10). Across the proteomic datasets five proteins had previously been annotated as PfNMT substrates via direct (GAP45, CDPK1) or indirect (GRASP113, Armadillo repeats-only protein ARO33, ARF130) experimental evidence. New proteins were identified that are N-myristoylated in other eukaryotes, including several ARF/ARL proteins and the 26S proteasome regulatory subunit 4 (P26s4), which is highly enriched in YnMyr-tagged samples34. Interestingly, studies in yeast have shown that loss of N-myristoylation of the yeast P26s4 homologue results in aberrant localization of the entire proteasome complex34 and the proteasome has recently been described as a promising drug target in P. falciparum35. Other substrates with important functions include Protein Kinase A regulatory subunit PKAr36, the small GTPase Rab5b37 and CDPK4, a kinase essential in gametogenesis and disease transmission38. We also identified the small inner-membrane complex (IMC) proteins ISP1 and ISP3; along with GAP45 these proteins have important roles in regulating formation of the IMC, a subcellular organelle that is critical for assembling the molecular motor apparatus required for RBC invasion39.
Identification and structural characterization of potent small molecule inhibitors of P. falciparum and P. vivax NMTs
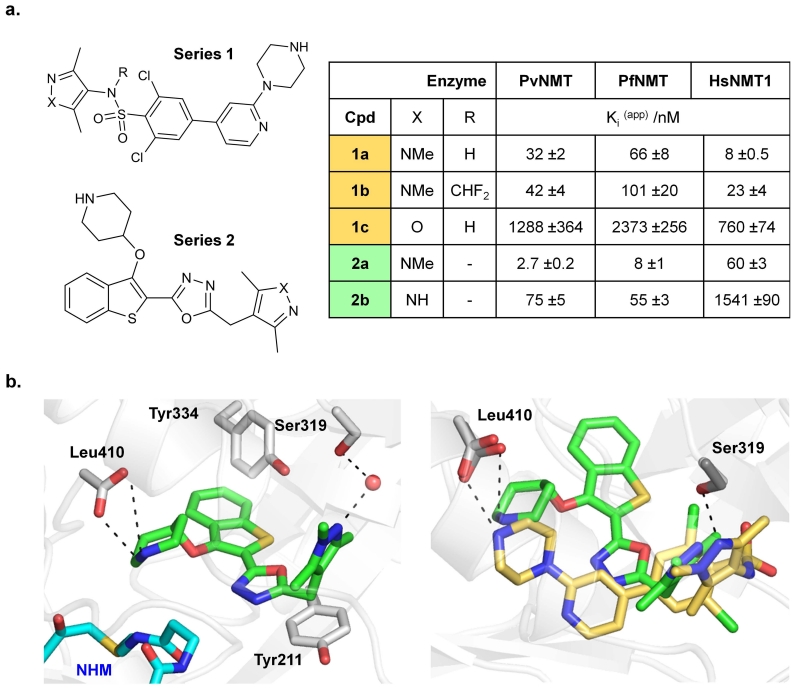
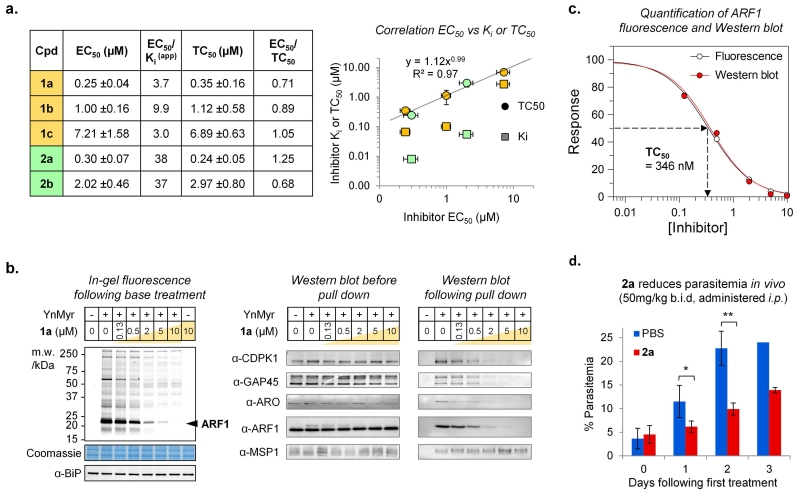
The diversity of function of the identified N-myristoylated proteome in P. falciparum indicates that specific NMT inhibition could substantially reduce parasite viability. To support this hypothesis, we sought to identify potent chemical inhibitors to enable selective and tunable chemical genetic knockdown of NMT activity in live parasites. As noted above, such tools are particularly powerful when applied to P. falciparum since there is currently no proven genetic method for inducible knockdown of an essential gene in this parasite. Through further development of our recently-reported Plasmodium NMT inhibitors40 combined with screening known inhibitors of NMT from other organisms we identified two distinct PfNMT/PvNMT inhibitor series 1 and 2 (Fig. 4a). Series 1 compounds were first reported as inhibitors of T. brucei NMT, and compound 1a is effective on trypanosomes in vitro and in vivo6; however, the causative agents of malaria and trypanosomiasis have entirely different mechanisms of infection and disease pathology. Series 2 is derived from a P. falciparum NMT inhibitor based on a benzothiophene scaffold,40 the structure-guided development of which will be reported elsewhere in due course. X-ray crystallography was used to determine the binding mode of the most potent compounds 1a and 2a in ternary complexes with PvNMT and a non-hydrolyzable myristoyl-CoA analogue (PDB 2YND and 2YNE; Supplementary Table S2). Both inhibitors bind at the peptide binding pocket, with 1a exhibiting a similar binding mode to that observed in NMTs from other species (Fig. 4b and Supplementary Fig. S11). A salt bridge is formed between the positively charged piperazine (1a) or piperidine (2a) nitrogen and the C-terminal carboxylate of the enzyme, which plays an essential catalytic role during myristate transfer4. Despite this common point of contact, the trajectory of these two chemotypes through the binding site is distinct. Substitution of the trimethyl pyrazole (1c, 2b) substantially reduces potency, likely due to reduced H-bonding capacity, whilst sulfonamide alkylation in 1b yielded a more subtle decrease (Fig. 4a). Although 1a-c41 are more potent against Homo sapiens NMT (HsNMT1) than against the Plasmodium enzymes, RBCs lack de novo protein synthesis and do not express NMT, suggesting that in vitro culture would not be compromised42. In contrast, 2a-b possess promising selectivity over HsNMT1, with 2a being the most potent PfNMT and PvNMT inhibitor reported to date.
Figure 4. Compounds 1a-c and 2a-b bind in the Plasmodium NMT peptide binding pocket and inhibit the recombinant enzyme.
(a) Structures and Ki(app) (apparent inhibitor dissociation constant) against recombinant PfNMT and PvNMT, and the host enzyme HsNMT1, for series 1a-c and 2a-b; IC50 values were determined by a fluorogenic assay51 and differences in substrate Km accounted for using the Cheng-Prusoff equation to give a value for apparent Ki. Errors are quoted ± standard error (n ≥ 3). (b) Structures for 1a and 2a in PvNMT were determined by X-ray crystallography with a non-hydrolyzable myristoyl-CoA (NHM) cofactor to 1.89 or 1.72 Å resolution (PDB 2YND and 2YNE), respectively. Left: interactions made by 2a (green) in the peptide binding site showing a water-mediated hydrogen bond to Ser319, an ionic bond to the C-terminal acid (Leu410) and interactions with aromatic residues Tyr334 and Tyr211. NHM is shown in cyan. Right: overlay of binding mode of 1a (yellow) and 2a (green) in PvNMT, following alignment of the protein backbone in PyMOL (DeLano Scientific).
NMT inhibitors act on-target to kill P. falciparum blood stage parasites and reduce parasite burden in an in vivo model of malaria
With inhibitor and substrate chemical probes for PfNMT characterized at the structural, biochemical and proteomic level, we sought to determine whether 1a-c and 2a-b act on-target in parasites in culture and to assess NMT as a potential drug target in blood stage P. falciparum. Analysis of EC50 values for parasite growth inhibition in synchronized intraerythrocytic P. falciparum cultures revealed that translation of potency from enzyme to cell (EC50/Ki(app)) varied significantly across series (Fig. 5a). Uninfected RBCs remained morphologically normal (Supplementary Fig. S12). Since protein N-myristoylation occurs co-translationally we reasoned that inhibition of NMT in parasites could be assessed directly by treatment with inhibitor combined with pulse tagging with YnMyr. When tagging experiments were repeated in the presence of 1a a highly specific dose-dependent depletion of fluorescence was observed for all bands previously identified as NMT substrates, with no change in GPI-anchored proteins (Fig. 5b; Supplementary Figs. S13 and S14). Parallel experiments with the tagged methionine surrogate azidohomoalanine (AHA43, Supplementary Fig. S3) showed that 1a does not directly affect protein synthesis (Supplementary Fig. S15). Selective inhibition was further confirmed for specific substrate proteins by Western blot against CDPK1 and GAP45, and using novel antibodies directed against ARF1 and ARO (Fig. 5b; Supplementary Figs. S16 and S17). Total abundance of each protein analyzed was unaltered by inhibitor treatment over this short pulse tagging period; however, on pull-down the amount of each YnMyr-tagged substrate recovered was dose-dependently inhibited, whilst MSP1 remained unaffected. Quantification of the 20 kDa ARF1 band by fluorescence intensity or Western blot gave 346 nM as the inhibitor concentration required for 50% reduction in YnMyr tagging (here termed TC50) for 1a, closely matching the TC50 values calculated for other specific substrates by quantification of Western blots (Fig. 5c and Supplementary Fig. S18).
Figure 5. Inhibition of NMT in P. falciparum blood stage parasites leads to anti-parasitic activity, and reduces parasitemia in an in vivo model of malaria.
(a) Left: EC50 (inhibitor concentration required for 50% reduction in parasitemia), EC50/Ki(app), TC50 (inhibitor concentration required for 50% reduction in YnMyr tagging) and EC50/TC50 for 1a-c and 2a-b against P. falciparum. Right: correlation between EC50 and TC50 for 1a-c and 2a-b. Ki is also plotted. Horizontal and vertical error bars show ± standard error (n ≥ 2). (b) Quantification of NMT inhibition in live parasites. Schizonts were treated with 1a (0.13-10 μM illustrated by gold shaded gradient, or DMSO) followed by tagging with YnMyr (or DMSO). Left: analysis by in-gel fluorescence following base treatment to remove GPI-anchored proteins. ARF1 band (marked) was used to calculate TC50; Coomassie and α-BiP are loading controls for gel and Western blots, respectively. Right: Analysis by Western blot before and after pull-down of YnMyr-tagged; all NMT substrates show dose-dependent reduction in enrichment, whilst MSP1 (GPI-anchored) does not. Full blots in Supplementary Information. (c) Quantification of normalized fluorescence and Western blot for ARF1, using GraFit 7.0 (Erithacus Software Ltd, UK). (d) Compound 2a dosed at 50 mg/kg b.i.d. by intraperitoneal injection (i.p.) (n = 3 animals in inhibitor-treated group; n = 2 in control group) reduces parasitemia in the P. berghei mouse model of malaria (one-tailed t-test; * p<0.05, ** p<0.01). An error bar is missing in the 3 day control because one animal was euthanized due to severe symptoms of malaria.
Series 1 and 2 were compared for their capacity to knock down NMT activity in parasites with the aim of rationalizing their diverse enzyme to cell translation (Fig. 5a and Supplementary Figs. S19 and S20). An excellent correlation was observed between TC50 and parasite viability (EC50) across all compounds tested (R2 = 0.97, Fig. 5a), establishing a direct link between in-cell NMT inhibition and efficacy, and resolving the differences in translation between members of series 1 and 2. In particular, compounds 1a and 2a have very similar effects on protein myristoylation in the cell despite their significantly different Ki values, resulting in similar EC50 values for parasite killing (Fig. 5a). These data demonstrate that 1a-c and 2a-b possess the same on-target mode of action, and establish the chemical tractability of NMT in blood stage human malaria parasites.
We also examined the impact of NMT inhibition on parasite burden in vivo in the rodent malaria parasite P. berghei. Both 1a and 2a maintained reasonable potency against recombinant PbNMT (Ki = 90 and 14 nM, respectively), but initial experiments found that 1a induced toxicity in vivo at sub-therapeutic concentrations. However, when 2a was administered to infected mice a significant (p ≤ 0.01, n = 3) reduction in parasitemia was observed compared to controls (PBS vehicle) over the course of 3 days treatment (50 mg/kg intraperitoneally, twice daily) (Fig. 5d). Furthermore, 2a caused no acute toxicity and animals survived without visible symptoms past day 3, whereas all control mice needed to be euthanized due to acute symptoms of malaria. This first demonstration of in vivo activity of a chemical inhibitor of Plasmodium NMT is encouraging, particularly given that the potency and pharmacokinetic properties of 2a are suboptimal (Supplementary Table S8).
NMT inhibition results in formation of non-viable parasites lacking critical components required for development and RBC invasion
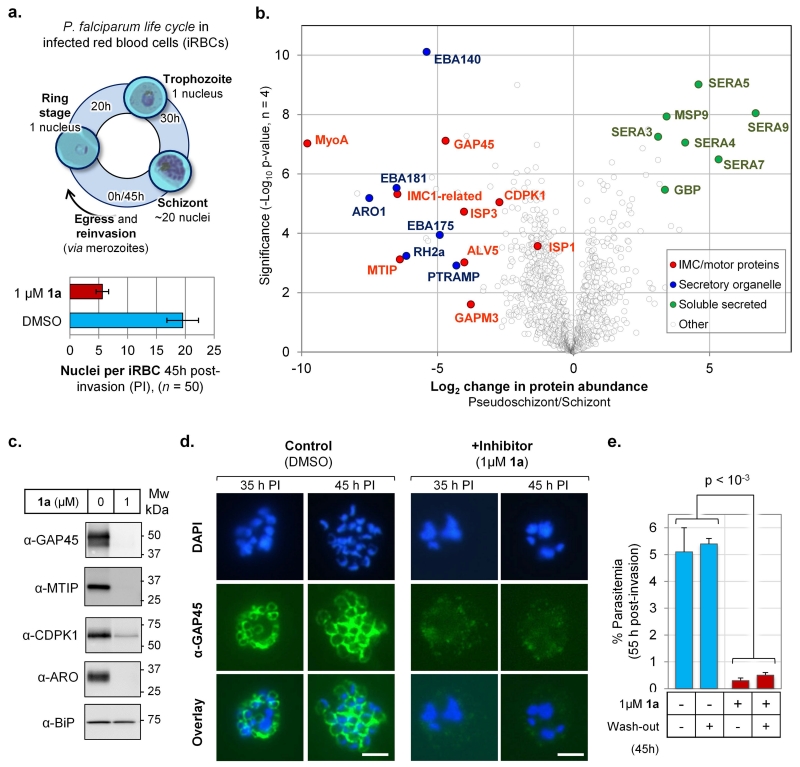
Having confirmed on-target action of NMT inhibitors in parasites, we sought to use these tools to investigate how NMT inhibition leads to parasite cell death. During asexual reproduction, P. falciparum undergoes a 45-48 hour life cycle within infected RBCs (iRBCs), maturing from ring stage, to trophozoite, and then schizont, before differentiation into merozoites that egress and reinvade RBCs (Fig. 6a). We examined the effects of NMT inhibition on life stage progression, monitoring growth at 1 μM 1a, a dose that causes approximately 80% inhibition of protein myristoylation in parasites (Fig. 5). NMT inhibitor or DMSO vehicle (control) was added to highly synchronized ring stage parasites 2 hours post-invasion (PI), and the parasite population was monitored up to 45 hours PI. Whilst control parasites progressed as expected over 45 hours to form multinucleate schizonts, 1a-treated parasites became trapped in a morphologically distinct life stage (here termed a ‘pseudoschizont’). Nuclear staining (DAPI) revealed that at 45 hours all (n = 50, p < 10−30) multinucleate parasites contained 4-7 (5.6 ± 1.1) nuclei in the presence of 1a, compared to ca. 20 (19.6 ± 2.7) nuclei in control schizonts (Fig. 6a and Supplementary Fig. S21). The clarity and severity of this phenotype is striking given that 1a was applied at a concentration that does not completely eliminate NMT activity in cells, and is consistent with the observed high sensitivity of parasites towards NMT inhibition (Fig. 5). At 66 hours PI, DMSO-treated parasites had egressed and reinvaded RBCs as expected, developing into new ring stages and resulting in an increase in parasitemia. However, in the presence of 1a parasitemia dropped by >85%, showing that NMT inhibition results in a substantial loss of parasite viability (Supplementary Fig. S22).
Figure 6. NMT inhibition results in loss of the inner membrane complex and irreversible loss of parasite viability prior to parasite egress.
(a) Top: P. falciparum intraerythrocytic life cycle showing approximate timings of life stages; parasite egress occurs ~45h post-invasion (PI). Bottom: mean number of nuclei per infected red blood cell (iRBC) 45h PI, +/− inhibitor 1a; error bars show ± standard deviation from mean (n = 50). (b) Label-free quantification (LFQ) of change in protein abundance plotted against significance of change for 1181 proteins in pseudoschizonts (NMT inhibition) vs. schizonts (DMSO), 45h PI; see Supplementary Table S9 for dataset; groups of proteins are highlighted by function. (c) Western blot of parasite proteins 45h PI, cultured in the presence of NMT inhibitor (1 μM 1a) or DMSO vehicle, showing depletion of IMC and motor proteins; BiP: loading control. (d) Indirect immunofluorescence microscopy of parasites 35h or 45h PI, cultured in presence of NMT inhibitor (1 μM 1a) or DMSO; green: secondary antibody fluorescence; blue: DAPI (nuclear stain); all images at identical magnification, white bar = 5 μm. (e) Synchronized parasites were cultured with NMT inhibitor (1 μM 1a) or DMSO vehicle to 45 h PI, whereupon medium was exchanged for medium containing either NMT inhibitor (1 μM 1a) or DMSO (45h wash-out), and parasite life stages analyzed at 55 h PI (p = 0.00081, two-tailed t-test; n = 3).
In order to characterize the distinguishing features of pseudoschizonts, we performed a quantitative proteomic analysis in pseudoschizonts (inhibitor-treated) and schizonts (control) at 45 hours PI. The relative abundance of 1181 parasite proteins was determined through label-free quantification, revealing a conspicuous clustering amongst strongly affected proteins with known function (Fig. 6b and Supplementary Table S9). Multiple markers of the Inner Membrane Complex (IMC) were greatly reduced in abundance in inhibited cells (e.g. MyoA, MTIP, GAP45, ISP3), as were a set of late-expressed proteins specific to apical (secretory) organelles in developing merozoites; these included ARO, PTRAMP and key RBC binding proteins and antigens (e.g. EBA140/175/181, RH2a). Western blot analyses of GAP45, MTIP, CDPK1 and ARO further confirmed marked depletion of these proteins in pseudoschizonts (Fig. 6c). Interestingly, there was relative accumulation of a set of soluble secreted proteins, including several members of the serine repeat antigen family (SERA) which under normal conditions are exported and processed during schizont development.
The IMC is comprised of flattened membrane compartments lying directly under the plasma membrane, where one of its roles is assembly of the molecular motor complex (including MTIP, MyoA and GAP45) that drives invasion of RBCs. In the intracellular parasite, assembly of the IMC occurs during schizogony and can be visualised with protein markers such as GAP4544. In Plasmodium nuclear division precedes cell division, and the IMC initially appears as small ring-like structures, which then extend around developing merozoites during schizont segmentation (cell division). This transition was recapitulated in immunofluorescence images of DMSO-treated parasites at 35 and 45 hours PI, whereas in the inhibitor treated parasites the IMC structure was not visible at either time point (Fig. 6d). Given the established importance of the IMC in parasite development and invasion, we hypothesized that NMT inhibition-induced loss of the IMC would have catastrophic and irreversible consequences for parasite viability. Indeed, when inhibited parasites (1 μM 1a) were analyzed 10 hours later (at 55 hours PI), iRBCs bearing pseudoschizonts were largely eliminated even when inhibitor was removed at 45 hours PI, whereas control parasites egressed and reinvaded new RBCs to form new ring stages over the same timeframe (Fig. 6e).
Discussion
Here we have established a novel chemical approach to study N-myristoylation and inhibition of NMT in the most important human malarial parasite, and demonstrated that NMT is an essential and chemically tractable target in P. falciparum. Metabolic chemical tagging has enabled the first proteome-wide survey of both N-myristoylated and GPI-anchored proteins in P. falciparum. Previously, identification of PfNMT substrates relied on painstaking generation of mutant parasites or specific antibodies and radiolabeling that are impractical for high-throughput studies. In contrast, our chemical approach identifies substrates directly across the proteome in their natural context in live parasites, without the need for genetic manipulation and substrate overexpression. In combination with a new reagent for the identification of protein lipidation sites by metabolic tagging, this technology provides high confidence assignment of over 30 NMT substrates; the majority are identified for the first time in this study, and have roles in a wide range of essential processes. These include protein trafficking and secretion (ARF1, Rab5b, ARO), ion channel regulation (PKAr), organelle biogenesis (GRASP1), motility (CDPK1, GAP45), inner membrane complex (IMC) integrity (ISP1, ISP3, GAP45), development (CDPK4), and protein homeostasis (P26s4). Combining PfNMT inhibitors with a chemical proteomic toolbox (YnMyr, AHA, AzKTB) backed up by extensive characterization (biochemical, structural, in vitro, in vivo) provides a powerful and self-consistent validation of on-target specificity. This study is also the first to apply AHA to track de novo protein synthesis in Plasmodium; notably, metabolic tagging proceeds efficiently without the need for defined media, and we anticipate that AHA will be a powerful alternative to radiolabeling for monitoring protein synthesis in malaria parasites.
Whilst work on fungi and trypanosomes suggests NMT may be an interesting potential target in infectious disease, Plasmodium parasites are evolutionarily specialized, and infect their host and mosquito vector through unique and highly complex mechanisms; the specific consequences of PfNMT inhibition were thus unknown prior to the work reported here. Our chemical approach offers a unique opportunity to examine the phenotype arising from perturbation of the pathways in which NMT substrates are involved, particularly since knockout of an essential gene such as PfNMT is currently intractable in human malaria parasites. In particular, we discovered that PfNMT inhibition induced an unexpected, irreversible and catastrophic failure to assemble the IMC during early schizogony in asexual blood stage parasites. This was accompanied by loss of essential components of the myosin motor that drives RBC invasion (MTIP, MyoA), as well as the absence of proteins critical for parasite engagement of new RBCs (EBA140/175/181, RH2a). A strong reduction in proteins synthesized late in intracellular development, including characteristic markers of organelles assembled at the apical end of developing merozoites (rhoptries/ARO, micronemes/EBA family), provides strong evidence that inhibited parasites are non-infectious33. Marked retention and failed processing of key proteins exported during late schizogony (SERA family, GBP, MSP9) further illustrates a failure to progress to merozoites. Interestingly, parasites commit to early rounds of nuclear division but are unable to proceed through cell division, supporting recent evidence for a key role of the IMC in development in addition to invasion44.
GAP45, ISP1 and ISP3 are NMT substrates identified here that are candidates for driving this IMC-related phenotype. N-Myristoylated GAP45 acts as a scaffold that spans the space between the plasma membrane and the IMC45; it also has an essential role in organizing the acto-myosin motor that drives invasion of RBCs44. We recently demonstrated that P. berghei ISP1 and ISP3 are highly expressed in invasive stages, and localize to the anterior apical end of the parasite where IMC organization is initiated39. Whilst deletion of ISP3 in P. berghei is compensated by up-regulation of ISP1, NMT inhibition may result in simultaneous inactivation of both proteins through loss of the membrane anchor. Loss of the IMC is sufficient to explain the irreversible and cidal mode of action of PfNMT inhibition during early schizogony, and failure to develop into infective merozoites. However, the comparative whole-proteome dataset obtained here shows that this is far from the only phenotype arising from PfNMT inhibition, and provides a rich resource that can be mined to initiate future studies of IMC formation, protein trafficking and assembly of the invasion apparatus.
Chemical knockdown of PfNMT effectively combines the benefits of a selective enzyme inhibitor with a defined multi-target, pleiotropic downstream mode of action. We have shown here that P. falciparum is highly sensitive to a global drop in N-myristoylation, and structural characterization of the binding modes of inhibitors at the protein substrate binding pocket of Plasmodium NMT provides the potential for future improvements in potency and selectivity through structure-guided design. Remarkably, despite unoptimized pharmacokinetic (PK) properties and modest in vitro potency, NMT inhibitor 2a significantly reduced parasitemia in the rodent model P. berghei with no acute toxicity, providing further evidence for the druggability of NMT in malaria. This in vivo proof of principle suggests that compounds with improved potency and PK properties could form the basis for an antimalarial drug discovery program46.
In future studies, the chemical tools described here may be applied to examine the role and druggability of PfNMT and N-myristoylation in other stages of the parasite, including in the sexual blood stages and liver stages that are important targets for blocking transmission. Our data suggest that NMT inhibitors could be highly effective against these life stages; for example CDPK4, identified here as an NMT substrate, is a promising target for kinase inhibitors that block development of sexual stage parasites38, whilst invasion of liver cells requires an intact IMC47. The chemical tools described here also provide a novel approach to study the role of the IMC in parasite development, given the striking effects of NMT inhibition on this structure. Future studies could usefully focus on proteins lacking functional annotation that are strongly changed in abundance in pseudoschizonts. Finally, contribution to parasite cell death could be explored for substrates identified here that have no annotated function, and our chemical tools will facilitate discovery of novel biology underlying these proteins.
Taken together, our results provide new insights into the scope and role of protein N-myristoylation in Plasmodium, and show for the first time that NMT is a chemically tractable target in the symptomatic stage of the most important human malarial parasite.
Methods
This section contains key experiments only. An extended experimental section is provided in the Supplementary Methods. Procedures used in this study were approved by the United Kingdom Home Office and carried out in compliance with European Directive 86/609/EEC for the protection of animals used for experimental purposes and in accordance with the United Kingdom “Animals (Scientific Procedures) Act 1986”.
Chemical probes
Synthesis of YnMyr, YnPal, AzTB and YnTB has been described previously20, 27. AHA was purchased from Iris Biotech. Synthesis of other compounds is described in the Supplementary Methods.
Parasite culture
Parasites were cultured as described previously and schizont stages purified using magnetic columns (MACS),48 before returning to culture for 2-4 h and synchronizing the newly invaded ring-stage parasites with 5 % D-sorbitol,49 one cycle prior to use.
YnMyr tagging and labeling
For tagging, assays were set up using parasites at 40 h post invasion (based on initial Giemsa-stained smear observations) and diluted to ~10 % parasitemia. A haematocrit of 4 % was used to increase the total number of parasites harvested. To each culture the required volume of YnMyr, YnPal, AHA or CHX was added from stocks, along with any inhibitors. DMSO controls were set up in parallel. Cultures were re-incubated (37 °C) for a further 5 h. Parasite cultures were pelleted and the red cells lysed in 1 × culture volume of 0.15 % saponin in PBS. Pellets were washed twice in the same buffer to remove red cell proteins. Parasite proteins were extracted using 1 % Triton X-100 in 10 mM Na2PO4 pH 8.2 with protease inhibitors (EDTA-free, Roche) and incubated overnight at 4 °C with rotation. Extracts were pelleted and the concentration of protein in the supernatant determined by DC protein assay (Bio-Rad).
CuAAC bioorthogonal ligation and pull-down of labeled proteins was carried out essentially as previously described21; full details are given in Supplementary Methods, along with details of in-gel fluorescence imaging and immunoblot analysis.
Proteomic identification of YnMyr tagged proteins by LC-MS/MS
Following CuAAC ligation and pull-down, samples were reduced, alkylated and digested on-bead with trypsin, as described in the Supplementary Methods. LC-MS/MS was carried out using an RSLCnano HPLC system (Dionex, UK) and an LTQ-Orbitrap-Velos mass spectrometer (Thermo Scientific). Data were further processed using Mascot (version 2.2.04, Matrix Science Ltd.) and Scaffold (version 3.6.02, Proteome Software). Protein identifications were required to contain at least 2 peptides exceeding 95 % probability giving a calculated protein false discovery rate of 0.0 %. Further experimental details and a description of data analysis are given in the Supplementary Methods.
Proteomics analysis of inhibited parasites
Equal protein quantities in whole cell lysate of parasites (45 h PI) cultured in presence of NMT inhibitor (1 μM 1a) or DMSO vehicle were processed by FASP™,50 subjected to tryptic digest and analyzed by nanoLC-MS/MS (n = 4). Protein identification (>99% confidence) and label-free quantification in MaxQuant produced a list of 1900 parasite proteins; data were filtered in Perseus v1.4.1.3 for completeness (minimum 3 quantifications per protein per group) yielding 1181 proteins for which relative abundance was determined using a two-tailed t-test (FDR = 0.05, 250 randomizations).
Supplementary Methods
Synthesis and characterization of compounds, protein crystallography, parasite assays, in vivo experiments, pharmacokinetics analysis, immunofluorescence and immunoblot analysis and further details of CuAAC labeling, gel analysis and LC-MS/MS proteomics is described in the Supplementary Methods. Substrate kinetic and inhibition assays for NMT were carried out essentially as described previously;25,51 details are given in the Supplementary Methods.
Supplementary Material
Acknowledgements
The authors acknowledge staff at the Diamond Light Source (Harwell, UK) for assistance with crystallography and Shirley Roberts for crystal handling. We are grateful to Lisa Haigh for assistance with mass spectrometry and to Paul Bowyer and members of the Tate group for critical reading of the manuscript, and also to Paul Wyatt for helpful discussions.
This work was supported by grants from the Institute of Chemical Biology (Imperial College London), the UK Engineering and Physical Sciences Research Council (Studentship awards and Doctoral Prize Fellowship awards to M.H.W. and M.D.R., grants EP/F500416/1 and EP/K039946/1), the UK Medical Research Council (grants G0900278, MR/K011782/1 and U117532067), the European Commission FP7 (2007-2013) (grant 242095), German Research Foundation (DFG, grant BR 4387/1-1), and the UK Biotechnology and Biological Sciences Research Council (grant BB/D02014X/1).
Footnotes
Accession codes
Primary accessions (Protein Data Bank): 2YNC (PvNMT-YnMyrCoA); 2YND (PvNMT-NHMCoA-1a); 2YNE (PvNMT-NHMCoA-2a).
Referenced accessions (Protein Data Bank): 1IID (ScNMT-NHM-GLYASKLA), 4A95 (PvNMT-NHM), 2WUU (LdNMT-NHM), 2P6E (ScNMT-MyrCoA), 2WSA (LmNMT-MyrCoA-1a), 3IWE (HsNMT1-MyrCoA-1a).
Additional information
Supplementary information and chemical compound information are available in the online version of the paper.
Competing Financial Interests Statement
The authors declare no competing financial interests.
References
- 1.Murray CJ, Rosenfeld LC, Lim SS, Andrews KG, Foreman KJ, Haring D, et al. Global malaria mortality between 1980 and 2010: a systematic analysis. Lancet. 2012;379(9814):413–431. doi: 10.1016/S0140-6736(12)60034-8. [DOI] [PubMed] [Google Scholar]
- 2.Price RN, Tjitra E, Guerra CA, Yeung S, White NJ, Anstey NM. Vivax malaria: neglected and not benign. Am J Trop Med Hyg. 2007;77(6 Suppl):79–87. [PMC free article] [PubMed] [Google Scholar]
- 3.Phyo AP, Nkhoma S, Stepniewska K, Ashley EA, Nair S, McGready R, et al. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet. 2012;379(9830):1960–1966. doi: 10.1016/S0140-6736(12)60484-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wright MH, Heal WP, Mann DJ, Tate EW. Protein myristoylation in health and disease. J Chem Biol. 2009;3(1):19–35. doi: 10.1007/s12154-009-0032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ebiike H, Masubuchi M, Liu P, Kawasaki K, Morikami K, Sogabe S, et al. Design and synthesis of novel benzofurans as a new class of antifungal agents targeting fungal N-myristoyltransferase. Part 2. Bioorg Med Chem Lett. 2002;12(4):607–610. doi: 10.1016/s0960-894x(01)00808-3. [DOI] [PubMed] [Google Scholar]
- 6.Frearson JA, Brand S, McElroy SP, Cleghorn LA, Smid O, Stojanovski L, et al. N-myristoyltransferase inhibitors as new leads to treat sleeping sickness. Nature. 2010;464(7289):728–732. doi: 10.1038/nature08893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tate EW, Bell AS, Rackham MD, Wright MH. N-Myristoyltransferase as a potential drug target in malaria and leishmaniasis. Parasitology. 2013 doi: 10.1017/S0031182013000450. 1-13. [DOI] [PubMed] [Google Scholar]
- 8.Traverso JA, Giglione C, Meinnel T. High-throughput profiling of N-Myristoylation substrate specificity across species including pathogens. Proteomics. 2013;13:25–36. doi: 10.1002/pmic.201200375. [DOI] [PubMed] [Google Scholar]
- 9.Moskes C, Burghaus PA, Wernli B, Sauder U, Durrenberger M, Kappes B. Export of Plasmodium falciparum calcium-dependent protein kinase 1 to the parasitophorous vacuole is dependent on three N-terminal membrane anchor motifs. Mol Microbiol. 2004;54(3):676–691. doi: 10.1111/j.1365-2958.2004.04313.x. [DOI] [PubMed] [Google Scholar]
- 10.Rees-Channer RR, Martin SR, Green JL, Bowyer PW, Grainger M, Molloy JE, et al. Dual acylation of the 45 kDa gliding-associated protein (GAP45) in Plasmodium falciparum merozoites. Mol Biochem Parasitol. 2006;149(1):113–116. doi: 10.1016/j.molbiopara.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Russo I, Oksman A, Goldberg DE. Fatty acid acylation regulates trafficking of the unusual Plasmodium falciparum calpain to the nucleolus. Mol Microbiol. 2009;72(1):229–245. doi: 10.1111/j.1365-2958.2009.06639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahlfs S, Koncarevic S, Iozef R, Mailu BM, Savvides SN, Schirmer RH, et al. Myristoylated adenylate kinase-2 of Plasmodium falciparum forms a heterodimer with myristoyltransferase. Mol Biochem Parasitol. 2009;163(2):77–84. doi: 10.1016/j.molbiopara.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Struck NS, Herrmann S, Langer C, Krueger A, Foth BJ, Engelberg K, et al. Plasmodium falciparum possesses two GRASP proteins that are differentially targeted to the Golgi complex via a higher- and lower-eukaryote-like mechanism. J Cell Sci. 2008;121(Pt 13):2123–2129. doi: 10.1242/jcs.021154. [DOI] [PubMed] [Google Scholar]
- 14.Resh MD. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat Chem Biol. 2006;2(11):584–590. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- 15.Gunaratne RS, Sajid M, Ling IT, Tripathi R, Pachebat JA, Holder AA. Characterization of N-myristoyltransferase from Plasmodium falciparum. Biochem J. 2000;348(Pt 2):459–463. [PMC free article] [PubMed] [Google Scholar]
- 16.Pino P, Sebastian S, Kim EA, Bush E, Brochet M, Volkmann K, et al. A tetracycline-repressible transactivator system to study essential genes in malaria parasites. Cell Host Microbe. 2012;12(6):824–834. doi: 10.1016/j.chom.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Limenitakis J, Soldati-Favre D. Functional genetics in Apicomplexa: potentials and limits. FEBS Lett. 2011;585(11):1579–1588. doi: 10.1016/j.febslet.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Heal WP, Tate EW. Getting a chemical handle on protein post-translational modification. Org Biomol Chem. 2010;8(4):731–738. doi: 10.1039/b917894e. [DOI] [PubMed] [Google Scholar]
- 19.Hang HC, Linder ME. Exploring protein lipidation with chemical biology. Chem Rev. 2011;111(10):6341–6358. doi: 10.1021/cr2001977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heal WP, Wickramasinghe SR, Leatherbarrow RJ, Tate EW. N-Myristoyl transferase-mediated protein labelling in vivo. Org Biomol Chem. 2008;6(13):2308–2315. doi: 10.1039/b803258k. [DOI] [PubMed] [Google Scholar]
- 21.Heal WP, Wright MH, Thinon E, Tate EW. Multifunctional protein labeling via enzymatic N-terminal tagging and elaboration by click chemistry. Nat Protoc. 2012;7(1):105–117. doi: 10.1038/nprot.2011.425. [DOI] [PubMed] [Google Scholar]
- 22.Charron G, Zhang MM, Yount JS, Wilson J, Raghavan AS, Shamir E, et al. Robust fluorescent detection of protein fatty-acylation with chemical reporters. J Am Chem Soc. 2009;131(13):4967–4975. doi: 10.1021/ja810122f. [DOI] [PubMed] [Google Scholar]
- 23.Hannoush RN, Arenas-Ramirez N. Imaging the lipidome: omega-alkynyl fatty acids for detection and cellular visualization of lipid-modified proteins. ACS Chem Biol. 2009;4(7):581–587. doi: 10.1021/cb900085z. [DOI] [PubMed] [Google Scholar]
- 24.Yap MC, Kostiuk MA, Martin DD, Perinpanayagam MA, Hak PG, Siddam A, et al. Rapid and selective detection of fatty acylated proteins using omega-alkynyl-fatty acids and click chemistry. J Lipid Res. 2010;51(6):1566–1580. doi: 10.1194/jlr.D002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goncalves V, Brannigan JA, Whalley D, Ansell KH, Saxty B, Holder AA, et al. Discovery of Plasmodium vivax N-myristoyltransferase inhibitors: screening, synthesis, and structural characterization of their binding mode. J Med Chem. 2012;55(7):3578–3582. doi: 10.1021/jm300040p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhatnagar RS, Futterer K, Farazi TA, Korolev S, Murray CL, Jackson-Machelski E, et al. Structure of N-myristoyltransferase with bound myristoylCoA and peptide substrate analogs. Nat Struct Biol. 1998;5(12):1091–1097. doi: 10.1038/4202. [DOI] [PubMed] [Google Scholar]
- 27.Heal WP, Jovanovic B, Bessin S, Wright MH, Magee AI, Tate EW. Bioorthogonal chemical tagging of protein cholesterylation in living cells. Chem Commun (Camb) 2011;47(14):4081–4083. doi: 10.1039/c0cc04710d. [DOI] [PubMed] [Google Scholar]
- 28.Green JL, Rees-Channer RR, Howell SA, Martin SR, Knuepfer E, Taylor HM, et al. The motor complex of Plasmodium falciparum: phosphorylation by a calcium-dependent protein kinase. J Biol Chem. 2008;283(45):30980–30989. doi: 10.1074/jbc.M803129200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerold P, Schofield L, Blackman MJ, Holder AA, Schwarz RT. Structural analysis of the glycosyl-phosphatidylinositol membrane anchor of the merozoite surface proteins-1 and -2 of Plasmodium falciparum. Mol Biochem Parasitol. 1996;75(2):131–143. doi: 10.1016/0166-6851(95)02518-9. [DOI] [PubMed] [Google Scholar]
- 30.Arumugam TU, Takeo S, Yamasaki T, Thonkukiatkul A, Miura K, Otsuki H, et al. Discovery of GAMA, a Plasmodium falciparum merozoite micronemal protein, as a novel blood-stage vaccine candidate antigen. Infect Immun. 2011;79(11):4523–4532. doi: 10.1128/IAI.05412-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilson PR, Nebl T, Vukcevic D, Moritz RL, Sargeant T, Speed TP, et al. Identification and stoichiometry of glycosylphosphatidylinositol-anchored membrane proteins of the human malaria parasite Plasmodium falciparum. Mol Cell Proteomics. 2006;5(7):1286–1299. doi: 10.1074/mcp.M600035-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Lin H, Su X, He B. Protein lysine acylation and cysteine succination by intermediates of energy metabolism. ACS Chem Biol. 2012;7(6):947–960. doi: 10.1021/cb3001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cabrera A, Herrmann S, Warszta D, Santos JM, John Peter AT, Kono M, et al. Dissection of minimal sequence requirements for rhoptry membrane targeting in the malaria parasite. Traffic. 2012;13:1335–1350. doi: 10.1111/j.1600-0854.2012.01394.x. [DOI] [PubMed] [Google Scholar]
- 34.Kimura A, Kato Y, Hirano H. N-Myristoylation of the Rpt2 subunit regulates intracellular localization of the yeast 26S proteasome. Biochemistry. 2012;51(44):8856–8866. doi: 10.1021/bi3007862. [DOI] [PubMed] [Google Scholar]
- 35.Li H, Ponder EL, Verdoes M, Asbjornsdottir KH, Deu E, Edgington LE, et al. Validation of the proteasome as a therapeutic target in Plasmodium using an epoxyketone inhibitor with parasite-specific toxicity. Chem Biol. 2012;19(12):1535–1545. doi: 10.1016/j.chembiol.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haste NM, Talabani H, Doo A, Merckx A, Langsley G, Taylor SS. Exploring the Plasmodium falciparum cyclic-adenosine monophosphate (cAMP)-dependent protein kinase (PfPKA) as a therapeutic target. Microbes Infect. 2012;14(10):838–850. doi: 10.1016/j.micinf.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rached FB, Ndjembo-Ezougou C, Chandran S, Talabani H, Yera H, Dandavate V, et al. Construction of a Plasmodium falciparum Rab-interactome identifies CK1 and PKA as Rab-effector kinases in malaria parasites. Biol Cell. 2012;104(1):34–47. doi: 10.1111/boc.201100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Billker O, Dechamps S, Tewari R, Wenig G, Franke-Fayard B, Brinkmann V. Calcium and a calcium-dependent protein kinase regulate gamete formation and mosquito transmission in a malaria parasite. Cell. 2004;117(4):503–514. doi: 10.1016/s0092-8674(04)00449-0. [DOI] [PubMed] [Google Scholar]
- 39.Poulin B, Patzewitz E-M, Brady D, Silvie O, Wright MH, Ferguson DJP, et al. Unique apicomplexan IMC sub-compartment proteins are early markers for apical polarity in the malaria parasite. Biol Open. 2013 doi: 10.1242/bio.20136163. in press; doi: 10.1242/bio.20136163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rackham MD, Brannigan JA, Moss DK, Yu Z, Wilkinson AJ, Holder AA, et al. Discovery of novel and ligand-efficient inhibitors of Plasmodium falciparum and Plasmodium vivax N-myristoyltransferase. J Med Chem. 2013;56(1):371–375. doi: 10.1021/jm301474t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brand S, Cleghorn LA, McElroy SP, Robinson DA, Smith VC, Hallyburton I, et al. Discovery of a novel class of orally active trypanocidal N-myristoyltransferase inhibitors. J Med Chem. 2011;55(1):140–152. doi: 10.1021/jm201091t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pasini EM, Kirkegaard M, Mortensen P, Lutz HU, Thomas AW, Mann M. In-depth analysis of the membrane and cytosolic proteome of red blood cells. Blood. 2006;108(3):791–801. doi: 10.1182/blood-2005-11-007799. [DOI] [PubMed] [Google Scholar]
- 43.Dieterich DC, Lee JJ, Link AJ, Graumann J, Tirrell DA, Schuman EM. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat Protoc. 2007;2(3):532–540. doi: 10.1038/nprot.2007.52. [DOI] [PubMed] [Google Scholar]
- 44.Ridzuan MA, Moon RW, Knuepfer E, Black S, Holder AA, Green JL. Subcellular location, phosphorylation and assembly into the motor complex of GAP45 during Plasmodium falciparum schizont development. PLoS One. 2012;7(3):e33845. doi: 10.1371/journal.pone.0033845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frenal K, Polonais V, Marq JB, Stratmann R, Limenitakis J, Soldati-Favre D. Functional dissection of the apicomplexan glideosome molecular architecture. Cell Host Microbe. 2010;8(4):343–357. doi: 10.1016/j.chom.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 46.Bell AS, Mills JE, Williams GP, Brannigan JA, Wilkinson AJ, Parkinson T, et al. Selective inhibitors of protozoan protein N-myristoyltransferases as starting points for tropical disease medicinal chemistry programs. PLoS Negl Trop Dis. 2012;6(4):e1625. doi: 10.1371/journal.pntd.0001625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bergman LW, Kaiser K, Fujioka H, Coppens I, Daly TM, Fox S, et al. Myosin A tail domain interacting protein (MTIP) localizes to the inner membrane complex of Plasmodium sporozoites. J Cell Sci. 2003;116(Pt 1):39–49. doi: 10.1242/jcs.00194. [DOI] [PubMed] [Google Scholar]
- 48.Ribaut C, Berry A, Chevalley S, Reybier K, Morlais I, Parzy D, et al. Concentration and purification by magnetic separation of the erythrocytic stages of all human Plasmodium species. Malar J. 2008;7:45. doi: 10.1186/1475-2875-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lambros C, Vanderberg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol. 1979;65(3):418–420. [PubMed] [Google Scholar]
- 50.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 51.Goncalves V, Brannigan JA, Thinon E, Olaleye TO, Serwa R, Lanzarone S, et al. A fluorescence-based assay for N-myristoyltransferase activity. Anal Biochem. 2012;421(1):342–344. doi: 10.1016/j.ab.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.