Abstract
Research in genetics of neurodevelopmental disorders such as autism suggests that several hundred genes are likely risk factors for these disorders. This heterogeneity presents a challenge and an opportunity at the same time. While the exact identity of many of the genes remains to be discovered, genes identified to date encode for proteins that play roles in certain conserved pathways: protein synthesis, transcriptional/epigenetic regulation and synaptic signaling. Next generation of research in neurodevelopmental disorders needs to address the neural circuitry underlying the behavioral symptoms and co-morbidities, the cell types playing critical roles in these circuits and common intercellular signaling pathways that link diverse genes. Results from clinical trials have been mixed so far. Only when we are able to leverage the heterogeneity of neurodevelopmental disorders into precision medicine, will the mechanism-based therapeutics for these disorders start to unlock success.
Neurodevelopmental disorders include a wide range of conditions such as epilepsy, intellectual disability and autism spectrum disorder (ASD). Patients with ASD exhibit early childhood onset of symptoms, first described over sixty years ago (1), that persist throughout life and produce significant impairments in social, communicative, cognitive and behavioral functioning (2). According to the Centers for Disease Control, ASD affects 1 in 68 children and 1 in 42 boys. ASD is a major public health problem that leads to significant disability and disrupts families, resulting in a total annual societal cost of ~$35 billion in the US alone (3).
ASD diagnosis is comprised of a constellation of behavioral symptoms as defined by a group of experts (DSM-5) and require persistent deficits in social communication and interaction across multiple contexts, as well as restricted, repetitive patterns of behavior, interests and activities. A key characteristic in ASD is its heterogeneity. Patients with ASD present with wide variation and levels of impairment with different co-morbidities, and the expression of these symptoms can change over time. Heterogeneity has been a huge obstacle in ASD research, but in recent years, researchers are starting to take advantage of the heterogeneity of ASD. Rather than focusing on “pure autism” (autism not confounded by intellectual disability) (4, 5), research has now opened up to examining genetic disorders with high penetrance of ASD, such as Fragile X syndrome, Rett syndrome and Tuberous Sclerosis Complex, which have now come to the forefront of translational efforts to find treatments for subsets of mechanism-based classification of ASD (6). Complementary to this effort, is the National Institute of Mental Health (NIMH) initiative to define psychiatric disorders according to mechanistic descriptions of symptom clusters rather than symptom inventories, also known as research domain criteria (7, 8). In ASD, the etiology seems to vary according to the individual’s genome and interaction with his/her environment. Genetic heterogeneity and overlap with other neuropsychiatric disorders makes it difficult to find unique risk factor for ASD. Improved understanding and classification of ASD based domains and levels of analysis could improve precision and treatment efficacy.
Here, we review research on neurodevelopmental disorders that spans genes, molecules, cells and circuits, as well as the whole individual and environment. We discuss current efforts and obstacles in clinical trials and offer recommendations for the future that lead towards precision medicine.
Genes
The genetic component of ASD susceptibility is evidenced by twin studies that demonstrated higher concordance of ASD among monozygotic than dizygotic twins, has benefitted from modern genome scanning initiatives to yield many new genes worthy of further study. Genome analysis revealed the association of copy number variants (such as 15q11–13, 16p11.2, and 22q11.2) and single nucleotide variants with ASD. Some of these variants are de novo (not found in either parent) and thus easier to deem as causal. Variants that are not de novo or sequencing variants that are not obviously deleterious are harder to evaluate. Several studies have used whole exome sequencing to reveal a number of ASD susceptibility genes, such as CHD8, GRIN2B, SCN2A etc. These studies estimate that 400–1,000 genes are involved in ASD susceptibility (9). The vast majority of ASD susceptibility genes have not yet been identified and will require much larger cohorts for adequate power, as was necessary for schizophrenia (10). Germ-line mutations are not the only contributor to brain disorders: somatic mutations that affect a subset of brain neurons can cause epilepsy, brain malformations, and quite possibly ASD (11). Somatic mosaicism affecting the brain will confound the genetic analysis of cohorts, which are almost always based on bulk DNA derived from the blood and intended to represent the inherited genome.
Along with larger cohort sizes, identifying many of the remaining hundreds of ASD susceptibility genes will require thoughtful and innovative study designs. One approach is to study families with consanguinity to reduce inherited variation and help identify rare recessive variants (12, 13). Another approach is study groups that are relatively protected from ASD. Since ASD is much more common among males than females, focusing on families with a history of severe autism among women appears to enrich for highly penetrant rare variants (14).
The estimated heritability of ASD is 0.7–0.8, which, while relatively high, leaves room for non-inherited factors, including de novo mutations and epigenetic and environmental factors, leading to a complex risk architecture. Environmental influences such as perinatal injury and maternal infection could play a significant role in the context of a susceptible genetic background and contribute to the development of ASD. For instance, premature infants with isolated cerebellar hemorrhage have a 30 fold higher incidence of ASDs compared to the general population (15, 16). Other epidemiological studies have implicated activation of the maternal immune system during gestation as a contributor to the development of various neuropsychiatric disorders (17–19) and more specifically in the development of autism (19–21). Maternal immune activation leads to region specific changes in brain cytokines (22) and neuropathological changes that can be detected even in nonhuman primates (23). Interestingly, maternal immune activation is implicated in the exacerbation of syndromic forms of ASD. For example, maternal immune activation has been shown to intensify social behavior deficits observed in Tsc2+/− mutant mice (24). Finally, the relationship between the gut microbiome and neurodevelopmental symptoms has attracted attention (25). Autism and accompanying gastrointestinal symptoms are associated with distinct gut microbial compositions (26). Furthermore, probiotic treatment can improve both the metabolite abnormalities and the behavioral deficits in a maternal immune activation mouse model, supporting a connection between gut microbiome and autism (27). Further studies are needed to test how robust these initial observations are and what cellular mechanisms mediate them.
Molecular and cellular pathways
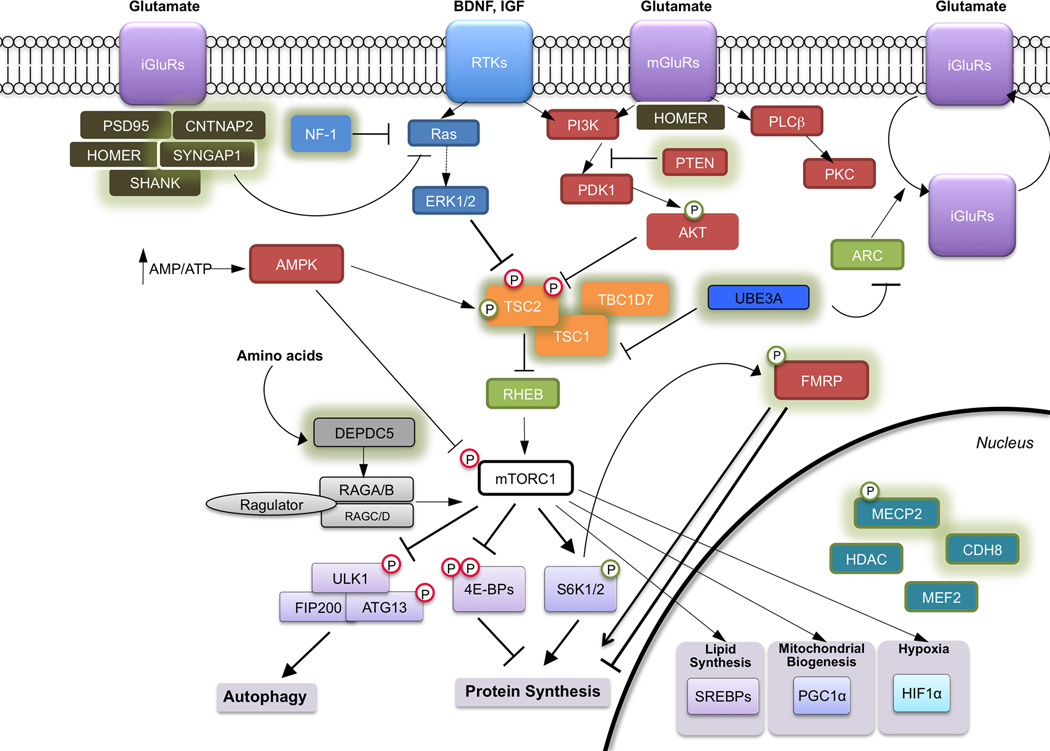
Every identified ASD susceptibility gene sheds new light on the cellular mechanisms underlying ASD. Many of the ASD-genes converge onto a few major signaling pathways: transcriptional control and chromatin remodeling, protein synthesis and cellular metabolism, and synapse development and function (6, 28–30). While many of these cellular processes are shared between neurons and non-neuronal cells, they appear to play roles particularly relevant to ASD in the brain (Figure 1).
Figure 1. Molecular pathways implicated in neurodevelopmental disorders.
Many of the genes mutated in individuals with ASD fall into several shared neuronal processes: transcriptional control and chromatin remodeling in the nucleus, protein synthesis, and synaptic structure. Proteins encoded by genes mutated in syndromes with high penetrance of ASD are shown with green outline. Many of the proteins (such as MECP2 and FMRP) have multiple functions and interactions in the cell, but are represented with the dominant functional role for the sake of clarity. Abbreviations not found in text include: RTKs = receptor tyrosine kinases; mGluRs = metabotropic glutamate receptors; iGluRs = metabotropic glutamate receptors; PGC-1α (Peroxisome proliferator-activated receptor gamma coactivator 1-alpha); SREBP = sterol-response binding proteins; HIF1α = hypoxia inducible factor 1 alpha; ULK1 = unc-51-like kinase 1; ARC = Activity-Regulated Cytoskeleton-Associated Protein; UBE3A = Ubiquitin Protein Ligase E3A.
Transcriptional control and chromatin remodeling
Several ASD genes influence transcription (31, 32), including those that are highly penetrant such as Rett Syndrome. MeCP2 (methyl CpG binding protein 2), which underlies Rett Syndrome, is a molecular multi-tasker that regulates gene expression by interacting with chromatin remodeling, transcription, and splicing (33). Initial findings suggested that MeCP2 binds to methylated CpG sites in the promoters of genes and associates with chromatin silencing complexes to repress gene expression (34–36). However, subsequent studies have demonstrated that MeCP2 also interacts with chromatin and transcriptional activators to activate gene expression (37, 38). MECP2 also mediates miRNA-mediated posttranscriptional control of gene expression (39, 40), as does FMRP (Fragile X Mental Retardation Protein) (41).
Signaling pathways and protein synthesis
One cellular process that has been implicated in multiple studies is that of mRNA synthesis and protein translation (28). Two key pathways of protein synthesis that contribute to synaptic function are the PI3K/mTOR pathway and the Ras-MAPK pathway. These pathways have been linked to neurodevelopmental disorders and to synaptic dysfunction. Due to the availability of specific and FDA-approved inhibitors, the mTOR pathway has been well characterized. Tuberous Sclerosis Complex (TSC) and PTEN Hamartoma Tumor Syndrome (PHTS) are two paradigmatic ‘mTORopathies’ such that loss of TSC1, TSC2 or PTEN function leads to activation of mTOR kinase activity and high incidence of intellectual disability, seizures and ASD (42). Other mutations in this pathway that present with ASD include the neurofibromin 1 (NF1) gene that result in neurofibromatosis type I. NF1 encodes a GTPase activating protein that suppresses the activity of the proto-oncogene Ras and also alter mTOR activity.
Dysregulation of protein synthesis is a prominent feature of several other neurodevelopmental disorders such as Fragile X syndrome (FXS) (43). FMRP is an mRNA binding protein that regulates the translation of mRNAs and is silenced in FXS, resulting in aberrant protein synthesis from key transcripts implicated in synaptic plasticity (44). Likewise, while MECP2 influences expression of several hundred genes (37), levels of BDNF (brain-derived neurotrophic factor) and IGF1 (insulin-like growth factor 1) are reduced in Mecp2 mutant mice, along with other molecules that cause both the PI3K/mTOR and ERK/MAPK pathways to be downregulated (37, 45–49). Treatment with recombinant human IGF1 upregulates these pathways in mice and iPSC-derived human neurons (50, 51), and ameliorates symptoms in mice (48). Preliminary results in human trials also appear promising (52). It is important to remember that PI3K/mTOR and ERK/MAPK pathways regulate a large number of cellular processes, including transcription, autophagy, metabolism, and organelle biogenesis and maintenance. The role of each of these cellular processes in the pathogenesis and therapeutics of ASD remain to be determined.
Disruptions of signaling pathways can change scaffolding of proteins at synapses. Such changes may cause neurodevelopmental disorders. PSD95 anchors NMDA/AMPA receptors at glutamatergic synapses. PSD95 expression is influenced by PI3K signaling; its levels, as well as excitatory synaptic transmission, are reduced in Mecp2 mutant mice (48) and rescued by IGF1 application. Similarly, SHANK3, which lies in the 22q13.3 deletion region associated with Phelan-McDermid Syndrome, encodes for a synaptic protein which regulates other protein partners such as PSD95; upregulation of the PI3K pathway by IGF1 rescues synaptic deficits in iPSC-derived human patient neurons (53) and Shank3 mutant mice (54), at least partly by upregulating PSD95.
Molecular convergence of pathways implicated by human genetics of ASD is apparent in studies of the Fmr1 knockout mouse. First, among the mRNA binding partners of FMRP are postsynaptic proteins such as SHANK3, and signaling proteins such as TSC2 and PTEN. Second, a number of studies in Fmr1 knockout mice indicate that interfering with protein synthesis in different ways can normalize the phenotype of the knockout mice. Knockout of S6 Kinase (55), Cebp (56), PI3 Kinase (57, 58) all are sufficient to ameliorate aspects of the Fmr1 knockout pathogenesis, raising the possibility of having multiple potential targets for intervening with loss of FMRP. Dysregulation of metabotropic glutamate receptor (mGluR) and aberrant mGluR-dependent long-term depression (LTD) has been reported first in FXS mouse models (59, 60), and several other ASD animal models including such as Nlgn3 (neuroligin 3) knockout and 16p11.2 knockout (61, 62). While we do not yet know if murine hippocampal LTD models the human ASD brain function, these findings raise the possibility that convergent cellular and molecular pathway targets exist in subsets of ASDs.
Brain regions and neural circuits
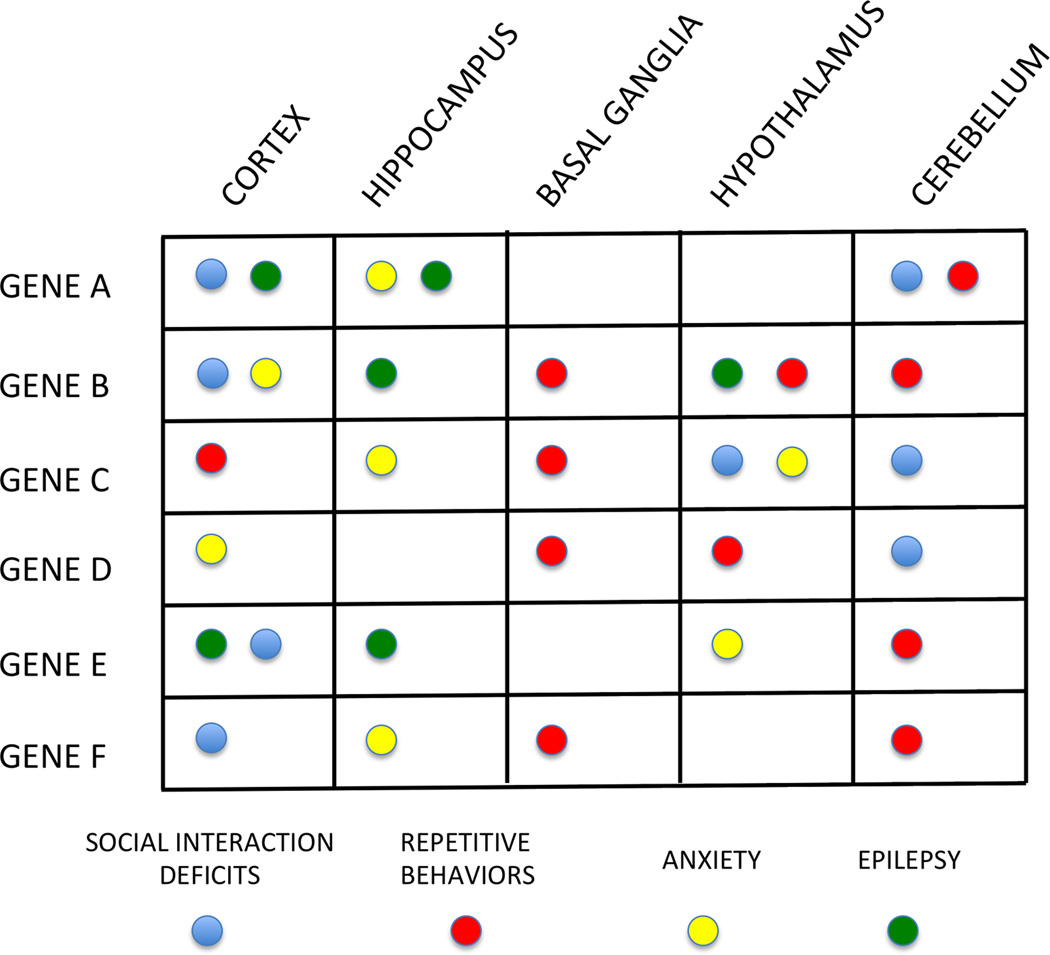
Molecular pathways in brain cells affect the function of neurons and synapses, and hence neuronal connectivity and circuits, to modify brain function. However, we lack insight about the brain regions and neuronal circuits underlying ASD. We do not yet know whether one cell type or circuit is crucial for the behavioral deficits observed in ASD patients. It is likely that different gene mutations perturb the neural circuitry underlying social interactions and repetitive behaviors at different nodes resulting in a complicated matrix of genes, brain regions and behavioral correlates (Figure 2).
Figure 2. Hypothetical matrix of genetic mutations and brain regions mapping onto behavioral profiles.
The approach to focus on mechanistic descriptions of symptom clusters rather than symptom inventories requires an understanding of the neural circuit(s) underlying these behavioral symptoms. One way to examine the neural circuits in animal models is to probe the relationship between a gene’s function in a certain brain region and the behavioral deficits in the animal. Use of conditional knockout mice has started to provide such information in certain genetic diseases such as TSC and RTT (93, 118). This matrix represents a hypothetical framework, which needs to be populated by future experimentation. One concrete example of this approach is currently in effect in epilepsy. Absence seizures are thought to arise from voltage-gated calcium channel dysfunction in the thalamus and respond best to ethosuximide treatment. In contrast, complex partial seizures occur due to increased excitation or decreased inhibition and thus respond to glutamate antagonists or GABAergic agonists. Such delineation of genetic, cellular and circuit defects may prove helpful in treating behavioral deficits associated with ASD with better precision as well.
Histopathological and imaging-based evidence that implicates specific brain regions and circuits underlying ASD is limited. The pathological studies are hampered by small sample size, and thus there is an urgent need for systematic and widespread collection of pathological specimens from those affected with a wide range of ASD. Imaging studies have been mostly performed on those with high functioning ASD because patients must be able to tolerate and comply with MRI protocols. Thus, while the functional MRI studies performed to date implicate certain areas of the brain in the “high functioning” ASD group, it is not clear whether the same circuits are involved in those who have more severe cognitive deficits. It is also possible that differences between ASD and control groups identified in such studies do not represent the aberrant circuits that are causally related to the behavioral abnormalities, but instead represent the activation of other brain regions that compensate for the neural circuitry abnormalities. Clinical protocols that enable MRI studies in ASD patients with intellectual disability and those who are younger will enhance our understanding of ASD and its associated neural circuitry. Such studies performed in individuals with genetically identified subsets of ASD may also shed light on genotype-phenotype correlations (63, 64).
In addition to functional MRI, complementary techniques to interrogate neuronal connectivity such as structural MRI, diffusion tensor imaging tractography, near-infrared spectroscopy, magnetoencephalography, and EEG can contribute to our understanding of brain connectivity at different time scales and with different spatial resolutions. It is likely that we will need to corroborate the findings from one modality with that from others to determine the most robust connectivity abnormalities in ASD. Some of these techniques may be more amenable for individuals with at different ages and at different functional levels.
Studies in genetic mouse models of ASD suggest abnormalities in both specific brain regions as well as in certain cell types. Two studies analyzed the co-expression patterns of a number of ASD genes in the human brain (65, 66). One study found enrichment in mid-gestation layer 5/6 cortical projection neurons and the other found enrichment in superficial cortical layers and glutamatergic projection neurons. While the exact layers of the cortex involved were different in the two analyses, the fact that cortical projection neurons were indicated in both studies is potentially significant.
In addition to cortical projection neurons, there is increasing evidence for the role of other neuronal subtypes in the pathogenesis of ASD. Multiple mouse models of ASD display reduction in parvalbumin (PV)-cells density in the neocortex (67). PV knockout mice display behavioral phenotypes with relevance to the core symptoms present in human ASD patients (68). In contrast, other groups have reported a selective increase in PV-immunopositive interneurons in the CA1 and CA3 subfields and calretinin-immunopositive neurons in CA1 in patients with ASD (69). Loss of PTEN in mice results in a preferential loss of a different subtype of GABAergic neurons, somatostatin (SST) interneurons (70). Interneuron specific deletions of ASD-related genes results in neurodevelopmental deficits in mice. For example, loss of MeCP2 from GABAergic interneurons leads to autistic-like repetitive movements, seizures and deficits in auditory event-related potentials (71, 72). Deficits in inhibitory neurotransmission, along with altered balance of excitation and inhibition (73) have been consistently observed in cortical and hippocampal neurons and circuits in diverse mouse models (74–77). In addition, the reversal potential of GABA may not mature fully when specific ASD genes are mutated, causing GABA to be depolarizing rather than hyperpolarizing (78). Consistent with these findings, a propensity for seizures is a major phenotype of ASDs. Taken together, these findings make a compelling case for dysregulation of inhibition as having a major role in neurodevelopmental disorders. More generally, cell type-specific and brain region-specific deletion of ASD genes is crucial for dissecting the circuit pathophysiology of ASD and in tying it to distinct symptom domains.
Connections between basal ganglia and cortex may underlie certain aspects of ASD. Neuroligin1 knockout mice exhibit ASD-like repetitive behaviors and abnormal corticostriatal synapses (79). Neuroligin3 mutations have similar abnormalities, but the defect appears to be due to a selective synaptic impairment in the nucleus accumbens/ventral striatum (80). SHANK3 is expressed in the basal ganglia, and Shank3 knockout mice exhibit repetitive grooming behavior, abnormal social interactions and changes at corticostriatal synapses (81).
The cerebellum is implicated in the pathogenesis of ASD via histopathology, imaging and epidemiological studies of injury. First, neuropathological studies demonstrate loss of cerebellar Purkinje cells in individuals diagnosed with ASD versus typically developing controls (82–85). Second, imaging studies of patients diagnosed with ASD indicate gray and white matter abnormalities in the cerebellum, dating to early childhood (86–90). Premature infants with isolated cerebellar hemorrhage have a higher incidence of ASDs, suggesting that cerebellar dysfunction early in life contributes to the pathogenesis of autism (15, 16). The developmental vulnerability of this circuit is further illustrated by study of genetic syndromes associated with ASD. Positron emission tomography (PET) studies in pediatric TSC patients with ASD demonstrate hyper-metabolism in the cerebellar nuclei, the output of the cerebellar cortex, in TSC patients with ASD, but not in TSC patients without ASD (91). The selective loss of Tsc1 or Tsc2 genes in the output cells of the cerebellum, the Purkinje neurons, appears to be sufficient to lead to an autistic-like phenotype in the two mouse models of TSC (92, 93). These findings suggest that abnormal cerebellar function contributes to ASD.
Non-neuronal cells in the brain such as astrocytes and microglia have also been implicated in the pathogenesis of neurodevelopmental disorders (94). Astrocyte processes extend into excitatory synapses, and they influence synaptic development (95) and synaptic transmission via uptake of glutamate (96) as well as by calcium-mediated alterations in synaptic function and plasticity (97, 98). ASD genes such as Fmr1 and Mecp2 are now known to influence astrocyte function (99). Astrocytes express mGluRs, providing a pathway for mGluR signaling to influence Fragile X pathophysiology (100). Astrocyte-specific restoration of Mecp2 in Mecp2 mutant mice restores function (101). Microglia also shape neuronal development and plasticity, and modulate synaptic transmission in the adult brain, via cytokine and chemokine release as well as phagocytosis (102, 103). Transplantation of wildtype microglia has been reported as reversing symptoms in a mouse model of Rett Syndrome, though the interpretation of these findings remains controversial (104, 105).
Treatments
Despite the many discoveries in basic neuroscience and human genetics, the number of drugs approved by the Food and Drug Administration for ASD patients is limited to risperidone (a dopamine antagonist) and aripiprazole (a dopamine agonist), which are both aimed at treating irritability and not the core features of ASD. Given the large number of genes that potentially confer ASD risk, the genetic heterogeneity of ASD presents a substantial obstacle to development of one-size fits all therapies. One can imagine several scenarios. It would be ideal to have one treatment for all causes of ASD. This seems rather unlikely; ASD is not one disease, and some genetic causes of ASD appear to have diametrically opposite manifestations at the synaptic level (106). It is also equally unlikely that different interventions can be developed for every genetic cause of ASD. So, the most realistic (and hopeful) scenario is that there will be a convergence upon a few molecular and circuit pathways that can be targeted by a limited number of interventions. Current focus is on the genetic syndromes with high penetrance of ASD symptoms, often caused by single-gene mutations (Table 1). The fact that mouse models of many of the syndromes associated with ASD respond positively to treatment, even in adulthood (107–109), has further bolstered optimism about the utility of pharmacological treatments in these disorders.
One of the first attempts at testing mechanism-based therapies in ASD was performed in FXS. The mGluR theory of Fragile X predicted that many symptoms of FXS are due to exaggerated responses to activation of mGluRs. This was demonstrated to be true in many animal models of FXS (110). Nonetheless, two mGluR antagonists (one made by Roche and another by Novartis) failed to show efficacy in Phase II trials (111). These negative results highlight the difficulties associated with clinical trials in neurodevelopmental disorders: Did the drugs engage their targets in the central nervous system? Were the endpoints chosen dynamic within the duration of the trial? Was the placebo effect too large? Was the right group of patients (e.g., patients at an appropriate stage of symptoms, or a subset with a particular genotype) chosen for enrollment?
Another important issue raised by these studies is how to best utilize animal models for developing therapies. Physiological and behavioral analyses in mice have been crucial for advancing our understanding of circuitry underlying social interactions and repetitive behaviors. However, a “good” mouse model needs to have both construct and face validity (112). Even more importantly, the circuit that is being interrogated needs to have some direct relevance to outcome measures in humans. Only then can the pharmacological interventions that modulate that circuit be translated effectively from mice to humans.
It is surprising that relatively few pharmacokinetic and pharmacodynamic (PK/PD) relationships are tested in preclinical studies in mouse models of neurodevelopmental disorders. While the pharmacokinetics will not be the same for a compound in mice and humans, understanding how much of the target needs to be engaged and over what period of time it must be engaged to achieve efficacy is crucial to interpret the preclinical data correctly and translate to clinical studies. To identify the correct target, more detailed preclinical studies will be necessary going forward. In terms of early stage clinical trials, many interventions look promising in open label studies but fail to show efficacy when compared to placebo. Thus, more placebo-controlled Phase II trials will be needed.
Biomarkers can be crucial for predicting subjects most likely to respond, confirm target engagement, and detect early signals of efficacy. Finding biomarkers that will segregate similarly diagnosed ASD patients into subsets of biologically more homogenous populations is a critical feature of good clinical trial design. A ‘stratification biomarker’ can be a biochemical measure from patient samples, a structural feature of a human imaging study, or a functional feature of an imaging or electrophysiological study. Aside from stratification, biomarkers can also be helpful in early diagnosis, assessing phenotype and severity, as well as measuring PK/PD in drug studies. Given that ASD represents circuit dysfunction, biomarkers that allow us to interrogate ASD-related circuits are likely to be most relevant. Especially, translatable biomarkers that can be employed in both mouse models and human subjects can be particularly powerful (e.g., EEG, MRI, visual or auditory evoked potentials, eye blink conditioning, etc. (64, 113–115)). Similarly, outcome measures that are circuit-based may be more fruitful in detecting efficacy rather than measurement of global functioning. A panel of relevant biomarkers, which together provide a unique profile of a patient, may be a crucial component of precision trial design in the future.
One of the most important questions about treatments is when mechanism-based treatments need to be initiated. Since the behavioral manifestations of ASD are quite early, one may have to intervene before symptoms arise. Animal models of syndromic ASD indicate that restoring function well into adulthood can rescue some of the symptoms of the disease (op. cit.). It is not yet clear if the same is true in humans and what exactly the critical windows for treatment are. However, regardless of age at treatment onset, relevant biomarkers and efficacy measures would be important for establishing the effectiveness of treatment.
One potential new tool to identity subjects who are likely to respond to a test drug is induced pluripotent stem cell (iPSC) derived neurons. This technology allows the possibility of testing the effects of a compound on a patient’s neurons first before giving it to the patient. Modeling the effects of mutations in iPSC-derived neurons can be informative about the molecular and cellular defects, but is unlikely with the current technology to provide insights on the emergent dysfunctions at the level of neuronal circuits. Nonetheless, a preliminary testing of efficacy in a patient’s iPSC-derived neurons should be a vital component of trial design for precision medicine.
Since many of the disease being targeted in these initial trials are genetic, they may be amenable to gene therapy in theory. Gene therapy using viral vectors is undergoing a renaissance and may be particularly applicable to diseases that arise from loss of function of a particular gene such as MECP2 or CDKL5 (cyclin-dependent kinase-like 5). Aside from the delivery issues, one needs to pay close attention to the dosage effects, since many of the genes that result in an ASD-related phenotype also have deleterious effects when they are expressed at high dosage. Thus, expression of the exogenous genes may have to be regulated tightly in spatial and temporal terms as well as levels of expression.
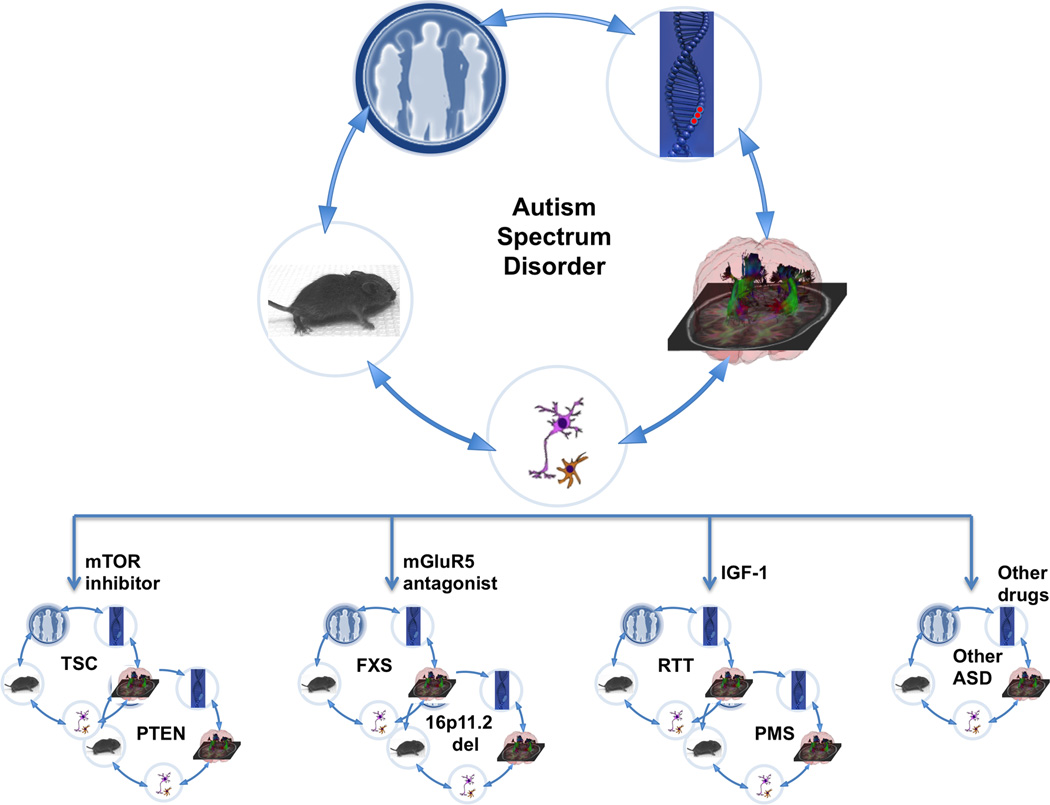
If we reach success in single genetically defined syndromes, there will be two new roadblocks in generalizing these findings to the larger ASD population. First, such an extension will require a comparative analysis of the different genetically defined causes of ASD to determine whether effective treatments in one may also be effective in another (Figure 3). Such comparative understanding of the genetic etiologies underlying ASD is in its infancy (106). A second and more difficult hurdle will be applying these findings to the “idiopathic” genetically unknown/undefined ASD population. Currently, we do not have an analytical tool to determine if an individual with ASD would benefit from a treatment that is effective, for example, in TSC or one effective in FXS. A marker to classify patients according to genetic, biochemical or circuit abnormalities does not yet exist. However, even single gene conditions involve multiple potential targets, and combination therapies are likely to be more effective than single drugs for single targets (47). The success of targeted pharmacological interventions would require integration of multiple kinds of data: knowledge of the genetic mutation and its signaling pathways and synaptic molecules, effectiveness of the therapy on neuronal and synaptic phenotypes in patient-derived neurons and non-neuronal cells in culture, and even analysis of transplanted human neurons in mice.
Figure 3. Translational research and clinical trials in ASD.
Translational studies in ASD have gained momentum from genetically defined causes such as FXS, TSC and RTT. The patients with these disorders are phenotyped in detail using advanced imaging and electrophysiology studies, with the aim of identifying potential biomarkers. There are cell-based models (both rodent and human) as well as mouse models of these syndromes enabling preclinical trials. Together, these efforts have led to clinical trials in some of these disorders. Based on the preclinical trials, the hypothesis is that different etiologies of ASD will respond to different therapies such as mTOR inhibitors (TSC and PTEN), mGluR5 antagonists (FXS and 16p11.2 deletion) and IGF-1 (RTT and PMS). Subsets of non-syndromic ASD patients may also benefit from one of these therapies, but further studies will be required to provide the tools and methods to stratify the individuals with non-syndromic ASD into treatment groups. It is important to remember that the discovery cycle will likely take more than one round in order to achieve safe and effective therapies for these disorders.
While pharmacological treatments may normalize neuronal and synaptic abnormalities, cognitive function is still dependent on complex circuits and interaction of the individual with his/her environment. Thus, pharmacological treatments alone may not be sufficient to reach the optimal outcome without behavioral treatments. Behavioral interventions appear promising in mouse models (116, 117) and could be combined with pharmacological interventions in future clinical trials. While simply correcting the synaptic abnormality using a pharmacological agent may not be sufficient to affect behavioral changes, it could accelerate the rate of learning and sociability in the setting of behavioral interventions. Although combining treatments adds complexity to the trial design, a few such trials are in the planning stages. Trials based on a mechanistic understanding of the disease, performed on a well defined group of subjects, with evidence of target engagement and supportive biomarkers, are the most likely to succeed. Once such trials prove effective in the highly penetrant genetic syndromes, the next challenge will be to identify patients with idiopathic autism who may benefit from the same treatment. Such an approach will finally realize the notion of precision medicine for autism and related neurodevelopmental disorders.
Acknowledgments
We thank Robin Kleiman, Annapurna Poduri and Kira Dies for critically reviewing the manuscript. Due to limited space we have not quoted all literature in the field, and we apologize to those whose articles are not referenced. Research in Mriganka Sur’s laboratory is supported by NIH grants MH085802 and EY007023 and the Simons Foundation Autism Research Initiative. Research in Mustafa Sahin’s laboratory is supported by the NIH (U01 NS082320, P20 NS080199, P30 HD018655), Department of Defense (W81XWH-13-1-0040, W81XWH-15-1-0189), Tuberous Sclerosis Alliance, Autism Speaks, Nancy Lurie Marks Family Foundation, Simons Foundation, Boston Children’s Hospital Translational Research Program, and Novartis, and Shire. The Developmental Synaptopathies Consortium (U54NS092090) is a part of the NCATS Rare Diseases Clinical Research Network (RDCRN). RDCRN is an initiative of the Office of Rare Diseases Research (ORDR), NCATS, funded through collaboration between NCATS, NIMH, NINDS and NICHD.
Contributor Information
Mustafa Sahin, Email: mustafa.sahin@childrens.harvard.edu.
Mriganka Sur, Email: msur@mit.edu.
References and Notes
- 1.Kanner L. Autistic disturbances of affective contact. Nervous Child. 1943;2:217–250. [PubMed] [Google Scholar]
- 2.Lord C. In: Understanding Autism: From Basic Neuroscience to Treatment. Moldin SO, Rubenstein JLR, editors. Boca Raton, FL: Taylor & Francis; 2006. pp. 1–23. [Google Scholar]
- 3.Ganz M. In: Understanding Autism: From Basic Neuroscience to Treatment. Moldin SO, Rubenstein JLR, editors. Boca Raton, FL: Taylor & Francis; 2006. pp. 475–502. [Google Scholar]
- 4.Vivanti G, Barbaro J, Hudry K, Dissanayake C, Prior M. Intellectual development in autism spectrum disorders: new insights from longitudinal studies. Front Hum Neurosci. 2013;7:354. doi: 10.3389/fnhum.2013.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillberg C, Fernell E. Autism plus versus autism pure. J Autism Dev Disord. 2014;44:3274–3276. doi: 10.1007/s10803-014-2163-1. [DOI] [PubMed] [Google Scholar]
- 6.Ebrahimi-Fakhari D, Sahin M. Autism and the synapse: emerging mechanisms and mechanism-based therapies. Curr Opin Neurol. 2015;28:91–102. doi: 10.1097/WCO.0000000000000186. [DOI] [PubMed] [Google Scholar]
- 7.Casey BJ, Oliveri ME, Insel T. A neurodevelopmental perspective on the research domain criteria (RDoC) framework. Biol Psychiatry. 2014;76:350–353. doi: 10.1016/j.biopsych.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Insel T, et al. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am J Psychiatry. 2010;167:748–751. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- 9.Geschwind DH, State MW. Gene hunting in autism spectrum disorder: on the path to precision medicine. Lancet Neurol. 2015 doi: 10.1016/S1474-4422(15)00044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schizophrenia C. Working Group of the Psychiatric Genomics, Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poduri A, Evrony GD, Cai X, Walsh CA. Somatic mutation, genomic variation, and neurological disease. Science. 2013;341:1237758. doi: 10.1126/science.1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu TW, et al. Using whole-exome sequencing to identify inherited causes of autism. Neuron. 2013;77:259–273. doi: 10.1016/j.neuron.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morrow EM, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turner TN, et al. Loss of delta-catenin function in severe autism. Nature. 2015;520:51–56. doi: 10.1038/nature14186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Limperopoulos C, et al. Does cerebellar injury in premature infants contribute to the high prevalence of long-term cognitive, learning, and behavioral disability in survivors? Pediatrics. 2007;120:584–593. doi: 10.1542/peds.2007-1041. [DOI] [PubMed] [Google Scholar]
- 16.Limperopoulos C, Chilingaryan G, Guizard N, Robertson RL, Du Plessis AJ. Cerebellar injury in the premature infant is associated with impaired growth of specific cerebral regions. Pediatric research. 2010;68:145–150. doi: 10.1203/PDR.0b013e3181e1d032. [DOI] [PubMed] [Google Scholar]
- 17.Shi L, Fatemi SH, Sidwell RW, Patterson PH. Maternal Influenza Infection Causes Marked Behavioral and Pharmacological Changes in the Offspring. J Neurosci. 2003;23:297–302. doi: 10.1523/JNEUROSCI.23-01-00297.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patterson PH. Immune involvement in schizophrenia and autism: Etiology, pathology and animal models. Behav Brain Res. 2009;204:313–321. doi: 10.1016/j.bbr.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 19.Garay PA, McAllister AK. Novel roles for immune molecules in neural development: implications for neurodevelopmental disorders. Front Synaptic Neurosci. 2010;2:136. doi: 10.3389/fnsyn.2010.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garbett K, et al. Immune transcriptome alterations in the temporal cortex of subjects with autism. Neurobiol Dis. 2008;30:303–311. doi: 10.1016/j.nbd.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vargas DL, Nascimbene C, Krishnan C, Zimmerman AW, Pardo CA. Neuroglial activation and neuroinflammation in the brain of patients with autism. Ann Neurol. 2005;57:67–81. doi: 10.1002/ana.20315. [DOI] [PubMed] [Google Scholar]
- 22.Garay PA, Hsiao EY, Patterson PH, McAllister AK. Maternal immune activation causes age- and region-specific changes in brain cytokines in offspring throughout development. Brain Behav Immun. 2013;31:54–68. doi: 10.1016/j.bbi.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weir RK, et al. Preliminary evidence of neuropathology in nonhuman primates prenatally exposed to maternal immune activation. Brain Behav Immun. 2015;48:139–146. doi: 10.1016/j.bbi.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ehninger D, et al. Gestational immune activation and Tsc2 haploinsufficiency cooperate to disrupt fetal survival and may perturb social behavior in adult mice. Mol Psychiatry. 2012;17:62–70. doi: 10.1038/mp.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mulle JG, Sharp WG, Cubells JF. The gut microbiome: a new frontier in autism research. Curr Psychiatry Rep. 2013;15:337. doi: 10.1007/s11920-012-0337-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang DW, et al. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS One. 2013;8:e68322. doi: 10.1371/journal.pone.0068322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsiao EY, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelleher RJ, 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 29.Zoghbi HY, Bear MF. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a009886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sudhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iossifov I, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernier R, et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158:263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castro J, Mellios N, Sur M. Mechanisms and therapeutic challenges in autism spectrum disorders: insights from Rett syndrome. Curr Opin Neurol. 2013;26:154–159. doi: 10.1097/WCO.0b013e32835f19a7. [DOI] [PubMed] [Google Scholar]
- 34.Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 35.Meehan RR, Lewis JD, Bird AP. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992;20:5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones PL, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 37.Chahrour M, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ben-Shachar S, Chahrour M, Thaller C, Shaw CA, Zoghbi HY. Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus. Hum Mol Genet. 2009;18:2431–2442. doi: 10.1093/hmg/ddp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Urdinguio RG, et al. Disrupted microRNA expression caused by Mecp2 loss in a mouse model of Rett syndrome. Epigenetics. 2010;5:656–663. doi: 10.4161/epi.5.7.13055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu H, et al. Genome-wide analysis reveals methyl-CpG-binding protein 2-dependent regulation of microRNAs in a mouse model of Rett syndrome. Proc Natl Acad Sci U S A. 2010;107:18161–18166. doi: 10.1073/pnas.1005595107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lipton JO, Sahin M. The neurology of mTOR. Neuron. 2014;84:275–291. doi: 10.1016/j.neuron.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qin M, et al. Altered cerebral protein synthesis in fragile X syndrome: studies in human subjects and knockout mice. J Cereb Blood Flow Metab. 2013;33:499–507. doi: 10.1038/jcbfm.2012.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang Q, Khare G, Dani V, Nelson S, Jaenisch R. The disease progression of Mecp2 mutant mice is affected by the level of BDNF expression. Neuron. 2006;49:341–348. doi: 10.1016/j.neuron.2005.12.027. [DOI] [PubMed] [Google Scholar]
- 46.Tropea D, et al. Partial reversal of Rett Syndrome-like symptoms in MeCP2 mutant mice. Proc Natl Acad Sci U S A. 2009;106:2029–2034. doi: 10.1073/pnas.0812394106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mellios N, et al. beta2-Adrenergic receptor agonist ameliorates phenotypes and corrects microRNA-mediated IGF1 deficits in a mouse model of Rett syndrome. Proc Natl Acad Sci U S A. 2014;111:9947–9952. doi: 10.1073/pnas.1309426111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Castro J, et al. Functional recovery with recombinant human IGF1 treatment in a mouse model of Rett Syndrome. Proc Natl Acad Sci U S A. 2014;111:9941–9946. doi: 10.1073/pnas.1311685111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ricciardi S, et al. Reduced AKT/mTOR signaling and protein synthesis dysregulation in a Rett syndrome animal model. Hum Mol Genet. 2011;20:1182–1196. doi: 10.1093/hmg/ddq563. [DOI] [PubMed] [Google Scholar]
- 50.Marchetto MC, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y, et al. Global transcriptional and translational repression in human-embryonic-stem-cell-derived Rett syndrome neurons. Cell Stem Cell. 2013;13:446–458. doi: 10.1016/j.stem.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khwaja OS, et al. Safety, pharmacokinetics, and preliminary assessment of efficacy of mecasermin (recombinant human IGF-1) for the treatment of Rett syndrome. Proc Natl Acad Sci U S A. 2014;111:4596–4601. doi: 10.1073/pnas.1311141111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shcheglovitov A, et al. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature. 2013;503:267–271. doi: 10.1038/nature12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bozdagi O, Tavassoli T, Buxbaum JD. Insulin-like growth factor-1 rescues synaptic and motor deficits in a mouse model of autism and developmental delay. Molecular autism. 2013;4:9. doi: 10.1186/2040-2392-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhattacharya A, et al. Genetic removal of p70 S6 kinase 1 corrects molecular, synaptic, and behavioral phenotypes in fragile X syndrome mice. Neuron. 2012;76:325–337. doi: 10.1016/j.neuron.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Udagawa T, et al. Genetic and acute CPEB1 depletion ameliorate fragile X pathophysiology. Nat Med. 2013;19:1473–1477. doi: 10.1038/nm.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gross C, et al. Selective role of the catalytic PI3K subunit p110beta in impaired higher order cognition in fragile X syndrome. Cell Rep. 2015;11:681–688. doi: 10.1016/j.celrep.2015.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gross C, et al. Increased expression of the PI3K enhancer PIKE mediates deficits in synaptic plasticity and behavior in fragile X syndrome. Cell Rep. 2015;11:727–736. doi: 10.1016/j.celrep.2015.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huber KM. Role for Rapid Dendritic Protein Synthesis in Hippocampal mGluR-Dependent Long-Term Depression. Science. 2000;288:1254–1256. doi: 10.1126/science.288.5469.1254. [DOI] [PubMed] [Google Scholar]
- 60.Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci U S A. 2002;99:7746–7750. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tian D, et al. Contribution of mGluR5 to pathophysiology in a mouse model of human chromosome 16p11.2 microdeletion. Nat Neurosci. 2015;18:182–184. doi: 10.1038/nn.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baudouin SJ, et al. Shared synaptic pathophysiology in syndromic and nonsyndromic rodent models of autism. Science. 2012;338:128–132. doi: 10.1126/science.1224159. [DOI] [PubMed] [Google Scholar]
- 63.Scott-Van Zeeland AA, et al. Altered functional connectivity in frontal lobe circuits is associated with variation in the autism risk gene CNTNAP2. Science translational medicine. 2010;2:56ra80. doi: 10.1126/scitranslmed.3001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peters JM, et al. Loss of white matter microstructural integrity is associated with adverse neurological outcome in tuberous sclerosis complex. Acad Radiol. 2012;19:17–25. doi: 10.1016/j.acra.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Willsey AJ, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parikshak NN, et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gogolla N, et al. Common circuit defect of excitatory-inhibitory balance in mouse models of autism. J Neurodev Disord. 2009;1:172–181. doi: 10.1007/s11689-009-9023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wohr M, et al. Lack of parvalbumin in mice leads to behavioral deficits relevant to all human autism core symptoms and related neural morphofunctional abnormalities. Transl Psychiatry. 2015;5:e525. doi: 10.1038/tp.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lawrence YA, Kemper TL, Bauman ML, Blatt GJ. Parvalbumin-, calbindin-, and calretinin-immunoreactive hippocampal interneuron density in autism. Acta Neurol Scand. 2010;121:99–108. doi: 10.1111/j.1600-0404.2009.01234.x. [DOI] [PubMed] [Google Scholar]
- 70.Vogt D, Cho KK, Lee AT, Sohal VS, Rubenstein JL. The Parvalbumin/Somatostatin Ratio Is Increased in Pten Mutant Mice and by Human PTEN ASD Alleles. Cell Rep. 2015;11:944–956. doi: 10.1016/j.celrep.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goffin D, Brodkin ES, Blendy JA, Siegel SJ, Zhou Z. Cellular origins of auditory event-related potential deficits in Rett syndrome. Nat Neurosci. 2014;17:804–806. doi: 10.1038/nn.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chao HT, et al. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263–269. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rubenstein JL, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2003;2:255–267. doi: 10.1034/j.1601-183x.2003.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Calfa G, Li W, Rutherford JM, Pozzo-Miller L. Excitation/inhibition imbalance and impaired synaptic inhibition in hippocampal area CA3 of Mecp2 knockout mice. Hippocampus. 2015;25:159–168. doi: 10.1002/hipo.22360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dani VS, et al. Reduced cortical activity due to a shift in the balance between excitation and inhibition in a mouse model of Rett syndrome. Proc Natl Acad Sci U S A. 2005;102:12560–12565. doi: 10.1073/pnas.0506071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chao HT, Zoghbi HY, Rosenmund C. MeCP2 controls excitatory synaptic strength by regulating glutamatergic synapse number. Neuron. 2007;56:58–65. doi: 10.1016/j.neuron.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wood L, Gray NW, Zhou Z, Greenberg ME, Shepherd GM. Synaptic circuit abnormalities of motor-frontal layer 2/3 pyramidal neurons in an RNA interference model of methyl-CpG-binding protein 2 deficiency. J Neurosci. 2009;29:12440–12448. doi: 10.1523/JNEUROSCI.3321-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ben-Ari Y, Gaiarsa JL, Tyzio R, Khazipov R. GABA: a pioneer transmitter that excites immature neurons and generates primitive oscillations. Physiol Rev. 2007;87:1215–1284. doi: 10.1152/physrev.00017.2006. [DOI] [PubMed] [Google Scholar]
- 79.Blundell J, et al. Neuroligin-1 deletion results in impaired spatial memory and increased repetitive behavior. J Neurosci. 2010;30:2115–2129. doi: 10.1523/JNEUROSCI.4517-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rothwell PE, et al. Autism-associated neuroligin-3 mutations commonly impair striatal circuits to boost repetitive behaviors. Cell. 2014;158:198–212. doi: 10.1016/j.cell.2014.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Peca J, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bauman ML, Kemper TL. Neuroanatomic observations of the brain in autism: a review and future directions. Int J Dev Neurosci. 2005;23:183–187. doi: 10.1016/j.ijdevneu.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 83.Casanova MF. The neuropathology of autism. Brain Pathol. 2007;17:422–433. doi: 10.1111/j.1750-3639.2007.00100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fatemi SH, et al. Consensus paper: pathological role of the cerebellum in autism. Cerebellum. 2012;11:777–807. doi: 10.1007/s12311-012-0355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Whitney ER, Kemper TL, Bauman ML, Rosene DL, Blatt GJ. Cerebellar Purkinje cells are reduced in a subpopulation of autistic brains: a stereological experiment using calbindin-D28k. Cerebellum. 2008;7:406–416. doi: 10.1007/s12311-008-0043-y. [DOI] [PubMed] [Google Scholar]
- 86.Courchesne E, Yeung-Courchesne R, Press GA, Hesselink JR, Jernigan TL. Hypoplasia of cerebellar vermal lobules VI and VII in autism. N Engl J Med. 1988;318:1349–1354. doi: 10.1056/NEJM198805263182102. [DOI] [PubMed] [Google Scholar]
- 87.Amaral DG, Schumann CM, Nordahl CW. Neuroanatomy of autism. Trends Neurosci. 2008;31:137–145. doi: 10.1016/j.tins.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 88.Kates WR, et al. Neuroanatomic variation in monozygotic twin pairs discordant for the narrow phenotype for autism. Am J Psychiatry. 2004;161:539–546. doi: 10.1176/appi.ajp.161.3.539. [DOI] [PubMed] [Google Scholar]
- 89.Akshoomoff N, et al. Outcome classification of preschool children with autism spectrum disorders using MRI brain measures. J Am Acad Child Adolesc Psychiatry. 2004;43:349–357. doi: 10.1097/00004583-200403000-00018. [DOI] [PubMed] [Google Scholar]
- 90.Allen G, Courchesne E. Differential effects of developmental cerebellar abnormality on cognitive and motor functions in the cerebellum: an fMRI study of autism. Am J Psychiatry. 2003;160:262–273. doi: 10.1176/appi.ajp.160.2.262. [DOI] [PubMed] [Google Scholar]
- 91.Asano E, et al. Autism in tuberous sclerosis complex is related to both cortical and subcortical dysfunction. Neurology. 2001;57:1269–1277. doi: 10.1212/wnl.57.7.1269. [DOI] [PubMed] [Google Scholar]
- 92.Reith RM, et al. Loss of Tsc2 in Purkinje cells is associated with autistic-like behavior in a mouse model of tuberous sclerosis complex. Neurobiol Dis. 2013;51:93–103. doi: 10.1016/j.nbd.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 93.Tsai PT, et al. Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature. 2012;488:647–651. doi: 10.1038/nature11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Molofsky AV, et al. Astrocytes and disease: a neurodevelopmental perspective. Genes Dev. 2012;26:891–907. doi: 10.1101/gad.188326.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Eroglu C, Barres BA. Regulation of synaptic connectivity by glia. Nature. 2010;468:223–231. doi: 10.1038/nature09612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schummers J, Yu H, Sur M. Tuned responses of astrocytes and their influence on hemodynamic signals in the visual cortex. Science. 2008;320:1638–1643. doi: 10.1126/science.1156120. [DOI] [PubMed] [Google Scholar]
- 97.Chen N, et al. Nucleus basalis-enabled stimulus-specific plasticity in the visual cortex is mediated by astrocytes. Proc Natl Acad Sci U S A. 2012;109:E2832–E2841. doi: 10.1073/pnas.1206557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Haydon PG, Nedergaard M. How do astrocytes participate in neural plasticity? Cold Spring Harb Perspect Biol. 2015;7:a020438. doi: 10.1101/cshperspect.a020438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yasui DH, et al. MeCP2 modulates gene expression pathways in astrocytes. Molecular autism. 2013;4:3. doi: 10.1186/2040-2392-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Higashimori H, et al. Astroglial FMRP-dependent translational down-regulation of mGluR5 underlies glutamate transporter GLT1 dysregulation in the fragile X mouse. Hum Mol Genet. 2013;22:2041–2054. doi: 10.1093/hmg/ddt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lioy DT, et al. A role for glia in the progression of Rett's syndrome. Nature. 2011;475:497–500. doi: 10.1038/nature10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xavier AL, Menezes JR, Goldman SA, Nedergaard M. Fine-tuning the central nervous system: microglial modelling of cells and synapses. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130593. doi: 10.1098/rstb.2013.0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schafer DP, Lehrman EK, Stevens B. The "quad-partite" synapse: microglia-synapse interactions in the developing and mature CNS. Glia. 2013;61:24–36. doi: 10.1002/glia.22389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Derecki NC, et al. Wild-type microglia arrest pathology in a mouse model of Rett syndrome. Nature. 2012;484:105–109. doi: 10.1038/nature10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang J, et al. Wild-type microglia do not reverse pathology in mouse models of Rett syndrome. Nature. 2015;521:E1–E4. doi: 10.1038/nature14444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480:63–68. doi: 10.1038/nature10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Michalon A, et al. Chronic pharmacological mGlu5 inhibition corrects fragile X in adult mice. Neuron. 2012;74:49–56. doi: 10.1016/j.neuron.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ehninger D, et al. Reversal of learning deficits in a Tsc2+/- mouse model of tuberous sclerosis. Nat Med. 2008;14:843–848. doi: 10.1038/nm1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Krueger DD, Bear MF. Toward fulfilling the promise of molecular medicine in fragile X syndrome. Annu Rev Med. 2011;62:411–429. doi: 10.1146/annurev-med-061109-134644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Scharf SH, Jaeschke G, Wettstein JG, Lindemann L. Metabotropic glutamate receptor 5 as drug target for Fragile X syndrome. Curr Opin Pharmacol. 2015;20:124–134. doi: 10.1016/j.coph.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 112.Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci. 2010;11:490–502. doi: 10.1038/nrn2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Reeb-Sutherland BC, Fox NA. Eyeblink conditioning: a non-invasive biomarker for neurodevelopmental disorders. J Autism Dev Disord. 2015;45:376–394. doi: 10.1007/s10803-013-1905-9. [DOI] [PubMed] [Google Scholar]
- 114.Gandal MJ, et al. Validating gamma oscillations and delayed auditory responses as translational biomarkers of autism. Biol Psychiatry. 2010;68:1100–1106. doi: 10.1016/j.biopsych.2010.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Durand S, et al. NMDA receptor regulation prevents regression of visual cortical function in the absence of Mecp2. Neuron. 2012;76:1078–1090. doi: 10.1016/j.neuron.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kondo M, et al. Environmental enrichment ameliorates a motor coordination deficit in a mouse model of Rett syndrome--Mecp2 gene dosage effects and BDNF expression. Eur J Neurosci. 2008;27:3342–3350. doi: 10.1111/j.1460-9568.2008.06305.x. [DOI] [PubMed] [Google Scholar]
- 117.Restivo L, et al. Enriched environment promotes behavioral and morphological recovery in a mouse model for the fragile X syndrome. Proc Natl Acad Sci U S A. 2005;102:11557–11562. doi: 10.1073/pnas.0504984102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fyffe SL, et al. Deletion of Mecp2 in Sim1-expressing neurons reveals a critical role for MeCP2 in feeding behavior, aggression, and the response to stress. Neuron. 2008;59:947–958. doi: 10.1016/j.neuron.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]