Abstract
The amphipathic helix 8 in the membrane-proximal C-terminus is a structurally conserved feature of class A seven transmembrane-spanning G protein-coupled receptors (GPCRs). Mutations of this helical motif often cause receptor misfolding, defective cell surface transport and dysfunction. Surprisingly, we demonstrated here that a single point mutation at Lys308 in helix 8 markedly enhanced the steady-state surface density of the angiotensin II type 1a receptor (AT1aR). Consistent with the enhanced cell surface expression, Lys308 mutation significantly augmented AT1aR-mediated mitogen-activated protein kinase ERK1/2 activation, inositol phosphate production, and vascular smooth muscle cell migration. This mutation also increased the overall expression of AT1aR without altering receptor degradation. More interestingly, Lys308 mutation abolished AT1aR interaction with β-COP, a component of COPI transport vesicles, and impaired AT1aR responsiveness to the inhibition of Rab6 GTPase involved in the Golgi-to-ER retrograde pathway. Furthermore, these functions of Lys308 were largely dependent on its positively charged property. These data reveal previously unappreciated functions of helix 8 and novel mechanisms governing the cell surface transport and function of AT1aR.
1. Introduction
G protein-coupled receptors (GPCRs) constitute a superfamily of cell surface receptors, which regulate a variety of cell functions. All GPCRs share common structural features characterized by a hydrophobic core of seven transmembrane-spanning α-helices, three intracellular loops, three extracellular loops, an extracellular N-terminus, and an intracellular C-terminus. Another important feature of the rhodopsin-like class A GPCRs is the formation of an additional amphipathic α-helix (helix 8) in the membrane-proximal C-termini. Helix 8 contains highly concentrated, positively charged residues and mutation of this region often causes receptor misfolding, defective cell surface transport and dysfunction [1–5].
The precise function of GPCRs is regulated by their intracellular trafficking, including cell surface export, internalization, recycling and degradation. Among these trafficking processes, the molecular mechanisms underlying the cell surface export of newly synthesized receptors are not well understood. GPCRs are synthesized in the endoplasmic reticulum (ER). After being synthesized, properly folded receptors are transported to the cell surface en route through the ER-Golgi intermediate complex (ERGIC), the Golgi apparatus and the trans-Golgi network (TGN), during which the receptors undergo post-translational modifications (e.g. glycosylation) to attain mature status. At the same time, the receptors at each intermediate compartment may be transported back to the previous compartment. It has been well defined that COPI-, COPII- and clathrin-coated vesicles mediate the transport between these intracellular compartments. COPI vesicles are involved in both anterograde and retrograde transport between different Golgi stacks and from the Golgi to the ER. COPII vesicles exclusively transport nascent cargos from the ER to the Golgi. Clathrin-coated vesicles mediate vesicular transport between the plasma membrane, the TGN and the endosomal compartment. Therefore, the steady state cell surface receptor expression is a balance of vesicle-mediated anterograde and retrograde transport through a series of intracellular compartments.
Angiotensin II (Ang II) plays an important role in the physiological function of virtually all organs and is involved in the development of many diseases, including diabetes, hypertension, myocardial infarction, congestive heart failure, stroke and cancer. There are two major Ang II receptors: type 1 receptor (AT1R) and type 2 receptor (AT2R), both are prototypic GPCRs. As demonstrated in many studies, AT1R mediates the most physiological actions of Ang II. AT1R mainly couples to the Gq family G proteins and activates a variety of signal transduction pathways, including the activation of mitogen-activated protein kinases (MAPK) and phospholipase C [6–8].
The C-terminus is a very important domain in the regulation of AT1R function, including G protein coupling, signaling, trafficking, phosphorylation and interaction with cytosolic proteins [9–18]. Based on the newly published crystal structure, the helix 8 of AT1R runs away from the membrane [19] which is in contrast to other GPCRs in which helix 8 is parallel to the membrane bilayer. This helix 8 forms high affinity interaction with the negatively charged lipids of the plasma membrane [20–22]. A number of studies have demonstrated that small GTPases and interacting proteins are involved in the regulation of AT1R transport to the cell surface [23–30]. We have previously shown that AT1aR cell surface transport depends on Sar1, a crucial regulator in the formation of COPII vesicles, and on ARF1, which is required for the function of both COPI- and clathrin-coated vesicles, suggesting an important role of small coated vesicles in the cell surface targeting of AT1R. We have also demonstrated that the di-acidic motif and dileucine motif in the C-terminus are essential for AT1aR cell surface transport [31–33]. Here, we report a surprising finding that a single point mutation at position Lys308 in helix 8 markedly enhances AT1aR cell surface export and function which is likely mediated via regulating overall expression and retrograde transport pathway of the receptor.
2. Experimental procedures
2.1. Materials
Rat AT1aR in vector pCDM8 was kindly provided by Dr. Kenneth E. Bernstein (Department of Pathology, Emory University, Atlanta, GA). Antibodies against GFP and phospho-ERK1/2 were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Antibodies against ERK1/2 were from Cell Signaling Technology, Inc. (Beverly, MA). Anti-β-COP polyclonal antibody was from Thermo Scientific. High affinity anti-HA-fluorescein (3F10) was from Roche Molecular Biochemicals (Mannheim, Germany). Ang II was purchased from Calbiochem (San Diego, CA). Myo-[3H]-inositol was from Perkin Elmer Life Sciences. Dowex AG1-X8 was from Bio-Rad (Hercules, CA). Rat aortic smooth muscle cells (RASMCs) were purchase from Cell Applications, Inc. (San Diego, CA). All other materials were obtained as described elsewhere [2, 34].
2.2. Plasmid construction
To generate the pcDNA3.1(−) vector containing three HA at the XbaI and XhoI restriction sites, two primers (forward primer: 5′–CTAGAATGTACCCATACGATGTTCCAGAT TACGCTTACCCATACGATGTTCCAGATTA CGCTTACCCATACGATGTTCCAGATTACG CTGATC-3′; reverse primer: 5′–TCGAGATCAGCGTA ATCTGGAACATCGTATGGGTAAGCGTAAT CTGGAACATCGTATGGGTAAGCGTAATCT GGAACATCGTATGGGTACATT-3′) encoding three YPYDVPDYA and containing XbaI and XhoI restriction sites were synthesized, annealed and ligated into the pcDNA3.1(−) vector (Invitrogen Life Technologies, Carlsbad, CA), which was digested with XbaI and XhoI. To generate AT1aR tagged with three HA at its N-terminus (HA-AT1aR), the full-length AT1aR was amplified by polymerase chain reaction (PCR). The PCR product was digested with XhoI and HindIII, purified and ligated to HA-tagged pcDNA3.1(−) vector, which was digested with XhoI and HindIII. AT1aR tagged with GFP at its C-termini (AT1aR-GFP) was generated as described previously [23, 34]. The GFP and HA epitopes have been used to label GPCRs including AT1aR, resulting in receptors with similar characteristics to the wild-type (WT) receptors. GST fusion protein constructs coding the C-terminus of AT1aR were generated in the pGEX-4T-1 vector as described previously [3]. The constructs of small GTPases (Rab1N124I, Rab6Q72L, Sar1H79G and ARF1N126I) were generated as described previously [23–27]. All mutants were made with the Quick Change site-directed mutagenesis kit. The sequence of each construct used in this study was verified by restriction mapping and nucleotide sequence analysis.
2.3. Cell culture and transient transfection
HEK293 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 units/ml streptomycin. RASMCs were cultured in Rat Smooth Muscle Cell Growth Medium (Cell Applications, Inc.). Transient transfection of the cells was carried out using Lipofectamine 2000 reagent (Invitrogen Carlsbad, CA).
2.4. Flow cytometric analysis of receptor expression
For measurement of the cell surface expression of AT1aR, HEK293 cells were cultured on 6-well dishes and transfected with 1 μg of HA-AT1aR for 36 h. The cells were collected, suspended in phosphate-buffered saline (PBS) containing 1% fetal calf serum (FCS) at a density of 1 × 107 cells/ml and incubated with high affinity anti-HA-fluorescein (3F10) at a final concentration of 2 μg/ml for 30 min at 4°C. After washing twice with 0.5 ml of PBS/1% FCS, the cells were resuspended and the fluorescence was analyzed on a flow cytometer (Dickinson FACSCalibur) as described previously [23]. Since the staining with the HA antibodies was carried out in the unpermeabilized cells and only those receptors expressed at the cell surface were accessible to the HA antibodies, the fluorescence measurement reflected the amount of the receptors at the cell surface. For measurement of total receptor expression, HEK293 cells were transfected with AT1aR-GFP for 36 h. The cells were collected, washed twice with PBS and resuspended in PBS containing 1% FCS at a density of 8 × 106 cells/ml. Total receptor expression was determined by measuring the GFP fluorescence. To determine the effect of drug treatment on receptor expression, HEK293 cells were incubated with MG132 (20 μM), chloroquine (100 μM) for 6 h at 37C°. To determine the effect of small GTPases, HEK293 cells were cultured on 6-well dishes and transfected with 200 ng of AT1aR plasmid together with 800 ng of individual small GTPase plasmids.
2.5. Biotinylation of the cell surface AT1aR
In addition to flow cytometry the cell surface AT1aR expression was also measured by immunoblotting following biotinylation and isolation of cell surface proteins by using the Pierce Cell Surface Protein Isolation Kit (Thermo Scientific). Briefly, HEK293 cells were cultured on 10-cm dishes and transfected with 4 μg of the pEGFP-N1 vector, AT1aR-GFP or K308E-GFP for 36–48 h. After washing, the cells were incubated with EZ-Link Sulfo-NHS-SS-Biotin for 30 min at 4 °C with gentle rotation. After the action was quenched, the cells were collected, lysed with lysis buffer containing protease inhibitors (Roche Molecular Biochemicals) and sonicated. The cell lysate was then centrifuged and the biotinylated proteins were isolated by using the NeutrAvidin Agarose column following the manufacturer’s instructions. The biotinylated cell surface AT1aR were measured by immunoblotting using GFP antibodies.
2.6. Fluorescence microscopy
Subcellular distribution of AT1aR was analyzed by fluorescence microscopy as described previously [33, 35]. Briefly, HEK293 cells were grown on coverslips pre-coated with poly-L-lysine in 6-well plates and transfected with 100 ng of AT1aR-GFP for 36 h. The cells were fixed with 4% paraformaldehyde-4% sucrose mixture in PBS for 15 min. The coverslips were mounted with prolong antifade reagent (Invitrogen) and images were captured using a Zeiss LSM780 confocal microscope equipped with a 63× objective. The cell surface expression of AT1aR was quantified by using ImageJ software.
2.7. Measurement of ERK1/2 activation
HEK293 cells were cultured on 6-well dishes and transfected with 1.2 μg of AT1aR or its mutant. At 6–8 h after transfection, the cells were split and cultured for additional 36 h. The cells were starved for 5 h and then stimulated with different concentrations of Ang II (0–100 nM) for 3 min at 37 °C. Stimulation was terminated by addition of 350 μl of 1 × SDS gel loading buffer. After solubilizing the cells, 20 μl of total cell lysates were separated by 12% SDS-PAGE. ERK1/2 activation was determined by measuring the levels of phosphorylation of ERK1/2 with phospho-specific ERK1/2 antibodies by immunoblotting as described previously [23].
2.8. Measurement of inositol phosphate (IP) production
AT1aR-stimulated IP accumulation was measured as described previously [23]. Briefly, HEK293 cells were cultured on 100-mm dishes and transfected with AT1aR or its mutant. At 24 h after transfection, the cells were labeled with fresh medium supplemented with myo-3H-inositol (2 μCi/ml) for 24 h. The cells were washed with 2 ml of buffer containing 1.26 mM Ca2+ and 0.89 mM Mg2+ three times and incubated with 2 ml of buffer containing 10 mM LiCl for 20 min. The cells were then stimulated with Ang II at a concentration of 1 μM for 45 min at 37 °C. The reactions were terminated with 1 ml of ice-cold 10% trichloroacetic acid. The cells were frozen at −80°C for 10 min, collected and mixed with 0.5 ml mixture of 0.72 M KOH/0.6 M KHCO3. After centrifugation at 3,000 rpm for 5 min to remove the precipitated proteins, the supernatants were applied to Dowex AG1-X8 anion exchange columns (1.0 ml) equilibrated with 0.1 M formic acid. The columns were washed with 4 X 4 ml 0.1 M formic acid to remove free myo-[3H]-inositol and inositol phosphates were eluted with 3 X 2.0 ml 0.1 M formic acid containing 1 M ammonium formate and counted by liquid scintillation spectrometry in 18 ml of Ecoscint A scintillation solution (National Diagnostics, Atlanta, GA).
2.9. Vascular smooth muscle cell migration assay
Migration assays were performed by using 24-well cell culture inserts with 8.0 μm pore size transwell culture chambers (Corning, NY, USA). RASMCs were cultured in 10-cm dishes and transfected with the control vector pEGFP-N1, AT1aR-GFP or K308E-GFP for 24 h. The cell were then seeded into chamber inserts at a density of 4x104 per well with Rat Smooth Muscle Cell Basal Medium (Cell Applications, Inc.) containing 05% FBS and stimulated with Ang II at a concentration of 100 nM for 7 h at 37 °C. The cell that did not migrate were removed from the upper surface of the membrane, the migrated cells on the lower surface were then fixed with 4% paraformaldehyde and stained with crystal violet solution for 30 min. Bright-field images were randomly taken with a light microscope at ×200 magnification and the numbers of migrated cells were counted.
2.10. Measurement of AT1aR ubiquitination
Ubiquitination of AT1aR was measured as described previously for AT2R [36]. HEK293 cells were cultured on 100-mm dishes and transfected with 3 μg of AT1aR-GFP and 3 μg of HA-ubiquitin. After transfection for 36 h, the cells were treated with MG132, a proteasome inhibitor at a concentration of 10 μM for 12 h. The cells were then washed with PBS and incubated with 500 μl of buffer containing 50 mM Tris, pH7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxychalate, 0.1% EDTA and protease inhibitors at 4 °C for 1 h with gentle rotation. After centrifugation for 15 min at 14,000 × g, 20 μl of protein G PLUS-Agarose beads was added into the supernatant for 1 h to remove unspecific binding proteins. The pre-cleared supernatant was then incubated with 2 μg of anti-GFP antibodies and rotated overnight at 4 °C followed by incubation with 12.5 μl of protein G-Agarose beads for 4 h at 4 °C. The beads were then washed twice with buffer and the bound proteins eluted with 2 × SDS-gel loading buffer. AT1aR-GFP and HA-ubiquitin in the immunoprecipitates were detected by immunoblotting using anti-GFP and HA antibodies, respectively.
2.11. GST fusion protein pulldown assays
GST fusion proteins were expressed in BL21 bacteria and purified using a glutathione affinity matrix as described previously [35, 37]. GST fusion proteins immobilized on the glutathione resin were either used immediately or stored at 4 °C for no longer than 3 days. Each batch of fusion proteins used in experiments was first analyzed by Coomassie Brilliant Blue R250 staining following SDS-PAGE. GST fusion proteins tethered to the glutathione resin were incubated with total HEK293 cell lysates in 1 ml of binding buffer (20 mM Tris-HCl, pH 7.5, 2% NP-40, 140 mM NaCl) at 4 °C overnight. The resin was washed four times with 1 ml of binding buffer, and the retained proteins were solubilized in 1 × SDS gel loading buffer and separated by SDS-PAGE. Bound β-COP were detected by immunoblotting.
2.12. Statistical analysis
Differences were evaluated using Student’s t test, and p < 0.05 was considered as statistically significant. Data are expressed as the mean ± S.E.
3. Results
3.1. Single mutation at Lys308 in helix 8 enhances the cell surface expression of AT1aR
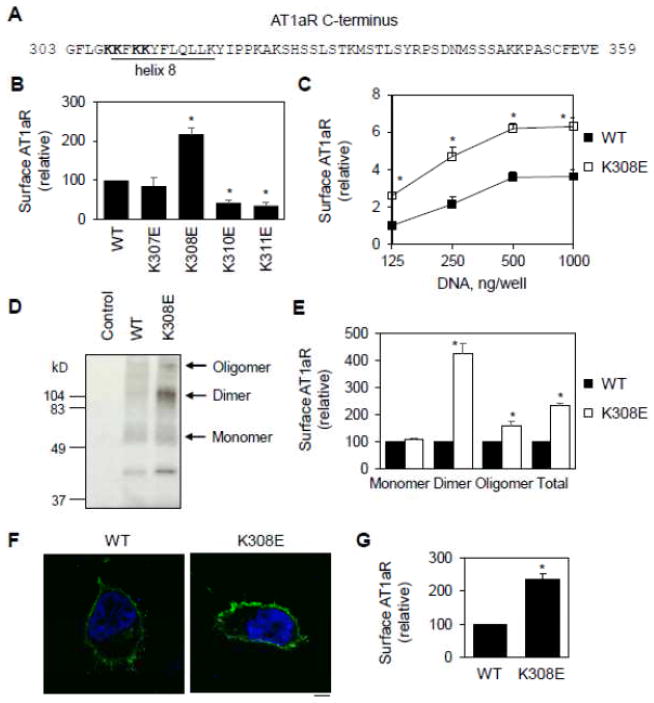
Positively charged residues are highly conserved in the helix 8 of family A GPCRs [2]. We and others have shown that mutating these residues markedly attenuated the cell surface expression of the receptors [2–5]. Here, we sought to further characterize the function of Lys residues in helix 8 (Fig. 1A) in the cell surface transport of AT1aR. In the first series of experiments, Wild-type (WT) AT1aR and its mutants were tagged with HA at the N-termini (HA-AT1aR) and their cell surface expression was quantified by flow cytometry following staining with anti-HA antibodies in non-permeabilized cells. Mutating Lys310 and Lys311 to Glu dramatically inhibited the cell surface expression of AT1aR. Surprisingly, Lys308 mutation produced a marked opposite effect and enhanced the cell surface expression of AT1aR by 2.2 folds. In contrast, Lys307 mutation did not have obvious effects (Fig. 1B).
Fig. 1.
Lys308 mutation to Glu enhances the cell surface expression of AT1aR. A, Sequence of the C-terminus of AT1aR. Residues in helix 8 are underlined and four Lys residues at positions 307, 308, 310 and 311 which were mutated to Glu individually are bolded. B, Effect of mutating Lys residues on the cell surface expression of AT1aR. HA-tagged AT1aR and its mutants were transfected in HEK293 cells and their cell surface expression was measured by flow cytometry following staining with HA antibodies in non-permeabilized cells (n=4–6). *, p < 0.05 versus WT. C, Effect of increasing amounts of plasmids (from 125 to 1000 ng per well) on the cell surface expression of AT1aR and K308E. The data are expressed as -folds relative to WT AT1aR expression in cells transfected with 125 ng plasmids (n=3). D, Effect of Lys308 mutation on the cell surface AT1aR expression measured in biotinylation assays. HEK293 cells were transfected with the pEGFP-N1 vector (control), AT1aR-GFP or K308E-GFP. The cell surface expression of the receptors was determined by Western blotting using GFP antibodies following biotinylation and isolation of the cell surface proteins. E, Quantitative data shown in D (n=3). F, Representative images showing subcellular distribution of AT1aR and K308E. HEK293 cells cultured on coverslips were transfected with AT1aR-GFP or K308E-GFP and the subcellular localization of the receptors was revealed by fluorescence microscopy. Green, GFP-tagged AT1aR; blue, DNA staining by 4,6-diamidino-2-phenylindole (nuclei). Scale bar, 10 μm. G, Quantitative data of cell surface expression shown in f (n=4). *, p < 0.05 versus respective WT.
We next determined the effect of transfection with different concentrations of plasmids on the cell surface expression of the receptors. The effect of increasing plasmid concentrations on the cell surface expression of the receptor was very much the same between WT AT1aR and its mutant K308E (Fig. 1C). However, at each plasmid concentration, the cell surface expression of the mutant K308E was significantly higher than its WT counterpart (Fig. 1C).
In the second series of experiments, we used the cell surface biotinylation method to determine the effect of Lys308 mutation on the cell surface expression of AT1aR. In these experiments, AT1aR and its mutant K308E were tagged with green fluorescent protein (GFP) at the C-termini and their cell surface expression levels were quantified by immunoblotting using anti-GFP antibodies after biotinylation and isolation of the cell surface proteins. Multiple immunoreactive bands were observed in the immunoprecipitates from cells expressing either AT1aR or the mutant K308R, but not from cells expressing GFP alone (Fig. 1D). Based on their apparent molecular weights, these bands likely represented the monomers, dimers and oligomers of AT1aR (Fig. 1D). The total cell surface AT1aR (Fig. 1D and 1E) was significantly augmented in cells expressing the mutant K308E as compared with cells expressing its WT AT1aR. Interestingly, increase in AT1aR dimers was much higher than its monomers and oligomers (Fig. 1D and 1E). These data are consistent with previous studies showing that AT1R undergoes dimerization [38–40].
In the third series of experiments, confocal microscopy was utilized to directly visualize the subcellular distribution of GFP-tagged AT1aR and its mutant K308E. As expected, AT1aR-GFP was robustly expressed at the cell surface (Fig. 1F). Consistent with the data obtained from flow cytometry, the cell surface localization of K308E-GFP was apparently stronger than that of AT1aR-GFP (Fig. 1F) and quantitative data showed that the cell surface AT1aR increased by 2.2 folds in cells transfected with the mutant K308E as compared to cells expressing WT AT1aR (Fig. 1F). Altogether, these data demonstrate that single point mutation at Lys308 in helix 8 markedly enhances the cell surface expression of AT1aR.
3.2. Lys308 mutation augments AT1aR-mediated signaling and function
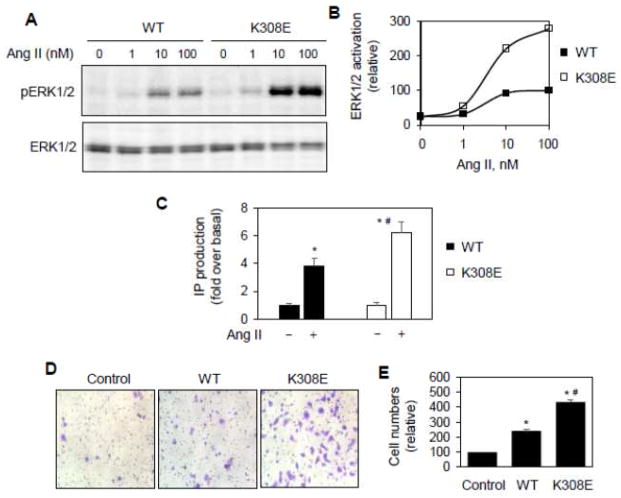
To define if Lys308 mutation could enhance AT1aR-mediated signaling, the MAPK ERK1/2 activation and IP production were compared in cells expressing WT AT1aR and its mutant K308E. Consistent with the effect on cell surface AT1aR expression, AT1aR-mediated ERK1/2 activation in response to Ang II stimulation was significantly greater in cells expressing the mutant K308E than in cells expressing WT AT1aR (Fig. 2A and 2B). Similarly, IP production was also higher in cells expressing K308E than in cells expressing WT AT1aR (Fig. 2C). The basal levels of ERK1/2 activation and IP production were almost the same in cells expressing AT1aR and its mutant K308E (Fig. 2A–2C), suggesting that the mutation did not induce constitutive activation of the receptor. These data indicate that Lys308 mutation enhances not only AT1aR cell surface expression but also AT1aR-mediated signaling.
Fig. 2.
Effect of Lys308 mutation on AT1aR-mediated ERK1/2 activation and IP production. A, Effect of Ly308 mutation on AT1aR-stimulated ERK1/2 activation. HEK293 cells cultured on 6-well dishes were transfected with AT1aR-GFP or K308E-GFP and then stimulated with increasing concentrations of Ang II (0–100 nM) for 3 min. ERK1/2 activation was determined by Western blot analysis using phospho-specific ERK1/2 antibodies. B, Quantitative data shown in A. The data are the percentage of ERK1/2 activation in cells expressing AT1aR-GFP and stimulated with Ang II at 100 nM. Similar results were obtained in 4 individual experiments. C, Effect of Lys308 mutation on AT1aR-mediated IP accumulation. HEK293 cells were transfected with AT1aR or its mutant K308E, incubated with myo-3H-inositol and stimulated with Ang II at 1 μM for 45 min. The data shown are the fold increase over respective basal levels of IP production in response to Ang II stimulation (n=3). *, p < 0.05 versus the cells without Ang II stimulation and #, p < 0.05 versus the cells transfected with WT AT1aR and stimulation with Ang II. D, Effect of Lys308 mutation on AT1aR-mediated migration. RASMCs were transfected with the control vector pEGFP-N1, AT1aR-GFP or K308E-GFP and seeded on transwell culture chambers. The cells were stimulated with Ang II at 100 nM for 7 h at 37 °C. After removal of non-migrating cells, the migrated cells on the lower surface were fixed and stained. Bright-field images were randomly taken with a light microscope at ×200 magnification. E, Quantitative data of migrated cells shown in D. The data are percentages of migrated RASMCs transfected with the control vector (n= 9–20 images from 3 individual experiments). *, p < 0.05 versus control and #, p < 0.05 versus WT AT1aR.
The activation of AT1R has been well defined to be important for the vascular smooth muscle migration [41, 42]. To further determine if Lys308 mutation could augment AT1R-mediated function, we determined the effect of transient expression of AT1aR and its mutant K308E on the migration of RASMCs in response to Ang II stimulation. Consistent with the role of AT1aR activation in promoting vascular smooth muscle cell migration, the number of migrated RASMCs was significantly higher in cells expressing WT AT1aR than control cells upon stimulation with Ang II (Fig. 2D and 2E). The migrated RASMCs were greater in cells expressing K308E than in cells expressing WT AT1aR and control cells (Fig. 2D and 2E). In contrast, the migration of RASMCs was about the same in cells expressing the control vector, WT AT1aR and K308E mutant in the absence of Ang II stimulation (data not shown). These data further indicate that Lys308 mutation augments the function of AT1aR.
3.3. Lys308 mutation increases overall expression of AT1aR
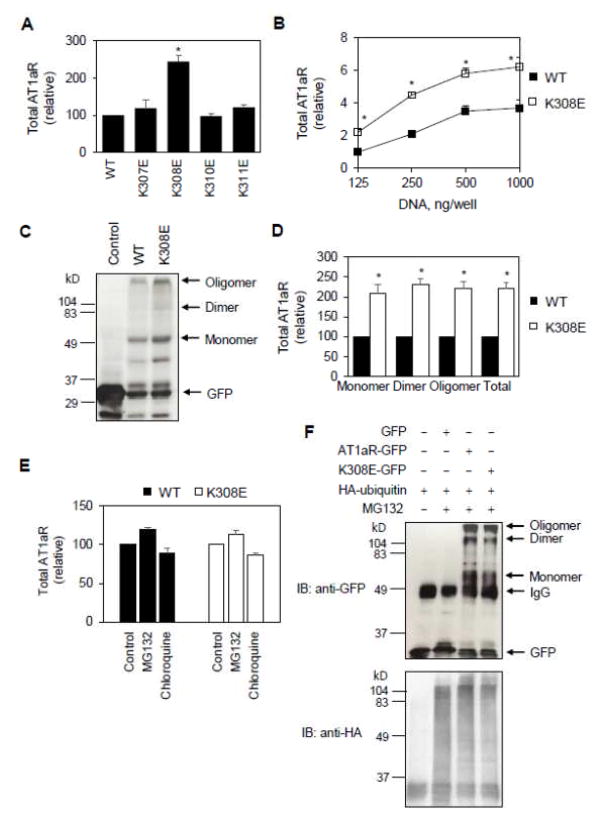
To explore the possible mechanisms of how Lys308 mutation increased the cell surface AT1aR expression, we determined if the mutation could alter overall AT1aR synthesis. AT1aR and its mutants were tagged GFP at their C-termini (AT1aR-GFP) and their total expression levels were measured by flow cytometry measuring total GFP signal. Similar to the effect on the cell surface expression, Lys308 mutation to Glu increased total AT1aR expression by 2.4 folds, whereas mutating Lys307, Lys310 and Lys311 did not have any clear effect (Fig. 3A). A significant increase in overall AT1aR expression by Lys308 mutation was also observed in cells transfected with different plasmid concentrations (Fig. 3B).
Fig. 3.
Effect of Lys308 mutation on AT1aR expression, degradation and ubiquitination. A, HEK293 ells were transfected with GFP-tagged AT1aR and its mutants and total receptor expression was determined by flow cytometry detecting the GFP signal (n=4–6). *, p < 0.05 versus WT. B, Effect of increasing plasmid concentrations on total expression of AT1aR and its mutant K308E. The data are expressed as -folds relative to WT AT1aR expression in cells transfected with 125 ng plasmids (n=5–7). C, HEK293 cells were transfected with the pEGFP-N1 vector (control), AT1aR-GFP or K308E-GFP. Total cell lysates were separated by SDS-PAGE followed by Western blotting using GFP antibodies. D, Quantitative data shown in C (n=3). E, Effect of the treatment of MG132 and chloroquine on total AT1aR expression. HEK293 cells were transfected with AT1aR-GFP or K308E-GFP and then treated with DMSO (control), GM132 (20 μM) or chloroquine (100 μM) for 6 h at 37C° (n=3). F, Effect of Lys308 mutation on AT1aR ubiquitination. HEK293 cells were transfected with AT1aR-GFP or K308E-GFP together with HA-ubiquitin and treated with MG132. Following immunoprecipitation with anti-GFP antibodies, AT1aR-GFP and HA-ubiquitin were detected by immunoblotting using anti-GFP (upper panel) and anti-HA (lower panel) antibodies, respectively. Similar results were obtained in 3 separate experiments. *, p < 0.05 versus respective WT.
We next measured total expression of AT1aR by immunoblotting of cell lysates using GFP antibodies. Total AT1aR was augmented in cells expressing the mutant K308E as compared with cells expressing WT AT1aR, and Lys308 mutation similarly enhanced the expression of AT1aR monomers, dimers and oligomers (Fig. 3C and 3D). These data suggest that Lys308 mutation also enhances total expression of AT1aR.
To test the possibility that increased total AT1aR expression by Lys308 mutation was due to reduced receptor targeting to the degradation pathways, we determined the effects of MG132, a proteasome degradation pathway inhibitor, and chloroquine, a lysosomal degradation pathway inhibitor, on total AT1aR expression. The treatments with GM132 and chloroquine only slightly affected total AT1aR expression and their effects were similar between AT1aR and the mutant K308E (Fig. 3E).
We further determined if AT1aR was ubiquitinated as well as the effect of Lys308 mutation on the ubiquitination. For this purpose, GFP-tagged AT1aR and HA-tagged ubiquitin were co-expressed in HEK293 cells and AT1aR was immunoprecipitated using GFP antibodies. The levels of AT1aR and ubiquitin in the immunoprecipitates were determined by immunoblotting using GFP and HA antibodies, respectively. Although both WT AT1aR and the mutant K308E were found in the anti-GFP immunoprecitates, no specific HA-ubiqintin bands were detected in the immunoprecipitates from cells expressing AT1aR and the mutant K308E, suggesting that AT1aR and the mutant K308E are not conjugated with ubiqintin (Fig. 3F). These data are consistent with a previous report showing that AT1R does not undergo ubiquitination under normal conditions [43]. Similar amounts of AT1aR and K308E were found in the immunoprecipitates which was likely due to the limited amount of GFP antibodies used in the immunoprecipitation experiments (Fig. 3F). Altogether, these data suggest that AT1aR degradation is unlikely a major cause for the increased synthesis of AT1aR induced by Lys308 mutation.
3.4. Lys308 mutation abolishes COPI interaction and disrupts Rab6-dependent transport of AT1aR
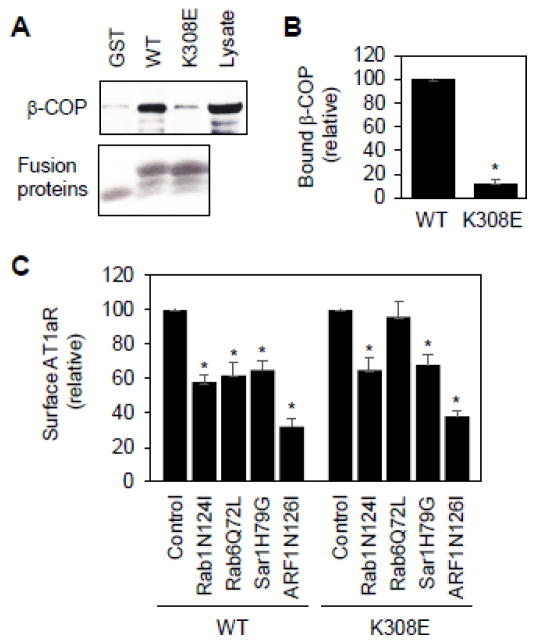
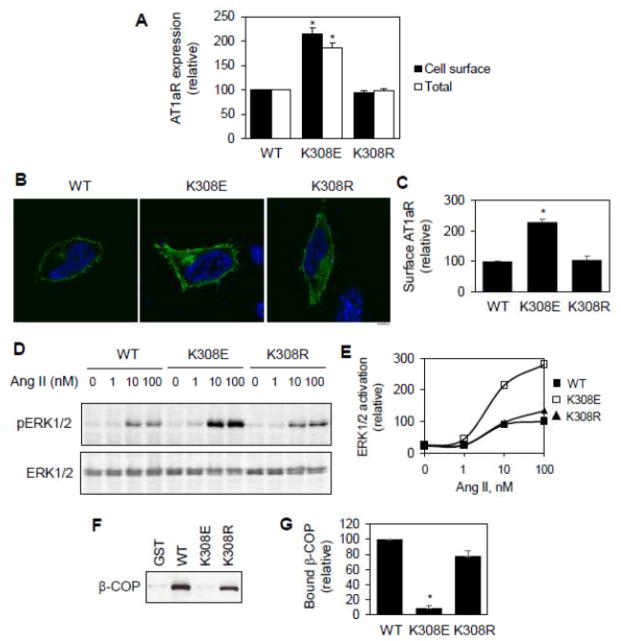
It has been well established that retrograde transport from the Golgi to the ER and between the Golgi stacks is mediated through the COPI vesicles and several motifs, such as the dilysine motifs, have been identified to directly interact with the COPI vesicles [44–47]. Therefore, we determined if Lys308 mediated AT1aR interaction with the COPI vesicles. The AT1aR C-terminus and its mutant K308E were generated as glutathione S-transferase (GST) fusion proteins and their interaction with β-COP, a component of COPI vesicles, was determined in GST fusion protein pulldown assays. The GST-fusion proteins containing the C-terminus, but not GST alone, strongly interacted with β-COP. Lys308 muation abolished the interaction of the C-terminus with β-COP (Fig. 4A and 4B). These data suggest that the AT1aR C-terminus physically associates with the COPI vesicles which is mediated via Lys308.
Fig. 4.
Effect of Lys308 mutation on AT1aR interaction with β-COP and responsiveness to small GTPase inhibition. A, Effect of Lys308 mutation on the interaction of the AT1aR C-terminus with β-COP. The AT1aR C-terminus and its mutant K308E were generated as GST fusion proteins and incubated with total cell lysates prepared from HEK293 cells. Bound β-COP was revealed by immunoblotting using anti-β-COP antibodies (upper panel). Lower panel shows Coomassie blue staining of purified GST fusion proteins. Lysate – 1% of total input. B, Quantitative data shown in A (n=3). *, p < 0.05 versus WT. C, Effect of transient expression of small GTPase mutants on the cell surface expression of AT1aR and its mutant K308E. HEK293 cells were transfected with HA-AT1aR or HA-K308E together with pcDNA3.1 (control) or individual small GTPase mutants. The cell surface AT1aR expression was quantitated by flow cytometry following incubation with anti-HA antibodies (n=3). *, p < 0.05 versus control.
We have previously shown that AT1aR cell surface transport depends on several small GTPases, including Rab1, Rab6, Sar1 and ARF1 [23–27]. Rab1 is well known to modulate the ER-to-Golgi transport, whereas Rab6 modulates retrograde transport from the late to the early Golgi cisternae or from the Golgi to the ER [48, 49]. Sar1 regulates the ER exit via controlling the formation of COPII vesicles [50]. ARF1 coordinates multiple pathways involved in both anterograde and retrograde transport [51]. To define if AT1aR and its mutant K308E used the same pathway to transport to the cell surface, we determined the effect of expressing these small GTPase mutants which were previously shown to inhibit AT1aR cell surface transport [23–27]. We found that the expression of the dominant-negative mutants Rab1N124I and ARF1N126I and the constitutively active mutant Sar1H79G significantly inhibited the cell surface expression of both AT1R and K308E (Fig. 4c). In contrast, the expression of the constitutively active mutant Rab6Q72L selectively attenuated the cell surface transport of AT1aR but had no effect on the transport of mutant K308E (Fig. 4C). These data suggest that Lys308 mutation may alter AT1aR transport via the Rab6-dependent retrograde pathway.
3.5. Lys308 function depends on its positively charged property
To further provide insights into how Lys308 modulates AT1aR cell surface transport, overall synthesis and interaction with COPI vesicles, we determined the effect of Lys308 mutation to Arg. In marked contrast to the effects of Lys308 mutation to Glu, Lys308 mutation to Arg did not significantly influence the cell surface and total expression of AT1aR (Fig. 5A). Confocal microscopic analysis of the subcellular distribution of AT1aR showed that the cell surface expression levels of AT1aR and the mutant K308R were similar which were significantly less than that of the mutant K308E (Fig. 5B and 5C). Lys308 mutation to Arg also did not alter the ability of AT1aR to activate the MAPK ERK1/2 in response to Ang II stimulation (Fig. 5D and 5E). In GST fusion protein assays, both WT C-terminus and its mutant K308R strongly bound to β-COP (Fig. 5F and 5G). These data demonstrate that Lys and Arg at position 308 of AT1aR are exchangeable. These data also suggest that the positively charged property of Lys308 is a main determinant of its functions in regulating AT1aR expression and interaction with β-COP.
Fig. 5.
Effect of Lys308 mutation on AT1R expression, interaction with β-COP and function. A, Effect of Lys308 mutation on the cell surface and total expression of AT1aR as described in legend of Fig. 1a and 3a. B, Representative images showing the subcellular distribution of AT1aR and its mutants revealed by fluorescence microscopy as described in legend of Fig. 1F. Green, AT1aR-GFP; blue, DNA staining by 4,6-diamidino-2-phenylindole (nuclei). Scale bar, 10 μm. C, Quantitative data of AT1aR cell surface expression shown in B (n=3). D, Effect of Ly308 mutation on AT1aR-stimulated ERK1/2 activation as described in legend of Fig 2A. E, Quantitative data shown in D. The data are the percentage of ERK1/2 activation in cells expressing AT1aR-GFP and stimulated with Ang II (0–100 nM) for 3 min. Similar results were obtained in 3 separate experiments. F, Effect of Lys308 mutation on the interaction of the AT1aR C-terminus with β-COP in GST fusion protein pulldown assays as described in legend of Fig. 4A. G, Quantitative data shown in F (n=3). *, p < 0.05 versus respective WT.
4. Discussion
The most important finding we have shown in this manuscript is that a single point mutation at Lys308 in the membrane-proximal C-terminal helix 8 significantly enhances the export of functional AT1aR to the cell surface, and as a result promotes receptor-mediated signaling and function. These functions of Lys308 are likely mediated through regulating overall AT1aR expression and AT1aR transport along the Rab6- and COPI-mediated retrograde pathway.
The amphipathic helix 8 is a structurally conserved motif in family A GPCRs. This helical region has been shown to be involved in the G protein activation, trafficking and function of the receptors [32, 52, 53]. Highly concentrated, positively charged residues in helix 8 have been defined to be essential for the cell surface expression of some GPCRs [2–5]. Here, we have demonstrated that the function of Lys308 in regulating AT1aR transport and function is in marked contrast to other positively charged residues located in the membrane-proximal C-terminal helix 8. Lys308 mutation to Glu markedly enhanced the cell surface expression of AT1aR as measured in three different assays: flow cytometry, biotinylation and confocal microscopy. Furthermore, Lys308 mutation also enhanced AT1aR-mediated ERK1/2 activation, IP production and vascular smooth muscle cell migration which are presumably caused by increased AT1aR cell surface expression. However, we cannot exclude the possibility that Lys308 mutation may alter the receptor’s abilities to bind to its ligands and to directly activate its downstream signaling molecules. As K308E did not alter basal ERK1/2 activation, IP production and cell migration in the absence of Ang II, it is not a constitutively active mutant.
Lys308 mutation enhanced the overall expression of AT1aR, suggesting that upregulated receptor expression is an important force that drives the cell surface transport of the mutated AT1aR. However, the mechanism of how Lys308 mutation enhances total AT1aR expression is unknown. The simplest explanation for this is that Lys308 is a cleavage site for some proteolytic digestive enzymes and its mutation reduces the degradation of AT1aR, resulting in the increase in total receptor numbers in the cell. As both Lys and Arg are the same cleavage sites for the enzymes, Lys308 mutation to Arg will not influence AT1aR degradation. However, AT1R cleavage has been demonstrated to occur at different residues at the C-terminus [54]. Our data have also shown that the amounts of the GFP fragment were about the same in total lysates prepared from cells expressing WT AT1aR and K308E (Fig. 2C). In addition, the treatment with the proteasomal inhibitor MG132 and the lysosomal inhibitor chloroquine only moderately altered total AT1aR synthesis. Furthermore, the functions of Lys308 can be replaced by Arg, suggesting it is unlikely involved in the regulation of AT1aR targeting to the ubiquitination and sumoylation pathways which are specifically mediated through Lys residues [55]. Indeed, consistent with other reports [43], our studies showed that AT1aR did not undergo ubiquitination under the condition that AT2R was ubiquitinated [36]. Altogether, these data suggest that Lys308 is unlikely involved in the AT1aR degradation processes.
Our studies suggest a possible role of retrograde transport in the regulation of AT1aR export trafficking to the cell surface. We demonstrated that the AT1aR C-terminus interacted with the COPI vesicle component β-COP and the interaction was abolished by Lys308 mutation. As the COPI vesicles are well demonstrated to mediate retrograde transport from the late to the early Golgi cisternae and from the Golgi to the ER, it is possible that, under the normal condition, AT1aR physically associates with the COPI vesicles via Lys308, leading to the enhancement of AT1aR recruitment onto the vesicles and subsequent transport from the Golgi to the ER. Lys308 mutation disrupts the interaction of AT1aR with the COPI vesicles, reducing receptor retrograde transport which presumably enhances receptor forward trafficking. This possibility is also supported by identification of the dilysine motifs that interact with COPI vesicles and function as retrieval signals recycling proteins from the Golgi back to the ER [44–47]. Furthermore, Rab6 is well characterized to be involved in regulation of protein retrograde transport [48, 49]. Consistent with our previous report, the expression of constitutively active mutant Rab6Q72L inhibited the transport of AT1aR, but not the mutant K308E. The lack of response of the AT1aR mutant K308E to Rab6 inhibition suggests that Lys308 may be involved in the regulation of AT1aR retrograde movement. It is also interesting to note that, in some GPCRs such as α2C-adrenergic and GABAB receptors, positively charged residues may function as ER retention motifs to block the receptor export from the ER [56, 57]. To the best of our knowledge, these data provide the first evidence implicating a role of retrograde transport in regulating GPCR forward trafficking. These data also suggest that the C-terminus-mediated specific interaction of AT1aR with the COPI vesicles is an important checkpoint for modulating AT1aR cell surface export and function.
The cell surface targeting of nascent AT1R is a crucial event in regulating its functionality under the physiological and pathological conditions. Abnormal expression of AT1R at the cell surface has been well described to significantly contribute to the development of many cardiovascular diseases and cancers and AT1R blockers are important drugs for the treatment of these diseases [6, 58, 59]. Although molecular mechanisms governing the cell surface export of AT1R are not well understood, recent studies have demonstrated that it is coordinated by many regulatory proteins, motif-mediated interaction with transport machineries and structural determinants [28, 30–33]. We have previously demonstrated that AT1aR may use export motifs, such as di-leucine and di-acidic motifs located in the C-terminus, to direct its anterograde transport [31–33]. As demonstrated in this study, Lys308 in helix 8 modulates AT1aR trafficking and function via different mechanisms. Because Lys308 is a highly conserved residue in many GPCRs [2], it will be very intriguing to determine if these mechanisms revealed by using AT1aR as a model can be applied to other GPCRs to regulate their trafficking and function.
Highlights.
A single mutation in helix 8 enhances the cell surface expression of AT1aR
The mutation augments AT1aR-mediated signaling and function
The mutation increases overall expression of AT1aR
The mutation abolishes COPI interaction and disrupts Rab6-dependent transport of AT1aR
Novel mechanisms that control AT1aR trafficking and signaling via helix 8 are proposed.
Acknowledgments
This work was supported by US National Institutes of Health Grant R01GM076167 (to G. W.) and National Natural Science Foundation of China Grant 31201069 (to H. W.).
The abbreviations used are
- GPCR
G protein-coupled receptor
- Ang II
angiotenin II
- AT1R
angiontensin II type 1 receptor
- AT1aR
angiontensin II type 1a receptor
- AT2R
angiontensin II type 2 receptor
- MAPK
mitogen-activated protein kinase
- ERK1/2
extracellular signal-regulated kinase 1 and 2
- WT
wild type
- IP
inositol phosphate
- GST
glutathione S-transferase
- GFP
green fluorescent protein
- HA
hemagglutinin
- RASMC
rat aortic smooth muscle cell
- DMEM
Dulbecco’s modified Eagle’s medium
- PBS
phosphate-buffered saline
- ER
endoplasmic reticulum
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gaborik Z, Mihalik B, Jayadev S, Jagadeesh G, Catt KJ, Hunyady L. FEBS Lett. 1998;428:147–151. doi: 10.1016/s0014-5793(98)00511-0. [DOI] [PubMed] [Google Scholar]
- 2.Duvernay MT, Wang H, Dong C, Guidry JJ, Sackett DL, Wu G. J Biol Chem. 2011;286:14080–14089. doi: 10.1074/jbc.M111.222323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang X, Wang H, Duvernay MT, Zhu S, Wu G. PLoS One. 2013;8:e57805. doi: 10.1371/journal.pone.0057805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tetsuka M, Saito Y, Imai K, Doi H, Maruyama K. Endocrinology. 2004;145:3712–3723. doi: 10.1210/en.2003-1638. [DOI] [PubMed] [Google Scholar]
- 5.Venkatesan S, Petrovic A, Locati M, Kim YO, Weissman D, Murphy PM. J Biol Chem. 2001;276:40133–40145. doi: 10.1074/jbc.M105722200. [DOI] [PubMed] [Google Scholar]
- 6.Hunyady L, Catt KJ. Mol Endocrinol. 2006;20:953–970. doi: 10.1210/me.2004-0536. [DOI] [PubMed] [Google Scholar]
- 7.Mehta PK, Griendling KK. Am J Physiol Cell Physiol. 2007;292:C82–97. doi: 10.1152/ajpcell.00287.2006. [DOI] [PubMed] [Google Scholar]
- 8.Oro C, Qian H, Thomas WG. Pharmacol Ther. 2007;113:210–226. doi: 10.1016/j.pharmthera.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huynh J, Thomas WG, Aguilar MI, Pattenden LK. Mol Cell Endocrinol. 2009;302:118–127. doi: 10.1016/j.mce.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Lee DK, Lanca AJ, Cheng R, Nguyen T, Ji XD, Gobeil F, Chemtob S, George SR, O’Dowd BF. Journal of Biological Chemistry. 2004;279:7901–7908. doi: 10.1074/jbc.M306377200. [DOI] [PubMed] [Google Scholar]
- 11.Ali MS, Sayeski PP, Dirksen LB, Hayzer DJ, Marrero MB, Bernstein KE. J Biol Chem. 1997;272:23382–23388. doi: 10.1074/jbc.272.37.23382. [DOI] [PubMed] [Google Scholar]
- 12.Morinelli TA, Raymond JR, Baldys A, Yang Q, Lee MH, Luttrell L, Ullian ME. Am J Physiol-Cell Ph. 2007;292:C1398–C1408. doi: 10.1152/ajpcell.00337.2006. [DOI] [PubMed] [Google Scholar]
- 13.Ju H, Venema VJ, Marrero MB, Venema RC. J Biol Chem. 1998;273:24025–24029. doi: 10.1074/jbc.273.37.24025. [DOI] [PubMed] [Google Scholar]
- 14.Thomas WG, Motel TJ, Kule CE, Karoor V, Baker KM. Mol Endocrinol. 1998;12:1513–1524. doi: 10.1210/mend.12.10.0179. [DOI] [PubMed] [Google Scholar]
- 15.Sano T, Ohyama K, Yamano Y, Nakagomi Y, Nakazawa S, Kikyo M, Shirai H, Blank JS, Exton JH, Inagami T. J Biol Chem. 1997;272:23631–23636. doi: 10.1074/jbc.272.38.23631. [DOI] [PubMed] [Google Scholar]
- 16.Becker BN, Cheng HF, Hammond TG, Harris RC. Mol Pharmacol. 2004;65:362–369. doi: 10.1124/mol.65.2.362. [DOI] [PubMed] [Google Scholar]
- 17.Thomas WG, Baker KM, Motel TJ, Thekkumkara TJ. J Biol Chem. 1995;270:22153–22159. doi: 10.1074/jbc.270.38.22153. [DOI] [PubMed] [Google Scholar]
- 18.Conchon S, Barrault MB, Miserey S, Corvol P, Clauser E. J Biol Chem. 1997;272:25566–25572. doi: 10.1074/jbc.272.41.25566. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Unal H, Gati C, Han GW, Liu W, Zatsepin NA, James D, Wang D, Nelson G, Weierstall U, Sawaya MR, Xu Q, Messerschmidt M, Williams GJ, Boutet S, Yefanov OM, White TA, Wang C, Ishchenko A, Tirupula KC, Desnoyer R, Coe J, Conrad CE, Fromme P, Stevens RC, Katritch V, Karnik SS, Cherezov V. Cell. 2015;161:833–844. doi: 10.1016/j.cell.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mozsolits H, Unabia S, Ahmad A, Morton CJ, Thomas WG, Aguilar MI. Biochemistry. 2002;41:7830–7840. doi: 10.1021/bi0121813. [DOI] [PubMed] [Google Scholar]
- 21.Kamimori H, Unabia S, Thomas WG, Aguilar MI. Anal Sci. 2005;21:171–174. doi: 10.2116/analsci.21.171. [DOI] [PubMed] [Google Scholar]
- 22.Hirst DJ, Lee TH, Pattenden LK, Thomas WG, Aguilar MI. Sci Rep. 2015;5:9972. doi: 10.1038/srep09972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu G, Zhao G, He Y. J Biol Chem. 2003;278:47062–47069. doi: 10.1074/jbc.M305707200. [DOI] [PubMed] [Google Scholar]
- 24.Dong C, Wu G. Cell Signal. 2007;19:2388–2399. doi: 10.1016/j.cellsig.2007.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong C, Zhou F, Fugetta EK, Filipeanu CM, Wu G. Cell Signal. 2008;20:1035–1043. doi: 10.1016/j.cellsig.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Filipeanu CM, Zhou F, Claycomb WC, Wu G. J Biol Chem. 2004;279:41077–41084. doi: 10.1074/jbc.M405988200. [DOI] [PubMed] [Google Scholar]
- 27.Dong C, Zhang X, Zhou F, Dou H, Duvernay MT, Zhang P, Wu G. J Pharmacol Exp Ther. 2010;333:174–183. doi: 10.1124/jpet.109.161489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cook JL, Re RN, deHaro DL, Abadie JM, Peters M, Alam J. Circ Res. 2008;102:1539–1547. doi: 10.1161/CIRCRESAHA.108.176594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin H, Li Q, Qian G, Wang Y, Li Y, Wu G, Wang G. Int J Biochem Cell Biol. 2011;43:401–408. doi: 10.1016/j.biocel.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Azuma K, Tamura K, Shigenaga A, Wakui H, Masuda S, Tsurumi-Ikeya Y, Tanaka Y, Sakai M, Matsuda M, Hashimoto T, Ishigami T, Lopez-Ilasaca M, Umemura S. Hypertension. 2007;50:926–932. doi: 10.1161/HYPERTENSIONAHA.107.096115. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Dong C, Wu QJ, Balch WE, Wu G. J Biol Chem. 2011;286:20525–20535. doi: 10.1074/jbc.M111.222034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duvernay MT, Zhou F, Wu G. J Biol Chem. 2004;279:30741–30750. doi: 10.1074/jbc.M313881200. [DOI] [PubMed] [Google Scholar]
- 33.Duvernay MT, Dong C, Zhang X, Zhou F, Nichols CD, Wu G. Mol Pharmacol. 2009;75:751–761. doi: 10.1124/mol.108.051623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duvernay MT, Dong C, Zhang X, Robitaille M, Hebert TE, Wu G. Traffic. 2009;10:552–566. doi: 10.1111/j.1600-0854.2009.00890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li C, Fan Y, Lan TH, Lambert NA, Wu G. J Biol Chem. 2012;287:42784–42794. doi: 10.1074/jbc.M112.410936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Wang G, Dupre DJ, Feng Y, Robitaille M, Lazartigues E, Feng YH, Hebert TE, Wu G. J Pharmacol Exp Ther. 2009;330:109–117. doi: 10.1124/jpet.109.153460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong C, Nichols CD, Guo J, Huang W, Lambert NA, Wu G. Traffic. 2012;13:857–868. doi: 10.1111/j.1600-0854.2012.01351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.AbdAlla S, Lother H, Langer A, el Faramawy Y, Quitterer U. Cell. 2004;119:343–354. doi: 10.1016/j.cell.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 39.Yamada M, Kushibiki M, Osanai T, Tomita H, Okumura K. Cardiovasc Res. 2008;79:169–178. doi: 10.1093/cvr/cvn064. [DOI] [PubMed] [Google Scholar]
- 40.AbdAlla S, Lother H, Quitterer U. Nature. 2000;407:94–98. doi: 10.1038/35024095. [DOI] [PubMed] [Google Scholar]
- 41.Dubey RK, Jackson EK, Luscher TF. J Clin Invest. 1995;96:141–149. doi: 10.1172/JCI118014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kohno M, Yokokawa K, Kano H, Yasunari K, Minami M, Hanehira T, Yoshikawa J. Hypertension. 1997;29:1309–1313. doi: 10.1161/01.hyp.29.6.1309. [DOI] [PubMed] [Google Scholar]
- 43.Li H, Armando I, Yu P, Escano C, Mueller SC, Asico L, Pascua A, Lu Q, Wang X, Villar VA, Jones JE, Wang Z, Periasamy A, Lau YS, Soares-da-Silva P, Creswell K, Guillemette G, Sibley DR, Eisner G, Gildea JJ, Felder RA, Jose PA. J Clin Invest. 2008;118:2180–2189. doi: 10.1172/JCI33637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andersson H, Kappeler F, Hauri HP. J Biol Chem. 1999;274:15080–15084. doi: 10.1074/jbc.274.21.15080. [DOI] [PubMed] [Google Scholar]
- 45.Harter C, Wieland FT. Proc Natl Acad Sci U S A. 1998;95:11649–11654. doi: 10.1073/pnas.95.20.11649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Letourneur F, Gaynor EC, Hennecke S, Demolliere C, Duden R, Emr SD, Riezman H, Cosson P. Cell. 1994;79:1199–1207. doi: 10.1016/0092-8674(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 47.Ma W, Goldberg J. EMBO J. 2013;32:926–937. doi: 10.1038/emboj.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez O, Goud B. Biochim Biophys Acta. 1998;1404:101–112. doi: 10.1016/s0167-4889(98)00050-0. [DOI] [PubMed] [Google Scholar]
- 49.White J, Johannes L, Mallard F, Girod A, Grill S, Reinsch S, Keller P, Tzschaschel B, Echard A, Goud B, Stelzer EH. J Cell Biol. 1999;147:743–760. doi: 10.1083/jcb.147.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuge O, Dascher C, Orci L, Rowe T, Amherdt M, Plutner H, Ravazzola M, Tanigawa G, Rothman JE, Balch WE. J Cell Biol. 1994;125:51–65. doi: 10.1083/jcb.125.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gillingham AK, Munro S. Annu Rev Cell Dev Biol. 2007;23:579–611. doi: 10.1146/annurev.cellbio.23.090506.123209. [DOI] [PubMed] [Google Scholar]
- 52.Kirchberg K, Kim TY, Moller M, Skegro D, Dasara Raju G, Granzin J, Buldt G, Schlesinger R, Alexiev U. Proc Natl Acad Sci U S A. 2011;108:18690–18695. doi: 10.1073/pnas.1015461108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaye RG, Saldanha JW, Lu ZL, Hulme EC. Mol Pharmacol. 2011;79:701–709. doi: 10.1124/mol.110.070177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cook JL, Singh A, DeHaro D, Alam J, Re RN. Am J Physiol Cell Physiol. 2011;301:C1175–1185. doi: 10.1152/ajpcell.00040.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dores MR, Trejo J. Mol Pharmacol. 2012;82:563–570. doi: 10.1124/mol.112.079418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Margeta-Mitrovic M, Jan YN, Jan LY. Neuron. 2000;27:97–106. doi: 10.1016/s0896-6273(00)00012-x. [DOI] [PubMed] [Google Scholar]
- 57.Filipeanu CM, Pullikuth AK, Guidry JJ. Mol Pharmacol. 2015;87:792–802. doi: 10.1124/mol.114.096198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arumugam S, Sreedhar R, Thandavarayan RA, Karuppagounder V, Krishnamurthy P, Suzuki K, Nakamura M, Watanabe K. Trends Cardiovasc Med. 2015 doi: 10.1016/j.tcm.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 59.Miyajima A, Kikuchi E, Kosaka T, Oya M. Rev Recent Clin Trials. 2009;4:75–78. doi: 10.2174/157488709788185996. [DOI] [PubMed] [Google Scholar]