The novel protein EHBP1L1 links Rab8 to Bin1and dynamin to regulate apical transport in epithelial cells.
Abstract
The highly conserved Rab guanosine triphosphatase (GTPase) Rab8 plays a role in exocytosis toward the polarized plasma membrane in eukaryotic cells. In murine Rab8-deficient small intestine cells, apical proteins are missorted into lysosomes. In this study, we identified a novel Rab8-interacting protein complex containing an EH domain–binding protein 1–like 1 (EHBP1L1), Bin1/amphiphysin II, and dynamin. Biochemical analyses showed that EHBP1L1 directly bound to GTP-loaded Rab8 and Bin1. The spatial dependency of these complexes at the endocytic recycling compartment (ERC) was demonstrated through overexpression and knockdown experiments. EHBP1L1- or Bin1-depleted or dynamin-inhibited small intestine organoids significantly accumulated apical membrane proteins but not basolateral membrane proteins in lysosomes. Furthermore, in EHBP1L1-deficient mice, small intestine cells displayed truncated and sparse microvilli, suggesting that EHBP1L1 maintains the apical plasma membrane by regulating apical transport. In summary, our data demonstrate that EHBP1L1 links Rab8 and the Bin1–dynamin complex, which generates membrane curvature and excises the vesicle at the ERC for apical transport.
Introduction
In polarized epithelial cells, the transport pathway is directed to the apical or basolateral plasma membrane, which differ in protein and lipid composition (Rodriguez-Boulan et al., 2005). Several findings suggest that newly synthesized protein exported from the TGN is delivered to the endocytic recycling compartment (ERC), which is regarded as a recycling endosome, and sorted to the apical or basolateral plasma membrane (Ang et al., 2004; Thuenauer et al., 2014).
Rab GTPases belong to the Ras small GTPase superfamily (Wennerberg et al., 2005). More than 60 mammalian Rab proteins define vesicle and organelle identity by recruiting various binding proteins to the membrane. The Rab protein acts upstream of SNARE-mediated fusion to the target membrane (Barr, 2013).
Rab8 is a highly conserved small GTPase in eukaryotic cells and regulates exocytic transport to a polarized plasma membrane (Peränen, 2011). The mammalian genome encodes two Rab8 isoforms: Rab8a and Rab8b. Small intestine cells in both Rab8a knockout (KO) and Rab8a/8b double-knockout (DKO) mice show accumulated apical cargo proteins in lysosomes, which suggests that Rab8 is involved in apical transport (Sato et al., 2007, 2014). Previous studies provide insight into the molecular mechanisms related to Rab8. In Saccharomyces cerevisiae, the Rab8 homolog Sec4 plays a role in exocytic vesicle motility with Myo2, an actin-based motor protein, and mediates vesicle docking to a polarized plasma membrane by binding to a tethering complex, the exocyst (Guo et al., 1999; He and Guo, 2009; Jin et al., 2011). Importantly, small intestines from microvillus inclusion disease patients with a gene mutation for Myo5B, a mammalian homolog of yeast Myo2, exhibit short microvilli and microvillus inclusion bodies, which are also observed in Rab8 KO mouse intestine cells (Sato et al., 2007; Ruemmele et al., 2010). Despite its role in exocytic vesicle motility and tethering, Rab8 is mainly localized to the ERC in mammals and Caenorhabditis elegans, indicating unknown functions of Rab8 at the ERC (Hattula et al., 2006; Sharma et al., 2009; Shi et al., 2010). Emerging evidence suggests that Rab proteins may coordinate with Bin/Amphiphysin/Rvs (BAR) domain proteins, which are membrane curvature sensing/generating proteins on the ERC (Pant et al., 2009; Giridharan et al., 2013).
In this study, we identified a novel Rab8-binding protein complex, EHBP1L1–Bin1–dynamin, and demonstrated its role in apical transport. Our results suggest that this complex localizes to the ERC and functions in the generation of vesicles/tubules containing apical cargo proteins. In small intestine organoids, such as in small intestines from Rab8a/b KO mice, depletion of EHBP1L1 and Bin1 or inhibition of dynamin GTPase activity caused missorting of the apical proteins.
Results and discussion
Identification of EHBP1L1 as a novel Rab8-binding protein
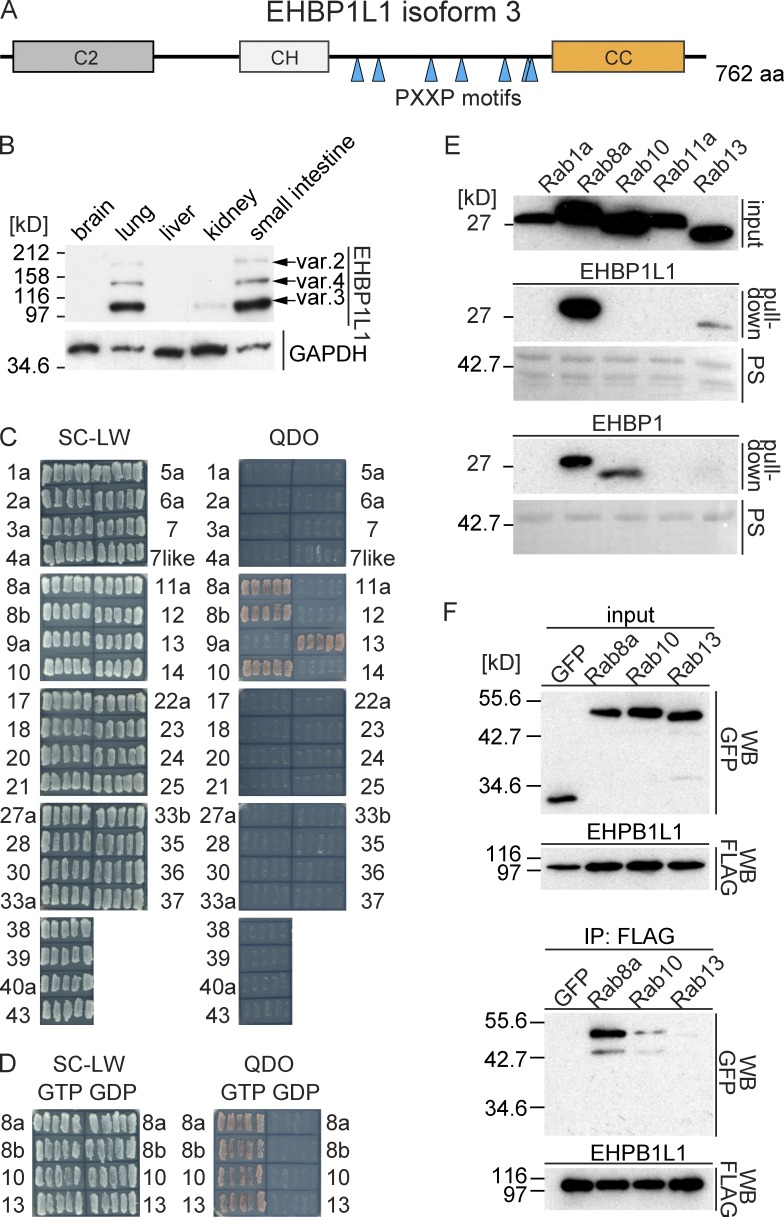
To identify novel proteins that bind Rab8 and that function predominantly in epithelial cells, we used yeast two-hybrid screening with a Rab8 GTP-restricted form against a mouse small intestine cDNA library. We obtained several positive clones from this screening. Most were previously reported, such as microtubule-associated monooxygenase, calponin, and LIM domain–containing 1 (MICAL 1), MICAL-like 1, MICAL-L2, and optineurin (Sahlender et al., 2005; Rahajeng et al., 2010). Seven clones represented the same uncharacterized gene that encoded the full-length EHBP1L1 isoform 3 (Fig. 1 A). Three EHBP1L1 isoforms were highly expressed in the lung and small intestine among several tissues (Fig. 1 B).
Figure 1.
EHBP1L1 is a novel Rab8-interacting protein. (A) Schematic representation of EHBP1L1. CH, calponin homology domain; CC, coiled-coil domain. Seven PXXP motifs are indicated by triangles. (B) Lysates from different mouse tissues were immunoblotted using an EHBP1L1 antibody. EHBP1L1 splicing isoforms based on the GenBank database are indicated by arrows. (C) A yeast two-hybrid assay between EHBP1L1 and the GTP-restricted forms of Rab proteins. The cells were grown on SC-LW plates, and five independent colonies were restreaked on QDO plates. (D) Yeast two-hybrid assay using EHBP1L1 and the GTP or GDP forms of Rab-8a, 8b, 10, and 13. (E) In vitro pull-down assay using recombinant GST-EHBP1L1-CC (top) or GST-EHBP1-CC (bottom) and the lysate from HEK293 cells expressing the FLAG-tagged GTP-form of Rab, as indicated. The transferred protein was stained with Ponceau S (PS) to detect GST-fusion proteins. The bound Rab proteins were detected by immunoblotting using a FLAG antibody. (F) HEK293 cell lysates coexpressing FLAG-tagged EHBP1L1 and EGFP-tagged Rab, or EGFP proteins were immunoprecipitated using a FLAG antibody.
EHBP1L1 includes three conserved domains: the C2 domain, the calponin homology (CH) domain, and the coiled-coil (CC) domain (Fig. 1 A). Previous studies show that the C2 domain binds phosphatidylserine and phosphatidylethanolamine (Lemmon, 2008). The CH domain has been observed in several F-actin–binding proteins (Sjöblom et al., 2008). The CC domain is conserved among many Rab-binding proteins, such as MICAL-1, MICAL-L1, MICAL-L2, and EHBP1 (Yamamura et al., 2008; Sharma et al., 2009; Giridharan et al., 2013). EHBP1 and EHBP1L1 in particular exhibit a similar domain composition and aa sequence identity (65% in C2, 75% in CH, and 71% in the CC domain). EHBP1L1 features a unique region between the CH and CC domains. The region includes a proline-rich (PR) sequence with seven PXXP motifs, which has been predicted to bind to SH3 domain–containing proteins (Fig. 1 A; Lee et al., 2002). EHBP1L1 bound a GTP-restricted form of Rab8a, Rab8b, Rab10, and Rab13 among 35 Rab proteins in a yeast two-hybrid assay. All of the bound Rab proteins are members of the Sec4/Rab8 subfamily (Fig. 1 C; Pereira-Leal and Seabra, 2001). The EHBP1L1 binding was specific for the GTP form but not for the GDP form (Fig. 1 D). These results suggest that EHBP1L1 specifically binds members of the Sec4/Rab8 subfamily.
We further confirmed the binding specificity using an in vitro pull-down assay between several GTP-loaded Rab proteins and a GST-fused CC domain of EHBP1L1 and EHBP1, as well as coimmunoprecipitation in HEK293 cells (Fig. 1, E and F). In the in vitro pull-down assay, EHBP1L1 specifically bound Rab8a and weakly bound Rab13. In contrast, EHBP1 bound Rab8a and Rab10 and faintly bound Rab13, but it did not bind Rab1a or Rab11a (Fig. 1 E). To examine in vivo binding, HEK293 cell lysates coexpressing FLAG-tagged EHBP1L1 and EGFP-tagged Rab proteins were immunoprecipitated using a FLAG antibody. Rab8a was most strongly coprecipitated among the Sec4/Rab8 subfamily (Fig. 1 F). Collectively, we conclude that EHBP1L1 is a specific binding protein for Rab8.
The EHBP1L1 PR domain binds Bin1
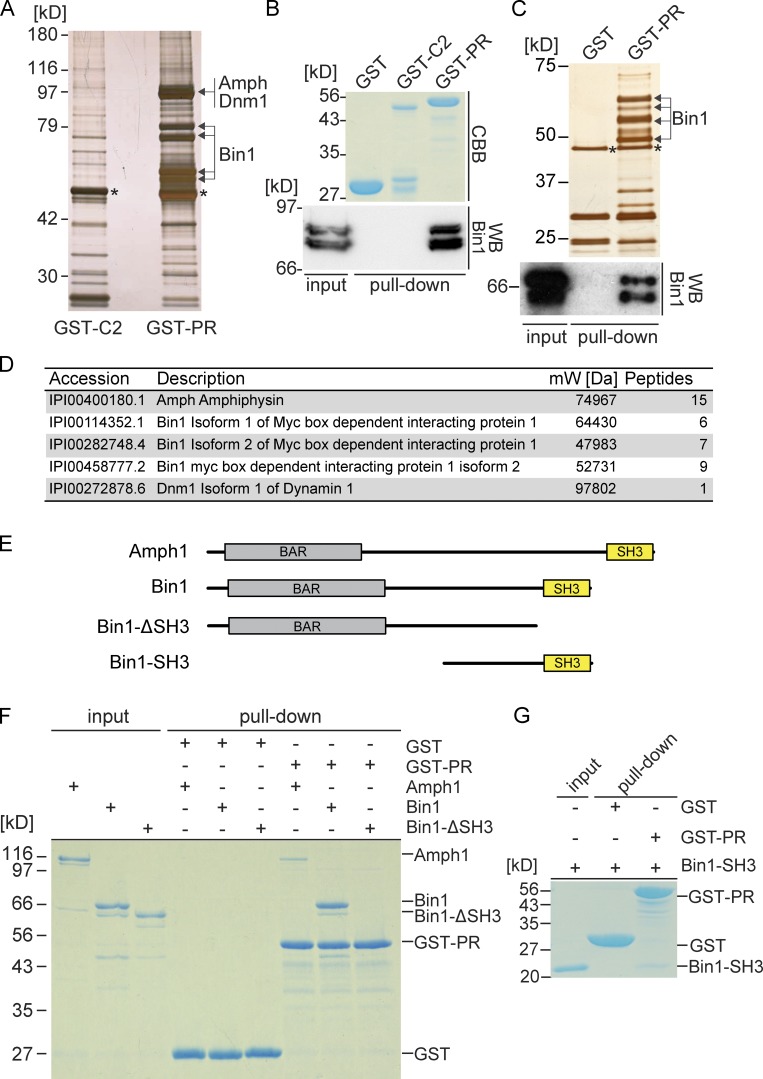
We took a biochemical approach to search for EHBP1L1-interacting proteins to gain further insight into EHBP1L1. Although EHBP1L1 is highly expressed in lung and small intestine (Fig. 1 B), these lysates were unsuitable for this approach because of low protein yields and high protease activity. Therefore, we used mouse brain or human Caco-2 cell lysates for a pull-down screen with GST-fusion proteins of the C2 domain and PR domain of EHBP1L1 (Fig. 2, A and C). From the brain lysate, mass spectrometry identified several proteins that were pulled down with the PR domain, including amphiphysin1 (Amph1), Bin1 (also termed as amphiphysin II), and dynamin1 (Fig. 2 D). Similarly, we also obtained Bin1 from Caco-2 lysate (Fig. 2 C). We confirmed an interaction between the PR domain and Bin1 through immunoblotting (Fig. 2, B and C). We demonstrated a direct interaction between the EHBP1L1 PR domain and full-length Amph1 and Bin1 using purified proteins (Fig. 2 F). Next, we examined the series of truncated Bin1 constructs shown in Fig. 2 E. Bin1 harboring an SH3 domain interacted with EHBP1L1, whereas the protein lacking the SH3 domain did not (Figs. 2, F and G). These data indicate that EHBP1L1 binds directly to Bin1 via its C-terminal SH3-containing region. Amph1 and Bin1 sense and generate membrane curvature via a BAR domain and couple the membrane scission (Meinecke et al., 2013; Daumke et al., 2014). Hereafter, we focused on Bin1, which is expressed ubiquitously, rather than Amph1, expressed specifically in the brain (De Camilli et al., 1993; Butler et al., 1997).
Figure 2.
EHBP1L1 proline-rich domain binds Bin1. (A) GST pull-down assay using mouse brain lysate and the GST-C2 domain or PR domain of EHBP1L1. The protein gel was visualized through silver staining. The bands with arrows or asterisks were digested and analyzed by mass spectrometry. The asterisks indicate α- and β-tubulin. (B) Pulled-down proteins were immunoblotted using a Bin1 antibody. Coomassie brilliant blue–stained gel shows the GST fusion protein input. (C) GST pull-down assay using Caco-2 lysate and a GST or GST-EHBP1L1 PR domain. Top, silver-stained gel; bottom, immunoblotting image using a Bin1 antibody; *, α- and β-tubulin. (D) List of proteins detected by mass spectrometry from the brain lysate. (E) Schematic representations showing amphiphysin1, Bin1, Bin1-SH3, and Bin1-ΔSH3. (F and G) In vitro pull-down assay using purified recombinant protein, Amph1, Bin1, and Bin1-ΔSH3 (F) or Bin1-SH3 (G) and either immobilized GST or the GST-tagged PR domain. Coomassie brilliant blue stain. We loaded 1 µg Amph1, Bin1, Bin1-ΔSH3, and Bin1-SH3 as input.
Rab8, EHBP1L1, and Bin1 stabilize each other’s localization to the ERC
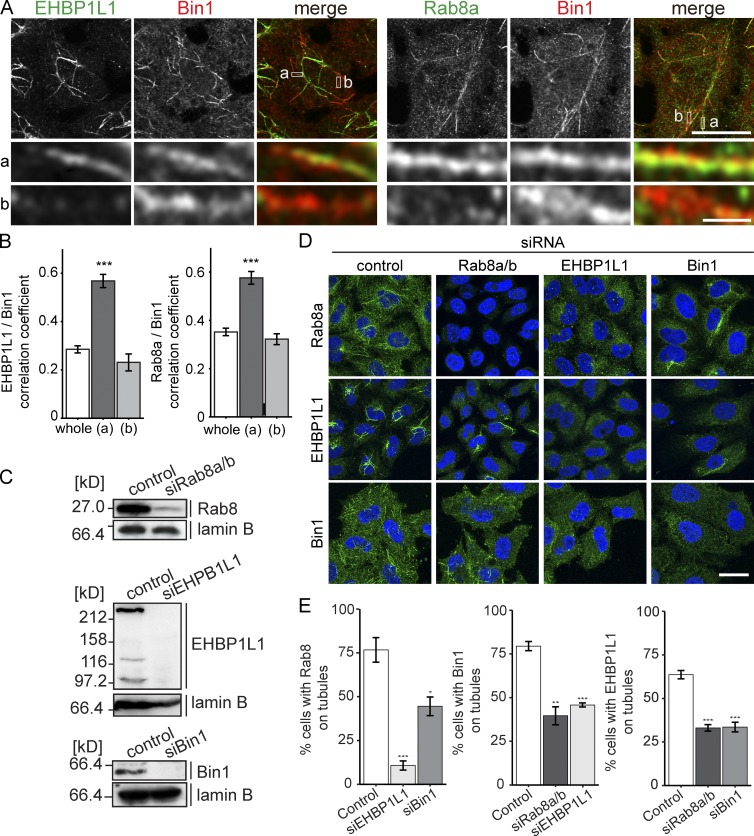
We then assessed the subcellular localization of Rab8, EHBP1L1, and Bin1 in HeLa cells (Fig. 3) and the mouse mammary epithelial cell line EpH4 (Figs. 4 and S2). In HeLa cells, Rab8 localizes to the prominent tubular structures that are referred to as the ERC (Maxfield and McGraw, 2004; Hattula et al., 2006). Although HeLa is a nonpolarized cell line, we took advantage of these structures to test the dependence of each protein there (Figs. 3 and S1). Bin1 and EHBP1L1 or Bin1 and Rab8a colocalized to the ERC. We also observed heterogeneous distribution between these proteins to some parts of the ERC (Fig. 3, A and B). Rab8a and Bin1 also colocalized with exogenously expressed FLAG-tagged EHBP1L1 in HeLa cells. In these cells, Rab8a and Bin1 exhibited strong recruitment to the ERC (Fig. S1 A). Next, each protein was depleted to examine the effect on the localization of the remaining proteins to tubular structures. Rab8a/8b, EHBP1L1, and Bin1 siRNA efficiently depleted the expression of each protein (Fig. 3 C). The immunofluorescence data showed that the loss of one protein altered the localization of the other proteins (Fig. 3, D and E). We also confirmed that the tubular ERC marked by CD147 was not altered in shape in these cells (Fig. S1 B). This result suggests that the three proteins stabilize each other’s localization.
Figure 3.
Rab8, EHBP1L1, and Bin1 stabilize each other on the tubules. (A) Immunofluorescence for EHBP1L1, Bin1, and Rab8a in HeLa cells. The enlarged views at the bottom show the overlap of the proteins on the tubular structures. Areas a and b are representative regions of the ERC depicting uniform intensity and heterogeneous intensity between the two proteins, respectively. Bars: (magnified views) 1 µm; (other views) 10 µm. (B) Correlation analysis comparing the distribution of EHBP1L1/Bin1 and Rab8/Bin1 in the whole cell (whole), the region positive for the two proteins (a), and the region positive for either protein (b). Data are mean ± SEM from at least 10 cells. ***, P < 0.001 relative to control; Student’s t test. (C) KD efficiency of Rab8, Bin1, and EHBP1L1 in HeLa cells. Lysates were analyzed by immunoblotting. Lamin B was used as a loading control. (D and E) HeLa cells transfected with siRNA for Rab8a and Rab8b, EHBP1L1, or Bin1 were stained with Rab8, Bin1, or EHBP1L1 antibody (green). Nuclei were indicated using DAPI (blue). Bar, 20 µm. (E) Quantification of the percentage of cells with tubular structures. Data are mean ± SEM from at least three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 relative to control; Student’s t test.
Figure 4.
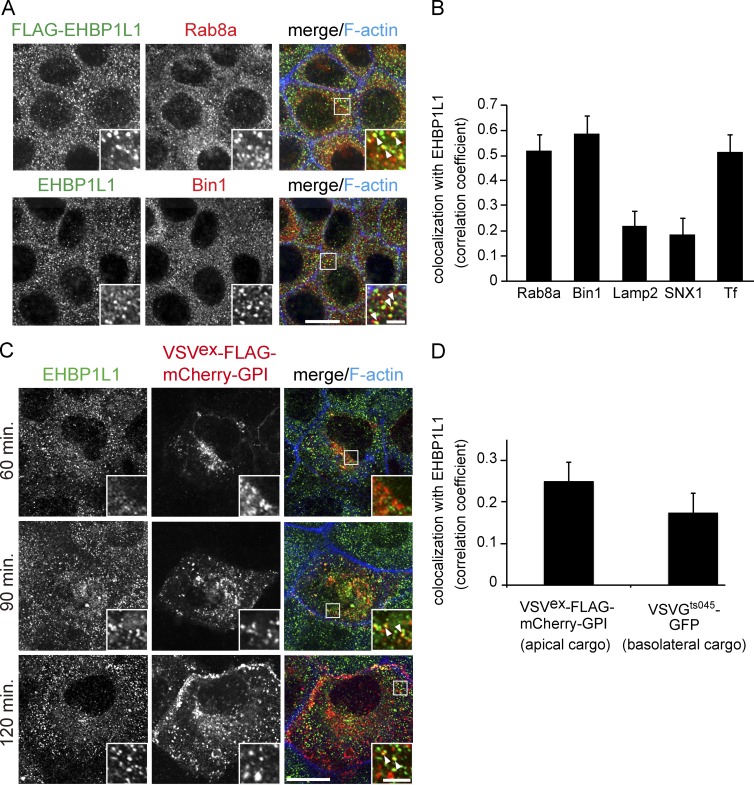
Apical cargo protein is transported through the ERC. (A) Subcellular localization of Rab8, EHBP1L1, FLAG-tagged EHBP1L1, and Bin1 in EpH4 cells was examined using immunofluorescence microscopy. (B) Colocalization was analyzed between EHBP1L1 and Rab8a, Bin1 (A), Lamp2, sorting nexin 1 (SNX1), and internalized transferrin (Tf; Fig. S2 B). (C) EpH4 cells expressing VSVex-FLAG-mCherry-GPI were cultured at 40°C and incubated for 60, 90, and 120 min at 32°C. The cells were fixed at each time point and stained using an EHBP1L1 antibody and Alexa Fluor 633 phalloidin. The insets show enlarged views. The arrowheads in the insets denote colocalization. Bars: (magnified views) 2 µm; (other views) 10 µm. (D) Colocalization was analyzed between EHBP1L1 and both VSVex-FLAG-mCherry-GPI (apical cargo) and VSVGts045-GFP (basolateral cargo; Fig. S2 C) after 90-min chase. Data are mean ± SEM from 10 cells; P < 0.01.
The apical cargo protein is transported through the ERC in polarized epithelial cells
Unlike in HeLa cells, EHBP1L1 localized to punctate structures throughout the cytoplasm in confluent monolayers of EpH4 cells (Fig. S2, A and B). EHBP1L1 overlapped with Bin1 and Rab8a on these structures (Fig. 4, A and B).
To define the EHBP1L1-positive punctate structures, the cells were costained for endosomal and lysosomal proteins. EHBP1L1 colocalized with the ERC, labeled with internalized transferrin, and partially overlapped with the sorting/early endosomal protein sorting nexin1 (SNX1) and the late endosomal/lysosomal protein Lamp2 (Figs. 4 B and S2 B). These data indicate that EHBP1L1 localizes to the ERC in epithelial cells as well as HeLa cells.
Next, we investigated whether the EHBP1L1-positive ERC in epithelial cells is a route for apical and basolateral cargo proteins, as previous studies suggested that the ERC is a sorting center for apical and basolateral destinations (Ang et al., 2004; Thuenauer et al., 2014). EpH4 cells expressing the apical model cargo protein VSVex-FLAG-mCherry-GPI were cultured at 40°C and incubated at 32°C to release the protein from the ER. After a 90-min chase, the protein was localized to punctate structures and colocalized with EHBP1L1 (Fig. 4, C and D). VSVGts045-GFP, a model cargo protein for the basolateral pathway, also colocalized with EHBP1L1-positive endosomes (Figs. 4 D and S2 C). These data suggest that the apical and basolateral cargo proteins traffic through the ERC in polarized epithelial cells.
EHBP1L1 and Bin1 depletion affects apical protein localization in polarized epithelial cells
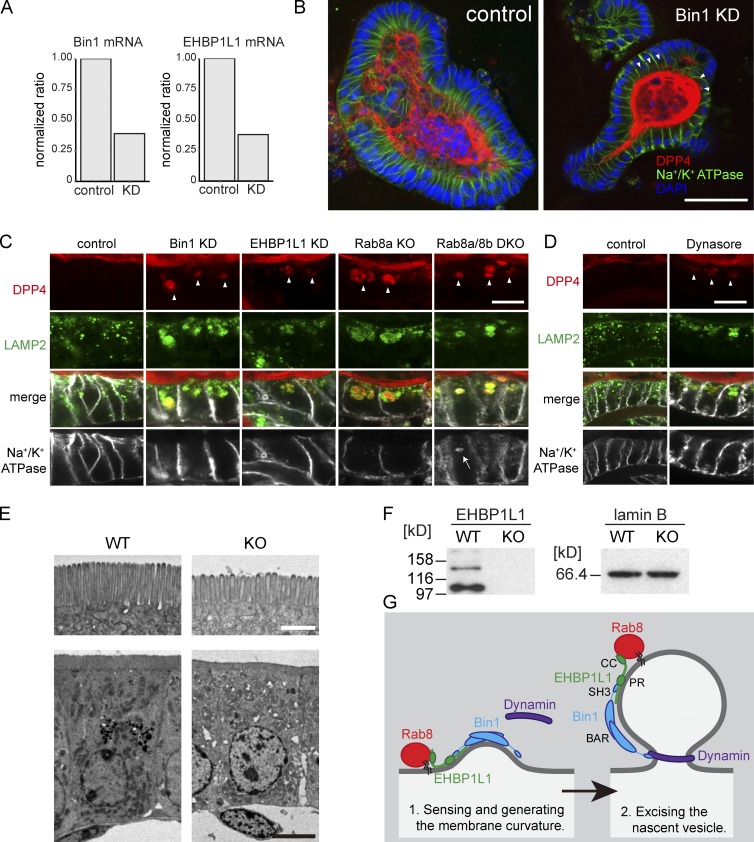
As previously reported, Rab8 deficiency induces the mislocalization of apical plasma membrane proteins to lysosomes in small intestine cells (Sato et al., 2014). We sought to determine whether EHBP1L1 and Bin1 also aid in transporting apical proteins in small intestine epithelial cells. EHBP1L1 and Bin1 were successfully depleted by lentivirus-based shRNA in mouse small intestine organoids (Sato et al., 2009). The depletion efficiency was determined using semiquantitative RT-PCR (Fig. 5 A). Localization of dipeptidyl peptidase 4 (DPP4) and Na+/K+ ATPase was demonstrated using immunofluorescence (Fig. 5 B). In the control organoids, DPP4 and Na+/K+ ATPase localized to the apical membrane and basolateral membrane, respectively, confirming successful epithelial polarization in this culture system. On the other hand, EHBP1L1-depleted organoids showed subapical punctate cytoplasmic localization of DPP4 in addition to apical plasma membrane localization. This cytoplasmic accumulation of DPP4 colocalized with Lamp2 (Fig. 5 C, EHBP1L1 KD). Similarly altered DPP4 localization was observed in the Bin1 knockdown (KD) organoids (Fig. 5, B and C, Bin1 KD) as well as in the organoids from the Rab8a KO and Rab8a/8b DKO mice (Fig. 5 C). In contrast, Na+/K+ ATPase localization remained intact in the Bin1-KD, EHBP1L1-KD, and Rab8a-KO organoids (Fig. 5 C). In the Rab8a/8b DKO organoids, Na+/K+ ATPase accumulated in the lysosome/late endosomes (Fig. 5 C, Rab8a/8b DKO, arrow). These data indicate that Rab8, EHBP1L1, and Bin1 play a role in apical protein transport and that Rab8 may play an additional role in basolateral transport in small intestine polarized epithelial cells.
Figure 5.
EHBP1L1 and Bin1 depletion induces the accumulation of apical but not basolateral membrane proteins in lysosomes. (A) EHBP1L1 and Bin1 mRNA levels in mouse small intestine organoids were analyzed using semiquantitative RT-PCR. The level of GAPDH mRNA was used for standardization. (B) Immunostaining for Na+/K+ ATPase (green) and DPP4 (red) in the mouse small intestine organoids. The nuclei were counterstained with DAPI (blue). The arrows indicate cytoplasmic DPP4 accumulation. Bar, 50 µm. (C) Magnified images of immunostaining for DPP4, Lamp2, and Na+/K+ ATPase in the small intestine Bin1 or EHBP1L1 KD organoids or organoids obtained from Rab8a or Rab8a/8b DKO mice. Arrowheads, DPP4 accumulation in lysosomes; arrow, Na+/K+ ATPase accumulation in lysosomes. Bar, 10 µm. (D) Immunostaining for DPP4 (red), Lamp2 (green), and Na+/K+ ATPase (white) in mouse small intestine organoids treated with DMSO (control) or 40 µM dynasore for 4 d. Bar, 10 µm. (E) Electron micrographs of epithelial cells in small intestines from EHBP1L1 KO mice and wild-type mice. Bars: (top) 1 µm; (bottom) 10 µm. (F) Immunoblot for intestine lysate from a WT or an EHBP1L1 KO mouse using an EHBP1L1 antibody. (G) Working model for Rab8, EHBP1L1, Bin1, and dynamin at the ERC. Bin1 is represented as a dimer (Owen et al., 1998).
The evidence that Bin1 senses membrane curvature and generates tubular structures raised an additional question regarding the mechanism of membrane scission machinery working with Rab8–EHBP1L1–Bin1. One candidate protein is dynamin, which excises the membrane in a GTPase-dependent manner (Takei et al., 1995). Our pull-down experiment using the EHBP1L1-PR domain revealed an association with Amph1, Bin1, and dynamin1 (Fig. 2 A). However, the Bin1 SH3 domain directly bound the EHBP1L1-PR domain (Fig. 2 F), and the binding between EHBP1L1-PR domain and dynamin1 appeared indirect, most likely via the Amph1 or Bin1 dimer (Owen et al., 1998; Meinecke et al., 2013; Neumann and Schmid, 2013). A recent study showed that inhibiting dynamin activity by either dominant-negative dynamin or the dynamin inhibitor dynasore in MDCK cells yielded the accumulation of an apical protein in a recycling endosome (Thuenauer et al., 2014). Therefore, we treated the small intestine organoids with dynasore. Recently, Park et al. (2013) reported that dynasore also inhibits fluid-phase endocytosis and membrane ruffling in addition to dynamin-dependent endocytosis, suggesting off-target effects. Thus, it is necessary to be careful in the interpretation of the data with dynasore. To diminish the off-target effect, we used 40 µM dynasore, half the concentration used by Park et al. This concentration is sufficient to inhibit the dynamin GTPase (Macia et al., 2006). After 4-d incubation with dynasore, DPP4 accumulated in lysosomes, but Na+/K+ ATPase did not (Fig. 5 D). Although we are not able to entirely exclude the possibility of off-target effects of dynasore, this result suggests that dynamin is likely to be involved in apical transport with the Rab8–EHBP1L1–Bin1 complex.
Small intestine in EHBP1L1-deficient mice exhibits truncated and sparse microvilli
We created EHBP1L1 KO mice using the CRISPR/Cas9 system (Fig. 5, E and F; Cong et al., 2013). The mice died within a day after birth. At that time, apical cargo proteins do not yet accumulate in lysosomes, even in Rab8a/8b DKO mice, which also exhibit defects in apical transport (Sato et al., 2014). Therefore, we could not detect accumulated apical cargo proteins in lysosomes from EHBP1L1 KO mice as in EHBP1L1-KD organoids (Fig. 5 C). Instead, the microvilli length (wild-type [WT]: mean ± SD 1.16 ± 0.11 µm measured on 42 cells; KO: 0.93 ± 0.11 µm [n = 23]; P < 0.0001; Student’s t test) and density (WT: 7.26 ± 0.35 µm−1 [n = 24]; KO: 6.15 ± 0.31 µm−1 [n = 38]; P < 0.0001) in the small intestines from EHBP1L1 KO mice were reduced (Fig. 5 E), as seen in Rab8a KO and Rab8a/8b DKO mice. These data indicate that EHBP1L1 maintains apical plasma membrane integrity by regulating apical transport.
In summary, our data indicate that the Rab8–EHBP1L1–Bin1 complex senses and generates membrane tubules to transport protein cargos to the apical plasma membrane, which is coupled with membrane scission by dynamin (Fig. 5 G). In polarized epithelial cells deficient in Rab8, EHBP1L1, Bin1, or dynamin, the cargo proteins eventually accumulated in lysosomes (Fig. 5, B–D). The proteins may have accumulated because the ERC contained unsorted apical proteins that directly fuse with lysosomes or change to lysosomes by maturation. In fact, a certain population of ERC proteins, including EHBP1L1, also partially localize to late endosome/lysosomes (Fig. S2; Yoshimura et al., 2010; Kanerva et al., 2013), which indicates spatial and functional relationships between the ERC and lysosomes.
Materials and methods
Plasmid construction
The mouse EHBP1L1 isoform C (PDB accession number NP_001108067.1), EHBP1, AMPH1, and BIN1 were amplified using PCR and KOD-Plus polymerase (Toyobo) with the Mouse 17-d Embryo Marathon-Ready cDNA library (Clontech). The cloned cDNA was subcloned into the mammalian expression plasmid pcDNA5/FRT/TO FLAG A or the yeast two-hybrid plasmids pACT2 or pFBT9.
The mammalian expression and yeast two-hybrid plasmids encoding the GTP-form and GDP-form Rab cDNAs were generated as previously described (Haas et al., 2005; Fuchs et al., 2007). The full-length AMPH1, BIN1, BIN1-ΔSH3 (1–448), BIN1-SH3 (391–521), EHBP1L1-C2 (1–185), and EHBP1L1-PR domain (442–595) were subcloned into the pQE32-TEV or pFAT2 vector for protein expression in Escherichia coli.
Protein production and purification from E. coli
Rosetta 2(DE3) pLysS cells (Novagen) expressing EHBP1L1-C2 or the EHBP1L1-PR domain in pFAT2 and the full-length AMPH1, BIN1, BIN1-ΔSH3, and BIN1-SH3 in pQE32-TEV were cultured in 2 l LB medium at 18°C for 16 h with 0.25 mM IPTG. The bacterial pellet was recovered by centrifugation, resuspended with IMAC20 (20 mM Tris-HCl, pH 8.0, 300 mM NaCl, and 20 mM imidazole), and sonicated. The lysate was centrifuged. The supernatant was mixed with 500 µl Ni-NTA Agarose (Qiagen). After incubating for 2 h at 4°C, the beads were washed three times in IMAC20, and the bound protein was eluted with IMAC200 (20 mM Tris-HCl, pH 8.0, 300 mM NaCl, and 200 mM imidazole). The eluted protein (5 ml) was dialyzed in PBS, and the aliquots were snap-frozen in liquid nitrogen and stored at −80°C until use.
Antibodies
A rabbit polyclonal anti-EHBP1L1 antibody was raised against a HisX6-GST-fused EHBP1L1-PR domain protein produced as described. The antiserum was affinity purified. Rabbit polyclonal anti-Rab8a antibody was prepared as described previously (Sato et al., 2007). The mouse monoclonal anti-BIN1 (clone 99D), mouse monoclonal anti-FLAG M2, rabbit monoclonal anti-DYKDDDDK, goat polyclonal DPP4, mouse monoclonal Na+/K+ ATPase, mouse monoclonal CD147 antibodies, and E-cadherin antibodies were purchased from Millipore, Sigma-Aldrich, Cell Signaling Technologies, R&D Systems, Upstate Biotechnology, eBioscience, and BD, respectively. Goat polyclonal anti-Lamin B (C-20) and goat polyclonal anti-SNX1 antibodies (T-19) were purchased from Santa Cruz Biotechnology. Rat monoclonal Lamp2 was obtained from the Developmental Studies Hybridoma Bank. Secondary antibodies Alexa Fluor 488 goat anti–rabbit IgG, donkey anti–rat IgG, Alexa Fluor 568 donkey anti–goat IgG, and Alexa Fluor 594 goat anti–mouse IgG were purchased from Life Technologies. Cyanine 5–conjugated donkey anti-rabbit antibody and horseradish peroxidase–conjugated antibodies for mouse, rabbit, and goat were purchased from Jackson ImmunoResearch Laboratories.
Yeast two-hybrid assay
The mouse small intestine library was generated using the Make Your Own “Mate & Plate” Library System (Clontech). In brief, 8-d-old mice were starved for 16 h and dissected to isolate the small intestines. We washed 2 cm of the small intestine in PBS and then quickly froze the sample in liquid nitrogen. The frozen intestine was minced, and total RNA was isolated using a NucleoSpin RNAII kit (Macherey-Nagel) in accordance with the manufacturer’s instructions. We applied 2 µg total RNA for reverse transcription using oligo-dT primer. The product was then amplified by PCR using Advantage 2 Polymerase Mix (Clontech). The cDNA was subjected to CHROMA SPIN+TE-400 Columns (Clontech) to exclude >200 bp DNA. We used 3 µg of purified cDNA and 0.5 µg pGADT7-Rec to transform a Y187 yeast strain, which was cultured in an SD/-Leu medium. The Y187 strain aliquots harboring the small intestine cDNA library were stored by freezing the medium at −80°C until use.
Yeast two-hybrid screening was performed essentially as described in the Matchmaker Gold Yeast Two-Hybrid System User Manual (Clontech). In brief, pFBT9 encoding GTP-restricted Rab8Q67L mutants were transformed into Y2HGold yeast, which were then mated with the Y187 yeast strain harboring a cDNA library in accordance with the Clontech manual and were grown on an SC-LW plate (SD/-Leu/-Trp) medium plate to calculate the number of screened colonies. The mated yeast strain was also grown on plates containing QDO (SD/-Ade/-His/-Leu/-Trp) medium and incubated for 10 d at 30°C. We collected 200 colonies from the QDO plate. The plasmids were then obtained from each colony through yeast DNA preparation and subsequently rescued through transforming the E. coli strain XL-1 Blue. The individual rescued plasmids and pGADT7 were retransformed into the PJ69-4A yeast strain and plated on SC-LW plates. The strain was then restreaked on QDO plates to verify self-activation. We collected 196 positive clones, and the sequences were analyzed.
Direct interaction was examined as described previously (Haas et al., 2005). In brief, the PJ69-4A strain was cotransformed with the AD plasmid (pACT2-EHBP1L1) and BD plasmid (pFBT9-Rabs). The transformed cells were grown on an SC-LW plate for 3 d at 30°C. Five independent colonies were restreaked on an SC-LW plate and QDO followed by incubation for 3 d at 30°C (Haas et al., 2005).
GST pull-down assay and mass spectrometry
We immobilized 500 µg of GST-fused EHBP1L1-C2-PR domain or GST protein expressed in E. coli on glutathione Sepharose 4B (GE Healthcare) and incubated them for 1 h at 4°C with 40 mg mouse brain lysate or human intestine cancer cell line Caco-2 lysate prepared using lysis buffer (20 mM Hepes-NaOH, pH 7.5, 100 mM NaCl, 5 mM MgCl2, and 0.1% Triton X-100) containing a 0.1% protease inhibitor mixture (Wako). The beads were washed three times in lysis buffer and once in buffer containing 20 mM Hepes-NaOH, pH 7.5, 200 mM NaCl, 20 mM EDTA, and 0.1% Triton X-100. The protein was eluted with 500 µl elution buffer (20 mM Hepes-NaOH, pH 7.5, 500 mM NaCl, 20 mM EDTA, and 0.1% Triton X-100). The eluted protein was precipitated using 5 µl of 1% (wt/vol) deoxycholic acid and 72 µl TCA for 30 min on ice and washed twice in ice-cold acetone. The precipitated protein was dissolved using 40 µl SDS-PAGE sample buffer.
For the mass spectrometric analyses, the proteins were separated using a NuPAGE Novex Bis-Tris Gel (Life Technologies) and silver-stained using the Silver Stain kit (Wako) as described in the manufacturer’s instructions. The specific bands were excised from the gel, treated with trypsin, and analyzed using a SYNAPT G2 mass spectrometer (Waters Corp.) at Osaka University Center for Medical Research and Education. Database searching was performed using ProteinLynx Global Server software, version 2.4 (Waters Corp.), and an International Protein Index (mouse, 3.77) database.
For the direct interaction between the bacterially expressed EHBP1L1 and full-length Bin1 or Amph1, Bin1-ΔSH3, or Bin1-SH3, we used 10 µg of proteins in an in vitro GST pull-down assay with 5 µg GST-EHBP1L1-PR domain as described. The protein was separated by SDS-PAGE and detected using Coomassie brilliant blue.
For the interaction between the CC domain of EHBP1L1 or EHBP1 and the Rab proteins, 5 µg of GST-fused EHBP1L1-CC or EHBP1-CC was used for 750 µg of lysate containing 5 µM Mg-GTP from HEK293 cells expressing the FLAG-tagged GTP form of Rab1a, 8a, 10, 11a, and 13. The protein was separated using SDS-PAGE, stained with Ponceau S, and analyzed by immunoblotting using a mouse anti-FLAG antibody.
Immunoprecipitation
HEK293 cells were cotransfected with 0.5 µg pcDNA5/FRT/TO FLAG A encoding EHBP1L1 isoform C and 0.5 µg of pEGFP-C2–encoding GTP-restricted Rab8a, 10, or 13. After 24 h of incubation, the cells were lysed in 1 ml NL100 buffer. The lysates were then preabsorbed with 30 µl of control agarose resin (Pierce) for 30 min at 4°C. The lysates were further incubated with FLAG-M2 agarose (Sigma-Aldrich) for 2 h. The beads were washed three times with 1 ml NL-100 buffer, and the proteins were eluted by 200 µg/ml FLAG peptide. The samples were analyzed by immunoblotting using an anti-FLAG antibody or GFP antibody.
Transfection and immunofluorescence in HeLa cells
HeLa cells were grown in high-glucose DMEM with l-glutamine, phenol red, and sodium pyruvate (Wako). The cells were transfected using a TransIT-LT1 Transfection Reagent (Mirus Bio) in accordance with the manufacturer’s instructions. Experiments were performed 24 h after transfection.
siRNA oligos for human EHBP1L1 (siRNA ID: SASI_Hs02_00320623) and Bin1 (siRNA ID: SASI_Hs01_00052359; Sigma-Aldrich) and Rab8a and Rab8b siRNA (SI02662254 and SI02662261; Qiagen) were transfected using Lipofectamine RNAiMAX Transfection Reagent (Life Technologies) as described in the manufacturer’s manual. The experiments were performed 72 h after transfection.
The EHBP1L1, Bin1, and Rab8a/b RNAi depletion efficiencies were validated through immunoblotting. Lamin B was also detected as a loading control.
For immunofluorescence, the cells on coverslips were fixed with 3% (wt/vol) paraformaldehyde in PBS, permeabilized with 0.2% (wt/vol) saponin in PBS, and incubated with primary antibodies for 1 h at RT. The cells were washed in PBS three times and incubated with the appropriate Alexa Fluor–conjugated secondary antibodies and DAPI for 1 h. The coverslips were mounted with Dako Fluorescence Mounting Medium.
All images were acquired using a FV1000D confocal microscope (Olympus) with UPLSAPO 100XO object glass (NA 1.40) or UPLSAPO 40× object glass (NA 0.95; Olympus) and FluoView software (Olympus). The percentage of cells with tubular structures was measured. A cell with no tubules was counted as negative, and a cell containing any tubules was considered positive. For each coverslip, at least 300 cells were counted. The results represent at least three independent experiments. Statistical analyses were performed using R software and ggplot2 (Wickham, 2009; R Core Team, 2015). The colocalizations between Bin1, EHBP1L1, and Rab8a were analyzed with Fiji software.
Transfection, infection, immunofluorescence, and transferrin uptake assay in EpH4 cells
Mouse mammary epithelial cells were maintained in high-glucose DMEM with l-glutamine, phenol red, and sodium pyruvate. The cells were transfected with FLAG-tagged EHBP1L1 using Lipofectamine 2000 (Life Technologies) into 50% confluent cells on coverslips. The cells were then incubated for 3 d at 37°C. For the apical cargo transport assay, VSVex-FLAG-mCherry-GPI (Maeda et al., 2008; Atik et al., 2014) was transfected into the cells as described, and the cells were incubated for 3 d at 40°C to allow the cells to accumulate VSVex-mCherry-GPI in the endoplasmic reticulum. The cells were subsequently incubated at 32°C and fixed after each time point, as indicated in Fig. 4 B. The cells were immunostained with the appropriate antibodies and Alexa Fluor 633 phalloidin. The infection of an adenovirus encoding VSVGts045-GFP was performed as previously described (Atik et al., 2014). The cells were incubated and fixed after each time point as in the apical transport assay. For transferrin uptake, the experiments were performed essentially as previously described (Zacchi et al., 1998). In brief, the cells were grown on Transwell filters (Corning) and preincubated with DMEM without FCS for 12 h. Next, the cells were incubated with DMEM without FCS containing 50 µg/ml Alexa Fluor 594–conjugated transferrin (Life Technologies) in DMEM and were incubated for 20 min. The cells were fixed and costained with an anti-EHBP1L1 antibody and DAPI.
The colocalization between FLAG-tagged EHBP1L1 and Rab8a and EHBP1L1 and Bin1, sorting nexin1, internalized transferrin, and Lamp2 were analyzed with Fiji software.
Small intestine organoids culture
Mouse small intestine organoid cultures were performed as previously described (Sato et al., 2009). In brief, the isolated crypts were derived from 2-wk-old C57BL/6J, Rab8a KO (Sato et al., 2007), or Rab8a/8b DKO mice (Sato et al., 2014) and mixed with 20 µl Matrigel (BD). The mixtures were then spotted in a single well of a 48-well plastic dish. The crypts were cultured in advanced DMEM/F12 (Life Technologies) containing penicillin/streptomycin (Wako), N2 supplement (Life Technologies), B27 supplement (Life Technologies), 2.5 mM N-acetylcysteine (Sigma-Aldrich), 100 ng/ml Noggin (Peprotech), 50 ng/ml EGF (Life Technologies), and 10% R-spondin conditioned medium (Kim et al., 2005) for 10–14 d until villi differentiation. The organoids were then used for the experiment.
For lentivirus production and organoid infection, lentivirus encoding EHBP1L1 and Bin1 shRNA were produced using a pLKO.1 plasmid (Addgene #10879) as described in the manual. The following oligos were designed for pLKO.1: 5′-CCGGGCAACACCTTCACAGTCAATTCTCGAGAATTGACTGTGAAGGTGTTGCTTTTTG-3′ (Bin1-shRNA-F), 5′-AATTCAAAAAGCAACACCTTCACAGTCAATTCTCGAGAATTGACTGTGAAGGTGTTGC-3′ (Bin1-shRNA-R), 5′-CCGGCATTGAAGAGTGGCAGAAATTCTCGAGAATTTCTGCCACTCTTCAATGTTTTTG-3′ (EHBP1L1-shRNA-F) and 5′-AATTCAAAAACATTGAAGAGTGGCAGAAATTCTCGAGAATTTCTGCCACTCTTCAATG-3′ (EHBP1L1-shRNA-R). The HEK293T cells were transfected with pLKO.1 plasmids encoding the shRNA, psPAX2 (12260; Addgene), and pMD2.G (12259; Addgene). The media containing virus were harvested for 2 d. The virus was concentrated as follows. A 20-ml volume of collected media was mixed with 6.7 ml of 4× PEG solution (32% PEG 6000, 0.4 M NaCl, and 40 mM Hepes-NaOH, pH 7.4) and incubated for 12 h at 4°C. The mixture was centrifuged at 1,500 g for 30 min. The pellet was resuspended with 250 µl of medium supplemented with 40% Wnt3a conditioned medium, 10 mM nicotinamide, 10 µM Y27632, and 8 µg/ml polybrene (Sigma-Aldrich). The trypsinized organoids from five wells were infected with the concentrated virus and selected by 2 µg/ml puromycin as previously described (Koo et al., 2012). For immunofluorescence, the organoids were mixed with 10 µl Matrigel and spotted on a coverslip, placed in a 48-well plate, and further cultured for 1–4 d. The organoids were fixed with ice-cold methanol for 30 min and washed three times in PBS. The organoids were incubated with 5% normal donkey serum for 30 min and immunostained with anti-Lamp2, DPP4, and Na+/K+ ATPase antibodies. In certain instances, DAPI was also used to stain the nuclei. The organoids were mounted and observed using a confocal microscope.
Semiquantitative RT-PCR
Total RNA was isolated from 20–30 cultured organoids in a single well of a 48-well plastic dish using NucleoSpin RNA XS (Macherey-Nagel). The reverse transcription product was obtained using PrimeScript Reverse transcription (Takara) as described in the manufacturer’s manual. EHBP1L1, Bin1, and GAPDH were amplified by PCR using the following primers: Mus musculus EHBP1L1, 5′-GTGTGCGCAGAGCTGCAAGCCCTG-3′ and 5′-CAGGGTACAGCGTTCCCGCCTGCT-3′; M. musculus Bin1, 5′-AGCTCTCTTCCGGCTGTGGTGGTG-3′ and 5′-CTGCACCCGCTCTGTAAAATTCTC-3′; and M. musculus GAPDH, 5′-ATGACATCAAGAAGGTGGTG-3′ and 5′-TGTCATACCAGGAAATGAGC-3′. The PCR products were subjected to an agarose gel, and the intensity of each band was quantified.
Generation of EHBP1L1 KO mice using the CRISPR/Cas9 system
Screening of the optimal small guide RNA (sgRNA) was performed in accordance with Mashiko et al. (2013). In brief, five sgRNA candidate sequences that targeted the first coding exon were selected using CRISPRdirect software (Naito et al., 2015), subcloned into pX330 (42230; Addgene), and cotransfected with pCAG-EGxxFP (Mashiko et al., 2013) into HEK293T cells. We used the sgRNA sequence (5′-CCCGCTTGCCAACCCGCTGC-3′) that yielded the highest GFP signal under the fluorescent microscope. In vitro transcribed 80-ng/µl NLS-Cas9 mRNA and 50-ng/µl Ehbp1l1 sgRNA in 10 mM Tris-HCl and 0.1 mM EDTA were injected into fertilized eggs as previously described (Yoshimi et al., 2014). EHBP1L1 mutations were verified by DNA sequencing, and the protein was analyzed by immunoblotting using an anti-EHBP1L1 antibody.
Analyses of EHBP1L1 KO mice
The KO mice were born alive but died within 1 d after birth. Their bodies and organs showed no overt abnormalities except severe anemia based on a histological examination after hematoxylin and eosin staining. For morphological analyses, the intestines were collected from unfixed P0 pups or P0 pups fixed by transcardial perfusion with 3% paraformaldehyde in 0.1 M of phosphate buffer, pH 7.4. Certain samples were further fixed in 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M of phosphate buffer for electron microscopy.
Online supplemental material
Fig. S1 confirmed the enhanced recruitment of Rab8a and Bin1 to the ERC tubules under EHBP1L1-overexpression condition. The figure also shows that ERC tubules were intact in Rab8a/b, EHBP1L1, or Bin1-depleted cells. Fig. S2 shows the subcellular localization of EHBP1L1 in polarized EpH4 cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201508086/DC1.
Supplementary Material
Acknowledgments
This work was supported by the Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (KAKENHI no. 24390046 to A. Harada and 25860140 and 24112512 to S. Yoshimura) and from the Mitsubishi Foundation and Takeda Science Foundation to A. Harada and S. Yoshimura. A. Nakajo is supported by the Osaka University Medical Doctor Scientist Training Program.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- BAR
- Bin/Amphiphysin/Rvs
- CC
- coiled coil
- DKO
- double knockout
- DPP
- dipeptidyl peptidase
- EHBP1L1
- EH domain–binding protein 1–like 1
- ERC
- endocytic recycling compartment
- KD
- knockdown
- KO
- knockout
- MICAL
- microtubule-associated monooxygenase, calponin, and LIM domain–containing
- PR
- proline-rich
- WT
- wild type
References
- Ang A.L., Taguchi T., Francis S., Fölsch H., Murrells L.J., Pypaert M., Warren G., and Mellman I.. 2004. Recycling endosomes can serve as intermediates during transport from the Golgi to the plasma membrane of MDCK cells. J. Cell Biol. 167:531–543. 10.1083/jcb.200408165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atik N., Kunii M., Avriyanti E., Furumoto N., Inami K., Yoshimura S., Harada R., and Harada A.. 2014. The role of PKD in cell polarity, biosynthetic pathways, and organelle/F-actin distribution. Cell Struct. Funct. 39:61–77. 10.1247/csf.13020 [DOI] [PubMed] [Google Scholar]
- Barr F.A. 2013. Rab GTPases and membrane identity: causal or inconsequential? J. Cell Biol. 202:191–199. 10.1083/jcb.201306010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler M.H., David C., Ochoa G.C., Freyberg Z., Daniell L., Grabs D., Cremona O., and De Camilli P.. 1997. Amphiphysin II (SH3P9; BIN1), a member of the amphiphysin/Rvs family, is concentrated in the cortical cytomatrix of axon initial segments and nodes of Ranvier in brain and around T tubules in skeletal muscle. J. Cell Biol. 137:1355–1367. 10.1083/jcb.137.6.1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., and Zhang F.. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science. 339:819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daumke O., Roux A., and Haucke V.. 2014. BAR domain scaffolds in dynamin-mediated membrane fission. Cell. 156:882–892. 10.1016/j.cell.2014.02.017 [DOI] [PubMed] [Google Scholar]
- De Camilli P., Thomas A., Cofiell R., Folli F., Lichte B., Piccolo G., Meinck H.M., Austoni M., Fassetta G., Bottazzo G., et al. 1993. The synaptic vesicle-associated protein amphiphysin is the 128-kD autoantigen of Stiff-Man syndrome with breast cancer. J. Exp. Med. 178:2219–2223. 10.1084/jem.178.6.2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E., Haas A.K., Spooner R.A., Yoshimura S., Lord J.M., and Barr F.A.. 2007. Specific Rab GTPase-activating proteins define the Shiga toxin and epidermal growth factor uptake pathways. J. Cell Biol. 177:1133–1143. 10.1083/jcb.200612068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giridharan S.S.P., Cai B., Vitale N., Naslavsky N., and Caplan S.. 2013. Cooperation of MICAL-L1, syndapin2, and phosphatidic acid in tubular recycling endosome biogenesis. Mol. Biol. Cell. 24:1776–1790: S1–S15. 10.1091/mbc.E13-01-0026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Roth D., Walch-Solimena C., and Novick P.. 1999. The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. EMBO J. 18:1071–1080. 10.1093/emboj/18.4.1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A.K., Fuchs E., Kopajtich R., and Barr F.A.. 2005. A GTPase-activating protein controls Rab5 function in endocytic trafficking. Nat. Cell Biol. 7:887–893. 10.1038/ncb1290 [DOI] [PubMed] [Google Scholar]
- Hattula K., Furuhjelm J., Tikkanen J., Tanhuanpää K., Laakkonen P., and Peränen J.. 2006. Characterization of the Rab8-specific membrane traffic route linked to protrusion formation. J. Cell Sci. 119:4866–4877. 10.1242/jcs.03275 [DOI] [PubMed] [Google Scholar]
- He B., and Guo W.. 2009. The exocyst complex in polarized exocytosis. Curr. Opin. Cell Biol. 21:537–542. 10.1016/j.ceb.2009.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Sultana A., Gandhi P., Franklin E., Hamamoto S., Khan A.R., Munson M., Schekman R., and Weisman L.S.. 2011. Myosin V transports secretory vesicles via a Rab GTPase cascade and interaction with the exocyst complex. Dev. Cell. 21:1156–1170. 10.1016/j.devcel.2011.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanerva K., Uronen R.L., Blom T., Li S., Bittman R., Lappalainen P., Peränen J., Raposo G., and Ikonen E.. 2013. LDL cholesterol recycles to the plasma membrane via a Rab8a-Myosin5b-actin-dependent membrane transport route. Dev. Cell. 27:249–262. 10.1016/j.devcel.2013.09.016 [DOI] [PubMed] [Google Scholar]
- Kim K.-A., Kakitani M., Zhao J., Oshima T., Tang T., Binnerts M., Liu Y., Boyle B., Park E., Emtage P., et al. 2005. Mitogenic influence of human R-spondin1 on the intestinal epithelium. Science. 309:1256–1259. 10.1126/science.1112521 [DOI] [PubMed] [Google Scholar]
- Koo B.-K., Stange D.E., Sato T., Karthaus W., Farin H.F., Huch M., van Es J.H., and Clevers H.. 2012. Controlled gene expression in primary Lgr5 organoid cultures. Nat. Methods. 9:81–83. 10.1038/nmeth.1802 [DOI] [PubMed] [Google Scholar]
- Lee E., Marcucci M., Daniell L., Pypaert M., Weisz O.A., Ochoa G.-C., Farsad K., Wenk M.R., and De Camilli P.. 2002. Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science. 297:1193–1196. 10.1126/science.1071362 [DOI] [PubMed] [Google Scholar]
- Lemmon M.A. 2008. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9:99–111. 10.1038/nrm2328 [DOI] [PubMed] [Google Scholar]
- Macia E., Ehrlich M., Massol R., Boucrot E., Brunner C., and Kirchhausen T.. 2006. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell. 10:839–850. 10.1016/j.devcel.2006.04.002 [DOI] [PubMed] [Google Scholar]
- Maeda Y., Ide T., Koike M., Uchiyama Y., and Kinoshita T.. 2008. GPHR is a novel anion channel critical for acidification and functions of the Golgi apparatus. Nat. Cell Biol. 10:1135–1145. 10.1038/ncb1773 [DOI] [PubMed] [Google Scholar]
- Mashiko D., Fujihara Y., Satouh Y., Miyata H., Isotani A., and Ikawa M.. 2013. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci. Rep. 3:3355 10.1038/srep03355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield F.R., and McGraw T.E.. 2004. Endocytic recycling. Nat. Rev. Mol. Cell Biol. 5:121–132. 10.1038/nrm1315 [DOI] [PubMed] [Google Scholar]
- Meinecke M., Boucrot E., Camdere G., Hon W.-C., Mittal R., and McMahon H.T.. 2013. Cooperative recruitment of dynamin and BIN/amphiphysin/Rvs (BAR) domain-containing proteins leads to GTP-dependent membrane scission. J. Biol. Chem. 288:6651–6661. 10.1074/jbc.M112.444869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y., Hino K., Bono H., and Ui-Tei K.. 2015. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 31:1120–1123. 10.1093/bioinformatics/btu743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann S., and Schmid S.L.. 2013. Dual role of BAR domain-containing proteins in regulating vesicle release catalyzed by the GTPase, dynamin-2. J. Biol. Chem. 288:25119–25128. 10.1074/jbc.M113.490474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D.J., Wigge P., Vallis Y., Moore J.D., Evans P.R., and McMahon H.T.. 1998. Crystal structure of the amphiphysin-2 SH3 domain and its role in the prevention of dynamin ring formation. EMBO J. 17:5273–5285. 10.1093/emboj/17.18.5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant S., Sharma M., Patel K., Caplan S., Carr C.M., and Grant B.D.. 2009. AMPH-1/Amphiphysin/Bin1 functions with RME-1/Ehd1 in endocytic recycling. Nat. Cell Biol. 11:1399–1410. 10.1038/ncb1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park R.J., Shen H., Liu L., Liu X., Ferguson S.M., and De Camilli P.. 2013. Dynamin triple knockout cells reveal off target effects of commonly used dynamin inhibitors. J. Cell Sci. 126:5305–5312. 10.1242/jcs.138578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peränen J. 2011. Rab8 GTPase as a regulator of cell shape. Cytoskeleton (Hoboken). 68:527–539. 10.1002/cm.20529 [DOI] [PubMed] [Google Scholar]
- Pereira-Leal J.B., and Seabra M.C.. 2001. Evolution of the Rab family of small GTP-binding proteins. J. Mol. Biol. 313:889–901. 10.1006/jmbi.2001.5072 [DOI] [PubMed] [Google Scholar]
- Rahajeng J., Giridharan S.S.P., Cai B., Naslavsky N., and Caplan S.. 2010. Important relationships between Rab and MICAL proteins in endocytic trafficking. World J. Biol. Chem. 1:254–264. 10.4331/wjbc.v1.i8.254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team 2015. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
- Rodriguez-Boulan E., Kreitzer G., and Müsch A.. 2005. Organization of vesicular trafficking in epithelia. Nat. Rev. Mol. Cell Biol. 6:233–247. 10.1038/nrm1593 [DOI] [PubMed] [Google Scholar]
- Ruemmele F.M., Müller T., Schiefermeier N., Ebner H.L., Lechner S., Pfaller K., Thöni C.E., Goulet O., Lacaille F., Schmitz J., et al. 2010. Loss-of-function of MYO5B is the main cause of microvillus inclusion disease: 15 novel mutations and a CaCo-2 RNAi cell model. Hum. Mutat. 31:544–551. 10.1002/humu.21224 [DOI] [PubMed] [Google Scholar]
- Sahlender D.A., Roberts R.C., Arden S.D., Spudich G., Taylor M.J., Luzio J.P., Kendrick-Jones J., and Buss F.. 2005. Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. J. Cell Biol. 169:285–295. 10.1083/jcb.200501162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Mushiake S., Kato Y., Sato K., Sato M., Takeda N., Ozono K., Miki K., Kubo Y., Tsuji A., et al. 2007. The Rab8 GTPase regulates apical protein localization in intestinal cells. Nature. 448:366–369. 10.1038/nature05929 [DOI] [PubMed] [Google Scholar]
- Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., and Clevers H.. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 459:262–265. 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- Sato T., Iwano T., Kunii M., Matsuda S., Mizuguchi R., Jung Y., Hagiwara H., Yoshihara Y., Yuzaki M., Harada R., and Harada A.. 2014. Rab8a and Rab8b are essential for several apical transport pathways but insufficient for ciliogenesis. J. Cell Sci. 127:422–431. 10.1242/jcs.136903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M., Giridharan S.S.P., Rahajeng J., Naslavsky N., and Caplan S.. 2009. MICAL-L1 links EHD1 to tubular recycling endosomes and regulates receptor recycling. Mol. Biol. Cell. 20:5181–5194. 10.1091/mbc.E09-06-0535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi A., Chen C.C., Banerjee R., Glodowski D., Audhya A., Rongo C., and Grant B.D.. 2010. EHBP-1 functions with RAB-10 during endocytic recycling in Caenorhabditis elegans. Mol. Biol. Cell. 21:2930–2943. 10.1091/mbc.E10-02-0149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöblom B., Ylänne J., and Djinović-Carugo K.. 2008. Novel structural insights into F-actin-binding and novel functions of calponin homology domains. Curr. Opin. Struct. Biol. 18:702–708. 10.1016/j.sbi.2008.10.003 [DOI] [PubMed] [Google Scholar]
- Takei K., McPherson P.S., Schmid S.L., and De Camilli P.. 1995. Tubular membrane invaginations coated by dynamin rings are induced by GTP-gamma S in nerve terminals. Nature. 374:186–190. 10.1038/374186a0 [DOI] [PubMed] [Google Scholar]
- Thuenauer R., Hsu Y.C., Carvajal-Gonzalez J.M., Deborde S., Chuang J.Z., Römer W., Sonnleitner A., Rodriguez-Boulan E., and Sung C.H.. 2014. Four-dimensional live imaging of apical biosynthetic trafficking reveals a post-Golgi sorting role of apical endosomal intermediates. Proc. Natl. Acad. Sci. USA. 111:4127–4132. 10.1073/pnas.1304168111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennerberg K., Rossman K.L., and Der C.J.. 2005. The Ras superfamily at a glance. J. Cell Sci. 118:843–846. 10.1242/jcs.01660 [DOI] [PubMed] [Google Scholar]
- Wickham H. 2009. ggplot2: Elegant Graphics for Data Analysis (Use R!). Springer, New York, NY. 213 pp. [Google Scholar]
- Yamamura R., Nishimura N., Nakatsuji H., Arase S., and Sasaki T.. 2008. The interaction of JRAB/MICAL-L2 with Rab8 and Rab13 coordinates the assembly of tight junctions and adherens junctions. Mol. Biol. Cell. 19:971–983. 10.1091/mbc.E07-06-0551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimi K., Kaneko T., Voigt B., and Mashimo T.. 2014. Allele-specific genome editing and correction of disease-associated phenotypes in rats using the CRISPR-Cas platform. Nat. Commun. 5:4240 10.1038/ncomms5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S., Gerondopoulos A., Linford A., Rigden D.J., and Barr F.A.. 2010. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J. Cell Biol. 191:367–381. 10.1083/jcb.201008051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacchi P., Stenmark H., Parton R.G., Orioli D., Lim F., Giner A., Mellman I., Zerial M., and Murphy C.. 1998. Rab17 regulates membrane trafficking through apical recycling endosomes in polarized epithelial cells. J. Cell Biol. 140:1039–1053. 10.1083/jcb.140.5.1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.