Summary
Amyloidogenic proteins, including prions, assemble into multiple forms of structurally distinct fibres. The [PSI+] prion, endogenous to the yeast Saccharomyces cerevisiae, is a dominantly inherited, epigenetic modifier of phenotypes. [PSI+] formation relies on the coexistence of another prion, [RNQ+]. Here, in order to better define the role of amyloid diversity on cellular phenotypes, we investigated how physiological and environmental changes impact the generation and propagation of diverse protein conformations from a single polypeptide. Utilizing the yeast model system, we defined extracellular factors that influence the formation of a spectrum of alternative self-propagating amyloid structures of the Sup35 protein, called [PSI+] variants. Strikingly, exposure to specific stressful environments dramatically altered the variants of [PSI+] that formed de novo. Additionally, we found that stress also influenced the association between the [PSI+] and [RNQ+] prions in a way that it superceded their typical relationship. Furthermore, changing the growth environment modified both the biochemical properties and [PSI+]-inducing capabilities of the [RNQ+] template. These data suggest that the cellular environment contributes to both the generation and the selective propagation of specific amyloid structures, providing insight into a key feature that impacts phenotypic diversity in yeast and the cross-species transmission barriers characteristic of prion diseases.
Introduction
A variety of progressive disorders including Alzheimer's disease (AD), Parkinson's disease (PD), frontotemporal dementia (FTD), amyotrophic lateral sclerosis (ALS) and prion diseases are characterized by the misfolding and accumulation of protein aggregates (Moreno-Gonzalez and Soto, 2011). In some manner, these protein aggregates have neurotoxic effects on the central nervous system of afflicted individuals. The proteins responsible for the degenerative nature of these diseases comprise a heterogeneous group in terms of primary structure, but they share the ability to form self-propagating, β sheet-rich structures that assemble into amyloid. Both genetic and environmental factors have been shown to contribute to amyloid formation; however, the number of sporadic cases greatly outweigh those associated with an identified underlying genetic cause, strongly implicating extracellular factors in disease pathogenesis (Holtzman et al., 2011; Klug et al., 2013; Prusiner, 2013). Recently, it has been proposed that several neurodegenerative disorders propagate in a prion-like fashion, in which the modified ‘prion’ form spreads along an axon and potentially throughout the brain (Miller, 2009; Eisenberg and Jucker, 2012).
Prion proteins have the ability to adopt more than one conformation. One conformer is a soluble, folded protein, and another is an insoluble, self-templating amyloid, but both originate from an identical polypeptide sequence (Prusiner, 1982; Stahl et al., 1993). Structurally distinct amyloid conformers of the mammalian prion protein (PrP), or prion ‘strains’, are characterized phenotypically by differences in disease progression, including dramatic differences in incubation time, and neuropathology (Dickinson et al., 1972; Safar et al., 1998). In fact, PrP has the ability to adopt more than 30 self-propagating structural variants (prion strains) (Safar, 2012). In a similar fashion, amyloidogenic proteins associated with other neurodegenerative diseases likely exist in a range of self-templating structural states, which may be responsible for the diversity in phenotypes; however, it is unknown how cellular environment contributes to this conformational diversity.
It has been a major challenge in the field to distinguish different amyloid conformers in complex biological systems and determine how they contribute to cellular phenotype, so researchers have utilized model systems as tools to investigate age-related protein aggregation. Similar to the phenomena that have been observed with variants of mammalian amyloidogenic proteins, yeast prion proteins have the ability to acquire multiple self-propagating structures (Derkatch et al., 1996; Schlumpberger et al., 2001; Bradley et al., 2002; Bradley and Liebman, 2003). Perhaps the best-characterized example of amyloid conformational variation is associated with the [PSI+] prion of Saccharomyces cerevisiae. [PSI+] is a self-replicating, aggregated form of the translation termination factor Sup35 (Chernoff et al., 1993; Patino et al., 1996; Paushkin et al., 1996). When Sup35 is in the soluble, non-prion form, called [psi−], translation termination is efficient (Frolova et al., 1994; Stansfield et al., 1995). Variability in the efficiency with which Sup35 joins aggregates in [PSI+] cells modulates the frequency of readthrough of stop codons in messenger RNA (Cox, 1965; Cox et al., 1988; Patino et al., 1996). [PSI+] variants can be distinguished phenotypically in yeast using sensitive reporters that monitor nonsense suppression, which is a read-out for the amount of ‘functional’ Sup35 in cells (Liebman and Derkatch, 1999; Zhou et al., 1999; Uptain et al., 2001). ‘Strong’ prion variants have a greater ability to recruit soluble, monomeric Sup35 protein into the aggregated prion state, and thereby generate a correspondingly strong nonsense suppression phenotype (Derkatch et al., 1996; Uptain et al., 2001; Tanaka et al., 2006). ‘Weak’ variants of [PSI+] maintain a higher steady-state level of soluble, functional Sup35, thereby exhibiting less nonsense suppression. Stronger variants also have higher mitotic stability than weaker variants, leading to less frequent spontaneous prion loss (Derkatch et al., 1996; 1997). Biochemically, [PSI+] variants differ both in the amount of soluble Sup35 (Kochneva-Pervukhova et al., 2001; Uptain et al., 2001) and in the relative size of the aggregate species (Kryndushkin et al., 2003). Similar to mammalian prions strains that are maintained upon serial passage between animals, once a [PSI+] variant is established in a cell, it typically propagates efficiently and is faithfully transmitted to progeny (Derkatch et al., 1996; Kochneva-Pervukhova et al., 2001).
The aggregation of prion proteins likely perpetuates through a mechanism involving a direct interaction between a normally folded protein and an aggregation-prone template or ‘seed’, resulting in the conformational conversion of the soluble monomer into the modified state. In addition to self-seeding reactions that increase amyloid formation, heterologous protein interactions can facilitate de novo amyloid formation in several protein misfolding disorders, resulting in the acceleration of the primary disease states and an increase in comorbidity of the diseases (Lundmark et al., 2005; Yan et al., 2007; Morales et al., 2009; 2010). The synergistic effects of these pathogenic proteins suggest that common molecular mechanisms underlie the amyloid formation and resultant pathology that is characteristic of several age-related protein-misfolding disorders. Interestingly, in the yeast model system, the de novo formation of [PSI+] appears to involve the cross-seeding of soluble Sup35 by non-Sup35 aggregates present in [psi−] cells (Derkatch et al., 2001; 2004; Patel and Liebman, 2007; Sharma and Liebman, 2013a). The predominant mediator of this heterologous cross-seeding is the yeast prion [RNQ+], which is formed by the ordered aggregation of the Rnq1 protein (Stein and True, 2011). This protein has no known function in its non-prion, [rnq−] state (Derkatch et al., 1997; 2001; Sondheimer and Lindquist, 2000; Strawn and True, 2006). Variability in Rnq1 protein structure gives rise to [RNQ+] variants that vary in their ability to induce [PSI+] formation (Bradley et al., 2002; Bradley and Liebman, 2003; Kalastavadi and True, 2010; Huang et al., 2013; Sharma and Liebman, 2013b; Westergard and True, 2014). These [RNQ+] variants have been characterized by their distinct biochemical properties, including the proportion of soluble Rnq1 (Bradley et al., 2002) and sensitivity of the protein aggregates to increased temperatures (Bagriantsev and Liebman, 2004). These collective differences suggest that these [RNQ+] variants represent different Rnq1 amyloid quaternary structures, which are hypothesized to interact with Sup35 differently and promote [PSI+] formation to different extents. The biological significance of the [PSI+]/[RNQ+] interaction is unknown. Previous research suggests that [PSI+] might be beneficial, or at least adaptive, in specific environments (Eaglestone et al., 1999; True and Lindquist, 2000; True et al., 2004; Chernoff, 2007; Newnam et al., 2011b; Holmes et al., 2013), and if so, [RNQ+] might serve as a regulatory effector of [PSI+] in these conditions.
Here, using the two-prion model system in yeast ([PSI+] and [RNQ+]), we investigate how environmental alterations, including treatment with stressors that perturb normal cell growth and metabolism, affect the formation and maintenance of prion variants. We demonstrate that the environment can have a striking impact on the prion structure that is formed. More surprisingly, the influence of the environment can alter typical variant formation even when the underlying mechanism may involve a direct physical interaction and cross-seeding on another amyloid. Furthermore, we demonstrate that transient environmental changes can have a surprising impact on the continued relationship between prion proteins, and can modify subsequent prion formation in long-lasting ways. Finally, these results may assist in our understanding of how specific cellular stressors affect the generation and propagation of amyloid structures. This will also facilitate our understanding of how structural differences in amyloid contribute to disease.
Results
[PSI+] induction rate corresponds to the strength of [PSI+] induced in cells harbouring different [RNQ+] variants
Amyloid-prone proteins most commonly convert into an aggregated form sporadically, without any known underlying genetic cause (Prusiner, 2013). However, in both yeast and mammalian cells, the propensity to form amyloid can be greatly accelerated by other aggregates present in the cell, upon which the soluble monomer can cross-seed (Derkatch et al., 2001; Vishveshwara and Liebman, 2009; Morales et al., 2010; Liebman and Chernoff, 2012). Because conformational variants of amyloid proteins impact disease progression, phenotype, and cross-species transmission in prion diseases, we sought to better understand the factors that influence the propensity to form amyloid using our model prion, [PSI+].
The [RNQ+] prion is the major facilitator of de novo [PSI+] formation (Derkatch et al., 1997; 2001). To explore the role of the pre-existing Rnq1 aggregates on [PSI+] induction, we took advantage of a set of isogenic yeast strains containing different [RNQ+] variants that were previously characterized and shown to have distinct biochemical, genetic and phenotypic properties, suggesting that they represent different, self-propagating structures (Bradley et al., 2002; Bradley and Liebman, 2003; Bagriantsev and Liebman, 2004). The most striking difference between these [RNQ+] variants is the rate at which they facilitate [PSI+] formation under normal growth conditions.
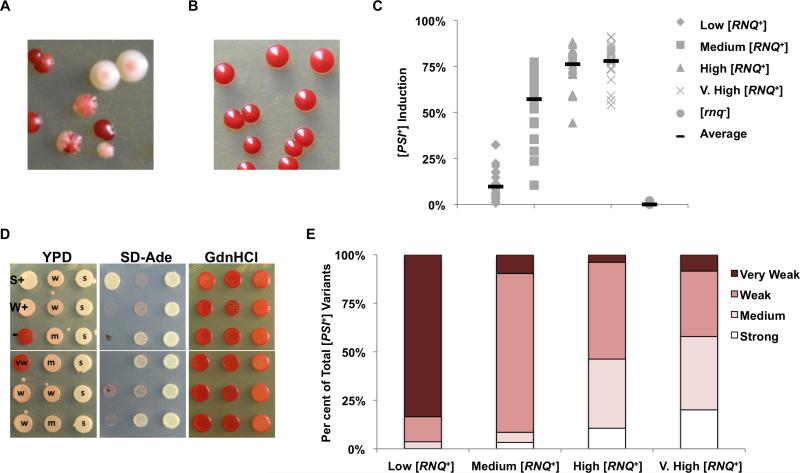
In order to establish baseline values for subsequent experiments, we utilized strains containing each of these four [RNQ+] variants to examine both [PSI+] induction frequency and [PSI+] variant formation upon continued over-expression of Sup35, which increases [PSI+] formation to more easily quantifiable levels (Chernoff et al., 1993). The 74-D694 yeast strain used in these studies contains a premature stop codon mutation in the ADE1 gene, which we use to track the [PSI+] state phenotypically (Chernoff et al., 1995). When Sup35 is in the functional, non-aggregated state in [psi−] cells, it efficiently terminates translation at the premature stop codon in the ade1-14 allele. This results in the inability of cells to grow on synthetic media lacking adenine (SD-Ade) and the accumulation of a red pigment when [psi−] adenine auxotrophs are grown on solid rich media (YPD). By contrast, when Sup35 is sequestered into prion aggregates in [PSI+] cells, its reduced functionality in translation termination causes readthrough of the ade1-14 premature stop codon. This results in the ability of [PSI+] cells to grow on SD-Ade, and form white or pink yeast colonies on YPD, depending on the [PSI+] variant.
For these control experiments, we assayed the proportion of cells containing white/pink sectored and completely white or pink yeast colonies upon overexpression of full-length Sup35 under normal growth conditions (Fig. 1A). Of note, we find that the population of transformants containing the plasmid, which expresses Sup35 under control of its endogenous promoter, is uniformly red on solid media that selects for the overexpression plasmid, suggesting that [PSI+] is not induced at this time. Using this colorimetric assay, we observed a [RNQ+]-dependent trend in [PSI+] induction frequency identical to the one previously reported (Bradley et al., 2002; Bradley and Liebman, 2003; Sharma and Liebman, 2013a), although our rates of [PSI+] formation greatly exceeded those that were previously published, likely due to the different Sup35 overexpression plasmid used. Additionally, we never observed [PSI+] formation in [rnq−] cells (Fig. 1B). [PSI+] formation was greatest in cells containing very high and high [RNQ+], followed by medium [RNQ+], and then low [RNQ+] (Fig. 1C). When [PSI+] cells were transferred to media containing 5-FOA, which selects against cells containing the URA3-marked plasmid, and back to rich media, they remained pink/white and were unable to grow on media lacking uracil (data not shown). This indicates that the [PSI+] phenotype was not dependent on nonspecific aggregation due to continued overexpression of Sup35 from the transformed plasmid. Notably, overexpression of Sup35 did not impact cell viability and there were no growth differences between cells containing the [RNQ+] variants and [rnq−] cells (data not shown).
Fig. 1.
[RNQ+] variants influence the rate of [PSI+] induction and [PSI+] variant formation.
A and B. Example of [PSI+] induction. 74-D694 yeast colonies containing the high [RNQ+] variant (A) or [rnq−] cells (B) were transformed with pSup2 and plated on solid YPD media, grown for 5 days at 30°C, and incubated overnight at 4°C for colour development.
C. 74-D694 yeast cells containing either the low, medium, high, or very high [RNQ+] variants, or [rnq−] 74-D694 cells were scored for [PSI+] status using the [PSI+] induction assay, as described in Experimental procedures. Colonies on YPD that were either white- or pink-sectored or completely white or pink colonies were scored as [PSI+]. For each [RNQ+] variant, at least 6000 total colonies from at least eight independent experiments were scored.
D. Colonies showing nonsense suppression were isolated and re-grown on YPD, SD-Ade, or YPD + 3 mM GdnHCl solid media, for verification of [PSI+] status and scoring of [PSI+] variants. This representative subset shows the spectrum of [PSI+] variants observed after re-spotting on YPD (left), SD-Ade (middle) and YPD + 3 mM GdnHCl (right). Each spot represents a different newly induced [PSI+] clone that was characterized according to our scoring protocol as very weak (vw), weak (w), medium (m) or strong (s). Controls are in the upper left: strong [PSI+] (S+), weak [PSI+] (W+) and [psi−] (−).
E. The number of GdnHCl-curable [PSI+] colonies scored according to growth on SD-Ade and plotted were as follows: low [RNQ+] 550, medium [RNQ+] 939, high [RNQ+] 800, very high [RNQ+] 747.
Next, we set out to confirm previous work that showed that the presence of a specific [RNQ+] variant influences the [PSI+] variants that are induced (Sharma and Liebman, 2013a). We re-grew [PSI+] clones on solid YPD media to distinguish colour, SD-Ade media to assess the strength of [PSI+], and rich media containing 3 mM guanidine hydro-chloride (GdnHCl) to confirm phenotype curability. GdnHCl inhibits the action of Hsp104, a chaperone which is required to propagate all known yeast prions (Ferreira et al., 2001; Jung and Masison, 2001; Liebman and Chernoff, 2012). [PSI+] variants were distinguished primarily by their growth on media lacking adenine and secondarily by their colour on rich media. These phenotypes differ depending on what [PSI+] variant is propagating, and presumably correlate to the efficiency of Sup35 aggregation. Stronger [PSI+] variants sequester the majority of Sup35, resulting in robust growth on SD-Ade and white colonies on rich media, whereas weaker [PSI+] variants retain a greater proportion of soluble Sup35, resulting in less growth on SD-Ade and darker pink colonies (Derkatch et al., 1996). We found that [RNQ+] variants not only influence the frequency of [PSI+] formation, but also biased the formation of particular [PSI+] variants in a very similar manner to what was recently reported, despite the fact that different methods were used to induce [PSI+] (Sharma and Liebman, 2013a). As represented in Fig. 1D, the continuum of [PSI+] variants we obtained exhibited a wide range of nonsense suppression, from strong to very weak, in agreement with previously published data (King, 2001; Kochneva-Pervukhova et al., 2001; Bateman and Wickner, 2013). The distribution of induced [PSI+] variants follows the distinct pattern that was previously reported: low [RNQ+] induces mostly very weak [PSI+], medium [RNQ+] induces mostly weak [PSI+], high [RNQ+] induces mostly medium and weak [PSI+], and very high [RNQ+] induces more medium and strong [PSI+] variants (Fig. 1E). In order to confirm our phenotypic assessment of the [PSI+] variant induced, we selected a subset of clones that we had categorized as ‘strong’, ‘medium’, ‘weak’ and ‘very weak’ [PSI+] and assayed their mitotic stability. When we assessed the [PSI+] status using our colorimetric assay, we found that prion loss was greatest in ‘weak’ and ‘very weak’ [PSI+] strains and rare in strains harbouring ‘strong’ or ‘medium’ [PSI+], in agreement with previous findings [(Derkatch et al., 1996), data not shown].
Extracellular environmental stress affects de novo amyloid formation and structure
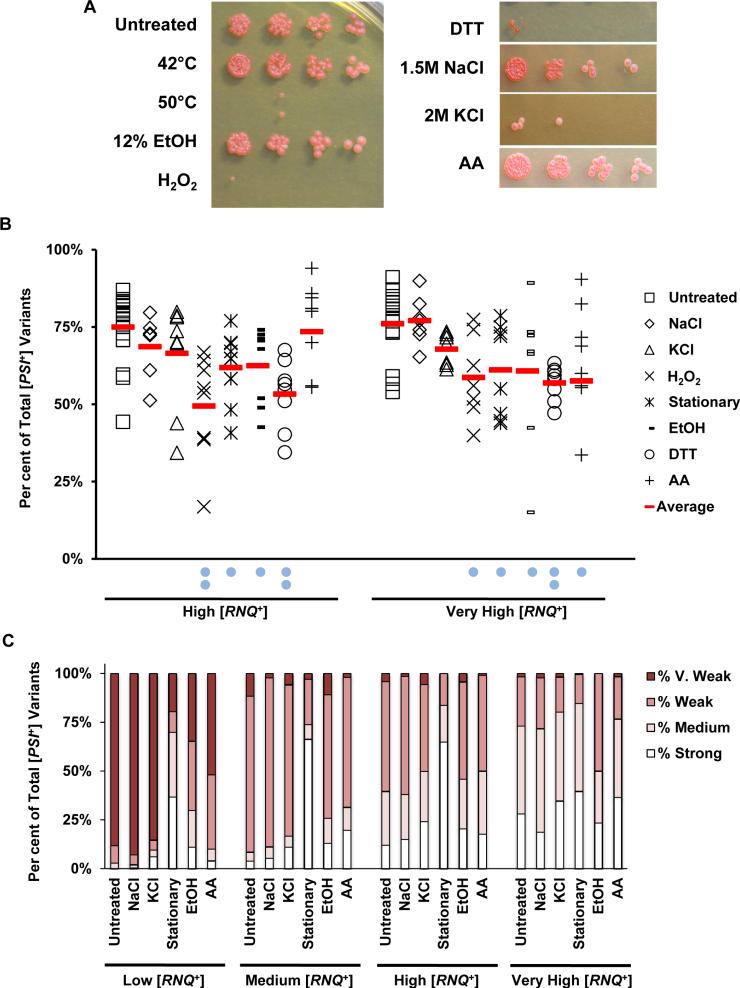
It was previously demonstrated that the extracellular environment influences both [PSI+] propagation (Newnam et al., 2011a) and spontaneous induction (Tyedmers et al., 2008). Our initial experiments confirmed previous reports that show that [RNQ+] variants induce [PSI+] at different frequencies and also differentially induce distinct [PSI+] variants in normal growth conditions (Bradley et al., 2002; Bradley and Liebman, 2003; Park et al., 2006; Sharma and Liebman, 2013a); therefore, we wanted to determine whether environmental stress would affect the ability of [RNQ+] to influence [PSI+] variant formation. We utilized conditions known to activate a range of different cellular stress response pathways and elicit varying degrees of toxicity (Fig. 2A, Supplemental Fig. S1), including the following: nutrient deprivation [incubation in stationary phase and in media lacking the amino acid histidine (AA)], oxidative stress [hydrogen peroxide (H2O2)], high salt [1.5 M sodium chloride (NaCl) and 2 M potassium chloride (KCl)], ethanol exposure (EtOH) and ER stress [treatment with dithiothreitol (DTT)]. Notably, the degree of cell toxicity was not influenced by the presence of [RNQ+] or [RNQ+] variant dependent (Supplemental Fig. S1; data not shown).
Fig. 2.
Extracellular environment influences [PSI+] induction and variant formation in a [RNQ+] variant-dependent manner.
A. 74-D694 yeast harbouring previously characterized [RNQ+] variants were normalized by OD600 and subjected to the indicated environmental stressors as described in Experimental procedures and Supplemental Table S1. Cells were serially diluted 5×, spotted on solid YPD rich media and grown for 3 days at 30°C. Representative plate of yeast harbouring the low [RNQ+] variant is shown.
B. 74-D694 yeast cells containing either the high or very high [RNQ+] variants were treated with or without environmental stress and scored for [PSI+] status using the [PSI+] induction assay, as described in Experimental procedures. At least 1500 colonies from at least three independent experiments were scored for each condition. Red lines denote the average of all experiments. Two blue dots below a condition indicate P < 0.0001 and one blue dot below a condition indicates P < 0.05 statistically significant difference from the untreated cells with the same [RNQ+] variant when analysed by two-tailed Student's t-test.
C. [PSI+] colonies were spotted on YPD, SD-Ade, and YPD + 3 mM GdnHCl media. GdnHCl-curable clones were assessed for [PSI+] variant based both on colour on YPD and on growth on SD-Ade. The number of colonies scored for each of the stress conditions ranged from 40 to 669 with an average number of colonies scored of ~ 300. Conditions that did not alter [PSI+] variants as compared to untreated controls were excluded from the chart. Percentage of total [PSI+] variants for each condition is plotted.
We continuously overexpressed Sup35 in cells containing one of each of the four [RNQ+] variants, or in [rnq−] cells. We found that the colony population was uniformly red on selection media after plasmid transformation, suggesting that [PSI+] is not induced at this stage (data not shown). We then treated the cells with acute (H2O2, EtOH, AA deprivation) or chronic stress (NaCl, KCl, DTT, incubation in stationary phase). After treatment with various stressors in liquid culture, we transferred cells from all conditions to solid YPD media in the absence of stress and scored for [PSI+] formation and [PSI+] variant formation, as above. Notably, with the exception of stationary phase, in which we observed a global decrease in protein expression, we did not observe any consistent, distinguishable changes in the steady-state level of Sup35 in the different conditions as compared to untreated cells (data not shown). Interestingly, we found that diverse stressors had differential effects on both the frequency of [PSI+] induction (Fig. 2B; Supplemental Fig. S2) and [PSI+] variant formation (Fig. 2C). These differences were dependent on the resident [RNQ+] variant, but did not correlate with the toxicity of the stressor. One of the most striking findings in these analyses was that, unlike in control unstressed cells harbouring the [RNQ+] variants, the increases in [PSI+] induction rate in the presence of external stress did not always correspond to an increase in the strength of the [PSI+] variant induced.
Incubation in 12% EtOH was one condition where the [PSI+] induction frequency and the strength of the [PSI+] variant induced diverged. We found a significant decrease in the frequency of [PSI+] induction in 12% EtOH in comparison to the untreated controls harbouring the same [RNQ+] variant (Fig. 2B, Supplemental Fig. S2; low [RNQ+] P = 0.03, medium [RNQ+] P < 0.0001, high [RNQ+] P = 0.01, very high [RNQ+] P = 0.03; Student's t-test). However, in low- and medium-[RNQ+]-containing cells, the resultant [PSI+] variants were generally stronger than those induced in the untreated cells harbouring the same [RNQ+] variants (Fig. 2C). For example, strong [PSI+] variants were never induced under normal growth conditions in cells harbouring low [RNQ+] but were induced approximately 10% of the time in cells treated with 12% EtOH. When we calculated the change in [PSI+] variants in cells propagating the low [RNQ+] variant using the cumulative induction of both medium and strong [PSI+], we found a 10.3-fold increase in medium/strong [PSI+] variants after 12% EtOH treatment when compared to untreated control cells that contain low [RNQ+].
Cells that were incubated for 7 days in stationary phase, or for 2 h without histidine, exhibited no change in [PSI+] induction rates with the low and medium [RNQ+] variants (Supplemental Fig. S2). However, cells containing both of these [RNQ+] variants showed an overall increase in the strength of [PSI+] variants induced (Fig. 2C). Stationary-phase incubation elicited the most obvious effect, including a 22.4-fold increase in medium/strong [PSI+] in cells containing the low [RNQ+] variant and a 17.1-fold increase in strong [PSI+] formation in the medium-[RNQ+]-containing cells (Fig. 2C). Conversely, stationary-phase cells containing either high and very high [RNQ+] exhibited statistically significantly lower [PSI+] induction rates, but did not modify the [PSI+] variants that were induced from the untreated controls. Thus, the [RNQ+] variants, and their interactions with Sup35, appear to be differentially impacted by stress. These results demonstrate that the extracellular environment influences both de novo amyloid formation and the generation of particular amyloid conformations, and these changes occur in a [RNQ+] variant-dependent manner.
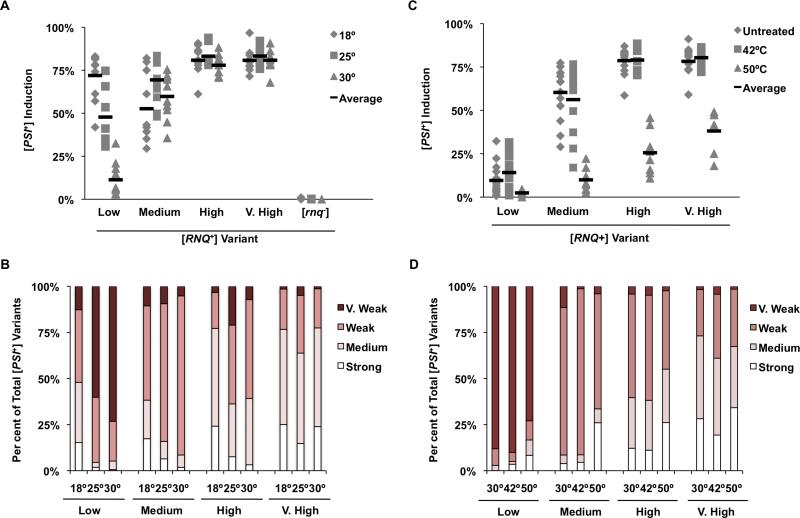
Extracellular growth temperature influences [PSI+] induction and variant formation
Exposure of cells to extreme conditions induces a protective stress response, which can include the induction of various heat shock proteins (Hsps), including: Hsp104, Hsp70s and Hsp40s (Morano et al., 2012). Increased expression of Hsps in S. cerevisiae occurs when the yeast are exposed to both low and high temperatures, and these temperature shifts can impact yeast prion formation. Therefore, we investigated the effect of prolonged growth at lower temperatures and transient heat shock, on [PSI+] induction and variant formation in vivo. We grew cells harbouring the different [RNQ+] variants at 18°C, 25°C or 30°C for the duration of the experiment, and examined them for [PSI+] induction and variant formation upon Sup35 overexpression. We found that growing yeast at 18°C or 25°C increased the rate of [PSI+] induction in a [RNQ+] variant-specific manner (Fig. 3A). Specifically, [PSI+] induction only increased in cells harbouring the low [RNQ+] variant (P < 0.0001; Student's t-test). Moreover, this increased induction rate corresponded with an increase in the strength of the [PSI+] variant induced in these cells (Fig. 3B). These effects were [RNQ+]-dependent, as [PSI+] was never induced in [rnq−] cells.
Fig. 3.
Temperature influences the rate of [PSI+] formation and is dependent on the resident [RNQ+] variant.
A. 74-D694 yeast cells containing either the low, medium, high, or very high [RNQ+] variants, or [rnq−] cells, were grown at either 18°C, 25°C or 30°C and scored for [PSI+] status as described in Experimental procedures.
B. Colonies showing nonsense suppression were picked and spotted on YPD, SD-Ade and YPD + 3 mM GdnHCl solid media and scored for curability and strength of [PSI+] by colour on YPD and growth on SD-Ade. Percentage of total [PSI+] variants for each condition are plotted. Each experiment was replicated at least three times and more than 300 colonies were scored for the presence of [PSI+] for each experiment. For each condition at least 300 colonies were selected and re-grown to score [PSI+] variant phenotype.
C. 74-D694 yeast cells containing either the low, medium, high, or very high [RNQ+] variants, or [rnq−] cells, were treated with either a 42°C or 50°C heat shock, or left at 30°C and scored for [PSI+] status as described in Experimental procedures.
D. [PSI+] colonies were picked and spotted on YPD, SD-Ade and YPD + 3 mM GdnHCl solid media and scored for curability and strength of [PSI+] as in panel B.
Conversely, cells that were subjected to a transient heat shock at 50°C exhibited a significant decrease in overall [PSI+] induction frequency in comparison to untreated controls harbouring the same [RNQ+] variant (Fig. 3C; medium, high and very high [RNQ+]: P < 0.0001, Student's t-test). However, in low and medium [RNQ+] cells, the resultant [PSI+] variants showed overall stronger phenotypes than those induced in the untreated cells (Fig. 3D). When we calculated the change in [PSI+] variants in cells propagating the low [RNQ+] variant, using the cumulative induction of both medium and strong [PSI+] as above, we found a 5.7-fold increase in medium/strong [PSI+] variants in the 50°C heat-shocked cells when compared to controls. Using this analysis method, we found that induction of strong [PSI+] increased 6.7-and 2.1-fold in cells harbouring the medium and high [RNQ+] variants, respectively. As we noted above with several of the other stressors, cells containing very high [RNQ+] that were stressed at 50°C did not show significant differences in the [PSI+] variants that were induced, further distinguishing the [RNQ+] variants from each other. Surprisingly, heat shock at 50°C and 42°C, which we found both elicit a similar increase in levels of Hsp104 (data not shown), had very different effects on [PSI+] formation. Incubation at 42°C, which was not toxic to the cells, did not change the [PSI+] induction rates nor the [PSI+] variants that were formed with any of the [RNQ+] variants. Cumulatively, these results demonstrate that both the mild stress of sustained growth at a low temperature and the extremely toxic stress of transient incubation at 50°C had dramatic effects on prion formation, in a [RNQ+] variant-specific manner. It is plausible that these environmental shifts modified the [RNQ+] prion such that it became a ‘stronger’ template that was more capable of inducing stronger variants of [PSI+].
Incubation in stationary phase impacts both de novo [PSI+] formation and [PSI+] loss
Following up on our observations that both the resident [RNQ+] variant and cellular growth conditions significantly affect [PSI+] induction, we investigated whether these factors would affect the rate and variants of [PSI+] formed spontaneously, without Sup35 overexpression. To analyse de novo [PSI+] formation, we incubated the cells in stationary phase, which showed the most dramatic changes to the [PSI+] variants formed relative to untreated controls upon Sup35 over expression. We grew cells to mid-log phase at 30°C (as the control), or agitated the cultures for 7 days at 30°C in saturation to achieve stationary phase. We selected for nonsense suppressors on synthetic solid medium lacking adenine and isolated all spontaneously formed colonies after 8 days of growth. When cells were grown for more than 8 days at 30°C on solid media lacking adenine (SD-Ade), non-curable adenine prototrophs obscured our analysis of true [PSI+] cells. We re-grew these clones, as above, on solid YPD media to confirm colour, SD-Ade media to assess the strength of [PSI+], and media containing GdnHCl to confirm [PSI+] curability. As an additional test of true [PSI+] formation, a subset of the spontaneously formed GdnHCl-curable adenine prototrophs were mated to [psi−] 74-D694 cells. We found that in all cases, the [PSI+] phenotype segregated 4:0, exhibiting the dominant non-Mendelian inheritance that defines yeast prions (Supplemental Fig. S3 and data not shown).
When we calculated the number of cells that spontaneously formed [PSI+], we found that, under normal growth conditions (overnight at 30°C), spontaneously acquired [PSI+] showed the same [RNQ+]-variant-dependent trend as observed by induction assays. The high and very high [RNQ+] variants spontaneously formed [PSI+] much more readily than cells containing the low [RNQ+] variant (Table 1). Interestingly, we found that modifying the growth environment by incubating cells in stationary phase increased spontaneous [PSI+] formation in a [RNQ+] variant-dependent manner. Stationary-phase growth caused dramatic increases in spontaneous [PSI+] formation, approximately three orders of magnitude, in both low-[RNQ+]- and high-[RNQ+]-containing cells. Surprisingly, incubation in stationary phase did not alter spontaneous [PSI+] formation in yeast harbouring very high [RNQ+]. For both conditions, cell harbouring the very high [RNQ+] variant spontaneously formed [PSI+] at a frequency of approximately 1 × 10−3, which is at least three to four orders of magnitude greater than rate of spontaneous [PSI+] formation that was previously reported in cells containing an unidentified [RNQ+] variant (Lancaster et al., 2010).
Table 1.
Incubation in stationary phase modifies spontaneous [PSI+] formation.
| Condition | [RNQ+] variant | [PSI+] rate | % Med/strong [PSI+] |
|---|---|---|---|
| Mid-log | Low | 2.2E-07 | 100 |
| Stationary | Low | 4.7E-04 | 85 |
| Mid-log | High | 5.0E-06 | 97 |
| Stationary | High | 1.1E-03 | 96 |
| Mid-log | Very high | 1.0E-03 | 93 |
| Stationary | Very high | 6.6E-03 | 96 |
Yeast containing the low, high or very high [RNQ+] variants or [rnq−] cells were grown either overnight in liquid YPD at 30°C or 7 days at 30°C. Diluted cells were spread on YPD plates and grown for 5 days at 30°C to assess total cell number. A dilution of the same culture was plated on SD-Ade plates and colonies that were confirmed to be curable by transient growth on media containing GdnHCl were scored as [PSI+]. The [rnq−] cells never showed the formation of any [PSI+] colonies for any condition. Colonies that were adenine prototrophs were picked and spotted on YPD, SD-Ade and YPD + 3 mM GdnHCl solid media and scored for curability and strength of [PSI+] by growth on SD-Ade. For column 5, the percentage of medium/strong [PSI+] was calculated using the sum of the number of medium and strong [PSI+] divided by the total number of curable [PSI+] clones, and is the cumulative percentage for all experiments combined. Each experiment was completed at least five times. Between 3 and 8 million cells were assessed for [PSI+] induction and an average of 150 colonies were scored for the strength of the [PSI+] variant, for each condition.
We characterized the strength of the [PSI+] variants that were formed after the overnight and week-long incubation conditions and observed a striking trend: the majority of [PSI+] variants formed in both conditions were either strong or medium (Table 1, Supplemental Fig. S4). Our results show that the cellular environment dramatically impacts the predominant amyloid structure that forms, thereby controlling the resultant phenotypes. We found that weak [PSI+] was rarely induced spontaneously in stationary phase; however, the possibility remained that weak [PSI+] formed, but was unstable, and therefore readily lost under these conditions. Because weak [PSI+] is spontaneously lost at a low frequency under normal growth conditions, we used cells harbouring a previously characterized weak [PSI+] variant to test whether long-term incubation in stationary phase would alter this mitotic loss. To assess the mitotic stability of weak [PSI+] in stationary phase, we grew cells harbouring the prion either overnight or for 7 days at 30°C, and scored colonies for [PSI+] loss (colonies that were red or contained red sectors). Interestingly, we found that weak [PSI+] is lost more readily during incubation in stationary phase than in an overnight culture (~ 5% versus ~ 1% loss: Supplemental Fig. S5). We propose that in weak [PSI+] cells, the Sup35 amyloid conformation is less compatible with the intracellular changes that occur during stationary phase, resulting in increased loss, and that the extracellular environment may serve as a selective pressure in maintaining particular prion conformers (Newnam et al., 2011b).
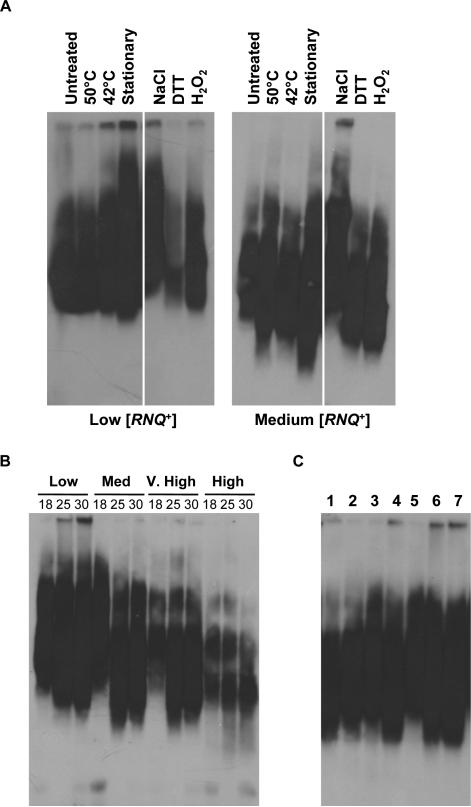
Extracellular growth environment modifies Rnq1 aggregates
As it was clear that cellular stress could impact [PSI+] variant formation, we wanted to explore the mechanistic basis underlying these changes in [PSI+] in different conditions. One plausible explanation may be that, in those conditions, the Rnq1 aggregate structure has changed in the [RNQ+] cells. If the structure of the [RNQ+] prion were altered, it could change the ability of Sup35 to cross-seed on the existing Rnq1 aggregates, perhaps by modifying the structure of the Sup35 protein, or the proteins’ interface. To investigate this possibility, we analysed the Rnq1 aggregates by SDD-AGE using lysates from [RNQ+] cells grown with the various environmental stressors (Fig. 4A) or at altered temperatures (Fig. 4B). Using this method, which allows intact aggregates to run through the large pores of an agarose gel with a low SDS concentration (Kryndushkin et al., 2003), we found that there were slight modifications to Rnq1 aggregate distribution in some of the conditions. For instance, cells incubated in stationary phase reproducibly expanded the distribution of Rnq1 aggregates as compared to the controls. In addition, aggregates of Rnq1 were reproducibly altered by cell growth at 18°C. Cells harbouring the low, medium and very high [RNQ+] variants showed a reduced population of lower aggregate species after growth at 18°C (Fig. 4B). Conversely, aggregates of the high [RNQ+] variant, which was previously shown to differ from the other variants when assayed by Rnq-GFP distribution and temperature sensitivity (Bradley and Liebman, 2003), were not altered by growth temperature (far right 3 lanes), indicating that it may not be affected by the environment in the same manner. These results indicate that the extracellular environment may modify the [RNQ+] prion structure. As such, the environmentally induced changes in [PSI+] formation may be due, at least in part, to modifications to the [RNQ+] template present in the cell.
Fig. 4.
Rnq1 aggregate distribution by SDD-AGE after treatment with stress.
A. Yeast cells harbouring the low or medium [RNQ+] variants were grown overnight at 30°C, subjected to the environmental changes indicated, and then lysates were analysed by SDD-AGE as detailed in Experimental procedures. The condition tested is indicated at the top of the panel.
B. Yeast strains containing the [RNQ+] variants were grown at 18°C, 25°C or 30°C. Cell lysates were subjected to SDD-AGE as in (A).
C. Cells harbouring the low [RNQ+] variant were grown at either: 18°C and then transferred to 30°C (lane 1), 30°C and then transferred to 18°C (lane 2), 18°C and then transferred to 18°C (lane 3), 30°C and then transferred to 30°C (lane 4), 18°C (lane 5), 25°C (lane 6), or 30°C (lane 7). Cells from each of the conditions were lysed and subjected to SDD-AGE as in (A).
Knowing the dynamic nature of prions in response to environmental changes, we next asked whether there were any noticeable changes to Rnq1 aggregates upon temperature shift. We examined the Rnq1 aggregate distribution in cells that were initially grown at 18°C and then moved to 30°C for subsequent growth, and vice versa. As mentioned above, when we grew cultures at 18°C, the majority of Rnq1 lower-molecular-weight aggregate species were lost when we analysed aggregate distribution by SDD-AGE. However, when we re-grew these cells at 30°C for all subsequent growth, the Rnq1 aggregate distribution returned to that seen in cells grown at 30°C (Fig. 4C). These results indicate that the changes to Rnq1 aggregate distribution, as observed by SDD-AGE, can be transiently modified by the extra-cellular environment.
Previous exposure to environmental stress can modify [RNQ+] and alter subsequent [PSI+] formation
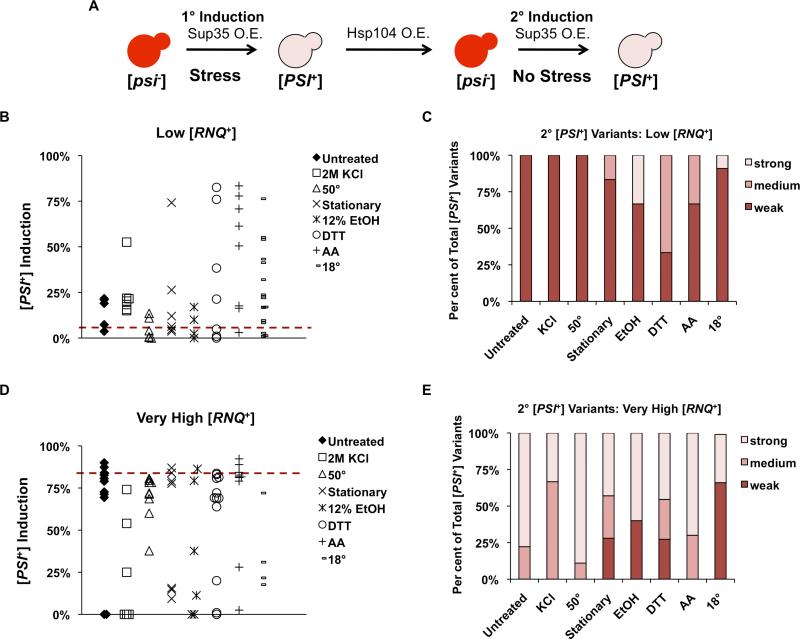
We have shown that incubation of cells in altered environments can have transient effects on Rnq1 aggregates in [RNQ+] cells when assessed biochemically. We were interested in whether environmental modifications could also have a long-lasting impact on [RNQ+] and whether these effects would influence the subsequent [PSI+]-inducing capabilities of the [RNQ+] variants. The ability to induce [PSI+] is the most sensitive assay to differentiate between structurally distinct [RNQ+] variants and illuminates subtle differences in prion variants that are impossible to see using current biochemical assays. Therefore, in order to complement our biochemical assays, we characterized the cells harbouring the [RNQ+] variants post-stress by analysing the re-formation of [PSI+] (See schematic in Fig. 5A). We selected clones harbouring an array of [PSI+] variants that were formed when [RNQ+] [psi−] cells were grown in a stressful environment (Figs 2 and 3). We then cured [PSI+] in these clones by transiently overexpressing Hsp104, which is known to cure [PSI+] but not [RNQ+] (Chernoff et al., 1995; Derkatch et al., 1997). Red colonies from the pGPD-Hsp104 transformation plates were selected, and the cells were re-plated on YPD media to confirm the red colour. Then, [PSI+] was re-induced in these [RNQ+] [psi−] cells under normal growth conditions, at 30°C. We selected phenotypically [psi−], red colonies from the pSup2 transformation plates and [PSI+] formation was assessed phenotypically as before.
Fig. 5.
Secondary (2°) [PSI+] induction rates and variants re-induced can be modified by previous exposure to stress.
A. Schematic depicting the protocol for the re-induction of [PSI+], as detailed in Experimental procedures.
B and D. [PSI+] clones induced under conditions of environmental stress were cured of [PSI+] by Hsp104 overexpression. [PSI+] was re-induced by overexpression of Sup35. Cells were scored for [PSI+] state using the [PSI+] induction assay described in the text and Experimental procedures. The dashed red line represents the initial level of [PSI+] induction in untreated control cells. Each symbol represents the [PSI+] induction rate of one re-induced clone.
C and E. The number of strong, medium and weak [PSI+] clones divided by the total number of [PSI+] clones analysed was calculated. For all panels, some conditions that yielded results similar to controls were excluded from the graphs. Each experiment was repeated, and all re-induced clones are combined in the graphs.
We were surprised to find that cells that had previously been [PSI+] and induced in stress, had changes in both the [PSI+] re-induction rates and the [PSI+] variants subsequently formed in normal growth conditions. For each condition and prion variant, at least four independent clones were cured and re-induced, and each symbol represents one of these clones (Fig. 5B and C). Yeast containing the low or very high [RNQ+] variants showed the most changes. Cells containing low [RNQ+] that were incubated during the initial induction with DTT, histidine deprivation, at 18°C, or in stationary phase, induced [PSI+] at rates similar to the ‘higher’ [RNQ+] variants in a subset of clones, upon [PSI+] re-induction (Fig. 5B). This was striking because, without stress, low-[RNQ+]-containing cells never induced [PSI+] at rates above 35% during the primary induction, but, post-stress several showed [PSI+] induced at rates of over 50%. Moreover, the [RNQ+] variant cells previously treated with stress were capable of generating [PSI+] variants that were much stronger than the [PSI+] variants that were formed in clones isolated from the controls (Fig. 5C). For the very high [RNQ+] variants, in a subset of clones, the post-stress [PSI+] re-induction rates were substantially lower than we ever observed for yeast containing the very high [RNQ+] variant in non-stressed conditions (Fig. 5D). These decreases in [PSI+] re-induction corresponded with an increase in the number of weak and medium [PSI+] variants induced (Fig. 5E).
Previous reports found incompatibilities between specific variants of [PSI+] and [RNQ+]: medium [RNQ+] was always lost upon induction of weak [PSI+], and both medium and very high [RNQ+] were sometimes lost upon induction of strong [PSI+] (Bradley and Liebman, 2003). We hypothesized that if the [RNQ+] structure was modified by extracellular environment, it may also affect the compatibilities of particular [PSI+] and [RNQ+] structures within a cell. Therefore, we assessed the [RNQ+] status in the clones that demonstrated atypically low rates of [PSI+] re-induction, or failed to re-induce [PSI+]. When we assayed the [RNQ+] status of cells that formed [PSI+] less than 5% of the time, by solubility assays using high-speed centrifugation, we initially saw similarities in incompatibilities to those previously reported (Bradley and Liebman, 2003). We found that, despite lacking the Rnq1 protein in the insoluble fraction, many of these strains re-induced [PSI+] after Hsp104 mediated curing, albeit at low levels; therefore, we analysed the [RNQ+] status by SDD-AGE. We found that these cells did indeed contain aggregates of the Rnq1 protein. As such, we failed to see incompatibilities between any [RNQ+] variants and [PSI+] in untreated, DTT, EtOH, and stationary-phase conditions. [RNQ+] was only lost in a proportion of cells grown in the presence of KCl or with histidine deprivation, after a brief shock at 50°C, or after growth at 18°C (Table 2). Therefore, we found that [PSI+] re-induction and SDD-AGE were more sensitive methods to detect [RNQ+], as compared to analysis of Rnq1 solubility or monitoring Rnq1 aggregate distribution using a Rnq1-GFP reporter protein (Sondheimer and Lindquist, 2000; Bradley and Liebman, 2003).
Table 2.
Rnq1 aggregates are lost in conditions of environmental stress.
| [RNQ+] variant | Per cent of clones that retain Rnq1 aggregates |
||||
|---|---|---|---|---|---|
| Untreated | KC1 | 50°C | AA | 18°C | |
| Low | 100% | 100% | 100% | 100% | 100% |
| Medium | 100% | 100% | 80% | 80% | 40% |
| High | 100% | 100% | 100% | 100% | 100% |
| Very high | 100% | 80% | 100% | 100% | 100% |
The [RNQ+] status was examined in clones containing [PSI+] variants that were induced under conditions of environmental stress but failed to re-induce [PSI+]. [PSI+] cells from the initial induction under stress, were grown overnight at 30°C without stress. Cell lysates were subjected to SDD-AGE and analysed as described in Experimental procedures. AA: amino acid deprivation.
Cumulatively, these results indicate that the changes to the [RNQ+] prion after stress are not necessarily transient. Furthermore, they suggest that the [RNQ+] variant may be modified by its environment after interacting with Sup35, during [PSI+] induction, in a way that altered the [RNQ+] structure or [PSI+]-inducing capabilities. These differences are not likely due to the effect of overexpression of Hsp104 on [RNQ+], because we did not see a similar degree of changes to [PSI+] re-induction rates when control unstressed cells were cured in the same fashion (Fig. 5B and D). Because [RNQ+] is the regulator of [PSI+] formation, in these experiments, this putative, ‘adapted’ [RNQ+] structure was capable of inducing [PSI+] at different frequencies and exhibited different compatibilities with [PSI+], presumably as a consequence of an altered ability to associate with Sup35.
Discussion
Using a yeast prion protein as a model for amyloid formation, we found that extracellular stress can have a predominant influence on both the rate of [PSI+] formation and the specific variants that are generated. The amyloid structure of Sup35 and the resultant [PSI+] phenotype are influenced by the external environment in a surprising way, such that it eclipses the typical interplay between the [RNQ+] template and Sup35. Interestingly, upon removal from the stressful condition, cells can either: retain the new, modified inter-prion relationship, form a new relationship, or revert to the initial one, suggesting that environmental modifications can have both transient and long-lasting affects. The effects of these stressors on prion formation may be due to an influence on amyloid formation itself, or on the selection and subsequent propagation of a certain amyloid structure. Indeed, it remains possible that a variety of prion structures form simultaneously in some conditions and, depending on the cellular environment, a subset are selectively propagated. Interestingly, the magnitude and scope of the effects of stress on [PSI+] variant formation could not be explained simply by the level or type of stress. The underlying mechanism that changes [PSI+] could result from a direct effect of the environmental condition on prion protein folding, or could be an indirect consequence of changes in proteostasis (including chaperone activity, translational fidelity, and protein turnover).
Interestingly, the ability of yeast to propagate a spectrum of prion structures from a single protein sequence creates the potential to encipher markedly diverse biological phenotypes from one genotype, plausibly providing a selective advantage to an organism that exists in varied cellular environments (Eaglestone et al., 2000; True and Lindquist, 2000; True et al., 2004; Chernoff, 2007; Halfmann et al., 2010; 2012; Newnam et al., 2011b; Holmes et al., 2013; Masel, 2013). We hypothesize that, in addition to the ability to be in the [PRION+] or [prion−] state, the specific conformation of each individual prion variant can impart an additional level of cellular control. Moreover, the resulting continuum of structural and phenotypic variation might impact cellular fitness in response to different environmental stressors to generate a range of novel phenotypes that could be adaptive (Kirschner, 2013). Several lines of evidence suggest that the ability to acquire the [PRION+] state is an evolutionarily conserved mechanism for increasing fitness, including: dominant inheritance, conservation of the prion forming domain (Ter-Avanesyan et al., 1993; 1994; Doel et al., 1994; Chernoff et al., 2000; True and Lindquist, 2000; Jensen et al., 2001; True et al., 2004), spontaneous formation in the wild (Alberti et al., 2009; Halfmann et al., 2012; Westergard and True, 2014), increase in frequency in response to changes in growth conditions (Derkatch et al., 2000; Chernoff, 2007; Tyedmers et al., 2008; Newnam et al., 2011b; Holmes et al., 2013), and benefits imparted in certain environments (Eaglestone et al., 1999; True and Lindquist, 2000). To add to this list, we found that in the context of different [RNQ+] variants, cells spontaneously formed a continuum of structural states of Sup35 to generate a multitude of [PSI+] variant-specific phenotypes (Fig. 1D and E). The generation of multiple conformers, adapted to modify particular physiological functions, may serve as a bet-hedging strategy by yeast to increase fitness by allowing access to complex phenotypes (Masel, 2013).
Similarly, when variants of the infectious form of PrP (PrPSc) are propagated in cell culture and then exposed to a toxic environment, the PrPSc variant population shifts from a sensitive conformer to one that increases cellular survival in the previously toxic conditions (Li et al., 2010; Mahal et al., 2010; Oelschlegel and Weissmann, 2013). Such selection of variants might occur via a mechanism that involves either the conversion of a sensitive variant to a more resilient conformation or the amplification of a species that previously persisted at low levels within a mixture of conformations (Weissmann, 2012). In fact, modifications to cellular environment, such as cross-species transmission, atypical inoculation route, preexisting peripheral inflammation and exposure to physical treatments that inactivate less resilient strains can dramatically alter the pathogenicity and infectivity of PrPSc variants that were previously bred true upon serial passage (Heikenwalder et al., 2005; Aguzzi et al., 2007; Collinge and Clarke, 2007; Ghaemmaghami et al., 2009; Shikiya et al., 2010). Uncovering the effects of these potential modifiers on the formation of amyloid is complicated in the context of mammalian systems: changes in cell physiology, cell-to-cell communication, the cell type, and the external environment may all play a role in the de novo generation and selective amplification of specific amyloid structures. In addition, the susceptibility of specific cell types for particular disease-associated amyloids (Eisenberg and Jucker, 2012) suggests that at least some of these factors play important roles in the generation and propagation of toxic conformers.
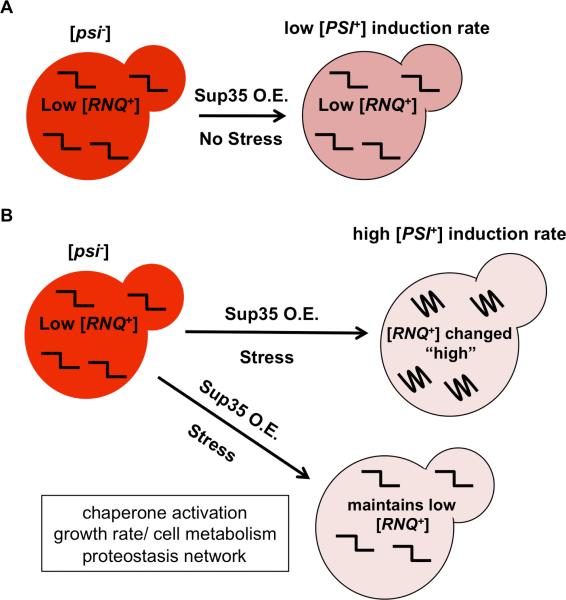
Our data suggest that environmental cues trigger yeast cells to selectively generate or amplify specific [PSI+] variants (Figs 2B and 3B and D; Supplemental Fig. S2). How might these changes in Sup35 protein structure arise? De novo [PSI+] induction occurs in a [RNQ+]-dependent manner. Some evidence suggests that the Rnq1 protein interacts with Sup35 via direct templating in [RNQ+] cells, perhaps inefficiently, but in doing so, promotes conversion of Sup35 to a prion state (Derkatch et al., 2000; 2004; Kulikov et al., 2001). Variants of [RNQ+] reproducibly facilitate [PSI+] induction at certain rates (Fig. 1C; Bradley et al., 2002; Bradley and Liebman, 2003; Sharma and Liebman, 2013a) under normal growth conditions. Because changes in the extracellular environment can elicit a range of intra-cellular effects, we reasoned that, either the structure of the [RNQ+] template, or the mechanism by which [RNQ+] interacts with Sup35, may be altered by the environmental changes. In fact, we found that changing environmental conditions can alter the templating process between [RNQ+] and Sup35, to induce [PSI+] (Figs 2A and 3A and C). Interestingly, the low and medium [RNQ+] variants were more sensitive to several stressors, and showed more dramatic changes to the [PSI+] variants that were induced. Moreover, cells containing very high [RNQ+] never followed this trend in the primary induction assays, but we did note changes to the very high [RNQ+] template by secondary [PSI+] induction (Fig. 5D and E) and biochemically (Fig. 4B). As compared to the [PSI+] variants formed in control conditions, those formed in stressed cells were generally strengthened. Because the very high [RNQ+] variant induces generally strong [PSI+] variants in normal growth environments, our assay was likely limited in detecting increases to this already strong range. However, we were able to elucidate alterations to the very high [RNQ+] template utilizing our other assays. In sum, our data (Figs 4 and 5) led us to hypothesize that the Rnq1 structure in [RNQ+] cells can be transiently or perhaps more permanently altered upon exposure to stressful growth conditions (Fig. 6) and these changes impact the subsequent [PSI+]-inducing capabilities of the [RNQ+] template. Because the only known function of [RNQ+] is to serve as a template on which to cross-seed [PSI+], the changes to Rnq1 aggregates that we observed may make a cell more poised to induce specific variants of [PSI+].
Fig. 6.
Model illustrating factors responsible for changes to [PSI+] variants and induction rate after stress exposure.
A. Under normal growth conditions, low [RNQ+] inefficiently cross-seeds Sup35 to generate [PSI+] cells. Sup35 overexpression (Sup35 O.E.) results in low [PSI+] induction rates and mostly weak [PSI+] colonies.
B. Under conditions of stress, various cellular modifications increase [PSI+] induction rates and/or the strength of the [PSI+] variants. This presumably occurs either by modifying the low [RNQ+] structure to become a template more like high [RNQ+] (top) or by affecting [PSI+] through cellular responses while maintaining the low [RNQ+] variant structure (bottom).
It was previously reported that when [psi−] cells were subjected to environmental stress, the toxicity of the stress corresponded with an increase in [PSI+] formation when an artificially expanded version of Sup35 was expressed in the cells (Tyedmers et al., 2008). We were unable to replicate these data using our methods, and, in fact, found that many conditions of stress actually decreased [PSI+] formation (Fig. 2B; Supplemental Fig. S2). In the previous work, however, [PSI+] formation was assayed by selecting adenine prototrophs. We hypothesize that the discrepancies between these data sets may be due to their method for assessing [PSI+] induction that limited detection to a subset of stronger [PSI+] variants. Similar to our experiments assessing spontaneous [PSI+] formation in stationary phase, without Sup35 overexpression, where it appeared that there was an increase in [PSI+] formation, when in actuality, it was likely an increase in the number of strong [PSI+] variants that formed or were selected.
Previous reports showed that an increase in the expression of the Hsp104 chaperone augmented the [RNQ+]-dependent induction of both [PSI+] and another yeast prion, [URE3] (Kryndushkin et al., 2011). Additionally, increasing Hsp104 levels by exposing cells to higher temperatures that are permissive for growth destabilizes the weak [PSI+] variant (Newnam et al., 2011b). In our experiments, exposure to conditions that were previously shown to increase Hsp104 expression, including: ethanol exposure, incubation in stationary phase, 50°C heat shock, and growth at low temperatures (Sanchez et al., 1992), increased the incidence of strong [PSI+] appearance. Moreover, while this manuscript was in preparation, it was reported that cells harbouring the high [RNQ+] variant preferentially formed stronger [PSI+] variants when they were grown at 4°C, as compared to those grown at 30°C, upon overexpression of a Sup35-GFP fusion protein (Sharma and Liebman, 2013b). It was also previously shown that strong [PSI+] is more resistant to changes in Hsp104 than weak [PSI+] (Kushnirov et al., 2000). Our data suggest that environmental stressors may modify the cellular environment in a manner that abolishes less resilient prion variants, allowing for the amplification of ‘resistant’ conformers. This could be due, in part, to changes in chaperone protein activity and resultant alterations in refolding or degradation of misfolded polypeptides and other general changes to the proteostasis network (PN) that become activated in response to the toxicity associated with several of these conditions. As the ‘normal laboratory conditions’ are quite distinct from the environment yeast typically encounter in the wild, these data potentially represent the relationship between, and range of structures acquired by, this two-prion system in natural habitats.
Organisms like yeast are exposed to diverse and ever-changing extracellular environments in their natural habitat. In the current study, we identified a mechanism by which the yeast S. cerevisiae epigenetically produces heritable phenotypic diversity through the generation and propagation of different amyloid structures in response to environmental changes, in a manner that could serve to increase adaptation and fitness in diverse biological niches. In normal growth conditions, both mammalian and yeast cells achieve protein quality control by utilizing the PN; however, ageing, extracellular stress, and protein aggregation can overwhelm the system, leading to cellular dysfunction. Our results suggest that the response of the PN to stress conditions, in the presence of prions, amyloids and other protein aggregates, could act as a mechanism to generate, alter, or select a specific prion structure (Fig. 6). As such, this response may impact any protein aggregation that may be subject to cross-seeding by altering ‘typical’ cross-seeding interactions. Heterologous protein interactions can also enhance the seeding in several neurodegenerative disorders (Gotz et al., 2001; Ferrari et al., 2003; Morales et al., 2010; Miller et al., 2011), resulting in the acceleration of the primary disease and an increase in co-morbidity of diseases. Environmental stressors, like those that occur with increased frequency in aged brain, may modify these interactions to alter the amyloid structures normally formed, or to overcome normal barriers to cross-seeding, thereby dramatically altering amyloid structure and resultant cellular toxicity.
Experimental procedures
Strains, plasmids and cultivation procedures
Yeast cells were grown and manipulated using standard techniques (Guthrie and Fink, 1991). The strain 74-D694 (MATA ade1-14 trp1–289 his3Δ-200 ura3-52 leu2-3112) of S. cerevisiae was used for all experiments (Chernoff et al., 1995). Isogenic strains harbouring the low, medium, high and very high [RNQ+] variants, as well as the [rnq−] control strain (Bradley et al., 2002; Bradley and Liebman, 2003) were kindly provided by Dr Susan Liebman.
Untransformed cells were grown in rich media: 1% yeast extract/2% peptone/2% dextrose (YPD). The plasmid pEMBL-SUP35 (pSup2) (Ter-Avanesyan et al., 1993) contains a URA3 marker, Sup35 under its native promoter and a 2μ origin of replication. A plasmid containing the coding region of HSP104 under control of the GPD promoter (pGPD-104) (Mumberg et al., 1995) with a URA3 selection marker was used to cure [PSI+] cells. Cells transformed with pSup2 or pGPD-104 were grown in synthetic media lacking uracil (SD-Ura) with 2% glucose, to select for cells containing the plasmid. YPD media containing 3 mM guanidine hydrochlo-ride (YPD + GdnHCl) was used to inhibit Hsp104 and cure prions (Ferreira et al., 2001; Jung and Masison, 2001). Cells were grown on synthetic media lacking adenine (SD-Ade) with 2% glucose to assess [PSI+] phenotype (nonsense suppression of ade1-14).
[PSI+] induction frequency under control conditions
Using the standard lithium acetate (LiOAc) protocol, cells containing each of the four [RNQ+] variants or the [rnq−] control strain were transformed with pSup2 [or with empty vector (pEMBL)], spread on SD-Ura solid medium and grown at 30°C for 4 days. A swipe of 5–10 URA+ colonies was used to inoculate 4mls of SD-Ura, which was then grown for 16 h at 30°C with agitation. Cultures were diluted 1:8000 in sterile water and 300 μl were spread onto 150 mm plates with YPD solid media using glass beads. Plates were incubated for 5 days at 30°C and then transferred to 4°C overnight for colour development. [PSI+] induction rate was quantified as was previously described (Bradley and Liebman, 2003). Briefly, for each experiment, at least 300 colonies for each strain were assessed for [PSI+] status using readthrough of the ade1-14 mutant allele, which prevents accumulation of a red pigmented intermediate, resulting in [PSI+] colonies to be white or pink. YPD rich media contains enough adenine to support yeast growth, but does not prevent accumulation of the red pigment. Petite yeast were easily distinguished using colony colour and were excluded from analyses and further characterization. Each marker represents one culture, for which at least 300 colonies were scored for [PSI+] phenotype. Data represent the cumulative results of at least three experiments.
Assessment of [PSI+] phenotypes: scoring of variants
Colonies containing white/pink sectors or completely white or pink colonies were selected in an unbiased manner, mixed in sterile water and replica plated to YPD, YPD + GdnHCl and SD-Ade solid medium using a 48-pin device, along with strong [PSI+], weak [PSI+] and [psi−] controls. YPD and YPD + GdnHCl plates were grown for 3 days and then transferred to 4°C for colour development. SD-Ade plates were grown for 6 days at 30°C. Clones that were curable (reverted to completely red) on YPD + GdnHCl, and remained red when removed from GdnHCl, were subsequently scored for strength of [PSI+] variant based on colour on YPD and degree of growth on SD-Ade. Less than 1% of colonies selected were non-GdnHCl curable spots in the pinnings. Therefore, other nonsense suppressors were readily distinguished phenotypically and not counted.
[PSI+] induction with environmental stress and during growth at low temperatures
Cells containing the [RNQ+] variants, or [rnq−] control cells, were transformed with pSup2 using LiOAc as described above. SD-Ura liquid selection media was inoculated with 5–10 colonies from the pSup2 transformation plate and cultures were grown at 30°C with agitation for approximately 16 h, or to an approximate OD600 of 1.0. Cells were normalized to OD600 of 1.0 and subjected to dilution and/or stress as described in Supplemental Table S1. Three hundred microlitres of diluted cells were spread onto 150 mm YPD plates using glass beads and were allowed to grow at 30°C for 5 days. After growth, plates were placed at 4°C for further colour development. A minimum of 300 colonies were counted for each condition, per experiment and each experiment was repeated at least three times.
Examination of [PSI+] induction frequency at different temperatures consisted of growing cells at either 30°C, 25°C or 18°C at all times (growth prior to pSup2 transformation through growth on YPD to analyse colour change due to [PSI+] induction). [PSI+] induction rates and [PSI+] variants induced were analysed as in control conditions.
Measurement of spontaneous [PSI+] loss or mitotic stability
Clones propagating different newly induced [PSI+] variants or an established weak [PSI+] variant were selected, grown overnight in liquid YPD media, spread on solid YPD media, and then the number of red colonies or colonies containing red sectors were counted.
Measurement of spontaneous [PSI+] induction frequency
Cultures inoculated with yeast containing the [RNQ+] variants were grown either 16 h (overnight control), or for 7 days at 30°C, with agitation. Undiluted cultures were plated on SD-Ade medium and incubated for 8 days at 30°C. A 1:20 000 dilution of cells was plated on YPD and grown for 3 days at 30°C to determine the total number of cells plated on SD-Ade. All colonies that grew on SD-Ade were selected and replated on YPD, SD-Ade and YPD + 3 mM GdnHCl. Clones that were curable on YPD + GdnHCl, were pink/white on YPD and grew on SD-Ade were scored as [PSI+]. The [PSI+] induction rate was calculated as curable [PSI+] colonies divided by the total number of cells plated on the SD-Ade plate. Data represent the cumulative results of three experiments.
SDD-AGE
Yeast containing the [RNQ+] variants were inoculated in YPD and grown at 30°C, 25°C or 18°C to an OD600 of 1.0. For analysis of Rnq1 aggregate distribution after treatment with environmental stress, cells were grown to an OD600 of 1.0 and subject to stress as detailed in Supplemental Table S1. Cells were lysed in a buffer containing 25 mM Tris (pH 7.5), 100 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 0.5 mM DTT, 3 mM phenylmethanesulphonylfluoride, 40 mM N-Ethylmaleimide, and 1 tablet of complete mini protease inhibitor (Roche). Cell lysates normalized for total protein using the Bradford Assay were subjected to a 7 min incubation at room temperature before loading on 1.5% agarose gel. Prion aggregates were separated by SDD-AGE as previously described (Bagriantsev and Liebman, 2004). Rnq1 protein was detected with a polyclonal anti-Rnq1 antibody.
[PSI+] re-induction
Newly induced [PSI+] colonies were replica plated from YPD to solid media containing 5-Fluoroorotic acid (5-FOA), which inhibits the growth of cells expressing URA3 or URA5 (Sikorski and Boeke, 1991), to select for cells that had lost the pSup2 plasmid. Colonies were selected from 5-FOA plates, and loss of the pSup2 plasmid was confirmed by lack of growth on SD-Ura solid media, and cells were grown overnight in liquid YPD at 30° on a rotator. Uracil auxotrophs were transformed with the pGPD-104 plasmid and selected on SD-Ura, to cure [PSI+] (Chernoff et al., 1995). Red, uracil prototrophs were selected from pGPD-104 transformation plates and struck on YPD. Red colonies were struck from YPD to 5-FOA to select for cells that had lost the pGPD-104 plasmid. Red, uracil auxotrophs were then transformed with pSup2 to re-induce [PSI+]. A swipe of 5–10 red colonies were inoculated from the pSup2 transformation plate, grown overnight at 30°C in liquid selection media. Diluted cells were spread on YPD solid media, grown at 30°C for 5 days and colonies were scored as [PSI+] by the presence of pink or white colour in the colony.
After analysing tens of thousands of colonies for [PSI+] status, we were able to reliably assess [PSI+] variants by examining the proportion and grade of pink in a single colony on the YPD plate spread plate, after [PSI+] induction. Bar graphs represent the number of strong, medium and weak [PSI+] clones divided by the total number of [PSI+] clones analysed/re-induced.
Supplementary Material
Acknowledgements
We thank members of the True lab for helpful discussions. We also thank Kevin Stein, Dr Daniel Summers, Dr Anil Cashikar, Dr Justin Fay, Dr Shannon Macauley-Rambach, Dr Olivia Mooren, Dr Isaac Soloman, and Dr Jessie Turnbaugh for comments on the manuscript. We thank Dr Susan Liebman and Dr Yuri Chernoff for yeast strains and plasmids.
Footnotes
Authors’ contributions
LW carried out all of the experiments. LW and HLT conceived and designed the study and drafted the manuscript. Both authors read and approved the final manuscript.
Conflict of interest
The authors declare that they have no conflict of interests.
Supporting information
Additional supporting information may be found in the online version of this article at the publisher's web-site.
References
- Aguzzi A, Heikenwalder M, Polymenidou M. Insights into prion strains and neurotoxicity. Nat Rev Mol Cell Biol. 2007;8:552–561. doi: 10.1038/nrm2204. [DOI] [PubMed] [Google Scholar]
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagriantsev S, Liebman SW. Specificity of prion assembly in vivo. [PSI+] and [PIN+] form separate structures in yeast. J Biol Chem. 2004;279:51042–51048. doi: 10.1074/jbc.M410611200. [DOI] [PubMed] [Google Scholar]
- Bateman DA, Wickner RB. The [PSI+] prion exists as a dynamic cloud of variants. PLoS Genet. 2013;9:e1003257. doi: 10.1371/journal.pgen.1003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley ME, Liebman SW. Destabilizing interactions among [PSI(+)] and [PIN(+)] yeast prion variants. Genetics. 2003;165:1675–1685. doi: 10.1093/genetics/165.4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion ‘strains’ in yeast. Proc Natl Acad Sci USA. 2002;30:30. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO. Stress and prions: lessons from the yeast model. FEBS Lett. 2007;581:3695–3701. doi: 10.1016/j.febslet.2007.04.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Derkach IL, Inge-Vechtomov SG. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr Genet. 1993;24:268–270. doi: 10.1007/BF00351802. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Galkin AP, Lewitin E, Chernova TA, Newnam GP, Belenkiy SM. Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Mol Microbiol. 2000;35:865–876. doi: 10.1046/j.1365-2958.2000.01761.x. [DOI] [PubMed] [Google Scholar]
- Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science. 2007;318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- Cox BS. PSI, a cytoplasmic suppressor of super-suppressor in yeast. Heredity. 1965;20:505–521. [Google Scholar]
- Cox BS, Tuite MF, McLaughlin CS. The psi factor of yeast: a problem in inheritance. Yeast. 1988;4:159–178. doi: 10.1002/yea.320040302. [DOI] [PubMed] [Google Scholar]
- Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Masse SV, Zadorsky SP, Polozkov GV, Inge-Vechtomov SG, Liebman SW. Dependence and independence of [PSI(+)] and [PIN(+)]: a two-prion system in yeast? EMBO J. 2000;19:1942–1952. doi: 10.1093/emboj/19.9.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)]. Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- Derkatch IL, Uptain SM, Outeiro TF, Krishnan R, Lindquist SL, Liebman SW. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Natl Acad Sci USA. 2004;101:12934–12939. doi: 10.1073/pnas.0404968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson AG, Fraser H, Meikle VM, Outram GW. Competition between different scrapie agents in mice. Nat New Biol. 1972;237:244–245. doi: 10.1038/newbio237244a0. [DOI] [PubMed] [Google Scholar]
- Doel SM, McCready SJ, Nierras CR, Cox BS. The dominant PNM2− mutation which eliminates the psi factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics. 1994;137:659–670. doi: 10.1093/genetics/137.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglestone SS, Cox BS, Tuite MF. Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. EMBO J. 1999;18:1974–1981. doi: 10.1093/emboj/18.7.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglestone SS, Ruddock LW, Cox BS, Tuite MF. Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI(+)] of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:240–244. doi: 10.1073/pnas.97.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari A, Hoerndli F, Baechi T, Nitsch RM, Gotz J. beta-Amyloid induces paired helical filament-like tau filaments in tissue culture. J Biol Chem. 2003;278:40162–40168. doi: 10.1074/jbc.M308243200. [DOI] [PubMed] [Google Scholar]
- Ferreira PC, Ness F, Edwards SR, Cox BS, Tuite MF. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol Microbiol. 2001;40:1357–1369. doi: 10.1046/j.1365-2958.2001.02478.x. [DOI] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, et al. A highly conserved eukaryotic protein family possessing properties of polypep-tide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Ahn M, Lessard P, Giles K, Legname G, DeArmond SJ, Prusiner SB. Continuous quinacrine treatment results in the formation of drug-resistant prions. PLoS Pathog. 2009;5:e1000673. doi: 10.1371/journal.ppat.1000673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz J, Chen F, van Dorpe J, Nitsch RM. Formation of neurofibrillary tangles in P301l tau transgenic mice induced by Abeta 42 fibrils. Science. 2001;293:1491–1495. doi: 10.1126/science.1062097. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink G. Guide to Yeast Genetics and Molecular Biology. Academic Press; San Diego: 1991. [Google Scholar]
- Halfmann R, Alberti S, Lindquist S. Prions, protein homeostasis, and phenotypic diversity. Trends Cell Biol. 2010;20:125–133. doi: 10.1016/j.tcb.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lindquist S. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature. 2012;482:363–368. doi: 10.1038/nature10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikenwalder M, Zeller N, Seeger H, Prinz M, Klohn PC, Schwarz P, et al. Chronic lymphocytic inflammation specifies the organ tropism of prions. Science. 2005;307:1107–1110. doi: 10.1126/science.1106460. [DOI] [PubMed] [Google Scholar]
- Holmes DL, Lancaster AK, Lindquist S, Halfmann R. Heritable remodeling of yeast multicellularity by an environmentally responsive prion. Cell. 2013;153:153–165. doi: 10.1016/j.cell.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DM, Morris JC, Goate AM. Alzheimer's disease: the challenge of the second century. Sci Transl Med. 2011;3:77sr71. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang VJ, Stein KC, True HL. Spontaneous variants of the [RNQ+] prion in yeast demonstrate the extensive conformational diversity possible with prion proteins. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0079582. doi:10.1371/journal.pone.0079582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MA, True HL, Chernoff YO, Lindquist S. Molecular population genetics and evolution of a prion-like protein in Saccharomyces cerevisiae. Genetics. 2001;159:527–535. doi: 10.1093/genetics/159.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Masison DC. Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr Microbiol. 2001;43:7–10. doi: 10.1007/s002840010251. [DOI] [PubMed] [Google Scholar]
- Kalastavadi T, True HL. Analysis of the [RNQ+] prion reveals stability of amyloid fibers as the key determinant of yeast prion variant propagation. J Biol Chem. 2010;285:20748–20755. doi: 10.1074/jbc.M110.115303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CY. Supporting the structural basis of prion strains: induction and identification of [PSI] variants. J Mol Biol. 2001;307:1247–1260. doi: 10.1006/jmbi.2001.4542. [DOI] [PubMed] [Google Scholar]
- Kirschner M. Beyond Darwin: evolvability and the generation of novelty. BMC Biol. 2013;11:110. doi: 10.1186/1741-7007-11-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug GM, Wand H, Simpson M, Boyd A, Law M, Masters CL, et al. Intensity of human prion disease surveillance predicts observed disease incidence. J Neurol Neurosurg Psychiatry. 2013;84:1372–1377. doi: 10.1136/jnnp-2012-304820. [DOI] [PubMed] [Google Scholar]
- Kochneva-Pervukhova NV, Chechenova MB, Valouev IA, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. [Psi(+)] prion generation in yeast: characterization of the ‘strain’ difference. Yeast. 2001;18:489–497. doi: 10.1002/yea.700. [DOI] [PubMed] [Google Scholar]
- Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- Kryndushkin DS, Wickner RB, Tycko R. The core of Ure2p prion fibrils is formed by the N-terminal segment in a parallel cross-beta structure: evidence from solid-state NMR. J Mol Biol. 2011;409:263–277. doi: 10.1016/j.jmb.2011.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulikov VV, Derkatch IL, Noskov VN, Tarunina OV, Chernoff YO, Rogozin IB, Pavlov YI. Mutagenic specificity of the base analog 6-N-hydroxylaminopurine in the LYS2 gene of yeast Saccharomyces cerevisiae. Mutat Res. 2001;473:151–161. doi: 10.1016/s0027-5107(00)00142-1. [DOI] [PubMed] [Google Scholar]
- Kushnirov VV, Kryndushkin DS, Boguta M, Smirnov VN, Ter-Avanesyan MD. Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr Biol. 2000;10:1443–1446. doi: 10.1016/s0960-9822(00)00802-2. [DOI] [PubMed] [Google Scholar]
- Lancaster AK, Bardill JP, True HL, Masel J. The spontaneous appearance rate of the yeast prion [PSI+] and its implications for the evolution of the evolvability properties of the [PSI+] system. Genetics. 2010;184:393–400. doi: 10.1534/genetics.109.110213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Browning S, Mahal SP, Oelschlegel AM, Weissmann C. Darwinian evolution of prions in cell culture. Science. 2010;327:869–872. doi: 10.1126/science.1183218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman SW, Chernoff YO. Prions in yeast. Genetics. 2012;191:1041–1072. doi: 10.1534/genetics.111.137760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman SW, Derkatch IL. The yeast [PSI+] prion: making sense of nonsense. J Biol Chem. 1999;274:1181–1184. doi: 10.1074/jbc.274.3.1181. [DOI] [PubMed] [Google Scholar]
- Lundmark K, Westermark GT, Olsen A, Westermark P. Protein fibrils in nature can enhance amyloid protein A amyloidosis in mice: cross-seeding as a disease mechanism. Proc Natl Acad Sci USA. 2005;102:6098–6102. doi: 10.1073/pnas.0501814102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahal SP, Browning S, Li J, Suponitsky-Kroyter I, Weissmann C. Transfer of a prion strain to different hosts leads to emergence of strain variants. Proc Natl Acad Sci USA. 2010;107:22653–22658. doi: 10.1073/pnas.1013014108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masel J. Q&A: evolutionary capacitance. BMC Biol. 2013;11:103. doi: 10.1186/1741-7007-11-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G. Could they all be prion diseases? Science. 2009;326:1337–1339. doi: 10.1126/science.326.5958.1337. [DOI] [PubMed] [Google Scholar]
- Miller Y, Ma B, Nussinov R. Synergistic interactions between repeats in tau protein and Abeta amyloids may be responsible for accelerated aggregation via polymorphic states. Biochemistry. 2011;50:5172–5181. doi: 10.1021/bi200400u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales R, Green KM, Soto C. Cross currents in protein misfolding disorders: interactions and therapy. CNS Neurol Disord Drug Targets. 2009;8:363–371. doi: 10.2174/187152709789541998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales R, Estrada LD, Diaz-Espinoza R, Morales-Scheihing D, Jara MC, Castilla J, Soto C. Molecular cross talk between misfolded proteins in animal models of Alzheimer's and prion diseases. J Neurosci. 2010;30:4528–4535. doi: 10.1523/JNEUROSCI.5924-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morano KA, Grant CM, Moye-Rowley WS. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics. 2012;190:1157–1195. doi: 10.1534/genetics.111.128033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Gonzalez I, Soto C. Misfolded protein aggregates: mechanisms, structures and potential for disease transmission. Semin Cell Dev Biol. 2011;22:482–487. doi: 10.1016/j.semcdb.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Newnam GP, Birchmore JL, Chernoff YO. Destabilization and recovery of a yeast prion after mild heat shock. J Mol Biol. 2011a;408:432–448. doi: 10.1016/j.jmb.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newnam GP, Birchmore JL, Chernoff YO. Destabilization and recovery of a yeast prion after mild heat shock. J Mol Biol. 2011b;408:432–448. doi: 10.1016/j.jmb.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelschlegel AM, Weissmann C. Acquisition of drug resistance and dependence by prions. PLoS Pathog. 2013;9:e1003158. doi: 10.1371/journal.ppat.1003158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KW, Hahn JS, Fan Q, Thiele DJ, Li L. De novo appearance and ‘strain’ formation of yeast prion [PSI+] are regulated by the heat-shock transcription factor. Genetics. 2006;173:35–47. doi: 10.1534/genetics.105.054221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel BK, Liebman SW. ‘Prion-proof’ for [PIN+]: infection with in vitro-made amyloid aggregates of Rnq1p-(132–405) induces [PIN+]. J Mol Biol. 2007;365:773–782. doi: 10.1016/j.jmb.2006.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino MM, Liu JJ, Glover JR, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Biology and genetics of prions causing neurodegeneration. Annu Rev Genet. 2013;47:601–623. doi: 10.1146/annurev-genet-110711-155524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, et al. Eight prion strains have PrP(Sc) molecules with different conformations. Nat Med. 1998;4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- Safar JG. Molecular pathogenesis of sporadic prion diseases in man. Prion. 2012;6:108–115. doi: 10.4161/pri.18666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Taulien J, Borkovich KA, Lindquist S. Hsp104 is required for tolerance to many forms of stress. EMBO J. 1992;11:2357–2364. doi: 10.1002/j.1460-2075.1992.tb05295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlumpberger M, Prusiner SB, Herskowitz I. Induction of distinct [URE3] yeast prion strains. Mol Cell Biol. 2001;21:7035–7046. doi: 10.1128/MCB.21.20.7035-7046.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma J, Liebman SW. Exploring the basis of [PIN(+)] variant differences in [PSI(+)] induction. J Mol Biol. 2013a;425:3046–3059. doi: 10.1016/j.jmb.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma J, Liebman SW. Variant-specific prion interactions. Cell Logistics. 2013b;3:e25698. doi: 10.4161/cl.25698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikiya RA, Ayers JI, Schutt CR, Kincaid AE, Bartz JC. Coinfecting prion strains compete for a limiting cellular resource. J Virol. 2010;84:5706–5714. doi: 10.1128/JVI.00243-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Boeke JD. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- Stahl N, Baldwin MA, Teplow DB, Hood L, Gibson BW, Burlingame AL, Prusiner SB. Structural studies of the scrapie prion protein using mass spec-trometry and amino acid sequencing. Biochemistry. 1993;32:1991–2002. doi: 10.1021/bi00059a016. [DOI] [PubMed] [Google Scholar]
- Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, et al. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein KC, True HL. The [RNQ+] prion: a model of both functional and pathological amyloid. Prion. 2011;5:291–298. doi: 10.4161/pri.5.4.18213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn LA, True HL. Deletion of RNQ1 gene reveals novel functional relationship between divergently transcribed Bik1p/CLIP-170 and Sfi1p in spindle pole body separation. Curr Genet. 2006;50:347–366. doi: 10.1007/s00294-006-0098-6. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Kushnirov VV, Dagkesamanskaya AR, Didichenko SA, Chernoff YO, Inge-Vechtomov SG, Smirnov VN. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol Microbiol. 1993;7:683–692. doi: 10.1111/j.1365-2958.1993.tb01159.x. [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Dagkesamanskaya AR, Kushnirov VV, Smirnov VN. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics. 1994;137:671–676. doi: 10.1093/genetics/137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- True HL, Berlin I, Lindquist SL. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431:184–187. doi: 10.1038/nature02885. Epub 15 Aug 2004. [DOI] [PubMed] [Google Scholar]
- Tyedmers J, Madariaga ML, Lindquist S. Prion switching in response to environmental stress. PLoS Biol. 2008;6:e294. doi: 10.1371/journal.pbio.0060294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uptain SM, Sawicki GJ, Caughey B, Lindquist S. Strains of [PSI(+)] are distinguished by their efficiencies of prion-mediated conformational conversion. EMBO J. 2001;20:6236–6245. doi: 10.1093/emboj/20.22.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishveshwara N, Liebman SW. Heterologous cross-seeding mimics cross-species prion conversion in a yeast model. BMC Biol. 2009;7:26. doi: 10.1186/1741-7007-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann C. Mutation and selection of prions. PLoS Pathog. 2012;8:e1002582. doi: 10.1371/journal.ppat.1002582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergard L, True HL. Wild yeast harbour a variety of distinct amyloid structures with strong prion-inducing capabilities. Mol Microbiol. 2014;92:183–193. doi: 10.1111/mmi.12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Fu X, Ge F, Zhang B, Yao J, Zhang H, et al. Cross-seeding and cross-competition in mouse apolipoprotein A-II amyloid fibrils and protein A amyloid fibrils. Am J Pathol. 2007;171:172–180. doi: 10.2353/ajpath.2007.060576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Derkatch IL, Uptain SM, Patino MM, Lindquist S, Liebman SW. The yeast non-Mendelian factor [ETA+] is a variant of [PSI+], a prion-like form of release factor eRF3. EMBO J. 1999;18:1182–1191. doi: 10.1093/emboj/18.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.