Abstract
Pharmacological inhibitors against the PI3K-AKT-mTOR pathway, a frequently deregulated signaling pathway in cancer, are clinically promising, but the development of drug resistance is a major limitation. We found that 4EBP1, the central inhibitor of cap-dependent translation, was a critical regulator of both prostate cancer initiation and maintenance downstream of mTOR signaling in a genetic mouse model. 4EBP1 abundance was distinctly different between the epithelial cell types of the normal prostate. Of tumor-prone prostate epithelial cell types, luminal epithelial cells exhibited the highest transcript and protein abundance of 4EBP1 and the lowest protein synthesis rates, which mediated resistance to the PI3K-AKT-mTOR pathway inhibitor MLN0128. Decreasing total 4EBP1 abundance reversed resistance in drug-sensitive cells. Increased 4EBP1 abundance was a common feature in prostate cancer patients that had been treated with the PI3K pathway inhibitor BKM120; thus 4EBP1 may be associated with drug resistance in human tumors. Our findings reveal a molecular program controlling cell type-specific 4EBP1 abundance coupled to the regulation of global protein synthesis rates that renders each epithelial cell type of the prostate uniquely sensitive or resistant to inhibitors of the PI3K-AKT-mTOR signaling pathway.
Introduction
The PI3K-AKT-mTOR signaling pathway is altered in 100% of advanced human prostate cancer patients, which is a disease that arises from the prostatic epithelium composed of two distinct epithelial cell types, luminal and basal epithelial cells (1). Both cell types can transform and develop into tumors in the context of various oncogenic stimuli. For example, loss of PTEN, the tumor suppressor and negative regulator of the PI3K-AKT-mTOR signaling pathway, leads to tumor development in either cell type in mouse models of prostate cancer (2). Others have shown that overexpression of the kinase AKT and the transcription factor MYC in normal basal epithelial cells leads to the formation of a luminal-like prostate cancer (3). Moreover, loss of PTEN within a prostate luminal epithelial stem cell population also leads to tumorigenesis in vivo (4). These findings demonstrate that multiple cancer initiating cell types exist within the prostate and that tumor initiation can be driven by oncogenic PI3K-AKT-mTOR activity. However, an important unanswered question is whether all prostate tumor epithelial cell types are equally sensitive to inhibitors of the PI3K pathway or specific cell types are primed for drug resistance. This is a critical question as an emerging problem shared by all PI3K pathway inhibitors is drug resistance, which is significantly stifling the clinical success of this class of therapeutic agents.
The kinase mTOR promotes mRNA translation by converging on the eIF4F cap-binding complex, which is a critical nexus that controls global protein synthesis as well as the translation of specific mRNA targets (5–7). All eIF4F complex members including the cap-binding protein and oncogene eIF4E (8, 9), the scaffolding molecule eIF4G (10), and the RNA helicase eIF4A (11) are required for cap-dependent translation. The eIF4F complex is negatively regulated by a critical interaction between eIF4E and the tumor suppressor eIF4E binding proteins (4EBPs), which are phosphorylated and inhibited by mTOR (6, 12). Using unique mouse models of prostate cancer, we addressed the important question of cell type specificity and translation control in tumor initiation, cancer progression, and drug resistance and found that 4EBP1 activity is not only a marker of PI3K-AKT-mTOR signaling, but is also critical for prostate cancer initiation and maintenance as well as the therapeutic response. We found that a specific population of tumor-forming luminal epithelial cells, which exhibit high transcript and protein levels of 4EBP1 and low protein synthesis rates, are remarkably resistant to inhibition of the PI3K-AKT-mTOR signaling pathway. Furthermore, we found that elevated 4EBP1 expression is necessary and sufficient for drug resistance. Importantly, utilizing patient samples acquired from a phase II clinical trial with the oral pan-PI3K inhibitor BKM120, we found that a high amount of 4EBP1 protein was a characteristic of post-treatment prostate cancer cells. Together, our findings reveal a normal cellular program characterized by high 4EBP1 abundance and low protein synthesis rates in luminal epithelial cells that can be exploited by prostate cancer to direct tumor growth in the context of PI3K pathway inhibition.
Results
Luminal epithelial cells with increased 4EBP1 abundance define a PI3K-AKT-mTOR pathway inhibitor-resistant cell type in vivo
PI3K-AKT-mTOR pathway inhibitors have demonstrated significant preclinical efficacy in prostate cancer preclinical trials; however, drug resistance inevitably develops (13). Multiple prostate epithelial cell types have been implicated in tumorigenesis, including luminal epithelial cells and basal epithelial cells (2), however, it is unknown if both cell types are equally sensitive to PI3K-AKT-mTOR pathway inhibition or if specific cell types are more resistant than others. We previously conducted a preclinical trial with the ATP site mTOR inhibitor MLN0128 (7) in mice that develop prostate cancer through loss of the tumor suppressor PTEN in both basal and luminal epithelial cells (herein referred to as PTENL/L) (14). Although we observed a decrease in the amount of prostate tumors, we also observed that a significant number of tumors remained despite a four-week therapeutic course with MLN0128 (7). To characterize the prostate cancer epithelial cell types prone to drug resistance, we quantified the number of basal epithelial cells and luminal epithelial cells that remained in PTENL/L mice treated with MLN0128 (7). After an eight-week treatment course with MLN0128 or vehicle, the tumors that remained in PTENL/L mice treated with MLN0128 were enriched for CK8+ luminal epithelial cells over CK5+ basal epithelial cells (Fig. 1A and fig. S1, A and B). These findings suggest that tumor-forming luminal epithelial cells are more resistant than basal epithelial cells to inhibition of the PI3K-AKT-mTOR signaling pathway.
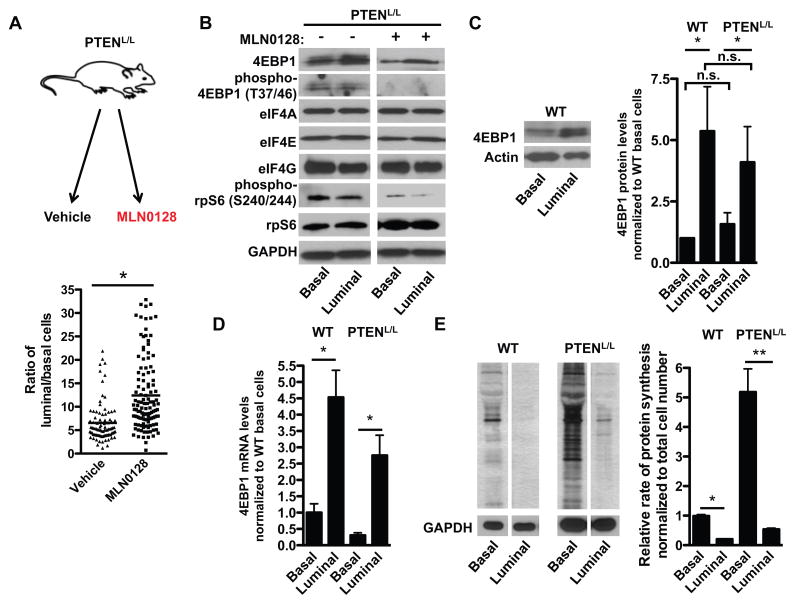
Figure 1. Luminal epithelial prostate cancer cells are enriched upon ATP site inhibition of mTOR and are characterized by increased 4EBP1 protein abundance and lower baseline protein synthesis rates in vivo.
(A) Schematic of MLN0128 preclinical trial. Age matched 9–12 month-old PTENL/L mice were treated with vehicle or MLN0128 (1 mg/kg) daily for 8 weeks (upper panel). Quantification for ratio of CK8+ luminal epithelial cells over CK5+ basal epithelial cells in vehicle or MLN0128 treated PTENL/L mice (lower panel, n = 6 mice/condition, *P<0.05, t-test). (B) Representative Western blots for 4EBP1, eIF4A, eIF4E, eIF4G, phospho-4EBP1 (Thr37/46), phospho-rpS6 (Ser240/244), and rpS6 in basal epithelial and luminal epithelial cells from PTENL/L mice treated with and without MLN0128 1mg/kg daily for 4 days by oral gavage. (C) Representative Western blot for 4EBP1 in basal epithelial and luminal epithelial cells from WT mice (left panel). Quantification of 4EBP1 densitometry in both wild-type (WT) and PTENL/L basal and luminal epithelial cells (right panel, n = 3 mice/genotype, *P = 0.04, t-test). (D) 4EBP1 mRNA expression in WT and PTENL/L basal and luminal epithelial cells (n = 2–3 mice/genotype, *P = 0.002, t-test). All samples were first normalized to actin. (E) Representative 35S-methionine incorporation run on the same gel in sorted basal and luminal epithelial cell types from WT and PTENL/L mice (left panel). Quantification of [35S]-methionine incorporation by densitometry in sorted basal and luminal epithelial cell types from WT and PTENL/L mice (right panel, n = 3 mice/genotype, *P<0.0001, **P = 0.002, t-test). Data are mean +/− S.E.M.
Next, we sought to determine the molecular features of resistant luminal epithelial cells that differentiate them from basal cells. The 4EBP1-eIF4E axis is a critical downstream target of mTOR that mediates the therapeutic efficacy of ATP site mTOR inhibitors (15). As such, we sought to determine the abundance of 4EBP1 and each of the eIF4F components (eIF4E, eIF4A, and eIF4G) in both luminal and basal epithelial cells. To this end, we sorted luminal and basal epithelial cells from wild-type and PTENL/L mice treated with and without MLN0128 and Western blotted for pathway components. We used the cell surface marker and integrin CD49f and the murine stem cell marker Sca-1 to distinguish luminal epithelial cells from basal epithelial cells (16). In particular, wild-type and PTENL/L basal epithelial cells are CD49f and Sca-1 high, and luminal epithelial cells are CD49f and Sca-1 low, which was confirmed by quantitative polymerase chain reaction (qPCR) analysis of their respective markers on sorted cell populations (fig. S2, A and B) (16). Loss of PTEN and hyper-phosphorylation of AKT were confirmed by qPCR, Western blot analysis, and immunofluorescence in PTENL/L basal and luminal epithelial cells (fig. S3, A to C). Amongst all the translation initiation components analyzed, only 4EBP1 protein levels were increased in PTENL/L luminal epithelial cells in both MLN0128 treated and untreated mice (Fig. 1, B and C). Notably, the phosphorylation of 4EBP1 was similar in basal and luminal epithelial cells and therefore do not contribute to the differences seen in total 4EBP1 abundance (Fig. 1B and fig. S3D). Moreover, we did not observe nuclear sequestration of 4EBP1 or cytoplasmic foci of 4EBP1 in luminal cells compared to basal cells (fig. S3E). The abundance of eIF4A and eIF4E was equivalent between treated and non-treated luminal and basal epithelial cells (Fig. 1B), whereas the abundance of eIF4G exhibited only a mild decrease in luminal epithelial cells compared to basal epithelial cells (Fig. 1B). Together, these findings reveal that luminal epithelial cells, which we found to be prone to drug resistance, are characterized by high abundance of the translational repressor 4EBP1.
Prostate epithelial cells have distinct amounts of 4EBP1 mRNA expression that underlies cell type-specific protein synthesis rates within the prostate
We next asked whether the higher amounts of 4EBP1 in prostate luminal epithelial cells were the result of PTEN loss or instead reflect a normal expression program. Unexpectedly, we found that 4EBP1 protein abundance was considerably higher in wild-type luminal epithelial cells compared to that in wild-type basal epithelial cells (Fig. 1C). To determine at what level of gene expression 4EBP1 is regulated in luminal and basal epithelial cells, we conducted quantitative PCR analysis of 4EBP1 expression in both cell types from wild-type and PTENL/L mice. 4EBP1 mRNA expression was 3–5 fold higher in normal and PTENL/L luminal epithelial cells compared to basal epithelial cells revealing differential transcriptional regulation of 4EBP1 in distinct tumor-prone prostate epithelial cell types (Fig. 1D).
Next we sought to determine the functional relevance of increased 4EBP1 transcript and protein amounts in luminal epithelial cells. 4EBP1 is a negative regulator of cap-dependent translation and has been shown to decrease the rate of protein synthesis (12). We therefore conducted [35S]-methionine incorporation assays on FACS sorted basal and luminal epithelial cells from wild-type and PTENL/L mice to determine the baseline mRNA translation rates in each cell type. The rates of de novo protein synthesis between the two cell types were significantly different, with basal epithelial cells exhibiting a markedly greater amount of protein synthesis than did luminal epithelial cells (Fig. 1E). To determine whether other translation components besides 4EBP1 may contribute to the difference in protein synthesis rates between basal and luminal epithelial cells, we conducted a candidate protein expression analysis of major regulators of translation, including the phosphorylation of eEF2, phosphorylation of eIF2α, phosphorylation of rpS6, and the abundance of the translation initiation inhibitors 4EBP2 and PDCD4. The phosphorylation of eEF2 and eIF2α was similar in both cell types in the PTENL/L with no changes in total abundance of eEF2 or eIF2α (fig. S4). Phosphorylation of rpS6, which has an inhibitory effect on protein synthesis (17), was decreased in luminal epithelial cells compared to basal epithelial cells (Fig. 1B). The abundance of both 4EBP2 and PDCD4 was decreased in PTENL/L luminal epithelial cells compared to that in basal epithelial cells (fig. S4). As such, the phosphorylation status of eEF2, eIF2α and rpS6, as well as protein abundance of 4EBP2 and PDCD4 cannot account for the difference in protein synthesis rates observed between PTENL/L basal and luminal epithelial cells.
Despite having similarly increased abundance of 4EBP1 as PTENL/L luminal epithelial cells, wild-type luminal epithelial cells displayed the lowest protein synthesis rates compared to all other cell types studied. This suggests that other factors besides 4EBP1 abundance may further limit wild-type luminal epithelial cell protein synthesis rates. For example, we observed that eIF2α phosphorylation was slightly increased in wild-type luminal epithelial cells compared to basal epithelial cells (fig. S4). This increase was not observed in PTENL/L luminal cells and therefore cannot account for drug resistance or the low protein synthesis rates in the transformed setting (fig. S4). Thus, prostate epithelial cell types were distinguished by their innate protein synthesis rates, which inversely correlated with 4EBP1 transcript and protein amounts and may underlie MLN0128 resistance. Moreover, these findings suggest that a transcriptional program, which dictates the cell type-specific expression of 4EBP1, may control the differential protein synthesis rates between basal and luminal epithelial cells. The molecular program that governs prostate epithelial cell-specific 4EBP1 expression remains a question for future investigation.
The 4EBP1-eIF4E axis is a critical driver of prostate cancer initiation and maintenance downstream of mTOR
Although in vitro work has demonstrated the importance of eIF4E activity in prostate cancer cell lines (18–20), little is known about the role of 4EBP1 in prostate cancer initiation and progression in vivo. Given the striking difference in 4EBP1 expression within the normal prostate, we next asked whether eIF4E hyperactivity is a critical driver of prostate cancer development as well as the therapeutic response or resistance of specific epithelial cell types. To directly address this question, we developed a genetic model in which eIF4E activity can be inhibited within all prostate epithelial cell types in an inducible fashion. In this model (Fig. 2A), the probasin promoter drives Cre recombinase expression (21) and prostate-specific recombination of a lox-stop-lox element, which precedes a reverse tetracycline-controlled transactivator gene (rtTA) within the Rosa26 locus (22). In the presence of the tetracycline (tet) analog doxycycline, the rtTA drives the expression of a tet responsive mutant form of 4EBP1 (TetO-4EBP1M) in which all the mTOR-sensitive phosphorylation sites have been mutated to alanine. We previously used the mTOR-insensitive mutant 4EBP1M to decrease eIF4E hyperactivation in vivo in hematologic cancers (15). Using this mouse model, we studied the effects of the TetO-4EBP1M in normal prostate epithelial cells. We induced the expression of 4EBP1M in PB-CreTg/+;Rosa-LSL-rtTAKI/KI;TetO-4EBP1M/M (herein referred to as 4EBP1M) mice at weaning before the onset of male puberty for 4–5 weeks with doxycycline (fig. S5A) (23). We verified transgene expression and found that the prostate glands of 4EBP1M-induced mice exhibited no histological differences compared to wild-type mice (Fig. 2B, fig. S5, B and C). Thus, 4EBP1M expression does not impact normal prostate gland development or histological features of a mature murine prostate.
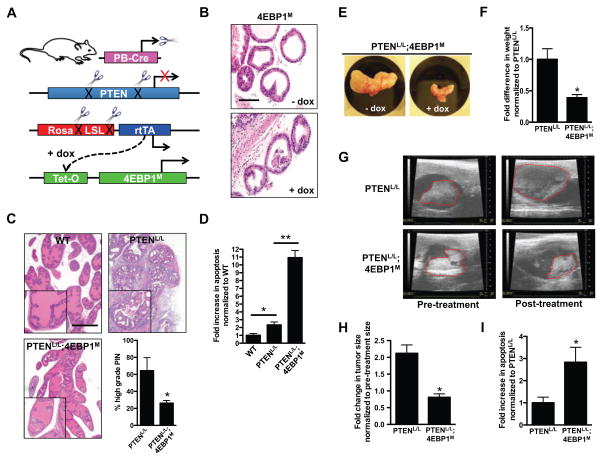
Figure 2. Inhibition of eIF4E activity does not affect wild-type prostate epithelial cell maintenance but suppresses prostate tumor initiation and progression in the setting of PTEN loss in vivo.
(A) Schematic representation of the prostate-specific and doxycycline-inducible PTENL/L;4EBP1M mouse model, in which the addition of doxycycline to the drinking water (at 2 g/L) induced the expression of the 4EBP1M transgene. (B) Representative H+E staining of ventral prostate glands from 4EBP1M mice after 4 weeks with or without doxycycline in their drinking water. Scale bar, 100 μm. (C) Representative H+E staining of wild-type (WT), PTENL/L, and PTENL/L;4EBP1M prostates after 4–5 weeks of exposure to doxycycline after weaning. Percent high grade prostatic intraepithelial neoplasia (PIN) positive glands in PTENL/L and PTENL/L;4EBP1M mice (n = 3 mice/genotype, *P = 0.03, t-test). Scale bar, 500 μm. (D) Fold change in TUNEL positive cells in WT, PTENL/L, and PTENL/L;4EBP1M prostates after 4–5 weeks of exposure to doxycycline after weaning (n = 3 mice/genotype, *P = 0.004, **P <0.0001, t-test). (E) Representative PTENL/L;4EBP1M prostate with or without exposure to doxycycline for 8 weeks starting at age 6–8 months. Mice were at 8–10 months of age at necropsy. (F) Quantification of mouse prostate weights between PTENL/L and PTENL/L;4EBP1M exposed to doxycycline for 8 weeks (n = 4–5 mice/genotype, *P = 0.02, t-test). (G) Representative ultrasounds of PTENL/L and PTENL/L;4EBP1M anterior prostates before and after 8 weeks exposure to doxycycline. (H) Quantification of tumor area in PTENL/L and PTENL/L;4EBP1M anterior prostates before and after 8 weeks exposure to doxycycline (n = 4–5 mice / genotype, *P= 0.003, t-test). (I) Fold change in TUNEL-positive (apoptotic) cells in PTENL/L and PTENL/L;4EBP1M (n = 3 mice/genotype, *P = 0.0006, t-test). Data are mean +/− S.E.M.
To genetically determine the role of the eIF4E hyperactivity toward prostate tumorigenesis and cancer progression, we developed the PB-CreTg/+;PTENL/L;Rosa-LSL-rtTAKI/KI;TetO-4EBP1M/M mouse model (here in referred to as PTENL/L;4EBP1M). In this system, loss of the PTEN tumor suppressor drives PI3K-AKT-mTOR signaling, hyperactivation of eIF4E, and subsequent development of pre-invasive prostatic intra-epithelial neoplasia (PIN) lesions by 10 weeks of age (14). Administration of doxycycline induces the expression of the 4EBP1M transgene (Fig. 2A), thereby decreasing eIF4E hyperactivation (15). Expression of 4EBP1M within the prostate did not affect the ability of mTOR to phosphorylate other downstream targets such as the p70S6K1 and p70S6K2 target ribosomal protein S6 (rpS6) (fig. S5D). PTENL/L and PTENL/L;4EBP1M mice were placed on doxycycline for 4–5 weeks immediately after weaning and analyzed for the effects on tumor initiation (fig. S5A). Loss of PTEN caused a two-fold increase in prostate size that was significantly blunted in PTENL/L;4EBP1M mice (fig. S5E). At a histological level, the 4EBP1M transgene markedly inhibited the development of PIN in PTENL/L;4EBP1M mice (Fig. 2C), which complements a previous finding that eIF4E phosphorylation is necessary for prostate cancer development (24). Next, we sought to delineate the cellular mechanism underlying the tumor suppressive effects of 4EBP1M expression. Confirming previously published work, PTENL/L mice exhibited increased baseline amounts of both proliferation and apoptosis compared to wild-type mice (Fig. 2D and fig. S5F) (14). However, inhibition of eIF4E hyperactivity substantially increased apoptosis by 5-fold (Fig. 2D), while having no effect on cell proliferation (fig. S5F), demonstrating that increased activity of eIF4E promotes a pro-survival program that is critical for tumor initiation.
Given the significant therapeutic potential of targeting eIF4E hyperactivity in established cancers, we used the inducible nature of the PTENL/L;4EBP1M mouse model to test the effect of eIF4E inhibition on tumor maintenance. PTENL/L and PTENL/L;4EBP1M mice were aged to 7–9 months, corresponding to the development of large prostate tumors that can be visualized by ultrasound. At that point, mice were imaged before and after 8 weeks of doxycycline administration (fig. S5G). By the end of the trial, PTENL/L;4EBP1M mouse prostates were 50% smaller than those of PTENL/L mice (Fig. 2, E and F). Moreover, whereas PTENL/L mice exhibited a 2-fold increase in tumor area, PTENL/L;4EBP1M tumors exhibited no growth, but instead a slight decline in tumor area (Fig. 2, G and H) which was associated with a 3-fold increase in apoptosis (Fig. 2I). Together, these findings provide in vivo evidence that eIF4E hyperactivity is critical for prostate cancer initiation and maintenance, and provide clinical precedence for therapeutically targeting upstream regulators of the translation initiation machinery, such as mTOR, in epithelial cancers.
PTENL/L luminal epithelial cells are resistant to inhibition of eIF4E activity
Our genetic studies reveal that despite the marked effect of restraining eIF4E hyperactivity on prostate cancer development, this effect is incomplete and certain prostate cancer cell populations continue to grow (Fig. 2G). This genetic effect was mirrored by what we observed after MLN0128 treatment, where a population of luminal epithelial cells was enriched after treatment (Fig. 1A). We therefore asked whether a specific population of prostate epithelial cells in PTENL/L;4EBP1M mice may be less sensitive to the expression of the 4EBP1M transgene. To address this question, we determined the change in the number of PTENL/L luminal epithelial cells compared to PTENL/L basal epithelial cells after the induction of the 4EBP1M by both immunofluorescence and FACS sorting. In the setting of PTEN loss (fig. S3, A and B), both basal and luminal epithelial cell populations increased in total number compared to wild-type cells and were characterized by increased entry into the cell-cycle (fig. S6A), demonstrating the mitogenic effects of increased PI3K-AKT-mTOR pathway activation (Fig. 3A). Strikingly, however, we found that luminal epithelial cells were specifically enriched upon inhibition of eIF4E by the 4EBP1M which phenocopied our pharmacological findings with MLN0128 (Figs. 1A and 3B; fig. S6, B to D). These findings suggest that eIF4E may be critical to transform basal epithelial cells, whereas luminal cells do not rely on increased eIF4E activation for their growth.
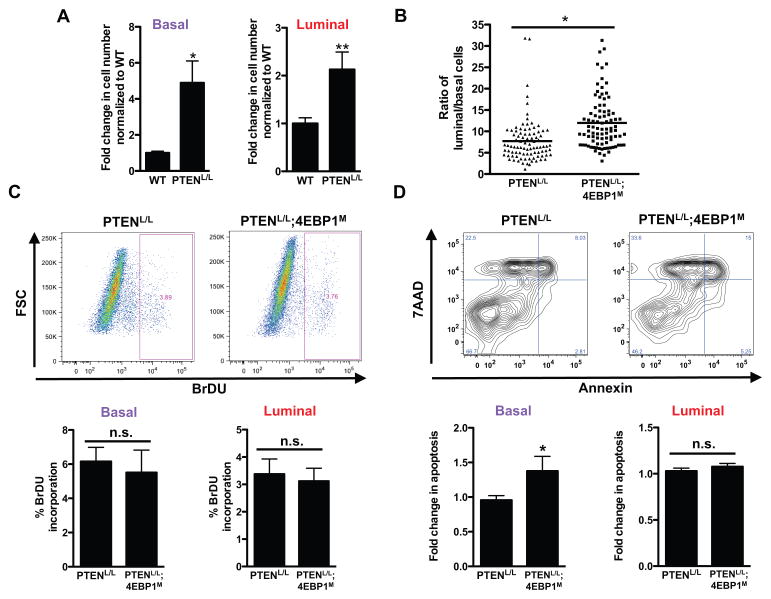
Figure 3. PTENL/L luminal epithelial cells are enriched upon expression of 4EBP1M.
(A) Fold change in basal and luminal epithelial cell numbers upon PTEN loss in wild-type (WT) and PTENL/L mice (n = 7–9 mice/genotype, *P = 0.005, **P = 0.01, t-test). (B) The ratio of CK8+ luminal epithelial cells over CK5+ basal epithelial cells in PTENL/L and PTENL/L;4EBP1M mice (n = 6 mice/condition,*P = 0.001, t-test). (C) Percent BrdU incorporation by fluorescence-activated cell sorting (FACS) of the basal and luminal epithelial cell types from mice exposed to doxycycline (administered in the drinking water at 2 g/L) at weaning for a total of 4–5 weeks. Upper panel: representative BrdU dot plot. Lower panel: quantification of percent BrdU positive basal and luminal cells in each mouse strain. FSC, forward scatter. (n = 5–6 mice/genotype, t-test). (D) 7AAD/Annexin FACS for apoptosis in basal and luminal epithelial cell types from mice exposed to the doxycycline regimen upon weaning for a total of 4–5 weeks. Upper panel: representative 7AAD/Annexin dot plot. Lower panel: quantification of 7AAD/Annexin double positive cells in basal or luminal cells in each mouse strain (n = 6–7 mice/genotype, *P = 0.03, t-test). All mice were at 8–10 weeks of age at necropsy. n.s. = not statistically significant. Data are mean +/− S.E.M.
To elucidate the cellular mechanism that enables this divergent response in basal and luminal epithelial cells to inhibition of eIF4E activity, we conducted in vivo analysis of cell proliferation and apoptosis. Induction of the TetO-4EBP1M transgene did not affect cell proliferation as determined by BrdU incorporation in any of the cell types in PTENL/L mice (Fig. 3C). However, 4EBP1M expression increased apoptosis in basal cells but did not affect luminal epithelial cells (Fig. 3D and fig. S6E). As such, despite having tumorigenic potential in the context of PTEN loss, luminal epithelial cells are not dependent on eIF4E activity for cell survival, which underscores a surprising specificity in cell identity and sensitivity to inhibition of eIF4E activity in prostate cancer. These findings are consistent with the overall marked increase in 4EBP1 transcript expression and decreased protein synthesis rates in luminal compared to basal epithelial cells. Together, these findings suggest that the high protein synthesis rates normally exhibited by prostate basal cells primes them to be more vulnerable to eIF4E down regulation in the oncogenic setting. These findings have further therapeutic implications, as cells that normally exhibit higher 4EBP1 transcript expression may be more resistant to pharmacological inhibition of the PI3K signaling pathway.
Increased 4EBP1 abundance controls the rate of protein synthesis and drives resistance to PI3K-AKT-mTOR pathway inhibition in human prostate cancer cells
To determine whether increased abundance of 4EBP1 primes cells for resistance to PI3K-AKT-mTOR pathway inhibition, we developed a human prostate cancer cell line characterized by knockdown of PTEN and overexpression of Large/Small T-antigen and hTERT in primary prostate epithelial cells (PTEN KD LHS PrEC) (fig. S7A) (25). These cells exhibit important similarities with human prostate cancer, including the expression of luminal epithelial markers and the ability to form colonies in clonigenic growth assays (fig. S7, A and B). Similar to our pharmacologic and genetic studies, these cells also exhibited a heterogeneous response to treatment with MLN0128 characterized by populations of sensitive and primary resistant cells. To determine whether 4EBP1 abundance defines the resistant cell type, single cell clones were isolated, characterized for their baseline 4EBP1 protein abundance, and treated with MLN0128 to determine the effects on cell survival by propidium iodide staining and annexin V analysis by FACS. We found that despite having similar loss of PTEN, individual clones had varying amounts of 4EBP1 mRNA and protein amounts (Fig. 4A and fig. S7C). Moreover, we observed that clones with lower 4EBP1 protein abundance were more sensitive to MLN0128 treatment, whereas those with high 4EBP1 abundance were resistant (Figs. 4, A and B). Next we sought to determine whether high 4EBP1 protein abundance primed cells for resistances to MLN0128. To this end, we knocked down 4EBP1 using pooled siRNAs to amounts comparable to those observed in the “low 4EBP1” clones (Fig. 4C). Remarkably, knockdown of 4EBP1 in resistant cells increased their sensitivity to MLN0128 (Fig. 4D). Moreover, enforced expression of wild-type 4EBP1 conferred resistance to a previously sensitive clone exhibiting low 4EBP1 abundance (figs. S7, D and E). At a molecular level, decreasing 4EBP1 mRNA and protein to amounts equivalent to those in sensitive cells appeared to increase de novo proteins synthesis (Fig. 4E) and increased the formation of the eIF4F complex (Fig. 4F). As such, high 4EBP1 abundance directly inhibited the activity of eIF4E by skewing the amount of eIF4G bound to eIF4E thereby lowering the amount of protein synthesis and conferring resistance to PI3K-AKT-mTOR pathway inhibitors. These findings implicate 4EBP1 as a marker and mediator of drug resistance.
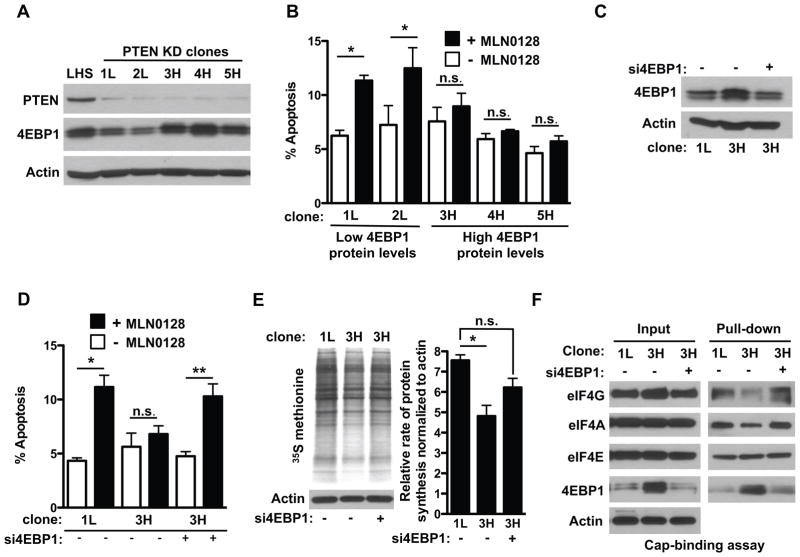
Figure 4. Increased abundance of 4EBP1 is required to maintain resistance to PI3K pathway inhibitors and is a marker of resistant cells in human prostate cancer.
(A) Representative Western blot analysis from 2 experiments for PTEN and 4EBP1 in PTEN KD LHS PrEC clones and control LHS cells. (B) Analysis of apoptosis by propidium iodide/annexin V staining in PTEN KD LHS PrEC cells after 12 hours’ exposure to MLN0128 or vehicle (–MLN0128). (n = 5 replicates in two independent experiments, *P<0.0001, **P = 0.04, t-test). (C) Representative Western blot of 4EBP1 in PTEN KD LHS PrEC cells with and without si4EBP1. (D) Analysis of apoptosis by propidium iodide/annexin V staining of PTEN-KD LHS PrEC cells after transfection with a 4EBP1-targeted siRNA pool (n = 8 replicates in two independent experiments *P<0.0001, **P = 0.005, t-test). (E) Representative autoradiograph (left) and quantification of S35-methionine incorporation assays in PTEN KD LHS PrEC clones upon silencing 4EBP1. (n = 3 independent experiments, *P < 0.05, ANOVA). (F) Representative cap-binding assay in PTEN knockdown LHS clones upon silencing 4EBP1. n.s. = not statistically significant. Data are mean ± S.E.M.
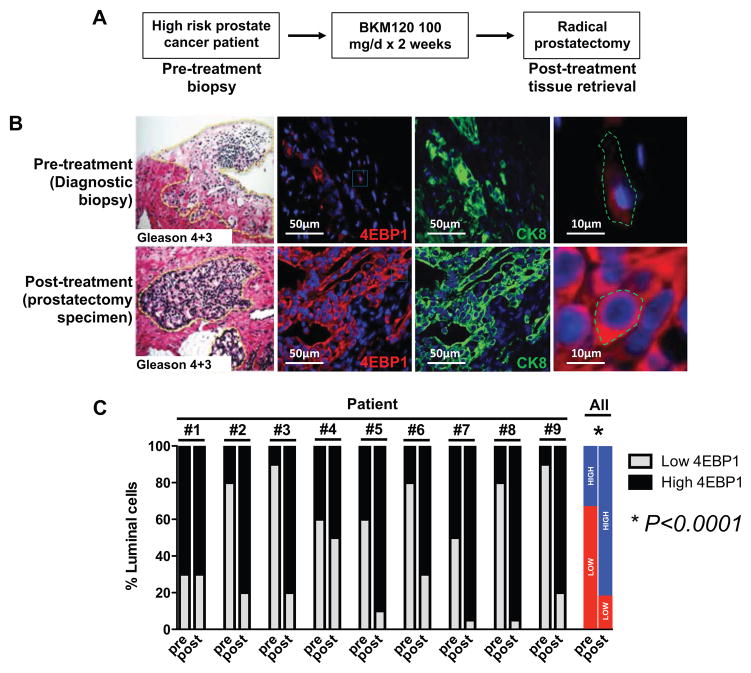
High 4EBP1 abundance is associated with resistance to PI3K inhibitors in prostate cancer patients
Next we sought to determine whether increased 4EBP1 abundance is a common feature of resistance to inhibitors of the PI3K-AKT-mTOR signaling pathway in prostate cancer patients. To this end, tissues obtained from prostate cancer patients enrolled in an ongoing, phase II clinical trial with the oral pan-PI3K inhibitor buparlisib (BKM120) were used to determine total abundance of 4EBP1 before and after PI3K-AKT-mTOR pathway inhibition (www.clinicaltrials.gov, NCT01695473). In this neoadjuvant trial, newly diagnosed prostate cancer patients are treated with buparlisib 100mg daily for 14 days prior to radial prostatectomy (Fig. 5A). Paired diagnostic prostate core biopsy and radical prostatectomy tissue specimens were collected from each patient for pharmacodynamic evaluation. PI3K pathway inhibition was confirmed by phosphorylated AKT immunohistochemistry in nearly all (7 out of 9) patients (fig. S8A). The two patients who did not exhibit significant differences had very low baseline phosphorylated AKT abundance in the pre-treatment setting (fig. S8A). Prostate specific antigen (PSA) is a biomarker commonly employed to monitor treatment response or disease progression. We found that no patients experienced an objective PSA response, defined as a >30% decrease in PSA serum concentration, to BKM120 during the course of the trial (fig. S8B), and this was associated with a significant enrichment of high-4EBP1-expressing luminal epithelial cells (CK8+) post-treatment compared to pre-treatment specimens (Figs. 5, B and C). Thus, increased 4EBP1 abundance may characterize a specific epithelial cell type with intrinsic resistance to PI3K-AKT-mTOR pathway inhibition in prostate cancer patients.
Figure 5. Increased 4EBP1 abundance is associated with drug resistance in prostate cancer patients.
(A) Schematic of the phase 2 neoadjuvant BKM120 clinical trial conducted at UCSF. (B) Representative H+E, CK8 and 4EBP1 immunofluorescence images of a patient tumor before and after treatment with the PI3K inhibitor BKM120. Far right: Magnified insets from the 4EBP1 images. Scale bars, 50 μm and 10 μm respectively. (C) Quantification of the percentage of luminal epithelial cells with low or high abundance of 4EBP1 for each patient as well as the average for all of the patients before (pre) and after (post) treatment with BKM120 (*P<0.0001, t-test).
Discussion
The PI3K pathway drives tumor growth in prostate cancer, but resistance is often observed to PI3K pathway inhibitors. We found a cell type-specific intrinsic resistance mechanism to PI3K pathway inhibitors in luminal epithelial prostate cells that was mediated by increased abundance of the translation repressor 4EBP1. Compared to luminal cells, basal epithelial cells in the prostate had considerably lower mRNA and protein abundance of 4EBP1, greater protein synthesis activity, and increased MLN0128 sensitivity. Thus, contrary to the expected outcome of increased PI3K signaling resulting in equivalent eIF4E activation across all cell prostate epithelial types, our data uncovered cell-type specificity in 4EBP1 transcript abundance that primes cells for drug sensitivity or resistance. In the future, it will be interesting to determine the molecular basis for the increased 4EBP1 expression in luminal epithelial cells compared to basal epithelial cells and whether these differences arise from transcription factor promoter binding (26, 27), chromatin remodeling, or mRNA degradation rates.
The clinical implication of these findings is the in vivo realization of a critical mode of resistance to PI3K pathway inhibitors other than those that have been previously proposed. For example, direct mutations of PI3K pathway components have been described to confer resistance to PI3K–mTOR inhibitors in prostate cancer (28), as have parallel mitogenic signaling pathways driven by the androgen receptor (29, 30). In addition to pathway-mediated mechanisms of resistance, the Brugge laboratory has demonstrated that the physical location of a cancer cell within a 3-dimensional culture can determine its sensitivity to PI3K pathway inhibitors (31). However, unlike these observations, which demonstrate adaptive resistance mechanisms, our findings illustrate the central importance of cell identity dictated by the status of the 4EBP1-eIF4E axis and protein synthesis rates to determine intrinsic resistance to PI3K-AKT-mTOR inhibition. This may be a cellular mechanism akin to oncogene addiction where the genetic make-up of the cell dictates its drug sensitivities (32).
Interestingly, loss of 4EBP1 or increased eIF4E, which is associated with increased protein synthesis, has been previously shown to confer drug resistant phenotypes in cell culture and xenografts (33–37). In the context of our in vivo studies, it is therefore intriguing to speculate as to why cancer-prone cells with high 4EBP1 abundance associated with lower protein synthesis rates escape target inhibition. Specific cell types, such as luminal epithelial cells with high 4EBP1 abundance, may harbor more quiescent features associated with lower protein synthesis and therefore may be less sensitive to therapeutic agents that impinge on protein synthesis control coupled to growth and survival. Together, our findings along with previously published reports (33–37) suggest that there is both a lower and upper threshold of protein synthesis rates that can prime cells for drug resistance. Thus, 4EBP1 abundance, which our study here indicated is cell type-specific (in the prostate) and correlated with mRNA translation, is a distinguishing factor that might be used to predict sensitivity or resistance to PI3K pathway inhibitors in patients.
Methods
Mice
PTENL/Land Rosa-LSL-rtTAKI/KI mice where obtained from Jackson Laboratories. PB-CreTg/+ mice were obtained from the Mouse Models of Human Cancers Consortium (MMHCC). All mice were maintained in the C57BL/6 background. The TetO-4EBP1M/M was generated as previously described (15). Mice were maintained under specific pathogen-free conditions, and experiments were performed in compliance with institutional guidelines as approved by the Institutional Animal Care and Use Committee of UCSF. Doxycycline (Sigma) was administered in the drinking water at 2 g/L.
Cell lines and reagents
The LHS PrEC cells were graciously provided by Dr. Phil Febbo (UCSF) and were cultured as previously described (25). A PTEN-targeted shRNA sequence 5′GACTTAGACTTGACCTATATT 3′ (Broad: TRCN0000355842) was cloned into the pLKO.1 vector and overexpressed using standard lentiviral packaging constructs. The PTEN shRNA virus was infected into LHS PrEC cells. Single cell clones were selected using trypsin soaked sterile cloning discs (Sigma). MLN0128 was generously provided by Dr. Kevan Shokat (UCSF) and used at 1 mg/mL in the preclinical trial and at 100 nM in the cell lines. 4EBP1 siRNAs were obtained from Thermo Scientific (ON-TARGET plus SMARTpool Human eIF4EBP1 siRNA: Cat# L-003005-00). Lipofectamine 2000 (Life Technologies) was used to transfect cancer cell lines with siRNA. The wild-type V5-tagged 4EBP1 construct (pLX304-4EBP1-V5) was generously provided by Dr. Patrick Paddison (Fred Hutchinson Cancer Research Center).
Clonigenic assay
In brief, five thousand cells PTEN-deficient LHS cells were plated on to a 35-mm six-well plate. The covering medium was changed every 2 or 3 days during culture. After 14 days, the cells were fixed and stained with crystal violet solution (10% acetic acid, 10% ethanol and 0.06% crystal) and then visualized. Colonies were enumerated and averaged over three independent experiments.
Prostate tissue processing
Whole mouse prostates were removed from wild-type (WT) and PTENL/L mice, microdissected, and frozen in liquid nitrogen. Frozen tissues were subsequently manually disassociated using a biopulverizer (Biospec) and additionally processed for protein and mRNA analysis as described below.
Prostate epithelium single cell dissociation
Wild-type, PTENL/L and PTENL/L;4EBP1M mouse prostates were retrieved, microdissected, and minced using a scalpel. Tissue chunks were incubated in 1 mg/mL Collagenase I (Life Technologies) in DMEM, 10% FBS, L-glutamine, and pen/strep for 1–2 hours at 37°C. This was followed by further dissociation with 0.05% trypsin, which was quenched using fully supplemented DMEM and 500 units of DNase (Roche). Cells were manually dissociated with an 18g followed by a 20g needle and syringe. Cells were counted using a hemocytometer.
Western blot analysis
Western blot analysis was performed as previously described (15) with antibodies specific to eIF4A (Cell Signaling), eIF4G (Cell Signaling), eIF4E (BD Biosciences), PTEN (Cell Signaling), 4EBP1 (Cell Signaling), phosphorylated 4EBP1 Thr37/46 (Cell Signaling), AKT (Cell Signaling), phosphorylated AKT Ser473 (Cell Signaling), rpS6 (Cell Signaling), phosphorylated rpS6 Ser240/244 (Cell Signaling), PTEN (Cell Signaling), GAPDH (Cell Signaling) and β-actin (Sigma), phosphorylated eEF2 Thr56 (Cell Signaling), eEF2 (Cell Signaling), PDCD4 (Cell Signaling), phosphorylated eIF2α Ser51 (Cell Signaling), eIF2α (Cell Signaling), 4EBP2 (Cell Signaling). Densitometry analysis was completed using Image J (http://imagej.nih.gov/ij/).
Quantitative PCR analysis
RNA was isolated using the manufacturer’s protocol for RNA extraction with TRIzol Reagent (Invitrogen) using the Pure Link RNA mini kit (Invitrogen). RNA was Dnase-treated with Pure Link Dnase (Invitrogen). Dnase- treated RNA was transcribed to cDNA with SuperScript III First-Strand Synthesis System for RT–PCR (Invitrogen), and 1 ml of cDNA was used to run a SYBR green detection qPCR assay (SYBR Green Supermix and MyiQ2, Biorad). Primers were used at 200 nM. Oligomer sequences are in table S1.
Fluorescence-activated cell sorting of distinct prostate epithelial populations
Live prostate epithelial cells were counted and labeled with CD49f-PE (eBioscience), Sca-1-PE-Cy7 (BioLegend), CD31-eFluor450 (eBioscience), CD45-eFluor450 (eBioscience), Ter119-eFluor450 (eBioscience). Data was acquired using a BD FACS Canto (BD Biosciences) and analyzed with FlowJo (v.9.4.10). In order to calculate the absolute number of cells per populations we multiplied the percentage of each cell populations with the total number of cells. To sort the basal and luminal populations, dissociated epithelial cells were stained as above and sorted on a BD FACS Aria III (BD Biosciences).
Prostate immunofluorescence and analysis
Prostates were dissected from mice and fixed in 10% formalin overnight at 4 °C. Tissues were subsequently dehydrated in ethanol (Sigma) at room temperature, mounted into paraffin blocks, and sectioned at 5 mm. Specimens were de-paraffinized and rehydrated using CitriSolv (Fisher) followed by serial ethanol washes. Antigen unmasking was performed on each section using Citrate pH 6 (Vector Labs) in a pressure cooker at 125 °C for 10–30 min. Sections were washed in distilled water followed by two washes in TBS. The sections were then incubated in 5% goat serum, 1% BSA in TBS for 1 h at room temperature. Various primary antibodies were used including those specific for cytokeratin 5 (Covance), cytokeratin 8 (Abcam), total 4EBP1 (Cell Signaling), phosphorylated 4EBP1 Thr37/46 (Cell Signaling), which were diluted in blocking solution and incubated on sections overnight at 4 °C. Specimens were then washed in TBS and incubated with the appropriate Alexa 488 and 594 labeled secondary (Invitrogen) at 1:500 for 2 h at room temperature. A final set of washes in TBS was completed at room temperature followed by mounting with DAPI Hardset Mounting Medium (Vector Lab). A Zeiss AxioImager M1 was used to image the sections. Individual prostate epithelial cells and cancer cells were quantified for mean fluorescence intensity (m.f.i.) using the Axiovision (Zeiss, Release 4.8) densitometric tool. To determine the ratio of luminal epithelial cells to basal epithelial cells, 10–15 images were taken for each pharmacologic or genetic manipulation and analyzed. Specifically, a mask for CK5+ basal epithelial cells and CK8+ luminal epithelial cells was drawn for each image and an area was calculated thereby providing a density of each cell type (ImageJ). To calculate the ratio, luminal epithelial areas were divided by basal epithelial areas and graphed (Graphpad Inc.)
Hematoxylin and eosin staining
Paraffin-embedded prostate specimens were deparaffinized and rehydrated as described above (see immunofluorescence section), stained with haematoxylin (Thermo Scientific), and washed with water. This was followed by a brief incubation in differentiation RTU (VWR) and two washes with water followed by two 70% ethanol washes. The samples were then stained with eosin (Thermo Scientific) and dehydrated with ethanol followed by CitriSolv (Fisher). Slides were mounted with Cytoseal XYL (Richard Allan Scientific).
Apoptosis analysis
TUNEL staining of OCT-embedded prostate was conducted per manufacturer’s protocol (Roche). Apoptosis analysis of live prostate epithelial cells was conducted by first labeling single cell prostate isolates with CD49f-PE (eBioscience), Sca-1-PE-Cy7 (BioLegend), CD31-eFluor450 (eBioscience), CD45-eFluor450 (eBioscience), Ter119-eFluor450 (eBioscience). This was followed by labeling with Annexin V-APC (BD Pharmingen) and 7-AAD (BD Pharmingen) following the manufacturer’s instructions (BD Biosciences). Data was acquired using a BD FACS Canto (BD Biosciences) and analyzed with FlowJo (v.9.4.10).
Cell proliferation analysis by BrdU incorporation in vivo
200 μL of 10 mg/mL BrdU (BD Pharmingen) was administered by intraperitoneal injection in mice 48 and 24 hours before they were euthanized. Prostates were retrieved and dissociated to single cells as above. Staining for BrdU was conducted using the manufacturer’s protocol (BD Pharmingen). In short, cells were labeled with CD49f-PE (eBioscience), Sca-1-PE-Cy7 (BioLegend), CD31-eFluor450 (eBioscience), CD45-eFluor450 (eBioscience), Ter119-eFluor450 (eBioscience) subsequently fixed an permeabilized with BD Cytofix/Cytopern buffer. Cells were treated with DNAse to expose the BrdU epitopes, and were stained with APC-anti-BrdU antibody. Data was acquired using a BD FACS Canto (BD Biosciences) and analyzed with FlowJo (v.9.4.10).
Ultrasound imaging of the mouse prostate
A Vevo 770 Ultrasound Imaging System (VisualSonic) was used to image PTENL/L and PTENL/L;4EBP1M anterior prostates before and after doxycycline treatment. Areas of interested were measured by assessing the largest area (height and width) of each mass within the anterior prostate.
Ex vivo [35S]-methionine labeling in primary prostate epithelial cells
Primary basal and luminal epithelial cells from wild-type and PTENL/L mice were dissociated and sorted as described above. For each cell type, 40,000 cells were incubated with 33 mCi of [35S]-methionine for 1.5 hrs in methionine-free DMEM (Invitrogen) plus 10% dialyzed FBS and L-glutamine. Cells were prepared using a standard protein lysate protocol, resolved on a 10% SDS polyacrylamide gel and transferred onto a PVDF membrane (BioRad). The membrane was exposed to autoradiography film (Denville) for 24 h and developed. Densitometry analysis was completed using Image J (http://imagej.nih.gov/ij/). For the LHS PTEN KD PrEC cells, 250,000 cells were plated and incubated as described above.
Neoadjuvant BKM120 prostate cancer clinical trial
A phase II prospective pharmacodynamic study of buparlisib (BKM120) (38), an oral pan-class PI3K inhibitor, in patients with high-risk localized prostate cancer was conducted at UCSF (NCT01695473). The primary study objective was to determine the proportion of men with downstream target inhibition of PI3K in prostate tumor tissue, as measured by immunohistochemistry. Eligible subjects had localized adenocarcinoma of the prostate that were candidates for and had selected radical prostatectomy as the primary treatment. Subjects had high-risk disease, defined as Gleason ≥8 and ≥2 discrete core biopsies containing ≥20% cancer, or Gleason 4+3 and ≥50% of core biopsies containing cancer. Patients were required to have adequate diagnostic core biopsy specimens for pharmacodynamic evaluation; patients who did not have adequate specimens for evaluation were required to undergo a pre-treatment biopsy for tissue acquisition. Patients received buparlisib 100 mg daily for the 14 days preceding radical prostatectomy, with the final dose taken on the night prior to surgery. The study was conducted in accordance with International Conference on Harmonization good clinical practice standards, with approval by the institutional review board at UCSF. All patients provided written informed consent to participate.
Cap-binding assay
Cells were lysed in buffer A (10mM Tris-HCL pH 7.6, 140mM KCl, 4mM MgCl2, 1mM DTT, 1mM EDTA and protease inhibitors) supplemented with 1% Nonidet P-40) and cell lysates (250 μg protein in 500 μl) were incubated overnight at 4 °C with 50 μl of the mRNA cap analogue m7GTP-sepharose (Jena Bioscience) in buffer A, under constant and gentle agitation. The protein complex-sepharose beads were washed with buffer A supplemented with 0.5 % NP-40 and the eIF4E-associated complex was resolved by SDS-PAGE and Western blotting.
Supplementary Material
Figure S1. Absolute quantification of basal epithelial cells and luminal epithelial cells after treatment with MLN0128.
Figure S2. qPCR phenotyping of distinct sorted epithelial cell populations in wild-type and PTENL/L mice.
Figure S3. Efficiency of PTEN deletion and the phosphorylation and localization of 4EBP1 in basal and luminal epithelial cells in vivo.
Figure S4. Western blot analysis of translation initiation factors and regulators in wild-type and PTENL/L basal and luminal epithelial cells.
Figure S5. 4EBP1M does not impact normal prostate homeostasis but impedes prostate cancer initiation and progression.
Figure S6. Effect of PTEN loss and 4EBP1M expression on absolute number of basal and luminal epithelial cells.
Figure S7. Expression of 4EBP1 endows resistance to MLN0128 in LHS PTEN knockdown cell lines.
Figure S8. Phosphorylated AKT (Ser473) immunohistochemistry of prostate tumors and serum PSA concentrations from patients before and after treatment with BKM120.
Table S1: qPCR oligonucleotide sequences.
Acknowledgments
We thank M. Barna for critical reading of the manuscript and J. Shen for editing; B. Hann of the UCSF Preclinical Therapeutics Core for technical advice regarding ultrasound imaging; J. Gordon of the UCSF Laboratory for Cell Analysis for technical advice with fluorescence activated cell sorting; M. Meng, M. Cooperberg for enrolling patients onto the BKM120 trial; A. Foye, A. Toschi, and J. Youngren for BKM120 clinical trial tissue acquisition, Goldberg-Benioff Program in Cancer Translational Biology for their support and commitment to prostate cancer research. L. Wang and Y. Liu for technical support. We also thank P. Paddison (Fred Hutchinson Cancer Research Center) for gifting the pLX304-4EBP1-V5 construct; P. Febbo (UCSF) for the LHS PrEC cells; and K. Shokat (UCSF) for MLN0128.
Funding: A.C.H. is supported by a Burroughs Wellcome Fund Career Award for Medical Scientists, by the Prostate Cancer Foundation, and by NIH 1K08CA175154. A.C.H. is a V Foundation for Cancer Research Scholar. D.R. is supported by NIH 1R01CA154916, 1R01CA184624, and a Stand Up to Cancer/Prostate Cancer Foundation Prostate Dream Team Translational Cancer Research Grant. This research grant is made possible by the generous support of the Movember Foundation. Stand Up to Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research. D.R. is a Leukemia and Lymphoma Society Research Scholar.
Footnotes
Author contributions: A.C.H and D.R. conceived of and designed all experiments. A.C.H., L.W., H.G.N., and M.P.E. conducted all experiments. W.K. was the lead investigator for the BKM120 Phase II clinical trial. P.R.C. provided prostate cancer specimens through the BKM120 clinical trial. A.C.H. and D.R. analyzed the data and wrote the manuscript. All authors discussed results and edited the manuscript.
Competing interests: The authors declare they have no competing interests.
References and Notes
- 1.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, Antipin Y, Mitsiades N, Landers T, Dolgalev I, Major JE, Wilson M, Socci ND, Lash AE, Heguy A, Eastham JA, Scher HI, Reuter VE, Scardino PT, Sander C, Sawyers CL, Gerald WL. Integrative genomic profiling of human prostate cancer. Cancer cell. 18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi N, Zhang B, Zhang L, Ittmann M, Xin L. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer cell. 2012;21:253–265. doi: 10.1016/j.ccr.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stoyanova T, Cooper AR, Drake JM, Liu X, Armstrong AJ, Pienta KJ, Zhang H, Kohn DB, Huang J, Witte ON, Goldstein AS. Prostate cancer originating in basal cells progresses to adenocarcinoma propagated by luminal-like cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:20111–20116. doi: 10.1073/pnas.1320565110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, Julio MK, Economides KD, Walker D, Yu H, Halili MV, Hu YP, Price SM, Abate-Shen C, Shen MM. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009 doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gingras AC, Raught B, Gygi SP, Niedzwiecka A, Miron M, Burley SK, Polakiewicz RD, Wyslouch-Cieszynska A, Aebersold R, Sonenberg N. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes & development. 2001;15:2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gingras AC, Kennedy SG, O’Leary MA, Sonenberg N, Hay N. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes & development. 1998;12:502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, Wang S, Ren P, Martin M, Jessen K, Feldman ME, Weissman JS, Shokat KM, Rommel C, Ruggero D. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012 doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, Pandolfi P. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nature medicine. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- 9.Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 10.Haghighat A, Sonenberg N. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. The Journal of biological chemistry. 1997;272:21677–21680. doi: 10.1074/jbc.272.35.21677. [DOI] [PubMed] [Google Scholar]
- 11.Rogers GW, Jr, Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. The Journal of biological chemistry. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 12.Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 13.Edlind MP, Hsieh AC. PI3K-AKT-mTOR signaling in prostate cancer progression and androgen deprivation therapy resistance. Asian journal of andrology. 2014;16:378–386. doi: 10.4103/1008-682X.122876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S, Gao J, Lei Q, Rozengurt N, Pritchard C, Jiao J, Thomas GV, Li G, Roy-Burman P, Nelson PS, Liu X, Wu H. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer cell. 2003;4:209–221. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh AC, Costa M, Zollo O, Davis C, Feldman M, Testa JR, Meyuhas O, Shokat K, Ruggero D. Genetic Dissection of the Oncogenic mTOR Pathway Reveals Druggable Addiction to Translational Control via 4EBP-eIF4E. Cancer cell. 2010;17:249–261. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawson D, Zong Y, Memarzadeh S, Xin L, Huang J, Witte O. Basal epithelial stem cells are efficient targets for prostate cancer initiation. Proceedings of the National Academy of Sciences. 2010;107:2610–2615. doi: 10.1073/pnas.0913873107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T, Dor Y, Zisman P, Meyuhas O. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes & development. 2005;19:2199–2211. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graff JR, Konicek BW, Lynch RL, Dumstorf CA, Dowless MS, McNulty AM, Parsons SH, Brail LH, Colligan BM, Koop JW, Hurst BM, Deddens JA, Neubauer BL, Stancato LF, Carter HW, Douglass LE, Carter JH. eIF4E activation is commonly elevated in advanced human prostate cancers and significantly related to reduced patient survival. Cancer research. 2009;69:3866–3873. doi: 10.1158/0008-5472.CAN-08-3472. [DOI] [PubMed] [Google Scholar]
- 19.Andrieu C, Taieb D, Baylot V, Ettinger S, Soubeyran P, De-Thonel A, Nelson C, Garrido C, So A, Fazli L, Bladou F, Gleave M, Iovanna JL, Rocchi P. Heat shock protein 27 confers resistance to androgen ablation and chemotherapy in prostate cancer cells through eIF4E. Oncogene. 2010;29:1883–1896. doi: 10.1038/onc.2009.479. [DOI] [PubMed] [Google Scholar]
- 20.Morad SA, Schmid M, Buchele B, Siehl HU, El Gafaary M, Lunov O, Syrovets T, Simmet T. A novel semisynthetic inhibitor of the FRB domain of mammalian target of rapamycin blocks proliferation and triggers apoptosis in chemoresistant prostate cancer cells. Molecular pharmacology. 2013;83:531–541. doi: 10.1124/mol.112.081349. [DOI] [PubMed] [Google Scholar]
- 21.Wu X, Wu J, Huang J, Powell WC, Zhang J, Matusik RJ, Sangiorgi FO, Maxson RE, Sucov HM, Roy-Burman P. Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissue-specific gene ablation. Mechanisms of development. 2001;101:61–69. doi: 10.1016/s0925-4773(00)00551-7. [DOI] [PubMed] [Google Scholar]
- 22.Belteki G, Haigh J, Kabacs N, Haigh K, Sison K, Costantini F, Whitsett J, Quaggin SE, Nagy A. Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic acids research. 2005;33:e51. doi: 10.1093/nar/gni051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinter O, Beda Z, Casaba Z, GI Differences in the onset of puberty in selected inbred mouse strains. Endocrine Abstracts. 2007;14:617. [Google Scholar]
- 24.Furic L, Rong L, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, Petroulakis E, Robichaud N, Pollak M, Gaboury LA, Pandolfi PP, Saad F, Sonenberg N. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proceedings of the National Academy of Sciences of the United States of America. 107:14134–14139. doi: 10.1073/pnas.1005320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berger R, Febbo PG, Majumder PK, Zhao JJ, Mukherjee S, Signoretti S, Campbell KT, Sellers WR, Roberts TM, Loda M, Golub TR, Hahn WC. Androgen-induced differentiation and tumorigenicity of human prostate epithelial cells. Cancer research. 2004;64:8867–8875. doi: 10.1158/0008-5472.CAN-04-2938. [DOI] [PubMed] [Google Scholar]
- 26.Azar R, Alard A, Susini C, Bousquet C, Pyronnet S. 4E-BP1 is a target of Smad4 essential for TGFbeta-mediated inhibition of cell proliferation. The EMBO journal. 2009;28:3514–3522. doi: 10.1038/emboj.2009.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Azar R, Lasfargues C, Bousquet C, Pyronnet S. Contribution of HIF-1alpha in 4E-BP1 gene expression. Molecular cancer research : MCR. 2013;11:54–61. doi: 10.1158/1541-7786.MCR-12-0095. [DOI] [PubMed] [Google Scholar]
- 28.Zunder ER, Knight ZA, Houseman BT, Apsel B, Shokat KM. Discovery of drug-resistant and drug-sensitizing mutations in the oncogenic PI3K isoform p110 alpha. Cancer cell. 2008;14:180–192. doi: 10.1016/j.ccr.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, Arora VK, Le C, Koutcher J, Scher H, Scardino PT, Rosen N, Sawyers CL. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer cell. 2011;19:575–586. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mulholland DJ, Tran LM, Li Y, Cai H, Morim A, Wang S, Plaisier S, Garraway IP, Huang J, Graeber TG, Wu H. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer cell. 2011;19:792–804. doi: 10.1016/j.ccr.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muranen T, Selfors LM, Worster DT, Iwanicki MP, Song L, Morales FC, Gao S, Mills GB, Brugge JS. Inhibition of PI3K/mTOR leads to adaptive resistance in matrix-attached cancer cells. Cancer cell. 2012;21:227–239. doi: 10.1016/j.ccr.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science (New York, NY. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 33.Boussemart L, Malka-Mahieu H, Girault I, Allard D, Hemmingsson O, Tomasic G, Thomas M, Basmadjian C, Ribeiro N, Thuaud F, Mateus C, Routier E, Kamsu-Kom N, Agoussi S, Eggermont AM, Desaubry L, Robert C, Vagner S. eIF4F is a nexus of resistance to anti-BRAF and anti-MEK cancer therapies. Nature. 2014 doi: 10.1038/nature13572. [DOI] [PubMed] [Google Scholar]
- 34.Mallya S, Fitch BA, Lee JS, So L, Janes MR, Fruman DA. Resistance to mTOR kinase inhibitors in lymphoma cells lacking 4EBP1. PloS one. 2014;9:e88865. doi: 10.1371/journal.pone.0088865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cope CL, Gilley R, Balmanno K, Sale MJ, Howarth KD, Hampson M, Smith PD, Guichard SM, Cook SJ. Adaptation to mTOR kinase inhibitors by amplification of eIF4E to maintain cap-dependent translation. Journal of cell science. 2014;127:788–800. doi: 10.1242/jcs.137588. [DOI] [PubMed] [Google Scholar]
- 36.Alain T, Morita M, Fonseca BD, Yanagiya A, Siddiqui N, Bhat M, Zammit D, Marcus V, Metrakos P, Voyer LA, Gandin V, Liu Y, Topisirovic I, Sonenberg N. eIF4E/4E-BP ratio predicts the efficacy of mTOR targeted therapies. Cancer research. 2012;72:6468–6476. doi: 10.1158/0008-5472.CAN-12-2395. [DOI] [PubMed] [Google Scholar]
- 37.Ilic N, Utermark T, Widlund HR, Roberts TM. PI3K-targeted therapy can be evaded by gene amplification along the MYC-eukaryotic translation initiation factor 4E (eIF4E) axis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E699–708. doi: 10.1073/pnas.1108237108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bendell JC, Rodon J, Burris HA, de Jonge M, Verweij J, Birle D, Demanse D, De Buck SS, Ru QC, Peters M, Goldbrunner M, Baselga J. Phase I, dose-escalation study of BKM120, an oral pan-Class I PI3K inhibitor, in patients with advanced solid tumors. J Clin Oncol. 2012;30:282–290. doi: 10.1200/JCO.2011.36.1360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Absolute quantification of basal epithelial cells and luminal epithelial cells after treatment with MLN0128.
Figure S2. qPCR phenotyping of distinct sorted epithelial cell populations in wild-type and PTENL/L mice.
Figure S3. Efficiency of PTEN deletion and the phosphorylation and localization of 4EBP1 in basal and luminal epithelial cells in vivo.
Figure S4. Western blot analysis of translation initiation factors and regulators in wild-type and PTENL/L basal and luminal epithelial cells.
Figure S5. 4EBP1M does not impact normal prostate homeostasis but impedes prostate cancer initiation and progression.
Figure S6. Effect of PTEN loss and 4EBP1M expression on absolute number of basal and luminal epithelial cells.
Figure S7. Expression of 4EBP1 endows resistance to MLN0128 in LHS PTEN knockdown cell lines.
Figure S8. Phosphorylated AKT (Ser473) immunohistochemistry of prostate tumors and serum PSA concentrations from patients before and after treatment with BKM120.
Table S1: qPCR oligonucleotide sequences.