Abstract
The nervous system consists of neurons and glial cells. Neurons generate and propagate electrical and chemical signals, whereas glia function mainly to modulate neuron function and signaling. Just as there are many different kinds of neurons with different roles, there are also many types of glia that perform diverse functions. For example, glia make myelin; modulate synapse formation, function, and elimination; regulate blood flow and metabolism; and maintain ionic and water homeostasis to name only a few. Although proteomic approaches have been used extensively to understand neurons, the same cannot be said for glia. Importantly, like neurons, glial cells have unique protein compositions that reflect their diverse functions, and these compositions can change depending on activity or disease. Here, I discuss the major classes and functions of glial cells in the central and peripheral nervous systems. I describe proteomic approaches that have been used to investigate glial cell function and composition and the experimental limitations faced by investigators working with glia.
The nervous system is composed of neurons and glial cells that function together to create complex behaviors. Traditionally, glia have been considered to be merely passive contributors to brain function, resulting in a pronounced neurocentric bias among neuroscientists. Some of this bias reflects a paucity of knowledge and tools available to study glia. However, this view is rapidly changing as new tools, model systems (culture and genetic), and technologies have permitted investigators to show that glia actively sculpt and modulate neuronal properties and functions in many ways. Glia have been thought to outnumber neurons by 10:1, although more recent studies suggest the ratio in the human brain is closer to 1:1 with region-specific differences (1). There are many different types of glia, some of which are specific to the central nervous system (CNS),1 whereas others are found only in the peripheral nervous system (PNS). The main types of CNS glia include astrocytes, oligodendrocytes, ependymal cells, radial glia, and microglia. In the PNS, the main glial cells are Schwann cells, satellite cells, and enteric glia. These cells differ and are classified according to their morphologies, distinct anatomical locations in the nervous system, functions, developmental origins, and unique molecular compositions. Among the different classes of glia there are additional subclasses that reflect further degrees of specialization. In this review, I will discuss the characteristics and functions of the major glial cell types including astrocytes, microglia, and the myelin-forming oligodendrocytes (CNS) and Schwann cells (PNS). Because of space limitations, it is impossible to give a complete accounting of all glia and what is known about each of these cell types. Therefore, I encourage the interested reader to refer to some of the many excellent reviews referenced below that focus on individual glial cell types. Finally, I will discuss proteomic studies of glial cell function and some of the unique challenges investigators face when working with these cells.
ASTROCYTES
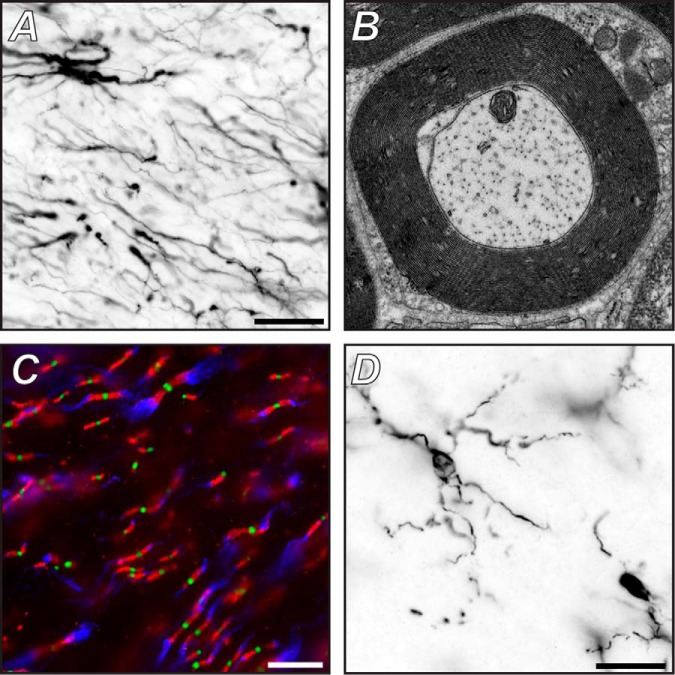
Astrocytes and neurons are derived from a common neuroepithelial precursor. These precursors undergo a gliogenic switch that depends on unique transcription factors regulated by both spatial and temporal elements. It has even been suggested that the spatial and temporal regulation of astrocyte development results in astrocyte heterogeneity, which in turn contributes to overall brain patterning (2). Astrocytes are the most numerous glial cell type in the central nervous system. In the cortex, most astrocytes are highly ramified with very fine processes that together define the domain of an individual astrocyte. In the healthy brain, adjacent astrocytes do not overlap, resulting in a “tiled” brain structure defined by astrocyte territory (3). Astrocytes are commonly identified in immunohistological experiments using antibodies against glial fibrillary acidic protein (Fig. 1A). More recently, methodologies for genetically labeling astrocytes using astrocyte-specific drivers (e.g. glial fibrillary acidic protein and Aldh1L1) have become available (4, 5) and have provided new insights into astrocyte development, structure, and function. These tools may also prove to be extremely useful in the application of proteomic approaches to understanding astrocyte function. The very fine processes of astrocytes form subcellular specializations that surround or contact neurons, especially at synapses, or endfeet that contact vasculature. Current estimates suggest that each astrocyte associates with about four neurons, 105 synapses, and one to two capillaries (6).
Fig. 1.
Glial cells and their specializations. A, astrocytes and their major processes immunolabeled using antibodies against glial fibrillary acidic protein. Scale bar, 20 μm. B, transmission electron micrograph of a myelin sheath made by a Schwann cell in the peripheral nervous system. C, central nervous system nodes (green), paranodes (red), and juxtaparanodes (blue) immunolabeled using antibodies against Na+ channels, contactin-associated protein, and Kv1.2 K+ channels, respectively. Scale bar, 10 μm. D, microglia and their branches immunolabeled using antibodies against IBA1. Scale bar, 20 μm.
The Neurovascular Unit
Astrocytes are essential elements of neural circuits because they are responsible for regulation of local blood flow as part of the neurovascular unit. Neuronal activity, reflected in the release of neurotransmitters from neurons, activates receptors on astrocytes. This in turn triggers a Ca2+ response that is thought to promote release of vasoactive substances in the astrocytic endfeet that contact blood vessels (7, 8). Astrocytes are proposed to not only promote increased blood flow and local oxygenation but also to take up glucose from the blood, convert it to lactate, and then release it to neurons for local energy use (9). Thus, astrocytes couple the demands of neuronal activity and metabolism to local blood flow.
Control of Ion and Water Homeostasis
Neuronal activity increases extracellular K+; if uncontrolled during high levels of activity, this can depolarize neuronal membrane potentials to pathological states. Astrocytes buffer extracellular K+ concentrations through Kir4.1 inwardly rectifying K+ channels (10). These channels take up excess K+ by passive diffusion down the K+ electrochemical potential gradient. Oligodendrocytes also express Kir4.1, and loss of Kir4.1 from both astrocytes and oligodendrocytes results in profound vacuolization of myelin, neuronal death, and premature lethality (11, 12). Astrocytes also regulate activity-dependent volume changes of the extracellular space by controlling water homeostasis through the aquaporin-4 water channel (13). These same water channels are the primary target of autoantibodies in neuromyelitis optica, an autoimmune astrocytopathy resulting in profound demyelination in the CNS. Thus, astrocytes play essential roles in controlling neuronal excitability and brain homeostasis by regulating brain volume and K+ concentrations.
Regulation of Synapse Formation, Function, and Plasticity
The main computational unit of the brain is the synapse. The discovery that astrocytes participate in many aspects of synapse function including their development, refinement, and modulation of activity has helped to firmly establish the view that astrocytes are not simply passive regulators of brain homeostasis but in fact are major contributors to cognition, learning, and memory. Some of the earliest studies on the role of astrocytes in synapse formation used highly purified cultures of retinal ganglion cells to show that the addition of astrocyte-conditioned medium induces the development of large numbers of synapses (14). These experiments supported the conclusion that astrocytes secrete factors that promote synaptogenesis and synapse function. This observation led to extensive searches for these factors, which are now known to contribute to structural assembly of synapses and modulation of pre- and postsynaptic function (15). For example, thrombospondins, which are large extracellular matrix molecules secreted by astrocytes, have a myriad of functions including the induction of glutamatergic synapse formation (16) and the inhibition of presynaptic release (17).
Not only do astrocytes promote synapse assembly, but recent studies of astrocyte gene expression suggested that they also participate in the refinement of neural circuits during development and in adult animals by activity-dependent synapse elimination. Specifically, astrocytes utilize MEGF10 and MERTK phagocytic pathways to promote synapse engulfment. Loss of these pathways results in failure to properly refine visual circuits and a greater number of weak synapses (18).
The close physical apposition of astrocytes to synapses suggested that astrocytes may directly participate in neurotransmission. Indeed, this close apposition has led to the idea of the so-called tripartite synapse where astrocytes affect synapse function (19). Many different laboratories have shown that astrocytes express neurotransmitter receptors, and the release of neurotransmitter can bidirectionally regulate astrocyte and neuron function (20). Astrocytes remove excess extracellular glutamate through excitatory amino acid transporters. Conversely, astrocytes can also release neurotransmitter to directly modify synapse properties. However, the concept of gliotransmission remains very controversial because one recent study claimed that the transgenic mouse (expressing a dominant-negative domain of the vesicular SNARE protein) originally used to discover gliotransmission was not specific to astrocytes (21).
Astrocytes in Disease
Astrocytes are prominent contributors not only to normal brain function but also play key roles in the brain's reaction to disease or injury. The response of astrocytes to injury or disease is called astrogliosis and may be defined by context-dependent morphological and molecular changes (22). Astrogliosis is a prominent feature of many diseases and injuries including Alzheimer disease, stroke, epilepsy, and traumatic brain and spinal cord injury (23–25). In response to a severe injury such as a spinal cord or traumatic brain injury, astrocytes become reactive and undergo a plethora of changes including cytoskeletal hypertrophy characterized by increased expression of glial fibrillary acidic protein and proliferation, and astrocytes begin to express many extracellular matrix molecules including chondroitin sulfate proteoglycans, laminins, and fibronectin (26). Together, these reactive astrocytes and the extracellular matrix form a dense glial scar that has both positive and negative consequences for nervous system repair. On the positive side, the glial scar promotes repair by limiting tissue damage due to inflammation (27); on the negative side, the extracellular matrix molecules comprising the glial scar are potent inhibitors of axon regeneration (28). Future studies will be required to separate the beneficial effects of astrogliosis from the detrimental effects of the glial scar on neural repair and regeneration. Malignant glioma can also result from the transformation of astrocytes into astrocytomas (29). Treatment of these cancers is especially difficult because of their resistance to chemotherapy and their highly invasive nature. Current efforts focus on defining the genetic changes in astrocytes that lead to transformation and the resulting phenotypic consequences of the mutations.
OLIGODENDROCYTES AND SCHWANN CELLS
Myelin is a key evolutionary adaptation unique to vertebrates that permitted the development of a complex nervous system. Myelin wraps axons as a multilamellar membrane sheath made by oligodendrocytes in the CNS and Schwann cells in the PNS (Fig. 1B). In the CNS, a single oligodendrocyte may contact and myelinate many axons, but in the PNS, a single Schwann cell myelinates only one axon. Thus, in the PNS, a myelinated axon consists of one axon ensheathed by many Schwann cells lined up one after another like very long beads on a string. During development, both oligodendrocytes and Schwann cells follow a well defined differentiation program that depends on distinct transcription factors and interactions with environmental cues (30, 31). In the brain and spinal cord, oligodendrocytes arise from precursors found in the subventricular zone and the ventral neural tube, respectively. In contrast, Schwann cells are neural crest derivatives.
The myelin sheath confers several important properties on axons that profoundly influence axon physiology: myelin reduces axonal membrane capacitance while simultaneously increasing the resistance to ion flux across the plasma membrane. This results in a decrease in the time constant and increase in length constant. In addition to these passive electrical properties, myelinating oligodendrocytes and Schwann cells also actively recruit and cluster ion channels (Na+ and K+ channels) at regularly spaced gaps in the myelin sheath called nodes of Ranvier (Fig. 1C). Clustering of ion channels restricts transmembrane ionic currents to the nodes. Together, the decrease in capacitance, increase in transverse membrane resistance, and clustering of ion channels dramatically increase the conduction velocity of axonal action potentials. The increase in conduction velocity means that axons can be smaller, resulting in dramatic space savings, which in turn allows for a more complex and interconnected nervous system. Finally, the decrease in axon diameter and restriction of transmembrane currents to the nodes of Ranvier also result in significant metabolic savings: far less energy must be expended to maintain the ionic gradients that underlie action potential generation and propagation.
Myelinating Glia Regulate Axonal Membrane Properties
Myelinating glia actively sculpt the functional organization of axons such that in both the PNS and CNS axons are subdivided into four major domains: nodes of Ranvier, paranodes, juxtaparanodes, and internodes (Fig. 1C). Nodes are characterized by the high density clustering of voltage-gated Na+ channels responsible for regeneration and propagation of the action potential. A complex set of glia-derived extracellular matrix molecules, axonal cell adhesion molecules, and cytoskeletal proteins function as one of two glia-dependent mechanisms that cluster Na+ channels (32, 33). Flanking the nodes are paranodal junctions where the myelin sheath attaches to the axon; here, paranodes form the largest known intercellular adhesive junction. Paranodes also have lipid raftlike properties that allow them to be purified and analyzed by mass spectrometry (34); we used this characteristic of the paranodal junctions to identify proteins enriched in this important domain. We found a specialized paranodal cytoskeleton consisting of spectrins in the axon (35) and ankyrins on the glial side of the paranode (36). Paranodal junctions and their associated cytoskeleton function both to isolate nodal currents from internodal regions and as the second glia-dependent mechanism for the clustering of axonal ion channels (37, 38). Adjacent to the paranodal junctions and beneath the myelin sheath is the juxtaparanode, a region defined by the clustering of Kv1 K+ channels. These K+ channels are also clustered through axon-glia interactions, but their physiological functions remain largely enigmatic (39). Finally, the vast majority of the myelinated axon consists of internode with very low densities of ion channels; it is this low density that confers an increase in membrane resistance on axons relative to nodes. Although these axonal domains consist of different sets of cell adhesion molecules, ion channels, and cytoskeletal proteins, all axonal domains are assembled and maintained by myelinating glia. Efforts to define the proteome of each domain have met with varying degrees of success. Because of their detergent insolubility and strong association with the axonal cytoskeleton, nodes and paranodes have proven to be resistant to typical immunoaffinity isolation of protein complexes. In contrast, juxtaparanodal protein complexes are readily solubilized and purified. We previously used immunoprecipitation followed by mass spectrometry to identify new components of juxtaparanodes (40).
Metabolism
Because myelinated axons are completely isolated from the extracellular space except at the nodes of Ranvier, myelinating glia must also provide metabolic support to axons. Current evidence suggests that oligodendrocytes and Schwann cells provide a variety of metabolites including cholesterol, lactate, and glycogen to axons (41). In support of the idea that myelinating glia provide lactate to axons for an energy source, loss of lactate transporters from oligodendrocytes results in axon damage and degeneration (42).
Myelin Plasticity
Recent experiments to determine the response of myelinating glia to activity or learning paradigms revealed that myelination is also plastic and can change in adults, not just during early development. For example, one study showed that development of new myelin sheaths is required for mice to learn how to run on a complex running wheel with irregularly spaced rungs (43). This remarkable result proves that some forms of learning require the development of new myelin segments in the adult brain and that this is an activity-dependent phenomenon. One can speculate that the addition of new myelin segments facilitates conduction velocity in specific circuits or alters the timing of action potential arrival. The signals that promote these plastic changes remain completely unknown.
Diseases of Myelin
The intimate interactions between myelinating glia and axons are also subject to disruption by disease or injury. De- or dysmyelination can result from genetic (e.g. Charcot-Marie-Tooth peripheral neuropathies or Pelizaeus-Merzbacher CNS hypomyelinating disease), autoimmune (e.g. PNS demyelinating Guillain-Barré syndrome or CNS demyelinating multiple sclerosis), metabolic (diabetic neuropathy), mechanical (spinal cord injury and carpal tunnel syndrome), or hypoxic-ischemic insults (stroke and neonatal hypoxia-ischemia). There are currently no cures for demyelinating diseases or injuries. A major consequence of demyelination is the loss of axonal support, which in turn leads to axon degeneration and permanent loss of function and disability (44). Thus, treatments for demyelinating diseases and injuries are an acute need and the subject of much current attention. Proteomic analyses of myelin from different demyelinating disease models may provide insights into pathogenesis and potential treatments for these diseases.
MICROGLIA
Microglia have traditionally been viewed primarily as the brain's resident immune cell, and their functions during disease and injury have been studied extensively (45). In their resting state, microglia are highly ramified cells with very elaborate thin processes that extend branches to surveil a defined territory (Fig. 1D). After injury, microglia undergo dramatic changes in shape and protein expression to protect the brain. Furthermore, microglia migrate to sites of injury where they release cytokines and phagocytose debris and dead and dying cells. After injury, microglia can strip dysfunctional synapses. Intriguingly, dysfunction or loss of microglia can also result in behavioral deficits and impaired learning-dependent synaptic plasticity (46). Recent studies have also shown that in the developing and adult brain microglia contact synapses and refine neural circuits by engulfment of some synapses (47). Together, these results suggest that microglia participate not only in defense mechanisms but also normal brain development and function. Microglia can be identified by a variety of cell surface antigens (e.g. IBA1; Fig. 1D), and mice with genetically labeled microglia (CX3CR1-GFP) have been widely used to study their functions in the healthy and diseased brain.
PROTEOMIC APPROACHES TO STUDYING GLIAL CELL FUNCTION
Recently, high throughput methods were developed to profile the RNAs expressed by thousands of individual cells (Drop-seq (48, 49)). This technology allows for unprecedented characterization of single cell gene expression among the cell types of the nervous system including glia. Despite the technical advances achieved in both resolution and sensitivity of mass spectrometry, single cell resolution of protein composition remains a distant goal. Even if sensitivity were high enough to permit single cell resolution, determining the proteome of a single astrocyte, neuron, microglial cell, or oligodendrocyte will likely remain impossible due to the experimental difficulty of isolating entire cells because of their extremely complex morphologies. Nevertheless, the use of mass spectrometry to dissect brain function has been wildly successful for neurons, and lessons learned in this context have been applied to the study of glia. Particularly notable examples include efforts to define the molecules involved in neurotransmission (synaptic vesicles and pre- and postsynaptic compartments (50–54)). The success of these experiments largely depended on the ability to biochemically isolate subcellular compartments or organelles of interest (55). For example, postsynaptic densities can be purified based on their detergent insolubility and their ability to partition into distinct fractions after gradient centrifugation. However, with the exception of oligodendrocytes and Schwann cells, analogous procedures for the fractionation and purification of subcellular compartments from glia have not been developed.
By weight, myelin is ∼80% lipid (mainly glycolipids) with the remainder being specific myelin proteins such as myelin basic protein and proteolipid protein. This unique biochemical composition has permitted both PNS and CNS myelin to be purified by biochemical fractionation procedures, which in turn has allowed this membrane specialization to be thoroughly characterized using mass spectrometry (56, 57). These studies revealed a previously unappreciated complexity to myelin and permitted follow-up studies on novel myelin proteins. For example, comparison of PNS and CNS myelin proteomes, together with unbiased immunostaining, revealed that N-ethylmaleimide-sensitive factor is highly enriched in Schwann cells surrounding nodes of Ranvier (56). Similarly, Septin 9, which is mutated in hereditary neuralgic amyotrophy, was identified as a PNS myelin protein that is dramatically increased in a mouse model of demyelinating neuropathy (57). Because of the ease of myelin purification, investigators have begun to analyze differences in myelin composition from models of nervous system diseases (e.g. fragile X, vanishing white matter disease, and multiple sclerosis (58–60)) and even between species. For example, a comparison between human and mouse myelin proteomes identified 678 proteins from human myelin and 515 proteins from mouse. Among the mouse proteins identified, 475 had clear human orthologues, and 308 were common between human and mouse proteomes (61). Because all proteomic results require validation, additional studies are necessary to confirm and determine the functions of mouse- or human-specific myelin proteins.
The challenge of glial proteomics is magnified by the highly complex morphologies of astrocytes and microglia. Thus, nearly all proteomic analyses of astrocytes and microglia have been performed on primary cultures or immortalized cell lines or were biased interrogations of proteins (and their interactors) thought to be specifically enriched in one glial cell type. Unfortunately, by definition, immortalized cell lines are unlike those found in the healthy brain, and cultured primary astrocytes or microglia are activated and have phenotypes that reflect a reactive rather than a native state.
The advent of new molecular and proteomic tools may begin to overcome some of these technical limitations. Genetic labeling strategies now permit isolation of specific brain cell types using fluorescence-activated cell sorting. This approach has the advantage of cellular specificity, but the disadvantage is that fine, thin complex cellular processes are still lost during the cell isolation procedure. Furthermore, these approaches result in a mixing of glial phenotypes; unless additional criteria for separation (e.g. brain region) are used, heterogeneity among glial types cannot be preserved. A combination of genetic and protein labeling approaches may overcome some of these limitations. For example, one can imagine combining Cre-loxP technology with protein labeling techniques such as BioID (62, 63). In this strategy, a promiscuous biotin ligase may be targeted to subcellular compartments (e.g. membrane, cytoplasm, or endoplasmic reticulum) but only in those cells where Cre recombinase is expressed. Thus, a membrane-targeted biotin ligase expressed only in astrocytes or microglia may allow for biotinylation of membrane proteins found not only on their cell bodies but also in their distal fine processes. Streptavidin-coated magnetic beads can then be used to “capture” these biotinylated proteins for subsequent analysis by mass spectrometry. It is easy to imagine variations on this approach by combining different Cre driver lines for different types of glia (with or without temporal control) with disease or injury models to determine the responses of glia. Alternatively, one could perform stereotactic injection of virally encoded Cre-dependent biotin ligases to investigate regional heterogeneity among glial cells. The spectrum of potential experiments may be further expanded by targeting the biotin ligase to other subcellular domains or compartments. Despite the technical limitations described above, comparison of glial proteomic data sets with the several excellent transcriptomics data sets of glia already available (64, 65) may provide powerful insights into regulation of glial protein expression and function.
In conclusion, the recent appreciation for and understanding of glia in the healthy and diseased nervous system has led to a rapid expansion in the tools available to study these important cell types. These tools coupled with advances in mass spectrometry and other protein analysis techniques make the field of glial proteomics ripe for the picking.
Footnotes
Author contributions: M.N.R. wrote the paper.
* This work was supported, in whole or in part, by National Institutes of Health Grants NS044916 and NS069688. This work was also supported by a grant from the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
1 The abbreviations used are:
- CNS
- central nervous system
- PNS
- peripheral nervous system.
REFERENCES
- 1.Azevedo F. A., Carvalho L. R., Grinberg L. T., Farfel J. M., Ferretti R. E., Leite R. E., Jacob Filho W., Lent R., and Herculano-Houzel S. (2009) Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J. Comp. Neurol. 513, 532–541 [DOI] [PubMed] [Google Scholar]
- 2.Rowitch D. H., and Kriegstein A. R. (2010) Developmental genetics of vertebrate glial-cell specification. Nature 468, 214–222 [DOI] [PubMed] [Google Scholar]
- 3.Bushong E. A., Martone M. E., Jones Y. Z., and Ellisman M. H. (2002) Protoplasmic astrocytes in CA1 stratum radiatum occupy separate anatomical domains. J. Neurosci. 22, 183–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsai H. H., Li H., Fuentealba L. C., Molofsky A. V., Taveira-Marques R., Zhuang H., Tenney A., Murnen A. T., Fancy S. P., Merkle F., Kessaris N., Alvarez-Buylla A., Richardson W. D., and Rowitch D. H. (2012) Regional astrocyte allocation regulates CNS synaptogenesis and repair. Science 337, 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim J. G., Suyama S., Koch M., Jin S., Argente-Arizon P., Argente J., Liu Z. W., Zimmer M. R., Jeong J. K., Szigeti-Buck K., Gao Y., Garcia-Caceres C., Yi C. X., Salmaso N., Vaccarino F. M., Chowen J., Diano S., Dietrich M. O., Tschöp M. H., and Horvath T. L. (2014) Leptin signaling in astrocytes regulates hypothalamic neuronal circuits and feeding. Nat. Neurosci. 17, 908–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halassa M. M., Fellin T., Takano H., Dong J. H., and Haydon P. G. (2007) Synaptic islands defined by the territory of a single astrocyte. J. Neurosci. 27, 6473–6477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filosa J. A., Morrison H. W., Iddings J. A., Du W., and Kim K. J. (2015) Beyond neurovascular coupling, role of astrocytes in the regulation of vascular tone. Neuroscience 10.1016/j.neuroscience.2015.03.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacVicar B. A., and Newman E. A. (2015) Astrocyte regulation of blood flow in the brain. Cold Spring Harb. Perspect. Biol. 7, a020388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magistretti P. J. (2006) Neuron-glia metabolic coupling and plasticity. J. Exp. Biol. 209, 2304–2311 [DOI] [PubMed] [Google Scholar]
- 10.Butt A. M., and Kalsi A. (2006) Inwardly rectifying potassium channels (Kir) in central nervous system glia: a special role for Kir4.1 in glial functions. J. Cell. Mol. Med. 10, 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neusch C., Papadopoulos N., Müller M., Maletzki I., Winter S. M., Hirrlinger J., Handschuh M., Bähr M., Richter D. W., Kirchhoff F., and Hülsmann S. (2006) Lack of the Kir4.1 channel subunit abolishes K+ buffering properties of astrocytes in the ventral respiratory group: impact on extracellular K+ regulation. J. Neurophysiol. 95, 1843–1852 [DOI] [PubMed] [Google Scholar]
- 12.Neusch C., Rozengurt N., Jacobs R. E., Lester H. A., and Kofuji P. (2001) Kir4.1 potassium channel subunit is crucial for oligodendrocyte development and in vivo myelination. J. Neurosci. 21, 5429–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagelhus E. A., Mathiisen T. M., and Ottersen O. P. (2004) Aquaporin-4 in the central nervous system: cellular and subcellular distribution and coexpression with KIR4.1. Neuroscience 129, 905–913 [DOI] [PubMed] [Google Scholar]
- 14.Ullian E. M., Sapperstein S. K., Christopherson K. S., and Barres B. A. (2001) Control of synapse number by glia. Science 291, 657–661 [DOI] [PubMed] [Google Scholar]
- 15.Chung W. S., Allen N. J., and Eroglu C. (2015) Astrocytes control synapse formation, function, and elimination. Cold Spring Harb. Perspect. Biol. 7, a020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christopherson K. S., Ullian E. M., Stokes C. C., Mullowney C. E., Hell J. W., Agah A., Lawler J., Mosher D. F., Bornstein P., and Barres B. A. (2005) Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell 120, 421–433 [DOI] [PubMed] [Google Scholar]
- 17.Crawford D. C., Jiang X., Taylor A., and Mennerick S. (2012) Astrocyte-derived thrombospondins mediate the development of hippocampal presynaptic plasticity in vitro. J. Neurosci. 32, 13100–13110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung W. S., Clarke L. E., Wang G. X., Stafford B. K., Sher A., Chakraborty C., Joung J., Foo L. C., Thompson A., Chen C., Smith S. J., and Barres B. A. (2013) Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature 504, 394–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haydon P. G., and Carmignoto G. (2006) Astrocyte control of synaptic transmission and neurovascular coupling. Physiol. Rev. 86, 1009–1031 [DOI] [PubMed] [Google Scholar]
- 20.Martín R., Bajo-Grañeras R., Moratalla R., Perea G., and Araque A. (2015) Circuit-specific signaling in astrocyte-neuron networks in basal ganglia pathways. Science 349, 730–734 [DOI] [PubMed] [Google Scholar]
- 21.Fujita T., Chen M. J., Li B., Smith N. A., Peng W., Sun W., Toner M. J., Kress B. T., Wang L., Benraiss A., Takano T., Wang S., and Nedergaard M. (2014) Neuronal transgene expression in dominant-negative SNARE mice. J. Neurosci. 34, 16594–16604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burda J. E., and Sofroniew M. V. (2014) Reactive gliosis and the multicellular response to CNS damage and disease. Neuron 81, 229–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Birch A. M., Katsouri L., and Sastre M. (2014) Modulation of inflammation in transgenic models of Alzheimer's disease. J. Neuroinflammation 11, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gleichman A. J., and Carmichael S. T. (2014) Astrocytic therapies for neuronal repair in stroke. Neurosci. Lett. 565, 47–52 [DOI] [PubMed] [Google Scholar]
- 25.Devinsky O., Vezzani A., Najjar S., De Lanerolle N. C., and Rogawski M. A. (2013) Glia and epilepsy: excitability and inflammation. Trends Neurosci. 36, 174–184 [DOI] [PubMed] [Google Scholar]
- 26.Yuan Y. M., and He C. (2013) The glial scar in spinal cord injury and repair. Neurosci. Bull. 29, 421–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sofroniew M. V. (2015) Astrocyte barriers to neurotoxic inflammation. Nat. Rev. Neurosci. 16, 249–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma K., Selzer M. E., and Li S. (2012) Scar-mediated inhibition and CSPG receptors in the CNS. Exp. Neurol. 237, 370–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zong H., Parada L. F., and Baker S. J. (2015) Cell of origin for malignant gliomas and its implication in therapeutic development. Cold Spring Harb. Perspect. Biol. 7, a020610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jessen K. R., and Mirsky R. (2005) The origin and development of glial cells in peripheral nerves. Nat. Rev. Neurosci. 6, 671–682 [DOI] [PubMed] [Google Scholar]
- 31.Gallo V., and Deneen B. (2014) Glial development: the crossroads of regeneration and repair in the CNS. Neuron 83, 283–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Susuki K., Chang K. J., Zollinger D. R., Liu Y., Ogawa Y., Eshed-Eisenbach Y., Dours-Zimmermann M. T., Oses-Prieto J. A., Burlingame A. L., Seidenbecher C. I., Zimmermann D. R., Oohashi T., Peles E., and Rasband M. N. (2013) Three mechanisms assemble central nervous system nodes of Ranvier. Neuron 78, 469–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feinberg K., Eshed-Eisenbach Y., Frechter S., Amor V., Salomon D., Sabanay H., Dupree J. L., Grumet M., Brophy P. J., Shrager P., and Peles E. (2010) A glial signal consisting of gliomedin and NrCAM clusters axonal Na+ channels during the formation of nodes of Ranvier. Neuron 65, 490–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schafer D. P., Bansal R., Hedstrom K. L., Pfeiffer S. E., and Rasband M. N. (2004) Does paranode formation and maintenance require partitioning of neurofascin 155 into lipid rafts? J. Neurosci. 24, 3176–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogawa Y., Schafer D. P., Horresh I., Bar V., Hales K., Yang Y., Susuki K., Peles E., Stankewich M. C., and Rasband M. N. (2006) Spectrins and ankyrinB constitute a specialized paranodal cytoskeleton. J. Neurosci. 26, 5230–5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang K. J., Zollinger D. R., Susuki K., Sherman D. L., Makara M. A., Brophy P. J., Cooper E. C., Bennett V., Mohler P. J., and Rasband M. N. (2014) Glial ankyrins facilitate paranodal axoglial junction assembly. Nat. Neurosci. 17, 1673–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang K. J., and Rasband M. N. (2013) Excitable domains of myelinated nerves: axon initial segments and nodes of Ranvier. Curr. Top. Membr. 72, 159–192 [DOI] [PubMed] [Google Scholar]
- 38.Zhang C., Susuki K., Zollinger D. R., Dupree J. L., and Rasband M. N. (2013) Membrane domain organization of myelinated axons requires βII spectrin. J. Cell Biol. 203, 437–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rasband M. N. (2004) It's ‘juxta’ potassium channel. J. Neurosci. Res. 76, 749–757 [DOI] [PubMed] [Google Scholar]
- 40.Ogawa Y., Oses-Prieto J., Kim M. Y., Horresh I., Peles E., Burlingame A. L., Trimmer J. S., Meijer D., and Rasband M. N. (2010) ADAM22, a Kv1 channel-interacting protein, recruits membrane-associated guanylate kinases to juxtaparanodes of myelinated axons. J. Neurosci. 30, 1038–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirrlinger J., and Nave K. A. (2014) Adapting brain metabolism to myelination and long-range signal transduction. Glia 62, 1749–1761 [DOI] [PubMed] [Google Scholar]
- 42.Lee Y., Morrison B. M., Li Y., Lengacher S., Farah M. H., Hoffman P. N., Liu Y., Tsingalia A., Jin L., Zhang P. W., Pellerin L., Magistretti P. J., and Rothstein J. D. (2012) Oligodendroglia metabolically support axons and contribute to neurodegeneration. Nature 487, 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKenzie I. A., Ohayon D., Li H., de Faria J. P., Emery B., Tohyama K., and Richardson W. D. (2014) Motor skill learning requires active central myelination. Science 346, 318–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trapp B. D., Peterson J., Ransohoff R. M., Rudick R., Mörk S., and Bö L. (1998) Axonal transection in the lesions of multiple sclerosis. N. Engl. J. Med. 338, 278–285 [DOI] [PubMed] [Google Scholar]
- 45.Hanisch U. K., and Kettenmann H. (2007) Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nat. Neurosci. 10, 1387–1394 [DOI] [PubMed] [Google Scholar]
- 46.Parkhurst C. N., Yang G., Ninan I., Savas J. N., Yates J. R. 3rd, Lafaille J. J., Hempstead B. L., Littman D. R., and Gan W. B. (2013) Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell 155, 1596–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schafer D. P., Lehrman E. K., Kautzman A. G., Koyama R., Mardinly A. R., Yamasaki R., Ransohoff R. M., Greenberg M. E., Barres B. A., and Stevens B. (2012) Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Macosko E. Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A. R., Kamitaki N., Martersteck E. M., Trombetta J. J., Weitz D. A., Sanes J. R., Shalek A. K., Regev A., and McCarroll S. A. (2015) Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeisel A., Muñoz-Manchado A. B., Codeluppi S., Lönnerberg P., La Manno G., Juréus A., Marques S., Munguba H., He L., Betsholtz C., Rolny C., Castelo-Branco G., Hjerling-Leffler J., and Linnarsson S. (2015) Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142 [DOI] [PubMed] [Google Scholar]
- 50.Takamori S., Holt M., Stenius K., Lemke E. A., Grønborg M., Riedel D., Urlaub H., Schenck S., Brügger B., Ringler P., Müller S. A., Rammner B., Gräter F., Hub J. S., De Groot B. L., Mieskes G., Moriyama Y., Klingauf J., Grubmüller H., Heuser J., Wieland F., and Jahn R. (2006) Molecular anatomy of a trafficking organelle. Cell 127, 831–846 [DOI] [PubMed] [Google Scholar]
- 51.Bayés A., van de Lagemaat L. N., Collins M. O., Croning M. D., Whittle I. R., Choudhary J. S., and Grant S. G. (2011) Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat. Neurosci. 14, 19–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trinidad J. C., Thalhammer A., Burlingame A. L., and Schoepfer R. (2013) Activity-dependent protein dynamics define interconnected cores of co-regulated postsynaptic proteins. Mol. Cell. Proteomics 12, 29–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trinidad J. C., Barkan D. T., Gulledge B. F., Thalhammer A., Sali A., Schoepfer R., and Burlingame A. L. (2012) Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol. Cell. Proteomics 11, 215–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Collins M. O., Yu L., Coba M. P., Husi H., Campuzano I., Blackstock W. P., Choudhary J. S., and Grant S. G. (2005) Proteomic analysis of in vivo phosphorylated synaptic proteins. J. Biol. Chem. 280, 5972–5982 [DOI] [PubMed] [Google Scholar]
- 55.Bergeron J. J., Au C. E., Desjardins M., McPherson P. S., and Nilsson T. (2010) Cell biology through proteomics—ad astra per alia porci. Trends Cell Biol. 20, 337–345 [DOI] [PubMed] [Google Scholar]
- 56.Taylor C. M., Marta C. B., Claycomb R. J., Han D. K., Rasband M. N., Coetzee T., and Pfeiffer S. E. (2004) Proteomic mapping provides powerful insights into functional myelin biology. Proc. Natl. Acad. Sci. U.S.A. 101, 4643–4648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patzig J., Jahn O., Tenzer S., Wichert S. P., de Monasterio-Schrader P., Rosfa S., Kuharev J., Yan K., Bormuth I., Bremer J., Aguzzi A., Orfaniotou F., Hesse D., Schwab M. H., Möbius W., Nave K. A., and Werner H. B. (2011) Quantitative and integrative proteome analysis of peripheral nerve myelin identifies novel myelin proteins and candidate neuropathy loci. J. Neurosci. 31, 16369–16386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Linker R. A., Brechlin P., Jesse S., Steinacker P., Lee D. H., Asif A. R., Jahn O., Tumani H., Gold R., and Otto M. (2009) Proteome profiling in murine models of multiple sclerosis: identification of stage specific markers and culprits for tissue damage. PLoS One 4, e7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gat-Viks I., Geiger T., Barbi M., Raini G., and Elroy-Stein O. (2015) Proteomics-level analysis of myelin formation and regeneration in a mouse model for vanishing white matter disease. J. Neurochem. 134, 513–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kalinowska M., Castillo C., and Francesconi A. (2015) Quantitative profiling of brain lipid raft proteome in a mouse model of fragile X syndrome. PLoS One 10, e0121464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ishii A., Dutta R., Wark G. M., Hwang S. I., Han D. K., Trapp B. D., Pfeiffer S. E., and Bansal R. (2009) Human myelin proteome and comparative analysis with mouse myelin. Proc. Natl. Acad. Sci. U.S.A. 106, 14605–14610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roux K. J., Kim D. I., and Burke B. (2013) BioID: a screen for protein-protein interactions. Curr. Protoc. Protein Sci. 74, Unit 19.23 [DOI] [PubMed] [Google Scholar]
- 63.Roux K. J., Kim D. I., Raida M., and Burke B. (2012) A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y., Chen K., Sloan S. A., Bennett M. L., Scholze A. R., O'Keeffe S., Phatnani H. P., Guarnieri P., Caneda C., Ruderisch N., Deng S., Liddelow S. A., Zhang C., Daneman R., Maniatis T., Barres B. A., and Wu J. Q. (2014) An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Doyle J. P., Dougherty J. D., Heiman M., Schmidt E. F., Stevens T. R., Ma G., Bupp S., Shrestha P., Shah R. D., Doughty M. L., Gong S., Greengard P., and Heintz N. (2008) Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135, 749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]