Abstract
Spatial memory depends on the hippocampus, which is particularly vulnerable to aging. This vulnerability has implications for the impairment of navigation capacities in older people, who may show a marked drop in performance of spatial tasks with advancing age. Contemporary understanding of long-term memory formation relies on molecular mechanisms underlying long-term synaptic plasticity. With memory acquisition, activity-dependent changes occurring in synapses initiate multiple signal transduction pathways enhancing protein turnover. This enhancement facilitates de novo synthesis of plasticity related proteins, crucial factors for establishing persistent long-term synaptic plasticity and forming memory engrams. Extensive studies have been performed to elucidate molecular mechanisms of memory traces formation; however, the identity of plasticity related proteins is still evasive. In this study, we investigated protein turnover in mouse hippocampus during long-term spatial memory formation using the reference memory version of radial arm maze (RAM) paradigm. We identified 1592 proteins, which exhibited a complex picture of expression changes during spatial memory formation. Variable linear decomposition reduced significantly data dimensionality and enriched three principal factors responsible for variance of memory-related protein levels at (1) the initial phase of memory acquisition (165 proteins), (2) during the steep learning improvement (148 proteins), and (3) the final phase of the learning curve (123 proteins). Gene ontology and signaling pathways analysis revealed a clear correlation between memory improvement and learning phase-curbed expression profiles of proteins belonging to specific functional categories. We found differential enrichment of (1) neurotrophic factors signaling pathways, proteins regulating synaptic transmission, and actin microfilament during the first day of the learning curve; (2) transcription and translation machinery, protein trafficking, enhancement of metabolic activity, and Wnt signaling pathway during the steep phase of memory formation; and (3) cytoskeleton organization proteins. Taken together, this study clearly demonstrates dynamic assembly and disassembly of protein-protein interaction networks depending on the stage of memory formation engrams.
Long-term synaptic plasticity is considered a cellular correlate of long-term memory (LTM)1. Contemporary understanding of memory formation is based on the initiation and maintenance of long-term synaptic plasticity (1–4), for which de novo protein synthesis is a vital requirement. De novo protein synthesis itself is secondary to activity-dependent changes in synapses that occur during learning processes. These activity changes trigger post-translational modifications of proteins initiating and sustaining multiple signal transduction pathways. In turn, these signaling pathways regulate changes in synaptic strength and connectivity by governing gene expression and protein translation (5–13). Depending on time elapsed since triggering of long-term synaptic plasticity, protein synthesis may be limited to the dendrites directly involved in the plasticity processes (14–18). Multiple synaptic activity-dependent signal transduction pathways (7–13, 19) orchestrate the regulation of synaptic plasticity on the translational level (for review see (20, 21)). Accumulated evidence shows that different types of LTM depend on protein synthesis, disregarding dependence on brain regions such as amygdala (22, 23), hippocampus (24–29), and medial prefrontal or insular cortex ((30–32); for review see (33)). However, LTM perseveres significantly longer than duration of translation-dependent long-term plasticity. Maintenance and persistence of LTM for days, months, or years requires replenishment of the mRNA pool coding for proteins necessary for memory consolidation. Moreover, importance of transcriptional regulation of LTM was demonstrated: several transcription factors (TFs), e.g. CREB, C/EBP, AP1, Egr, and Rel/NF-κB have been shown to be critical to synaptic plasticity, memory formation (for review see (34)), and regulation via multiple signal transduction pathways (34–36).
Protein degradation is another pole of protein turnover regulation. Studies over the last decade demonstrate strong links between maintenance of long-term potentiation (LTP, a type of long-term synaptic plasticity) and protein degradation ((37); for review, see (38)). It was recently shown that inhibition of the proteasome system may enhance LTP induction (39) because of prevention of translation activator targeting (40). Multiple behavioral studies have also confirmed the crucial role of the ubiquitin-proteasome system in memory consolidation in the amygdala (23, 41), hippocampus (24, 42), and prefrontal cortex (32).
In this study, we aimed to investigate protein turnover (expression term is henceforth used for simplicity) alteration in the hippocampus during long-term spatial memory formation. The hippocampus is known to be crucial for coding, consolidation, and reconsolidation of a wide variety of memory types, including spatial memory (for review, see (43)). The reference memory version of the radial arm maze (RAM) paradigm allows conduction for temporal tracking of protein expression changes occurring during memory acquisition.
The importance of protein turnover in memory consolidation and retrieval is indisputable. However, little is known about those proteins which undergo expression changes during memory formation and what are the dynamics of these changes. Although several transcriptomic studies were conducted on different types of learning (44–46), there is very limited proteomic information based on behavioral paradigms and temporal dynamics of memory acquisition. To our knowledge, there is only a single publication to date showing protein profile change during the Morris water maze paradigm, and this study was limited to the first 24 h of memory acquisition (47). The current study includes a comprehensive proteomic analysis of protein expression profiles occurring during the whole course of long-term spatial learning acquired by the RAM paradigm.
EXPERIMENTAL PROCEDURES
The Radial Arm Maze
Description
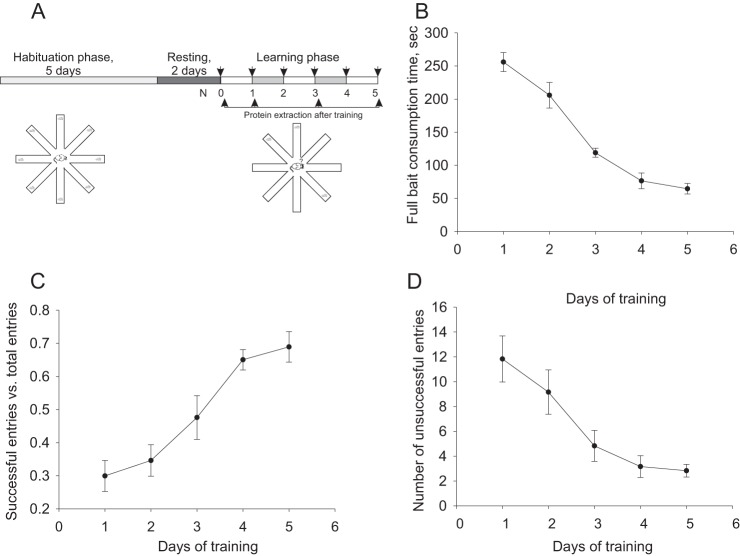
The RAM paradigm (48, 49) was conducted using a Plexiglass maze whose eight arms (35 cm × 8 cm × 8 cm) are connected by removable guillotine doors to a circular central chamber (21 cm diameter, Fig. 1A). At the end of each arm was a 3 cm dish in which bait (semi-soft cheese, 15% fat) was placed as needed. Four of the eight arms were marked with spatial cues for navigation purposes. Animals underwent 5 days of food deprivation (12 h daily without access to food and water accessible ad libitum) preceding the test, inducing a reduction in body weight of not more than 15%. RAM habituation phase began with a 3 day training period, during which each animal was placed in the maze for 8 min daily, with free access to 5 grams of bait located at the end of each arm. After the habituation period, the animals were left for 2 additional days without training. Learning (trial) phase: subsequently, animal learning ability was assessed daily during a 5 day testing period, in which three arms were baited. The baited arms differed from mouse to mouse, but remained constant for each individual mouse. In the beginning of the trial session, the mouse was placed in the center of the maze, with doors closed. Then the doors were opened, allowing the animals to freely enter the arms. Each mouse remained in the maze for 8 mins, or until all the bait was consumed. Each animal performed the task once per day. Animal navigation of the maze was recorded by EthoVision video tracking system (version 7.1, Noldus Information Technology, Wageningen, The Netherlands) and analyzed according to the following parameters: (1) Correct Entries: entries to baited arms as a portion of total arm entries; (2) Incomplete entries: entries to baited arms without consuming bait, as a portion of total entries to baited arms; (3) Latency, a time from trial start point to complete bait consumption; (4) Re-entries to formerly baited arms; (5) Distance traveled (cm); and (6) Velocity (cm/sec). The mice were deemed to have entered an arm when its center point was located in the arm. Animal learning was assessed as gradual elimination of randomness in animal navigation of the maze, reducing bait consumption time, re-entry, and incorrect entries. Furthermore, learning ability of the animals was validated using factor analysis.
Fig. 1.
Radial arm maze (RAM) paradigm used as a behavioral model of long-term reference memory. A, Experimental setup of RAM and its temporal relationship with protein extraction for proteomic study. B–D, Parameters used for learning evaluation. B, Improvement curve of time necessary to consume all baits. C, Rate of correct entries into the baited arms. D, Number of incorrect entries during each session.
Data Analysis
Animal velocities and traveled distances were evaluated to exclude changes in learning curve not related to differences in latency times. To evaluate reference memory performance, latency time, incorrect entry number, and a fraction of correct entries from the total entry number were calculated. A fraction of correct entry was deduced from the sum of correct, incorrect, incomplete entries and re-entries. Learning performance was evaluated using Kruskal-Willis one-way ANOVA on ranks test with subsequent post-hoc Dunn's analysis or one way ANOVA with Holm-Sidak post-hoc method, depending on Shapiro-Wilk normality test results.
Sample Preparation
Proteins were extracted from hippocampi of 5–10 mice per each group in each biological replicate. Each group was collected in three biological replicates, hence the total amount of mice analyzed per each group was about 30. Mice from three different generations were used as three biological replicates, overall one biological replicate per each generation. Protein extraction was performed as described previously (50–52). To be concise, hippocampi were homogenized in a Transport Buffer (20 mm Tris/HEPES; 110 mm Potassium acetate; 5 mm Magnesium acetate; 0.5 mm EGTA; 0.1 mm PMSF and 0.1% Triton X100) and titrated with KOH (pH 7.3) supplemented with Complete protease inhibitor (1:25, Roche, Cat#1838145). The homogenates of hippocampi were centrifuged and the extracted supernatants were subjected to protein denaturation, reduction, and alkylation procedures (in 6 m Guanidin-HCl and 105 mm TCEP, dissolved in 25 mm Ammonium bicarbonate, incubated for 1 h at 57 °C with subsequent incubation in 210 mm Iodacetamid, dissolved in 25 mm Ammonium bicarbonate for 45 min at room temperature, in dark). The obtained samples were diluted to 1 m Guanidine-HCl using 25 mm Ammonium bicarbonate. The pH of the diluted samples was adjusted to 8 by 1 m Ammonium bicarbonate and the samples were further subjected to proteolytic digestion by side-chain protected trypsin (Promega, Madison, WI) in ration 1:50 (protein:trypsin), overnight at 37 °C. Digestion was stopped by adjusting pH to 3 using 10% formic acid and frozen immediately using liquid nitrogen.
Liquid Chromatography
ULC/MS grade solvents were used for all chromatographic steps. Each sample was loaded using split-less nano-Ultra Performance Liquid Chromatography (10 kpsi nanoAcquity; Waters, Milford, MA) in high-pH/low-pH reversed phase (RP) 2 dimensional liquid chromatography mode. 20 μg of digested protein from each sample was loaded onto a C18 column (XBridge, 0.3 × 50 mm, 5 μm particles, Waters). The following two buffers were combined: (A) 20 mm ammonium formate, pH 10 and (B) acetonitrile (ACN). Peptides were released from the column using a step gradient: 10.8%B, 13.8%B, 15.8%B, 17.8%B, 20.1%B, 23.4%B, 65%B. Each fraction flowed directly to the second dimension of chromatography. The buffers used in the low pH RP were: (A) H2O + 0.1% formic acid and (B) ACN + 0.1% formic acid. Desalting of samples was performed online using a reverse-phase C18 trapping column (180 μm i.d., 20 mm length, 5 μm particle size, Waters). Then the peptides were separated using a C18 T3 HSS nano-column (75 μm i.d., 200 mm length, 1.8 μm particle size, Waters) run at 0.4 μl/minute. Finally, peptides were eluted from the column and loaded onto the mass spectrometer using the following protocol: 3 to 30%B over 60 min, 30to 95%B over 5min, 95% maintained for 7 min (and then back to initial conditions).
Mass Spectrometry
The nanoLC was coupled online through a nanoESI emitter (7 cm length, 10 mm tip; New Objective; Woburn, MA) to a quadrupole ion mobility time-of-flight mass spectrometer (Synapt G2 HDMS, Waters) tuned to 20,000 mass resolution (full width at half height). Data were acquired using Masslynx version 4.1 in data independent acquisition mode (DIA), HDMSE positive ion mode. The ions were separated in the T-Wave ion mobility chamber and transferred into the collision cell. Collision energy was alternated from low to high throughout the acquisition time. In low-energy (MS1) scans, the collision energy was set to 5 eV and this was ramped from 27 to 50 eV for high-energy scans. For both scans, the mass range was set to 50–2000 Da with a scan time set to 1 s. A reference compound (Glu-Fibrinopeptide B; Sigma) was infused continuously for external calibration using a LockSpray and scanned every 30 s.
Data Processing, Searching and Analysis
Raw data processing and database searching was performed using Proteinlynx Global Server (PLGS) version 2.5.2. Database searching was carried out using the Ion Accounting algorithm described by Li et al. (53). Data were searched against a combined target and reversed (decoy) mouse sequences in UniprotKB database and the CRAP list of common laboratory contaminants, version 2013_06 with 50,901 entries. Trypsin was set as the protease, and two missed cleavages were allowed. Carbamidomethylation was set as a fixed modification and oxidation of methionine as variable modification. Raw data were also imported into Rosetta Elucidator System, version 3.3 (Rosetta Biosoftware, Seattle, WA). Elucidator was used for alignment of raw MS1 data in RT and m/z dimensions as described (54). Aligned features were extracted and quantitative measurements obtained by integration of three-dimensional volumes (time, m/z, intensity) of each feature as detected in the MS1 scans. Search results were then imported directly from PLGS for annotation and the minimum identification score was set to achieve a maximum global false discovery rate of 1% at the protein level. Relative protein abundance was calculated using the Hi-3 method (55).
Data Acquisition and Peptide Identification Protein Abundancy Reconstruction
Median/standard deviation scaling was used for protein quantitative data reconstruction. The peptides were median-centered and then scaled by the raw of standard deviation. Protein abundance was obtained as the median of the abundances of the peptides in the group. Scaling was conducted on log2 transformed peptide abundance data. Outliers were removed using Grubb's test, and the minimum number of peptides per protein for Grubb's test was set to 6, to minimize multiple iteration related change of probability of outlier detection in InfernoRDN software (InfernoRDN, Richland, WA) (56). For proteins with the number of peptides less than six, we used the Tukey two-sided outlier test based on the data point location in regard to 25th (LV) and 75th (UV) percentiles: upper outlier > UV+OC*(UV-LV) and lower outlier < LV+OC*(UV-LV), where OC, the outlier coefficient was defined as 1.5.
Data Clustering
Cluster analysis was performed as described in (52) with several modifications. Briefly, prior cluster analysis log2 of protein expression change ratios between all the tested groups were calculated to minimize the impact of biological variability. Then the data was standardized using a z-score method. Hierarchic clustering was performed by evaluation of the Euclidean distances, and the distance matrix was linked using Ward's minimum variance linkage method (57, 58). Clustering was validated and the number of clusters was supervised using root mean square deviation at steps of clustering, pseudo-F ratio, pseudo T2 evaluation, and Dunn's cluster separation maximum group assessment approach. In addition, partitioning was visually evaluated by the amalgamation curves. Several types of nonhierarchic clustering were used. For k-mean cluster analysis the standardized data was subjected to exhaustive searching for the optimal cluster number using cubic clustering criterion (CCC) (59), as well as using silhouette plot (Matlab, Natick, MA). The maximal number of clusters for the search range was set based on the number of hierarchic clustering applied to the same data. The number of clusters was validated by v-fold cross-validation (Statsoft, Tulsa, OK) (57) and, in case of limited number of points, the data were simulated for 10,000 points per variable and reclustered. An expectation maximization approach was also utilized, where minimum increase of log likelihood was set to 0.001. Self-organizing maps (SOM) were used for nonhierarchic clustering of data filtered out by factor analysis (see below). The number of clusters was evaluated using CCC. As in the case of k-mean clustering, the maximal number of clusters was set in accordance to the number derived from hierarchic clustering analysis applied to the same data. Grid arrangement was also exhaustive allowing all possible columns and raw combinations for the range. Validity of the obtained clusters was verified by simulation of the data with at least 10,000 points per variable.
Data Dimensionality Reduction and Factor Extraction
Data dimensionality was reduced using principal component analysis applied to the log2-fold change ratios generated from protein abundance data. Multiple correlation matrices generated from comparison of all variables were used for estimation of eigenvalue to assess principal components. Squared cosines were calculated as the ratio of squared factor score per protein divided by squared distance of all the factor scores of this observation (60):
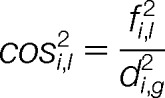 |
Proteins with larger squared cosine values related to the nonprincipal components with eigenvalue less than 1 were removed from the entire data set. Reduced data were subjected to factor analysis. Orthogonal factors were extracted by principal component method and verified by maximal likelihood method. Factors with eigenvalue higher than 1 were considered significant. The threshold for correlation between factor loadings and the variables was set to 0.7. The contribution of factors on variables was defined by evaluation of communalities. All orthogonal rotation methods were used for optimal principal axes alignment, though no significant differences were found among them, hence quartimax rotation was used as a default approach. Secondary factors were evaluated with hierarchic analysis of oblique rotation using oblimin method (61). The impact of factors on protein expression profiles was evaluated using factor scores analysis. Proteins expression profiles which correlated with a specific factor were extracted by projection of factor score coordinates in four-dimensional space onto six orthogonal planes. After projection on each plane, the azimuth angle was calculated for each projection point by arctan function. In each plane, one of the factors was assigned to x axis, and any points residing within the range of [-π/8, π/8] 200 [3π/8, 5π/8] were considered to be maximally correlated with a chosen factor. Obtained data sets were statistically evaluated for significance using Kruskal Walis one-way analysis of variance on rank with Dunn's posthoc test and p value cut-off set to 0.001. Proteins, which met these statistical criteria were further subjected to SOM clustering analysis (see Experimental Procedures, Data clustering).
Gene Ontology and Network Analysis
A protein data set correlating with a specific factor was subjected to visANT (integrative visual analysis tool for biological networks and pathways, (62)) versus mouse databases. The generated network was analyzed using network analysis tools implemented in the Cytoscape software package (63). Network clustering was evaluated using a fast agglomerative algorithm based on edge clustering (FAG-EC) implemented in ClusterViz plug-in of Cytoscape (64). The complex size threshold was set to 10. A network hub analysis was performed using node classification according to Guimera-Amaral functional cartography (65) based on spectral clustering implemented in GIANT plug-in of Cytoscape (65). Minimal hub criteria was set as within-module degree, z>2.5. Networks of proteins with strong expression level changes disregarding their clustering were assembled based on STRING10 database (66).
Gene ontology analysis was conducted using BiNGO plug-in of Cytoscape (67). A hypergeometric statistical test with Benjamini and Hochberg false discovery rate (FDR) correction was used. The significance level was set to 0.001. Data were analyzed versus the network generated in visANT (for Mus musculus database, see above) from the proteins detected in hippocampus during mass-spectrometry analysis for gene ontology (GO) containing all three ontological divisions. The obtained categories were further filtered to reduce redundancy of grouping into high hierarchical level categories using only GO categories with total frequency less than 5% and cluster frequency within the range of 5–15%. The obtained data was compared with GO clustering obtained from Functional annotation tool implemented in DAVID bioinformatics service (68). Finally, the obtained data was aligned against GO categories extracted from Ontologizer v.2 (69) using a topology-weighted algorithm corrected on Benjamin-Hochberg FDR. Only categories which were overlapped in two out of three methods were considered as enriched.
RESULTS
Radial Arm Maze Paradigm and Mass-spectrometry Analysis
Fifty mice per each biological replicates were subjected to the Radial arm maze paradigm. Initially, all animals were exposed to the habituation phase. Forty mice in each replicate were subjected to the learning phase and were tested during five consecutive days. Ten mice were used as a naïve control group (Fig. 1A). Trained animals exhibited significant improvement in learning curve expressed in reduction of time necessary to consume all the baits (p < 0.001, H = 34.173, ANOVA on rank; Fig. 1B), reduction of number of incorrect entries (p < 0.01, H = 16.675; Fig. 1C), and increase of fraction of correct entries (p < 0.01, F = 8.164, one way ANOVA; Fig. 1D).
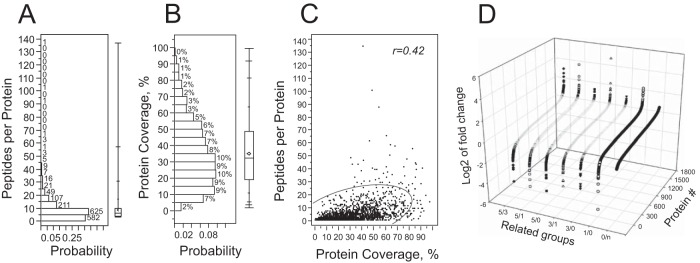
For the proteomic study, five experimental groups were generated based on memory acquisition time points and control, as follows: day 0, 1, 3, and 5 after the first training (0d, 1d, 3d, and 5d groups) and naïve group (N-group, Fig. 1A). At each time point, up to 10 mice were sacrificed for hippocampal protein extraction per each biological replicate. Mice for each biological replicate were collected from animals of different generation at the age of 3 months. Protein extracts obtained from hippocampi of the sacrificed animals were pooled per each time point in each biological replicate. Even though protein extracted from hippocampi of one animal would be sufficient for subsequent proteomic analysis, we took into consideration intrinsic variability of behavioral experiments. Hence, to minimize the impact on protein expression profiles of the fluctuations of individual animals in response to multiple external and nonspecific factors during learning: (1) we evaluated a higher number of animals than necessary based on power analysis (10 instead of 6 for power of 0.95) and (2) for each group we pooled a protein mix of individual mice. Importantly, no pooling was done on the animals of the same groups across the biological replicates. Protein mixes (7.5 μg) per each group of each replicate were used for total protein expression evaluation in label-free proteomic analysis (see Experimental Procedures). Tryptic digests of the protein extracts separated into five fractions per each group of each replicate were interrogated on the SynaptG2 instrument operating IMS-MS/MS mode. After removal of false positives by filtering against the UniProt decoy database and hits with less than 0.3% of FDR and with lowest minimal identification score at 5.8, the acquired spectra corresponded to 15245 unique peptides from 2256 unique proteins (supplemental Table S1; data stored on publically accessible server: http://www.ebi.ac.uk/pride/archive/projects/PXD002176), reconstituted from at least two peptides in all experimental groups from the three experiments from all the hippocampus, averaging 6.76 ± 0.17 peptides per identified protein, within a median number of 4. For quantitative analysis, we used 1592 unique proteins reconstituted from at least three unique peptides in each group of all biological replicates. For the quantitative case, we had 8.62 ± 0.22 peptides per protein (median = 6, 25th and 75th quartiles 4 and 10, respectively, Fig. 2A). The protein coverage level was about 35.10 ± 0.49% (median = 32.2%, with 25th and 75th quartiles: 19.2% and 48.5%, respectively) (Fig. 2B). A positive correlation trend was found between the peptide per protein number and protein sequence coverage (Fig. 2C). Despite a slight negative trend between protein sequence coverage and molecular masses of the proteins, we did not observe significant correlation (data not shown).
Fig. 2.
Statistical characteristics of data preprocessing used for protein quantitation. A, Histogram of peptides per protein distribution profile. Numbers above the histogram bars denote counts per bin. B, Distribution of sequence coverage of proteins reproduced from at least three peptides. Numbers above the histogram bars denote counts per bin. C, Scattered plot of all point correlation between Sequence coverage and peptides per proteins values. The “r” denotes correlation coefficient. Semieliptic line shows 95% confidence interval. D, Protein expression mean values distribution profile per each tested group averaged over all biological replicates. Protein # is unique for each expression profile in each group.
In order to analyze memory formation effect on protein expression, we further analyzed log2 of fold changes of protein expression between the tested groups. Fold changes per each protein were averaged over three biological replicates per each time point relation, for example, 0d training group versus naïve were presented as 0/n group, 5d versus 1d as 5/1 group. Analysis of log2 fold change of the tested groups revealed that only 15.2 ± 2.04% showed protein expression change larger by 50% (0.585 in log2 dimension) to either increase or decrease (Fig. 2D). However, this change was differently distributed in the tested groups (data not shown), hence the total number of proteins, expression profile of which underwent at least 50% change, was 1007, which is about 63.25% of all reconstituted proteins.
Protein Expression Profile Showed Significant Change During RAM Paradigm
Protein expression change may be the result of alteration in protein synthesis and/or protein degradation. Formation of long-term memory is dependent on both phenomena (38). The major changes in de novo protein synthesis appear hours after induction of long-term plasticity (15, 35). However, random fluctuation in protein expression level may occur within several minutes after behavioral paradigm triggering memory formation. These fluctuations cannot be correlated with memory formation. Hence, excluding proteins, which exhibited expression changes between the initial day of learning (0d) versus the animals, which were subjected only to the habituation phase, we filtered out the data not related to learning formation. Analysis revealed that protein expression changes were markedly larger in animals, which underwent RAM paradigm versus naïve. Namely, log2 of changes of protein expression 0d versus naïve group (0/n) was in the range of [-0.97; 0.99] (Fig. 2D). Although 147 proteins were found to be changed more than 1.5-fold in this group, neither of them showed twofold changes and only 13 proteins exhibited more than 90% change. In contrast, log2 of protein expression change observed between day 1 and day 0 of learning, group 1/0 (around 24 h after exposure to RAM paradigm), was in the range of [-5.26; 2.66] (Fig. 2D). Despite the number of proteins, which exhibited more than 1.5-fold change that was not marked different between 1/0 and 0/n groups (179 versus 147, respectively), the number of proteins with more than twofold change in the group 1/0 was 57, with none in 0/n group. A large number of proteins showed more than 2-fold changes at day 3 and day 5 of memory paradigm (56 and 89 proteins, respectively).
Despite clear evidence of protein expression change over time in the behavioral paradigm, individual protein profiles were very complex (supplemental Fig. S1A). Neither clustering method was optimal to recognize memory specific pattern change in protein expression profiles. Hierarchic cluster analysis converged on 22 clusters (supplemental Fig S1B). Despite the large number of clusters, protein expression profiles in each cluster showed complex behavior (supplemental Fig. S1C). The k-mean, expectation maximization (EM) clustering and self-organized maps (SOM) were not satisfactory. Using method v-fold cross-validation, k-mean clusters were optimized at 3 clusters, whereas EM clustering converged at 7 clusters. However, probability distributions and per cluster averages analysis revealed poor data separation (supplemental Fig. S1D–S1G). SOM algorithm failed to converge at any cluster number arrangement.
Enrichment Spatial Memory Formation Related Proteins
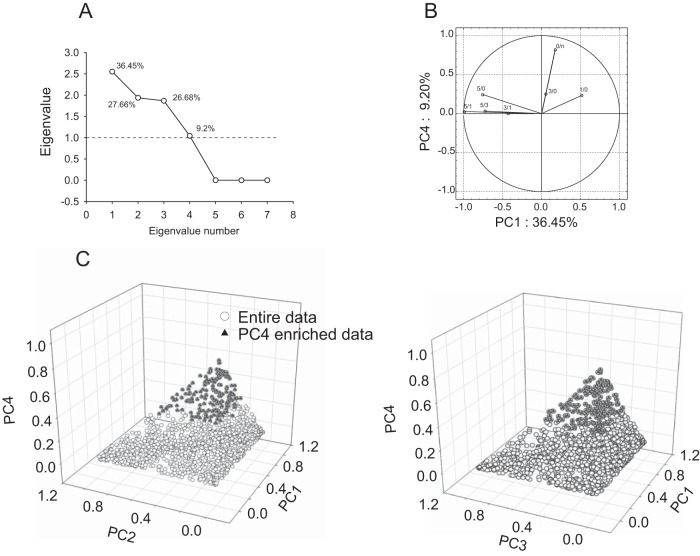
We reduced data complexity using advantages from our previous approach of linear decomposition of measured variables onto factors. According to the approach, variances of the variables measuring protein expression are determined by linear combination of numerous factors including the related memory formation (52). Initially, application of principal component analysis (PCA) to the entire data set revealed four principal components (PC) correlating with 99% of data (Fig. 3A). Factor loading analysis showed 81% correlation between group 0/n and PC4 (Fig. 3B). We considered PC4 as a memory nonrelated component. Using squared cosine data extracted from PCA analysis (see Experimental Procedures), 167 proteins highly correlating with PC4 were eliminated (Fig. 3C).
Fig. 3.
Removal RAM paradigm nonrelated proteins using Principal component analysis (PCA). A, Scree plot depicting eigenvalues distribution of PCA applied to variables representing protein expression ratio of all time point ratio groups averaged over three biological replicates. Percentage numbers above each eigenvalue coordinate depict data variance accounted for by that eigenvalue, hence being correlating with a specific factor. B, Scattered plot representing correlation between the variables and principal components, PC1 and PC4. Graph displays significant correlation between 0/n and PC4. C, Scattered plot of factor score coordinates of individual proteins on four dimensional space (four PCs) represented in two 3D plot. Extraction of PC4 correlating proteins (solid triangles) using squared cosine extraction method.
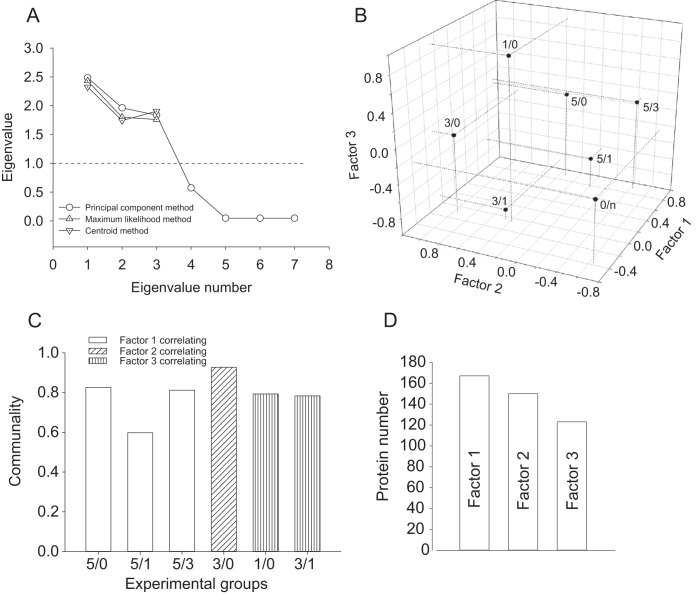
The enriched 1424 protein expression profiles were subjected to exploratory factor analysis. Factor extraction was conducted using three different approaches: (1) principal component, (2) maximum likelihood, and (3) principal factors/centroid based methods. All methods identified three factors although with slight differences in eigenvalues (Fig. 4A). Quartimax rotation was found as the best correlation fit of factor loadings on the variables. No factor interdependence and no secondary factors were detected upon application to the data of Oblimin rotation and hierarchic analysis (data not shown). The extracted orthogonal factors showed the following pattern of correlation: factor 1 strongly correlated with variable of the 5d versus other learning days (5/0, 5/1 and 5/3), factor 2 strongly correlated with variable 3/0 and factor 3 with variables 3/1 and 1/0 (Fig. 4B). Neither of the factors disregarding the method of extraction correlated with variable 0/n, indicating that preliminary PCA eliminated protein was unrelated to the RAM paradigm based spatial memory formation. Analysis of communalities showed that the extracted factors are capable to explain a majority of variance of the correlated variables (Fig. 4C). Analysis of factor scores resulted in total enrichment of 440 proteins, which were significantly affected by the correlating factor (Fig. 5D, supplemental Data S1). Quality of factor analysis was validated by support vector machine (SVM) algorithm, showing strong linear correlation of protein expression profiles and factor score based predicted variables as a result of factor analysis application (supplemental Fig. S2). Outlier proteins, which were enriched by factor analysis, however, were not within α = 0.95 range, as a result of SVM, and were removed.
Fig. 4.
Factor analysis of reference memory related protein expression profiles. A, Scree plot depicting eigenvalues distribution of principal factors extracted by three different methods, as denoted. B, 3D scatter plot representation of factor loading on variables with projections on three principal axes. C, Communality evaluation for each averaged variable showing contribution of factors in explanation of variance of a specific variable. D, Number of proteins correlating with the factors specified inside the bars.
Fig. 5.
Quantitative and functional characterization of proteins correlating with factor 1. A, Expression profiles of individual proteins enriched in specific clusters using hierarchic cluster analysis applied to proteins correlating with factor 1, as shown in Supplemental Fig. 3A. B, Expression landscape corresponding to variable 5/0 representing protein level changes days 5 versus 0 for the group correlating with factor 1. C, Average of each hierarchic cluster per most correlated variable for factor 1 (5/0). Box and Whisker representation: the boxes show the 25–75% range, and the inner square in each box is the median. Error bars denote nonoutlier region. D, GO annotation and KEGG and BioCarta signaling pathway database based functional clustering of proteins agglomerated in hierarchic cluster of data negatively correlating with factor 1. E, As in D for proteins positively correlating with factor 1.
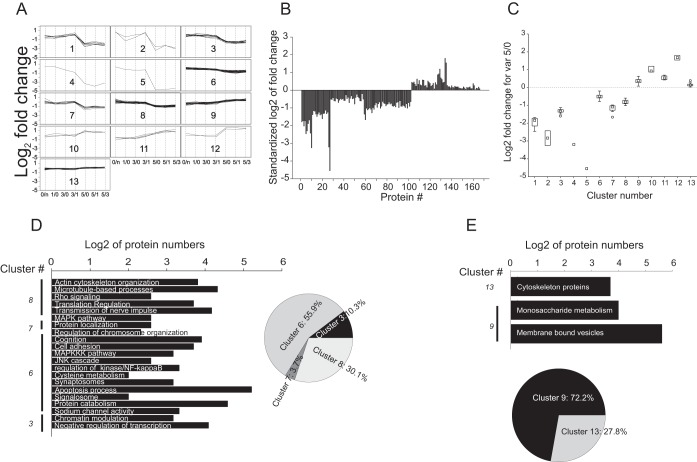
Proteins Correlating with Factor 1
Expression profile distribution of 165 proteins correlating with factor 1 showed a strong agglomeration pattern, which prevented appropriate partitioning by nonhierarchic clustering (data not shown). Hierarchic clustering partitioned the entire protein data set into 13 clusters (Fig. 5A; supplemental Fig. S3A; supplemental Data S1). Clusters 1–8 and 9–13 showed negative and positive correlation with factor 1, respectively. The expression profiles within the clusters did not show normal distribution (Shapiro-Wilk normality test failed, p < 0.05). Kruskal-Wallis one-way analysis of variance on ranks revealed statistically significant difference between the clusters (clusters 1–8: H = 85.755, p < 0.001 and Dunn's post-hoc analysis Q ∈[2.594; 6.867]; clusters 9–13: H = 39.113, p < 0.001 and Dunn's post-hoc analysis Q∈[3.731; 4.830]). Proteins correlating with factor 1 showed a significant change of expression pattern at day 5 in comparison to the previous days of the RAM paradigm (Fig. 5B; supplemental Fig. S3B). Comparison of distribution of medians of the clusters showed that a majority of changes occurred within 1.2–twofold range (Fig. 5C).
The clusters were subsequently subjected to functional categorization analysis based on GO categories and signal transduction pathways' enrichment. Three out of eight clusters negatively correlating with factor 1 were excluded from functional analysis there were less than five proteins per cluster. Protein sets of the clusters subjected to functional analysis were mapped onto a protein-protein interaction network using visANT software (see Experimental Procedures). Enriched first order interacting proteins were assembled into the protein-protein interaction networks for each cluster separately. The generated networks were further analyzed for GO categories and signal transduction pathway enrichment. A protein network generated from the whole proteomic data of all detected proteins was used as a background of the GO analysis. Proteins negatively correlating with factor 1 were significantly downregulated on day 5, a day of established spatial memory. Functional analysis of the networks of proteins negatively correlating with factor 1 revealed that upon completion of memory formation, transcriptional activity might be enhanced because of (1) enrichment of proteins negatively regulating transcription (GO: 0030163; p < 10−6, fdr<10−3) and (2) proteins involved in chromatin organization (GO: 0016568; p < 0.001; fdr<0.01). These GO categories were enriched in a generated network based on cluster 3, protein expression of which was downregulated more than twice (Fig. 5A, 5C, 5D). Indirectly, enrichment of programmed cell death regulation (GO: 0043067; p < 0.0001, fdr<0.01) and different pathways of proteolysis (GO: 0044257; p < 0.0001, fdr<0.01) including signalosome (GO: 0019717; p < 0.001, fdr<0.01) functional categories, also indicated reduction of protein degradation (because of negative correlation). The latter categories were enriched in cluster 6, which exhibited minor reduction in protein level in contrast to the data set of cluster 3 (Fig. 5A, 5C, 5D, supplemental Data S2). In addition, we observed enrichment of (1) proteins involved in action potential transmission (different types of voltage gated sodium channels (GO: 0005272; p < 0.0001, fdr<0.001)); (2) proteins associated with numerous signal transduction pathways, such as NF-κB (GO: 0043122; p < 0.001, fdr<0.01); MAPKKK (GO: 0043408; p < 0.001, fdr<0.01) and JNK (GO: 0046328; p < 0.001, fdr<0.01) cascades; (3) proteins associated with focal adhesion (GO: 0004707; p < 0.001, fdr<0.01). These categories were enriched in cluster 6 and 8, respectively, manifesting less than twofold expression changes (Fig. 5A, 5C, 5D, supplemental Data S2). Clusters 6 and 8 were also enriched for MAPK signaling pathways (GO: 0004707; p < 0.001, fdr<0.01, Fig. 5D, supplemental Data S2). Despite significant homogeneity of quantitative changes in cluster 6, FAG-EC analysis of the network assembled on cluster 6 revealed the existence of six network subclusters/domains (nc1 to nc6) exhibiting significant functional modularity (supplemental Data S2). Although three smaller network subclusters, nc3–5 were enriched for the protein degradation category, nc1 showed functional association with the voltage gated sodium channels. Network subclustering revealed hidden enrichment for actin cytoskeleton organization category (GO: 0030036; p < 0.0001, fdr<0.001). Heterogeneity of the network of cluster 8 was also reflected in MAPK activity, translation regulation (GO:0006417; p < 0.001; fdr<0.01), nerve impulse transmission regulation (GO: 0019226; p < 0.001, fdr<0.01), cytoskeletal process organization including microtubules (GO: 0007017; p < 0.001, fdr<0.01) and microfilaments (GO: 0030036; p < 0.001, fdr<0.01). Notably, the strongest expression level decrease was observed in single element clusters 4 and 5, containing Na/K-ATPase subunit beta1 and Rab11A protein, respectively.
Among clusters of the proteins positively correlated with factor 1, only cluster 9 and 13 were large enough for functional analysis. Protein-protein interaction networks generated by these data sets showed strong heterogeneity, which was confirmed by enrichment of a network hub using Guimera-Amaral's cartographic analysis. A 1.7-fold up-regulated CaMKIIa was found to serve as a network hub in the enriched data set (supplemental Fig. S3C). The assembled network was found to be enriched for metabolic processes and intracellular transport. Namely, among the most enriched categories were found proteins associated with membrane bound vesicles (GO: 0031988; p < 0.001, fdr<0.01) and intracellular vesicular transport (GO:0031988, p < 0.0001, fdr<0.01), proteins involved in monosaccharide metabolic enzymes and regulators (GO:0005996; p < 10−8, fdr<10−6). (Fig. 5E, Suppl. Data 2). Despite a small number of proteins, clusters 10–12 exhibited the strongest increase in expression profiles, especially for cytoskeleton regulation related proteins, ROCK2, Rho GEF7, Metastasis suppressor protein 1, and for neuronal adhesion protein NCAM1, with about 3.5-fold enhancement. Hence enhancement of cytoskeleton rearrangement and organization should not be excluded.
Proteins Correlating with Factor 2
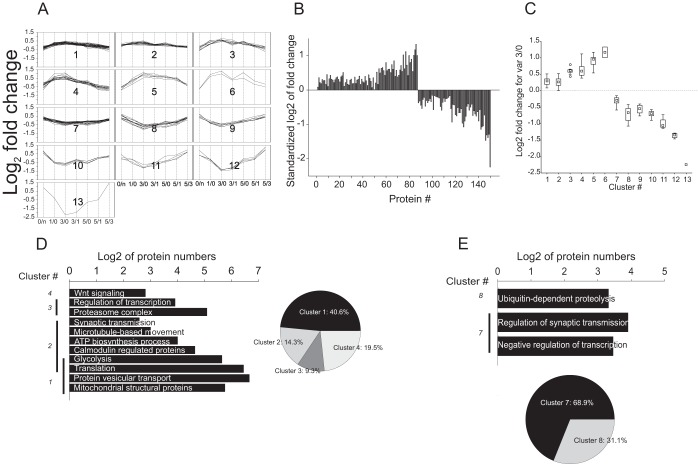
Factor 2 exhibited strong correlation with a variable 3/0 pointing to a factor associated with protein turnover changes occurring during memory engram formation process at the steep phase of the learning curve. Hierarchic analysis of this protein data set partitioned the data into 13 clusters containing 148 proteins. Clusters 1–6 and 7–13 were positively and negatively correlating with factor 2 (Fig. 6A; supplemental Fig. S4A; supplemental Data S1). Although most of the changes in protein expression occurred within 1.5–twofold range, a limited number of proteins (clusters 12 and 13) exhibited more than threefold change in the expression level (Fig. 6B, 6C; supplemental Fig. S4A; supplemental Data S1). The expression profiles within the clusters did not show normal distribution (Shapiro-Wilk normality test failed, p < 0.05). Kruskal-Wallis one way analysis of variance on ranks revealed statistically significant difference between the clusters (clusters 1–6: H = 57.034, p < 0.001 and Dunn's post-hoc analysis Q ∈[3.998; 5.058]; clusters 7–13: H = 46.150, p < 0.001 and Dunn's post-hoc analysis Q∈[3.199; 5.065]).
Fig. 6.
Quantitative and functional characterization of proteins correlating with factor 2. A–C, As in Fig. 5A–C. D, GO annotation and KEGG and BioCarta signaling pathway database based functional clustering of proteins agglomerated in hierarchic cluster of data positively correlating with factor 2. E, As in D for negatively correlating proteins.
Five out of six clusters showing positive correlation with factor 2 contained at least 5 proteins, sufficient for functional analysis. The assembled networks of protein-protein interaction exhibited a marked enhancement of biological processes associated with protein synthesis, metabolic processes necessary for its maintenance, and processes of protein intracellular transport and dynamics. Cluster 1 and 2 showing moderate expression profile changes contained different isoforms of proteins 14–3-3 (14–3-3 epsilon and 14–3-3 zeta/delta, respectively), which interact with a large number of proteins. Consequently, networks generated based on clusters 1 and 2 incorporated a significant number of nodes (3799 and 790, respectively) and exhibited a strong network heterogeneity with proteins 14–3-3 serving as a hub of the networks (supplemental Fig. S4B). Existence of a hub protein within the network led not only to topological, but also functional heterogeneity. The network of cluster 1 was enriched in three major GO categories: (1) protein translation (GO: 0006412; p < 0.001, fdr<0.01), (2) proteins populating mitochondrial matrix (GO: 0005759; p < 0.001, fdr<0.01); (3) proteins associated with vesicle-mediated transport (GO:0016192). Analysis of the networks of cluster 2 showed enrichment for translation category (GO: 0006412; p < 0.001, fdr<0.01) and proteins involved in bioenergetics processes necessary for protein anabolism, such as glycolysis (GO: 0006096; p < 10−13, fdr<10−11) and proteins of the ATP biosynthetic machinery (GO: 0005739; p < 0.001; p < 0.05) (Fig. 6D; supplemental Data S3). Despite differences of protein expression patterns between cluster 1 and 2, the networks of these clusters evidenced involvement in synergistic metabolic processes related to protein synthesis and its maintenance. One set of functional processes related to protein translation with subsequent protein movement and localization and another set of functional processes necessary for bioenergetics support of protein anabolism and transport. Notably, despite significant recruitment of proteins involved in gene expression and protein synthesis, the quantitative extent of these changes was limited to 22.3 ± 2.9% increase of protein levels (Fig. 6A–6C, supplemental Data S3), indicating an extensive enhancement of protein anabolism, though at relatively low expense of cellular resources. In addition to up-regulation of the core machinery of protein anabolism, we observed enrichment of proteins interacting with calmodulin (GO: 0005516; p < 0.0001, fdr<0.01), a key regulator protein of multiple signal transduction pathways in the network of cluster 2. The network of clusters 3–5, with 60 ± 2.6% averaged increase in protein expression profiles, exhibited functional enrichment consistent with the clusters 1 and 2 (Fig. 6A–6C; supplemental Data S3). A homogenous network of cluster 3, containing proteins such as SNAP25, dynactin 2, and tubulin was enriched for vesicular transport and exocytosis, including microtubule-based movement (GO: 007018, p < 10−7, fdr<10−5), proteins associated with synaptic vesicles (GO: 0031982, p < 10−11, fdr<10−10), and proteins regulating synaptic transmission (GO: 0007268, p < 10−8, fdr<10−6). Significant heterogeneity was observed in the network of cluster 4 being enriched for protein catabolism machinery (GO: 0009057, p < 0.001, fdr<0.01) and regulation of transcription (GO: 0006350, p < 0.001, fdr<0.01) with marked enhancement of the α5 subunit expression of ribosomal S6 kinase (Fig. 6D, supplemental Data. S1, S3). This significant diversity of functional roles was entailed because of subcluster/domains inside the network. FAG-EC analysis (see Experimental Procedures) partitioned the network into 3 network subclusters (nc) showing functional modularity. The network subcluster 1 (nc1), composed of 31 nodes, was enriched for proteasome core complex GO category. The nc2 including 31 nodes and a network hub protein PCBD1, was enriched for transcription regulation category. Interestingly, the partitioning of the network increased the resolution of functional analysis leading to enrichment of Wnt signaling pathway (GO: 0016055, p < 0.001, fdr<0.001) in nc3, which included 14 nodes (Fig. 6E, supplemental Data S3). No functional enrichment was observed for the network based on cluster 5.
Only three out of seven clusters negatively correlating with factor 2 (clusters 7–13, Fig. 6A–6C, supplemental Data S3) included a sufficient number of proteins to be subjected to functional analysis. The network of cluster 7 was enriched in two major functional groups involved in (1) negative regulation of transcription (GO: 0010629, p < 10−9, fdr<10−7) and (2) regulation of synaptic transmission (GO: 0010629; p < 10−12, fdr<10−9). The diversity within the cluster was explained by the network heterogeneity (Fig. 6E). FAG-EC analysis revealed four network clusters. Two network subclusters were found to be associated with transcriptional regulation (nc4), regulation of synaptic transmission, and long-term synaptic plasticity (nc1) (supplemental Data S3). Clusters 8 and 10 revealed a simple functional picture. The network of cluster 8 was enriched for ubiquitin dependent proteolysis (GO: 0051603, p < 0.001, fdr<0.01). Neither of clusters 9–13 bore sufficient proteins for their functional evaluation, although proteins of these clusters exhibited strong expression reduction (Fig. 6A–6C).
Proteins Correlating with Factor 3
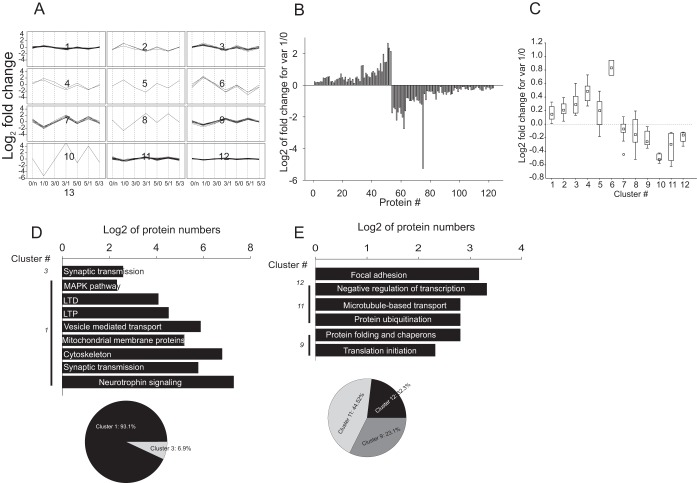
Factor 3 exhibited strong correlation with a variable 1/0 pointing to a factor associated with protein turnover changes occurring during the initial phase of memory acquisition and, potentially, upon consolidation of long-term synaptic plasticity. Hierarchic clustering partitioned the whole data set into 12 clusters containing 123 proteins: clusters 1–6 and 7–12 were positively and negatively correlating with factor 3 (Fig. 7A; supplemental Fig. S5A; supplemental Data S4). Overall, the up-regulation pattern prevailed in the data set, although the increase was up to 2-fold, at most (Fig. 7B, 7C).
Fig. 7.
Quantitative and functional characterization of proteins correlating with factor 3. A–C, As in Fig. 6A–C. D, GO annotation and KEGG and BioCarta signaling pathway database based functional clustering of proteins agglomerated in hierarchic cluster of data negatively correlating with factor 2. E, As in D for positively correlating proteins.
The clusters positively correlating with factor 1 showed that only two out of six, clusters 1 and 3, were useful to obtain function information. Cluster 1, containing the largest cluster of this group, included 32 proteins with a subtle changes in expression (Fig. 7A–7C, supplemental Data S4). The protein-protein interaction network generated based on this data set was found to be highly heterogeneous. Heterogeneity was related to the existence of four network subclusters/domains according to FAG-EC analysis and was enhanced with a hub protein, 14–3-3β/α (supplemental Data S4). Topological heterogeneity was accompanied by the functional diversity of the network. Three out of four network subclusters failed to be associated with specific GO categories. The network subcluster 4 (nc4), which was organized as a large network domain around the hub protein 14–3-3β/α, showed significant functional diversity. The nc4 was enriched for proteins involved in synaptic transmission (GO: 0007268; p < 0.0001, fdr<0.01), including both pre- and postsynaptic components (supplemental Data S4). In addition, nc4 was enriched for proteins of mitochondrial membranes (GO: 0031966; p < 0.01, fdr<0.05) and cytoskeleton associated proteins (GO:0005856; p < 0.0001, fdr<0.01), including the core components of microfilament networks, as well as multiple regulatory proteins and kinases, proteins of post-synaptic density, and motor proteins (supplemental Data S4). Another network domain, nc2, showed enrichment for neurotrophin (mmu04722; p < 0.001, fdr<0.05) and MAPK (p < 10−7, fdr<10−5) signaling pathways, as well as for pathways involved in long-term potentiation (mmu04720; p < 0.001; fdr<0.01) and long-term depression (mmu04730; p < 0.001, fdr<0.01; Fig. 7C, supplemental Data S4). Network domain, nc3, was enriched for neurotrophic factor signaling (p < 10−9, fdr<10−7; supplemental Data S4). Protein assembling networks of cluster 3 were associated with synaptic transmission, though because of the small number of proteins, a low level of enrichment was observed (Fig. 7D, supplemental Data S4). The strongest up-regulation pattern and the highest factor scores were observed for proteins of cluster 3–6. Using the STRING10 database (66) revealed two major domains of protein-protein interaction domains involved in protein processing assembled around the AKT hub protein (supplemental Fig. S5B). Significant enrichment was observed in protein degradation and translation initiation GO categories (p < 10−10, FDR<10−8). Applying a Markov clustering algorithm, MCL (70) to the combined network of the highly expressed proteins positively correlating with factor 3, revealed two major network clusters, protein turnover, and tyrosine kinase signaling, including ErbB (GO: 0038138; p < 10−13, fdr<10−11) pathway.
70 proteins distributed over 6 clusters negatively correlating with factor 3 showed a mild reduction of the expression level at day 1 versus day 0 (Fig. 7A–7C). Two major clusters, 11 and 12, showing more than 25% protein expression reduction were found to be functionally enriched (Fig. 7E, supplemental Data S4). Among enriched GO categories, we identified negative transcription regulation (GO: 0016481; p < 0.001, fdr<0.01), protein folding (GO: 0006457; p < 0.001, fdr<0.01), protein ubiquitination (GO: 0016567; p < 0.0001, fdr<0.01) and degradation (GO: 0030163; p < 0.001, fdr<0.01), endocytosis (GO: 0006897; p < 0.001, fdr<0.05), focal adhesion (mmu04510; p < 0.01, fdr<0.05) and (Fig. 7E, supplemental Data S4). Prominent diversity of expression patterns observed for this data set was related to network modularity linked to the existence of multiple subdomains identified by FAG-EC analysis (supplemental Data S4).
DISCUSSION
Proteomic study of memory related processes is a complicated endeavor. Learning and memory are complex processes incorporating multiple neuronal networks across different structures of the brain (71). These networks may elaborate a vast number of different types of neurons. Despite the abundant candidates involved even in a specific learning process, the actual number of neurons may be limited and a memory engram may be spatially curbed to specific subspaces of neurons. Learning is a dynamic process while protein expression and post-translational modification profiles may markedly change at different stages (33, 72, 73). Long-term synaptic plasticity is believed to be a cellular correlate of LTM. However, long-term potentiation (LTP), the most thoroughly studied form of long-term synaptic plasticity, is established within hours and may persist for hours, days, and even longer (74). In contrast, complex learning tasks, such as spatial memory, are formed during several days, require multiple recurrence of enforcement, and may elaborate interaction of different structures in and outside of the hippocampus (75). Hence, proteomic study of synaptic plasticity may not directly be extrapolated onto protein expression and post-translational modification changes occurring during formation of LTM. In this study, using label-free quantitative proteomics of the hippocampus, to our best knowledge for the first time, we tried to track down protein turnover changes occurring along the whole process of formation of reference spatial memory using the RAM paradigm. Appreciating dynamic profiles of about 1600 proteins, quantitative changes occurring during the RAM paradigm (supplemental Table S1), we found that the protein expression pattern and their functional categories are strictly related to temporal frames of memory formation.
The label-free quantitative proteomics was capable of estimating expression profiles of 1592 proteins (Fig. 2) reconstructed based on at least three peptides at all tested time points in all three biological replicates. A time-dependent study of memory formations may raise several critical issues capable of affecting noisiness of the proteomic data: (1) behavioral issue: individual learning ability of animals may markedly differ, potentially amplifying individual measurement related fluctuations in memory-dependent protein expression; (2) biological source issue: the RAM paradigm induces a spatial memory, which is hippocampal-dependent, however (1) the hippocampus is a complex and heterogeneous region of the brain and (2) spatial memory on its own is a heterogeneous phenomenon (75); and (3) biological replicates issue: learning ability and protein expression extent may be affected by the batches of used animals as well as by slight seasonal differences occurring during the learning process between different biological replicates. The behavioral issue was resolved by using animals which did not show existence of outliers in RAM measurement parameters as denoted by low values of standard deviations (Fig. 1). The biological source issue could not be resolved at the level of proteomic analysis; hence, it was inherently affecting protein expression at every measured time point, while averaging over three biological replicates and pooling of hippocampal extracts within the groups was supposed to suppress oscillation related to this issue. Use of a multivariate analysis approach including PCA and factor analysis, as well as validation with SVM on the averaged data per time point, allowed removal of proteins not-related to learning formation and enrichment of proteins differentially expressed during memory formation (Fig. 4 and supplemental Fig. S1, supplemental Data S1). An additional enrichment level was provided by subjection of proteomic data to protein–protein interaction network analysis.
Acquisition of memory is supposed to initiate activity-dependent changes in synapses leading emergence of long-term synaptic plasticity. At these stages numerous molecular and morphological changes occur on the synapses, including formation of new spines and reorganization of existing ones (76–79), as well as silent synapses activation (80–82). The early appearance of these changes should occur during 24 h from the memory acquisition initiation and correspond to the alteration of protein expression/degradation during the late phase LTP (14–16, 38, 83). In turn, protein turnover changes are dependent on enhancement of transport and metabolic activity and may lead to changes in the synaptic component. Factor 3 correlated with a variable, which corresponded to changes occurring during initiation of memory acquisition within 24 h after exposure of the positive reward. Proteins correlating with factor 3 showed strong functional association with the activity-dependent changes occurring in synapses. The eIF3d, Psma6, Ubxn6, and Usp9x, showing strong positive correlation with factor 3, were assembled into the protein–protein interaction network involved in protein synthesis and degradation (supplemental Fig. S5B) indicating enhancement of protein turnover integrity during memory consolidation. Moreover, protein synthesis requires involvement of MAPK pathway (84, 85), which was also found to correlate positively with factor 3 (Fig. 7D). Enrichment of neurotrophic factor signaling, necessary for late-phase LTP, such as BDNF activity (86), also supported a link between protein correlation with factor 3 and synaptic plasticity consolidation. Importance of ErbB signaling pathway enrichment in this group of proteins was also shown (87, 88). Other components of the networks of clusters positively correlating with factor 3 were aggregated into synaptic structural proteins, synaptic transmission categories, signaling pathways related to synaptic plasticity, LTP, and LTD (Fig. 7D, supplemental Data S4). This enrichment pattern is highly suitable to the processes occurring during late and persistent phases of long-term synaptic plasticity occurring during consolidation of memory (89). Previous studies already showed that expression and activity of the numerous proteins enriched in the network emerged from the proteins positively correlating with factor 3 to be essential for long-term memory. Expression of the presynaptic release machinery proteins, enriched in the network analysis, such as synapsins (90, 91), SNAP25(92), synaptotagmin (93), and syntaxin 1A(94, 95), was shown to be involved in regulation of the different forms of associative, punishment and pain-relief related memories, hippocampal-dependent long-term memory formation, and short- and long-term synaptic plasticity. Similarly, the enriched proteins of synaptic vesicle turnover, Rab3a (96, 97), piccolo (98) were also shown to participate in regulation of long-term memory including the reversal of spatial memory, as well as in regulation of synaptic plasticity observed in Mossie fibers. Strong up-regulated homer 3 (supplemental Data S1) belonging to homer family of post-synaptic density scaffold proteins, which plays an important role in mGluR1 signaling and regulation of LTP and LTD, was also shown to be transcriptionally regulated by synaptic activity (99, 100). Formation of new spines and change of their morphology would be impossible without changes in cytoskeletal components, actin microfilaments (e.g. actin-related proteins, cortactin), and motor proteins (kif5A, kif5B, dynein heavy chain), which were enriched in the network positively correlating with factor 3 (101). Enhancement of expression of actin related proteins, cortactin, and tubulin polymerization-promoting protein (supplemental Data S4) further supports the importance of cytoskeletal proteins in long-term memory consolidation. Motor proteins, found to correlate positively with factor 3, also play an active role in LTM formation (supplemental Data S1, S4). Kinesins were shown to be essential for delivery to the synapse of mRNA, necessary for local synthesis of synaptic proteins (reviewed in (102)). De novo protein synthesis, enhancement of synaptic release, and new spine structure formation because of cytoskeleton rearrangement are energy consuming processes. Our data showing enrichment of protein networks positively correlating with factor 3 and associated with mitochondrial metabolic activity (Fig. 7D) is consistent with the requirement of enhancement of metabolic activity to maintain changes associated with synaptic consolidation (103). It is not surprising that a limited number of proteins negatively correlating with factor 3 were also associated with the same GO categories as proteins positively correlating with factor 3. Uba2 is of a specific interest, acting as E1 ligase for SUMO 1–3 version of the protein degradation system (104, 105). Strong reduction of Uba2 expression accords well with the recent findings demonstrating reduction of SUMOyation of aggregated CPEB3, a prion-like protein, which promotes protein synthesis leading to maintenance of LTP and consolidation of long-term memory (106, 107). The role of CREB SUMOylation has been established in maintenance of long-term spatial memory (108). In light of these reports, negative correlation of factor 3 with the components of protein folding category does not seem very surprising. The components of protein folding are particularly important during de novo protein synthesis as occurs upon memory consolidation. However, the requirement for the protein aggregation upon formation of long-term memory (107) may justify reduction the levels of chaperones, as CCT6A and CCT2 (supplemental Data S1). Suppression of negative transcription regulation observed in this group of proteins may be the initial step of enhancement of transcriptional activity observed at the later stages of memory consolidation and requiring increase of mRNA pool (109, 110). Negative correlation of synaptojanin 1 and dynamin 1 with factor 3 is controversial, particularly in light of the recent publication showing a crucial role of dynamin 1 in LTP and memory modulation (111). However, these changes may be finely tuned depending on the stage of LTP and memory engram formation. The additional important outcome of evaluation of proteins correlating with factor 3 was very mild expression changes in protein levels. Remarkably, a local protein synthesis is assumed to be necessary for establishing late phase synaptic plasticity (14–18). Considering that most of the changes may occur in a very limited space, as claimed by the cluster plasticity hypothesis (16, 112), no dramatic increase of total protein level changes could be expected.
Factor 2 showed correlation with protein expression change during a steep phase of the learning curve. Presumably, during this phase of the learning curve consolidation and reconsolidation coincide (113, 114). LTM consolidation and reconsolidation require increase of mRNA pool and processing, necessary to sustain protein expression (34, 115) that was directly supported by up-regulation of mRNA processing proteins, such as, tRNA-splicing ligase RtcB homolog, hnRNPul2, and hnRNP-K (Suppl. Data. 1 and 3) among the factor 2-correlating group. Interestingly, recent studies have already shown hnRNP-K is required for LTP and dendritic spine development (116). Nonetheless, transcription factor level changes could be too subtle to be identified in the proteomic analysis. Definitely, the role of their post-translational modifications of the transcription may be of superior importance. Moreover, transcription factor up-regulation might occur in response to the first phase of transcription occurring shortly after induction of mechanisms leading to long-term synaptic plasticity (117). Contribution of transcription factors could be detected during the second transcription phase necessary for memory gene expression program, as it was recently shown for CREB transcription factor activity, a hallmark of LTM (118). This suggestion is suitable for the network assembled around the proteins positively correlating with factor 2, showing enrichment of transcription, mRNA processing, and translation related gene enrichment (Suppl. Data1 and 3). Hence, these findings indicate that, at this stage, formation of memory engram may be sustained and enforced not only by local protein synthesis, but also by increase of cellular translation and delivery of de novo synthesized proteins to the tagged synapses (119). Strong up-regulation of dynactin 2, SNAP25, SV2A, and Sorting nexin 2 during the steep phase of the learning curve led to enhancement of intracellular vesicle trafficking and microtubular transport (Fig. 6D, supplemental Data S3), which are processes necessary for synaptic plasticity. In line with protein synthesis, factors involved in protein degradation were also found to be enriched. Presumably, strict balance in protein synthesis and degradation should be preserved for maintenance of long-term memory. Enrichment of Wnt pathway is also compatible with previous findings of the involvement of this signaling in regulation of late phase LTP and LTM (120, 121). Enhancement of metabolic activity introduces strong demand in the energetic resources, which were found to be supplied by up-regulation of proteins involved in glycolysis and the ATP biosynthesis machinery (Fig. 6D, supplemental Data S3). The ensemble of proteins negatively correlating with factor 2 evidenced reduction of activity of the network involved in heterochromatin organization and negative regulation of transcription, such as down-regulation of TRIMM28. Weakening of these factors should be favorable for intensive protein anabolic activity occurring in neurons. More controversial were results related to negative correlation of some network components involving synaptic transmission, syntaxin 1B, synapsin 2, and syntaxin binding protein 1 found to be important for synaptic plasticity and memory formation. For example, previous studies had shown syntaxin 1B as an essential factor in LTP and memory induced synaptic plasticity (122, 123). Association with learning and age related cognitive impairment was found for synapsin 2 as well (124–126). A possible explanation could be related to differential modification of synaptic efficacy and memory engram formation may involve not only enhancement but also weakening of synaptic efficacy depending on specific neuronal networks and time elapsed since initiation of LTM.
Factor 1 correlated with variables characterizing changes occurring at the last day of the RAM paradigm showing near maximal improvement of reference memory. The most prominent outcome of expression profile positively correlating with factor 1 was related to the appearance of cytoskeleton organization proteins, such as profilin, fascin, coronin, and MTSS1. Multiple previous studies showed an essential role of profiling in long-term potentiation, associated cytoskeletal rearrangement, and importance in memory (127–129). Involvement of coronin in regulation of synaptic plasticity and cognition was also confirmed (130). Despite the robust network, observation of mild expression level changes indicate that during persistence of memory minor re-arrangement of cytoskeletal components of cellular and synaptic structures may still occur. As much can be indicated by moderate, though steady, co-incident increase in complexin 1 and 2 levels on the 5th versus all the previous days. Of interest, complexins were shown to be necessary for AMPARs exocytosis on the postsynapses, as well as needed on the presynapses, for LTP expression and proper cognitive functioning (131–133). Hence, slight increase in protein levels of the networks responsible for the intracellular vesicular transport also supports this notion.
Analysis of proteins negatively correlating with factor 1 provides unambiguous evidence that intensive anabolic processes are on their decline phase. Namely, down-regulation of signaling pathways such as MAPK, NF-κB and JNK, as well as ribosomal proteins and microtubular motor components, regulators of microfilament cytoskeleton, spine scaffolding proteins and cytoplasmic vesicle pools associated with NMDA, and insulin-like growth factor receptors. Together with the data of ErbB down-regulation pathway, this evidence indicates that no additional enhancement of synaptic efficacy is going to appear at this stage of memory formation, but all the processes are directed to maintenance of the formed memory engram. Nevertheless, some unexpected data was also observed such as down-regulation of voltage gated sodium channel that inevitably should lead to reduction of neuronal excitability (Fig. 5E, supplemental Data S2). Considering that LTP may lead to intrinsic excitability increase (134), reduction of sodium channel expression may be attributed to a compensatory mechanism preventing overexcitability. Because the changes occurring at this stage of learning have never been observed previously, as well as a lack of functional, electrophysiological data, further study is needed for better understanding of the occurring changes.
Taken together, proteomic study of temporal changes in protein expression profiles during acquisition of long-term spatial memory showed a clear correlation between behavioral changes and their molecular counterparts. Moreover, despite huge data complexity, the impact of multiple factors, including behavioral variability, a combinatorial analysis of the data enriched by multifactorial, network, and functional analysis draw a clear correlation between previous knowledge about protein expression related to synaptic plasticity and long-term memory. Moreover, this research clearly demonstrated dynamic assembly and disassembly of protein-protein interactions' functional network depending on the stage of formation of memory engram. Despite these interesting findings, current research is just the first step in understanding how and which proteins are necessary for memory engram formation. This study made only low resolution snapshots of protein expression changes during memory formation based on the whole neuronal extracts, being also contaminated by multiple glia cells. Although, the role of the glia cells cannot be ignored, particularly based on the recent reports showing their importance in regulation of long-term potentiation, long-term memory, and memory consolidation (135–139). Future directions of understanding protein behavior at different stages of memory formation should incorporate comparison of changes at the synaptic level versus the whole alteration, as well as analyze different regions of the brain and increase spatial resolution of study, namely locking down onto subregions, such CA1 or CA3 or dentate gyrus in the hippocampus. And, finally, understanding of post-translational modifications is the key component in the creation of complete pictures of “molecular memory.”
Supplementary Material
Footnotes
Author contributions: N.B., E.N., A.P., and I.M. designed research; N.B., E.N., Y.L., M.R., and I.M. performed research; Y.L. and I.M. analyzed data; N.B., E.N., M.R., A.P., and I.M. wrote the paper.
* This work was supported by a grant from the National Institute of Psychobiology in Israel and Recanati fund.
 This article contains supplemental Figs. S1 to S5, Data S1 to S4, and Table S1
This article contains supplemental Figs. S1 to S5, Data S1 to S4, and Table S1
1 The abbreviations used are:
- page: LTM
- long-term memory
- ANOVA
- analysis of variance
- AP1
- activating protein 1
- C/EBP
- cytosine-cytosine-adenosine-adenosine-thymidine box motif enhancer binding protein
- CA1
- Cornu Ammonis area 1, a region in hippocampus
- CA3
- Cornu Ammonis area 3, a region in hippocampus
- CaMKIIa
- calcium/calmodulin-dependent kinase IIa
- CCC
- cubic clustering criterion
- CREB
- cAMP response element-binding protein
- DIA
- data independent acquisition
- EM
- expectation maximization
- FAG-EC
- fast agglomerative algorithm based on edge clustering
- FDR
- false discovery rate
- GO
- gene ontology
- HDMSE
- high definition MSE
- IMS-MS/MS
- ion mobility spectroscopy with tandem mass-spectrometry
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- LTD
- long-term depression
- LTP
- long-term potentiation
- NanoESI
- nanoelectrospray ionization
- PC
- principal component
- PCA
- principal component analysis
- PCBD1
- pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1
- PLGS
- Proteinlynx Global Server
- PRP
- plasticity related protein
- RAM
- radial arm maze
- SOM
- self-organizing maps
- SVM
- support vector machine
- TF
- transcription factor
- ULC/MS
- ultraliquid chromatography/mass-spectrometry.
REFERENCES
- 1.Dudai Y. (2004) The neurobiology of consolidations, or, how stable is the engram? Annu. Rev. Psychol. 55, 51–86 [DOI] [PubMed] [Google Scholar]
- 2.Martin K. C., Casadio A., Zhu H., Yaping E., Rose J. C., Chen M., Bailey C. H., and Kandel E. R. (1997) Synapse-specific, long-term facilitation of aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell 91, 927–938 [DOI] [PubMed] [Google Scholar]
- 3.Schafe G. E., Nadel N. V., Sullivan G. M., Harris A., and LeDoux J. E. (1999) Memory consolidation for contextual and auditory fear conditioning is dependent on protein synthesis, PKA, and MAP kinase. Learn. Mem. 6, 97–110 [PMC free article] [PubMed] [Google Scholar]
- 4.Squire L. R., and Barondes S. H. (1972) Variable decay of memory and its recovery in cycloheximide-treated mice. Proc. Natl. Acad. Sci. U.S.A. 69, 1416–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bliss T. V., and Collingridge G. L. (1993) A synaptic model of memory: long-term potentiation in the hippocampus. Nature 361, 31–39 [DOI] [PubMed] [Google Scholar]
- 6.Malenka R. C., and Bear M. F. (2004) LTP and LTD: an embarrassment of riches. Neuron 44, 5–21 [DOI] [PubMed] [Google Scholar]
- 7.Barria A., Muller D., Derkach V., Griffith L. C., and Soderling T. R. (1997) Regulatory phosphorylation of AMPA-type glutamate receptors by CaM-KII during long-term potentiation. Science 276, 2042–2045 [DOI] [PubMed] [Google Scholar]
- 8.Esteban J. A., Shi S. H., Wilson C., Nuriya M., Huganir R. L., and Malinow R. (2003) PKA phosphorylation of AMPA receptor subunits controls synaptic trafficking underlying plasticity. Nat. Neurosci. 6, 136–143 [DOI] [PubMed] [Google Scholar]
- 9.Lee H. K., Barbarosie M., Kameyama K., Bear M. F., and Huganir R. L. (2000) Regulation of distinct AMPA receptor phosphorylation sites during bidirectional synaptic plasticity. Nature 405, 955–959 [DOI] [PubMed] [Google Scholar]
- 10.Costa-Mattioli M., Gobert D., Stern E., Gamache K., Colina R., Cuello C., Sossin W., Kaufman R., Pelletier J., Rosenblum K., Krnjevic K., Lacaille J. C., Nader K., and Sonenberg N. (2007) eIF2alpha phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell 129, 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dudai Y. (2002) Molecular bases of long-term memories: a question of persistence. Curr. Opin. Neurobiol. 12, 211–216 [DOI] [PubMed] [Google Scholar]
- 12.Dudai Y. (2009) Predicting not to predict too much: how the cellular machinery of memory anticipates the uncertain future. Philos. Trans. R Soc. Lond. B Biol. Sci. 364, 1255–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizuno M., Yamada K., Maekawa N., Saito K., Seishima M., and Nabeshima T. (2002) CREB phosphorylation as a molecular marker of memory processing in the hippocampus for spatial learning. Behav. Brain Res. 133, 135–141 [DOI] [PubMed] [Google Scholar]
- 14.Barco A., Lopez de Armentia M., and Alarcon J. M. (2008) Synapse-specific stabilization of plasticity processes: the synaptic tagging and capture hypothesis revisited 10 years later. Neurosci. Biobehav. Rev. 32, 831–851 [DOI] [PubMed] [Google Scholar]
- 15.Frey S., and Frey J. U. (2008) “Synaptic tagging” and “cross-tagging” and related associative reinforcement processes of functional plasticity as the cellular basis for memory formation. Prog. Brain Res. 169, 117–143 [DOI] [PubMed] [Google Scholar]
- 16.Govindarajan A., Kelleher R. J., and Tonegawa S. (2006) A clustered plasticity model of long-term memory engrams. Nat. Rev. Neurosci. 7, 575–583 [DOI] [PubMed] [Google Scholar]
- 17.Sajikumar S., and Frey J. U. (2004) Resetting of “synaptic tags” is time- and activity-dependent in rat hippocampal CA1 in vitro. Neuroscience 129, 503–507 [DOI] [PubMed] [Google Scholar]
- 18.Sajikumar S., Navakkode S., and Frey J. U. (2007) Identification of compartment- and process-specific molecules required for “synaptic tagging” during long-term potentiation and long-term depression in hippocampal CA1. J. Neurosci. 27, 5068–5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin H. H., Davis M. I., Ronesi J. A., and Lovinger D. M. (2006) The role of protein synthesis in striatal long-term depression. J. Neurosci. 26, 11811–11820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raymond C. R., and Redman S. J. (2006) Spatial segregation of neuronal calcium signals encodes different forms of LTP in rat hippocampus. J. Physiol. 570, 97–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reymann K. G., and Frey J. U. (2007) The late maintenance of hippocampal LTP: requirements, phases, “synaptic tagging,” “late-associativity,” and implications. Neuropharmacology 52, 24–40 [DOI] [PubMed] [Google Scholar]
- 22.Hoeffer C. A., Cowansage K. K., Arnold E. C., Banko J. L., Moerke N. J., Rodriguez R., Schmidt E. K., Klosi E., Chorev M., Lloyd R. E., Pierre P., Wagner G., LeDoux J. E., and Klann E. (2011) Inhibition of the interactions between eukaryotic initiation factors 4E and 4G impairs long-term associative memory consolidation but not reconsolidation. Proc. Natl. Acad. Sci. U.S.A. 108, 3383–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jarome T. J., Werner C. T., Kwapis J. L., and Helmstetter F. J. (2011) Activity dependent protein degradation is critical for the formation and stability of fear memory in the amygdala. PloS. One 6, e24349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Artinian J., McGauran A. M., De Jaeger X., Mouledous L., Frances B., and Roullet P. (2008) Protein degradation, as with protein synthesis, is required during not only long-term spatial memory consolidation but also reconsolidation. Eur. J. Neurosci. 27, 3009–3019 [DOI] [PubMed] [Google Scholar]
- 25.Bourtchouladze R., Abel T., Berman N., Gordon R., Lapidus K., and Kandel E. R. (1998) Different training procedures recruit either one or two critical periods for contextual memory consolidation, each of which requires protein synthesis and PKA. Learn. Mem. 5, 365–374 [PMC free article] [PubMed] [Google Scholar]
- 26.Da Silva W. C., Cardoso G., Bonini J. S., Benetti F., and Izquierdo I. (2013) Memory reconsolidation and its maintenance depend on L-voltage-dependent calcium channels and CaMKII functions regulating protein turnover in the hippocampus. Proc. Natl. Acad. Sci. U.S.A. 110, 6566–6570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossato J. I., Bevilaqua L. R., Myskiw J. C., Medina J. H., Izquierdo I., and Cammarota M. (2007) On the role of hippocampal protein synthesis in the consolidation and reconsolidation of object recognition memory. Learn. Mem. 14, 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taubenfeld S. M., Milekic M. H., Monti B., and Alberini C. M. (2001) The consolidation of new but not reactivated memory requires hippocampal C/EBPbeta. Nat. Neurosci. 4, 813–818 [DOI] [PubMed] [Google Scholar]
- 29.Warburton E. C., Barker G. R., and Brown M. W. (2013) Investigations into the involvement of NMDA mechanisms in recognition memory. Neuropharmacology 74, 41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blum S., Runyan J. D., and Dash P. K. (2006) Inhibition of prefrontal protein synthesis following recall does not disrupt memory for trace fear conditioning. BMC Neuroscience 7, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moguel-Gonzalez M., Gomez-Palacio-Schjetnan A., and Escobar M. L. (2008) BDNF reverses the CTA memory deficits produced by inhibition of protein synthesis. Neurobiol. Learn. Mem. 90, 584–587 [DOI] [PubMed] [Google Scholar]
- 32.Reis D. S., Jarome T. J., and Helmstetter F. J. (2013) Memory formation for trace fear conditioning requires ubiquitin-proteasome mediated protein degradation in the prefrontal cortex. Front. Behav. Neurosci. 7, 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenberg T., Gal-Ben-Ari S., Dieterich D. C., Kreutz M. R., Ziv N. E., Gundelfinger E. D., and Rosenblum K. (2014) The roles of protein expression in synaptic plasticity and memory consolidation. Front. Mol. Neurosci. 7, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alberini C. M. (2009) Transcription factors in long-term memory and synaptic plasticity. Physiol. Rev. 89, 121–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costa-Mattioli M., Sossin W. S., Klann E., and Sonenberg N. (2009) Translational control of long-lasting synaptic plasticity and memory. Neuron. 61, 10–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee Y. S., Bailey C. H., Kandel E. R., and Kaang B. K. (2008) Transcriptional regulation of long-term memory in the marine snail Aplysia. Mol. Brain 1, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fonseca R., Vabulas R. M., Hartl F. U., Bonhoeffer T., and Nagerl U. V. (2006) A balance of protein synthesis and proteasome-dependent degradation determines the maintenance of LTP. Neuron 52, 239–245 [DOI] [PubMed] [Google Scholar]
- 38.Hegde A. N. (2010) The ubiquitin-proteasome pathway and synaptic plasticity. Learn. Mem. 17, 314–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong C., Upadhya S. C., Ding L., Smith T. K., and Hegde A. N. (2008) Proteasome inhibition enhances the induction and impairs the maintenance of late-phase long-term potentiation. Learn. Mem. 15, 335–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong C., Bach S. V., Haynes K. A., and Hegde A. N. (2014) Proteasome modulates positive and negative translational regulators in long-term synaptic plasticity. J. Neurosci. 34, 3171–3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeh S. H., Mao S. C., Lin H. C., and Gean P. W. (2006) Synaptic expression of glutamate receptor after encoding of fear memory in the rat amygdala. Mol. Pharmacol. 69, 299–308 [DOI] [PubMed] [Google Scholar]
- 42.Lopez-Salon M., Alonso M., Vianna M. R., Viola H., Mello e Souza T., Izquierdo I., Pasquini J. M., and Medina J. H. (2001) The ubiquitin-proteasome cascade is required for mammalian long-term memory formation. Eur. J. Neurosci. 14, 1820–1826 [DOI] [PubMed] [Google Scholar]
- 43.Bird C. M., and Burgess N. (2008) The hippocampus and memory: insights from spatial processing. Nat. Rev. Neurosci. 9, 182–194 [DOI] [PubMed] [Google Scholar]
- 44.Keeley M. B., Wood M. A., Isiegas C., Stein J., Hellman K., Hannenhalli S., and Abel T. (2006) Differential transcriptional response to nonassociative and associative components of classical fear conditioning in the amygdala and hippocampus. Learn. Mem. 13, 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leil T. A., Ossadtchi A., Nichols T. E., Leahy R. M., and Smith D. J. (2003) Genes regulated by learning in the hippocampus. J. Neurosci. Res. 71, 763–768 [DOI] [PubMed] [Google Scholar]
- 46.O'Sullivan N. C., McGettigan P. A., Sheridan G. K., Pickering M., Conboy L., O'Connor J. J., Moynagh P. N., Higgins D. G., Regan C. M., and Murphy K. J. (2007) Temporal change in gene expression in the rat dentate gyrus following passive avoidance learning. J. Neurochem. 101, 1085–1098 [DOI] [PubMed] [Google Scholar]
- 47.Monopoli M. P., Raghnaill M. N., Loscher J. S., O'Sullivan N. C., Pangalos M. N., Ring R. H., von Schack D., Dunn M. J., Regan C. M., Pennington S., and Murphy K. J. (2011) Temporal proteomic profile of memory consolidation in the rat hippocampal dentate gyrus. Proteomics 11, 4189–4201 [DOI] [PubMed] [Google Scholar]
- 48.Brady M. L., Allan A. M., and Caldwell K. K. (2012) A limited access mouse model of prenatal alcohol exposure that produces long-lasting deficits in hippocampal-dependent learning and memory. Alcohol. Clin. Exp. Res. 36, 457–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng K. K., Yeung C. F., Ho S. W., Chow S. F., Chow A. H., and Baum L. (2013) Highly stabilized curcumin nanoparticles tested in an in vitro blood-brain barrier model and in Alzheimer's disease Tg2576 mice. AAPS J. 15, 324–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gross M., Sheinin A., Nesher E., Tikhonov T., Baranes D., Pinhasov A., and Michaelevski I. (2015) Early onset of cognitive impairment is associated with altered synaptic plasticity and enhanced hippocampal GluA1 expression in a mouse model of depression. Neurobiol. Aging. 36(5), 1938–1952 [DOI] [PubMed] [Google Scholar]
- 51.Michaelevski I., Segal-Ruder Y., Rozenbaum M., Medzihradszky K. F., Shalem O., Coppola G., Horn-Saban S., Ben-Yaakov K., Dagan S. Y., Rishal I., Geschwind D. H., Pilpel Y., Burlingame A. L., and Fainzilber M. (2010) Signaling to transcription networks in the neuronal retrograde injury response. Sci. Signal. 3, ra53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Michaelevski I., Medzihradszky K. F., Lynn A., Burlingame A. L., and Fainzilber M. (2010) Axonal transport proteomics reveals mobilization of translation machinery to the lesion site in injured sciatic nerve. Mol. Cell. Proteomics 9, 976–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li G. Z., Vissers J. P., Silva J. C., Golick D., Gorenstein M. V., and Geromanos S. J. (2009) Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics 9, 1696–1719 [DOI] [PubMed] [Google Scholar]
- 54.Levin Y., Hradetzky E., and Bahn S. (2011) Quantification of proteins using data-independent analysis (MSE) in simple and complex samples: a systematic evaluation. Proteomics 11, 3273–3287 [DOI] [PubMed] [Google Scholar]
- 55.Silva J. C., Gorenstein M. V., Li G. Z., Vissers J. P., and Geromanos S. J. (2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156 [DOI] [PubMed] [Google Scholar]
- 56.Polpitiya A. D., Qian W. J., Jaitly N., Petyuk V. A., Adkins J. N., Camp D. G. 2nd, Anderson G. A., and Smith R. D. (2008) DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics 24, 1556–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.MacKay D. (2004) Information theory, inference, and learning algorithms. Cambridge University Press, Cambridge, UK. [Google Scholar]
- 58.Salvador S., and Chan P. (2004) Determining the number of clusters/segments in hierarchical clustering/segmentation algorithms. Proceedings of the 16th IEEE International Conference on Tools with Artificial Intelligence, pp. 576–584, IEEE, Los Alamitos, CA [Google Scholar]
- 59.Sarle W. S. (1983) Cubic clustering criterion, SAS Institute, Cary, NC [Google Scholar]
- 60.Abdi H., and Williams L., J. (2010) Principal component analysis. Wiley Interdiscip. Rev. Comput. Stat. 2, 433–459 [Google Scholar]
- 61.Bernaards C. (2005) Gradient Projection Algorithms and Software for Arbitrary Rotation Criteria in Factor Analysis. Ed. Psychol. Meas. 65, 676–696 [Google Scholar]
- 62.Hu Z., Mellor J., Wu J., Yamada T., Holloway D., and Delisi C. (2005) VisANT: data-integrating visual framework for biological networks and modules. Nucleic Acids Res. 33, W352–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J., Zhong J., Chen G., Li M., Wu F., and Pan Y. (2014) ClusterViz: a Cytoscape APP for Clustering Analysis of Biological Network. Comput. Biol. Bioinform. PP, 1. [DOI] [PubMed] [Google Scholar]
- 65.Cumbo F., Paci P., Santoni D., Di Paola L., and Giuliani A. (2014) GIANT: a cytoscape plugin for modular networks. PloS One 9, e105001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., Jensen L. J., and von Mering C. (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated, and scored. Nucleic Acids Res. 39, D561–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maere S., Heymans K., and Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in biological networks. Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 68.Huang da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 69.Bauer S., Grossmann S., Vingron M., and Robinson P. N. (2008) Ontologizer 2.0 – a multifunctional tool for GO term enrichment analysis and data exploration. Bioinformatics 24, 1650–1651 [DOI] [PubMed] [Google Scholar]
- 70.Enright A. J., Van Dongen S., and Ouzounis C. A. (2002) An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30, 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Henke K. (2010) A model for memory systems based on processing modes rather than consciousness. Nat. Rev. Neurosci. 11, 523–532 [DOI] [PubMed] [Google Scholar]
- 72.Abel T., and Lattal K. M. (2001) Molecular mechanisms of memory acquisition, consolidation, and retrieval. Curr. Opin. Neurobiol. 11, 180–187 [DOI] [PubMed] [Google Scholar]
- 73.Santini E., Huynh T. N., and Klann E. (2014) Mechanisms of translation control underlying long-lasting synaptic plasticity and the consolidation of long-term memory. Prog. Mol. Boil. Transl. Sci. 122, 131–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raymond C. R. (2007) LTP forms 1, 2, and 3: different mechanisms for the “long” in long-term potentiation. Trends Neurosci. 30, 167–175 [DOI] [PubMed] [Google Scholar]
- 75.Bannerman D. M., Sprengel R., Sanderson D. J., McHugh S. B., Rawlins J. N., Monyer H., and Seeburg P. H. (2014) Hippocampal synaptic plasticity, spatial memory, and anxiety. Nat. Rev. Neurosci. 15, 181–192 [DOI] [PubMed] [Google Scholar]
- 76.Lai C. S., Franke T. F., and Gan W. B. (2012) Opposite effects of fear conditioning and extinction on dendritic spine remodeling. Nature 483, 87–91 [DOI] [PubMed] [Google Scholar]
- 77.Leuner B., Falduto J., and Shors T. J. (2003) Associative memory formation increases the observation of dendritic spines in the hippocampus. J. Neurosci. 23, 659–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moser M. B., Trommald M., and Andersen P. (1994) An increase in dendritic spine density on hippocampal CA1 pyramidal cells following spatial learning in adult rats suggests the formation of new synapses. Proc. Natl. Acad. Sci. U.S.A. 91, 12673–12675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang Y., Wang X. B., Frerking M., and Zhou Q. (2008) Spine expansion and stabilization associated with long-term potentiation. J. Neurosci. 28, 5740–5751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Atwood H. L., and Wojtowicz J. M. (1999) Silent synapses in neural plasticity: current evidence. Learn. Mem. 6, 542–571 [DOI] [PubMed] [Google Scholar]
- 81.Martin S. J., Grimwood P. D., and Morris R. G. (2000) Synaptic plasticity and memory: an evaluation of the hypothesis. Annu. Rev. Neurosci. 23, 649–711 [DOI] [PubMed] [Google Scholar]
- 82.Poncer J. C. (2003) Hippocampal long term potentiation: silent synapses and beyond. J. Physiol. 97, 415–422 [DOI] [PubMed] [Google Scholar]
- 83.Pang P. T., and Lu B. (2004) Regulation of late-phase LTP and long-term memory in normal and aging hippocampus: role of secreted proteins tPA and BDNF. Ageing Res. Rev. 3, 407–430 [DOI] [PubMed] [Google Scholar]
- 84.Rosenblum K., Futter M., Voss K., Erent M., Skehel P. A., French P., Obosi L., Jones M. W., and Bliss T. V. (2002) The role of extracellular regulated kinases I/II in late-phase long-term potentiation. J. Neurosci. 22, 5432–5441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schafe G. E., Swank M. W., Rodrigues S. M., Debiec J., and Doyere V. (2008) Phosphorylation of ERK/MAP kinase is required for long-term potentiation in anatomically restricted regions of the lateral amygdala in vivo. Learn. Mem. 15, 55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Messaoudi E., Ying S. W., Kanhema T., Croll S. D., and Bramham C. R. (2002) Brain-derived neurotrophic factor triggers transcription-dependent, late phase long-term potentiation in vivo. J. Neurosci. 22, 7453–7461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen Y. J., Zhang M., Yin D. M., Wen L., Ting A., Wang P., Lu Y. S., Zhu X. H., Li S. J., Wu C. Y., Wang X. M., Lai C., Xiong W. C., Mei L., and Gao T. M. (2010) ErbB4 in parvalbumin-positive interneurons is critical for neuregulin 1 regulation of long-term potentiation. Proc. Natl. Acad. Sci. U.S.A. 107, 21818–21823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Min S. S., An J., Lee J. H., Seol G. H., Im J. H., Kim H. S., Baik T. K., and Woo R. S. (2011) Neuregulin-1 prevents amyloid beta-induced impairment of long-term potentiation in hippocampal slices via ErbB4. Neurosci. Lett. 505, 6–9 [DOI] [PubMed] [Google Scholar]
- 89.Lynch G., Rex C. S., and Gall C. M. (2007) LTP consolidation: substrates, explanatory power, and functional significance. Neuropharmacology 52, 12–23 [DOI] [PubMed] [Google Scholar]
- 90.Kushner S. A., Elgersma Y., Murphy G. G., Jaarsma D., van Woerden G. M., Hojjati M. R., Cui Y., LeBoutillier J. C., Marrone D. F., Choi E. S., De Zeeuw C. I., Petit T. L., Pozzo-Miller L., and Silva A. J. (2005) Modulation of presynaptic plasticity and learning by the H-ras/extracellular signal-regulated kinase/synapsin I signaling pathway. J. Neurosci. 25, 9721–9734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Niewalda T., Michels B., Jungnickel R., Diegelmann S., Kleber J., Kahne T., and Gerber B. (2015) Synapsin determines memory strength after punishment- and relief-learning. J. Neurosci. 35, 7487–7502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hou Q. L., Gao X., Lu Q., Zhang X. H., Tu Y. Y., Jin M. L., Zhao G. P., Yu L., Jing N. H., and Li B. M. (2006) SNAP-25 in hippocampal CA3 region is required for long-term memory formation. Biochem. Biophys. Res. Commun. 347, 955–962 [DOI] [PubMed] [Google Scholar]
- 93.Ferguson G. D., Wang H., Herschman H. R., and Storm D. R. (2004) Altered hippocampal short-term plasticity and associative memory in synaptotagmin IV (-/-) mice. Hippocampus 14, 964–974 [DOI] [PubMed] [Google Scholar]
- 94.Guo C. H., Senzel A., Li K., and Feng Z. P. (2010) De novo protein synthesis of syntaxin-1 and dynamin-1 in long-term memory formation requires CREB1 gene transcription in Lymnaea stagnalis. Behav. Genet. 40, 680–693 [DOI] [PubMed] [Google Scholar]
- 95.Hu J. Y., Meng X., and Schacher S. (2003) Redistribution of syntaxin mRNA in neuronal cell bodies regulates protein expression and transport during synapse formation and long-term synaptic plasticity. J. Neurosci. 23, 1804–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Castillo P. E., Janz R., Sudhof T. C., Tzounopoulos T., Malenka R. C., and Nicoll R. A. (1997) Rab3A is essential for mossy fibre long-term potentiation in the hippocampus. Nature 388, 590–593 [DOI] [PubMed] [Google Scholar]
- 97.D'Adamo P., Wolfer D. P., Kopp C., Tobler I., Toniolo D., and Lipp H. P. (2004) Mice deficient for the synaptic vesicle protein Rab3a show impaired spatial reversal learning and increased explorative activity but none of the behavioral changes shown by mice deficient for the Rab3a regulator Gdi1. Eur. J. Neurosci. 19, 1895–1905 [DOI] [PubMed] [Google Scholar]
- 98.Ibi D., Nitta A., Ishige K., Cen X., Ohtakara T., Nabeshima T., and Ito Y. (2010) Piccolo knockdown-induced impairments of spatial learning and long-term potentiation in the hippocampal CA1 region. Neurochem. Int. 56, 77–83 [DOI] [PubMed] [Google Scholar]
- 99.Xiao B., Tu J. C., Petralia R. S., Yuan J. P., Doan A., Breder C. D., Ruggiero A., Lanahan A. A., Wenthold R. J., and Worley P. F. (1998) Homer regulates the association of group 1 metabotropic glutamate receptors with multivalent complexes of homer-related, synaptic proteins. Neuron 21, 707–716 [DOI] [PubMed] [Google Scholar]
- 100.Snyder E. M., Philpot B. D., Huber K. M., Dong X., Fallon J. R., and Bear M. F. (2001) Internalization of ionotropic glutamate receptors in response to mGluR activation. Nat. Neurosci. 4, 1079–1085 [DOI] [PubMed] [Google Scholar]
- 101.Bramham C. R. (2008) Local protein synthesis, actin dynamics, and LTP consolidation. Curr. Opin. Neurobiol. 18, 524–531 [DOI] [PubMed] [Google Scholar]
- 102.Puthanveettil S. V. (2013) RNA transport and long-term memory storage. RNA Biol. 10, 1765–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cheng A., Hou Y., and Mattson M. P. (2010) Mitochondria and neuroplasticity. ASN Neuro. 2, e00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Azuma Y., Tan S. H., Cavenagh M. M., Ainsztein A. M., Saitoh H., and Dasso M. (2001) Expression and regulation of the mammalian SUMO-1 E1 enzyme. FASEB J. 15, 1825–1827 [DOI] [PubMed] [Google Scholar]
- 105.Tatham M. H., Jaffray E., Vaughan O. A., Desterro J. M., Botting C. H., Naismith J. H., and Hay R. T. (2001) Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276, 35368–35374 [DOI] [PubMed] [Google Scholar]
- 106.Drisaldi B., Colnaghi L., Fioriti L., Rao N., Myers C., Snyder A. M., Metzger D. J., Tarasoff J., Konstantinov E., Fraser P. E., Manley J. L., and Kandel E. R. (2015) SUMOylation is an inhibitory constraint that regulates the prion-like aggregation and activity of CPEB3. Cell Rep. 11, 1694–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fioriti L., Myers C., Huang Y. Y., Li X., Stephan J. S., Trifilieff P., Colnaghi L., Kosmidis S., Drisaldi B., Pavlopoulos E., and Kandel E. R. (2015) The persistence of hippocampal-based memory requires protein synthesis mediated by the prion-like protein CPEB3. Neuron 86, 1433–1448 [DOI] [PubMed] [Google Scholar]
- 108.Chen Y. C., Hsu W. L., Ma Y. L., Tai D. J., and Lee E. H. (2014) CREB SUMOylation by the E3 ligase PIAS1 enhances spatial memory. J. Neurosci. 34, 9574–9589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cortes-Mendoza J., Diaz de Leon-Guerrero S., Pedraza-Alva G., and Perez-Martinez L. (2013) Shaping synaptic plasticity: the role of activity-mediated epigenetic regulation on gene transcription. Int. J. Dev. Neurosci. 31, 359–369 [DOI] [PubMed] [Google Scholar]
- 110.Lynch G., Cox C. D., and Gall C. M. (2014) Pharmacological enhancement of memory or cognition in normal subjects. Front. Sys. Neurosci. 8, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fa M., Staniszewski A., Saeed F., Francis Y. I., and Arancio O. (2014) Dynamin 1 is required for memory formation. PloS One 9, e91954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Govindarajan A., Israely I., Huang S. Y., and Tonegawa S. (2011) The dendritic branch is the preferred integrative unit for protein synthesis-dependent LTP. Neuron 69, 132–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sara S. J. (2000) Retrieval and reconsolidation: toward a neurobiology of remembering. Learn. Mem. 7, 73–84 [DOI] [PubMed] [Google Scholar]
- 114.Stickgold R., and Walker M. P. (2005) Memory consolidation and reconsolidation: what is the role of sleep? Trend Neurosci. 28, 408–415 [DOI] [PubMed] [Google Scholar]
- 115.Gebicke-Haerter P. J. (2014) Engram formation in psychiatric disorders. Front. Neurosci. 8, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Folci A., Mapelli L., Sassone J., Prestori F., D'Angelo E., Bassani S., and Passafaro M. (2014) Loss of hnRNP K impairs synaptic plasticity in hippocampal neurons. J. Neurosci. 34, 9088–9095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Loebrich S., and Nedivi E. (2009) The function of activity-regulated genes in the nervous system. Physiol. Rev. 89, 1079–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lakhina V., Arey R. N., Kaletsky R., Kauffman A., Stein G., Keyes W., Xu D., and Murphy C. T. (2015) Genome-wide functional analysis of CREB/long-term memory-dependent transcription reveals distinct basal and memory gene expression programs. Neuron 85, 330–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cohen S., and Greenberg M. E. (2008) Communication between the synapse and the nucleus in neuronal development, plasticity, and disease. Annu. Rev. Cell Dev. Biol. 24, 183–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Oliva C. A., Vargas J. Y., and Inestrosa N. C. (2013) Wnt signaling: role in LTP, neural networks and memory. Ageing Res. Rev. 12, 786–800 [DOI] [PubMed] [Google Scholar]
- 121.Tan Y., Yu D., Busto G. U., Wilson C., and Davis R. L. (2013) Wnt signaling is required for long-term memory formation. Cell Rep. 4, 1082–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Davis S., Rodger J., Hicks A., Mallet J., and Laroche S. (1996) Brain structure and task-specific increase in expression of the gene encoding syntaxin 1B during learning in the rat: a potential molecular marker for learning-induced synaptic plasticity in neural networks. Eur. J. Neurosci. 8, 2068–2074 [DOI] [PubMed] [Google Scholar]
- 123.Hicks A., Davis S., Rodger J., Helme-Guizon A., Laroche S., and Mallet J. (1997) Synapsin I and syntaxin 1B: key elements in the control of neurotransmitter release are regulated by neuronal activation and long-term potentiation in vivo. Neuroscience 79, 329–340 [DOI] [PubMed] [Google Scholar]
- 124.Corradi A., Zanardi A., Giacomini C., Onofri F., Valtorta F., Zoli M., and Benfenati F. (2008) Synapsin-I- and synapsin-II-null mice display an increased age-dependent cognitive impairment. J. Cell Sci. 121, 3042–3051 [DOI] [PubMed] [Google Scholar]
- 125.Nesher E., Koman I., Gross M., Tikhonov T., Bairachnaya M., Salmon-Divon M., Levin Y., Gerlitz G., Michaelevski I., Yadid G., and Pinhasov A. (2015) Synapsin IIb as a functional marker of submissive behavior. Sci. Rep. 5, 10287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Revest J. M., Kaouane N., Mondin M., Le Roux A., Rouge-Pont F., Vallee M., Barik J., Tronche F., Desmedt A., and Piazza P. V. (2010) The enhancement of stress-related memory by glucocorticoids depends on synapsin-Ia/Ib. Mol. Psychiatry 1125, 1140–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Birbach A. (2008) Profilin, a multimodal regulator of neuronal plasticity. BioEssays 30, 994–1002 [DOI] [PubMed] [Google Scholar]
- 128.Krucker T., Siggins G. R., and Halpain S. (2000) Dynamic actin filaments are required for stable long-term potentiation (LTP) in area CA1 of the hippocampus. Proc. Natl. Acad. Sci. U.S.A. 97, 6856–6861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lamprecht R., Farb C. R., Rodrigues S. M., and LeDoux J. E. (2006) Fear conditioning drives profilin into amygdala dendritic spines. Nat. Neurosci. 9, 481–483 [DOI] [PubMed] [Google Scholar]
- 130.Jayachandran R., Liu X., Bosedasgupta S., Muller P., Zhang C. L., Moshous D., Studer V., Schneider J., Genoud C., Fossoud C., Gambino F., Khelfaoui M., Muller C., Bartholdi D., Rossez H., Stiess M., Houbaert X., Jaussi R., Frey D., Kammerer R. A., Deupi X., de Villartay J. P., Luthi A., Humeau Y., and Pieters J. (2014) Coronin 1 regulates cognition and behavior through modulation of cAMP/protein kinase A signaling. PLoS Biol. 12, e1001820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ahmad M., Polepalli J. S., Goswami D., Yang X., Kaeser-Woo Y. J., Sudhof T. C., and Malenka R. C. (2012) Postsynaptic complexin controls AMPA receptor exocytosis during LTP. Neuron 73, 260–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sawada K., Barr A. M., Nakamura M., Arima K., Young C. E., Dwork A. J., Falkai P., Phillips A. G., and Honer W. G. (2005) Hippocampal complexin proteins and cognitive dysfunction in schizophrenia. Arch. Gen. Psychiatry 62, 263–272 [DOI] [PubMed] [Google Scholar]
- 133.Takahashi S., Ujihara H., Huang G. Z., Yagyu K. I., Sanbo M., Kaba H., and Yagi T. (1999) Reduced hippocampal LTP in mice lacking a presynaptic protein: complexin II. Eur. J. Neurosci. 11, 2359–2366 [DOI] [PubMed] [Google Scholar]
- 134.Xu J., Kang N., Jiang L., Nedergaard M., and Kang J. (2005) Activity-dependent long-term potentiation of intrinsic excitability in hippocampal CA1 pyramidal neurons. J. Neurosci. 25, 1750–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Babayan A. H., Kramar E. A., Barrett R. M., Jafari M., Haettig J., Chen L. Y., Rex C. S., Lauterborn J. C., Wood M. A., Gall C. M., and Lynch G. (2012) Integrin dynamics produce a delayed stage of long-term potentiation and memory consolidation. J. Neurosci. 32, 12854–12861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Han J., Kesner P., Metna-Laurent M., Duan T., Xu L., Georges F., Koehl M., Abrous D. N., Mendizabal-Zubiaga J., Grandes P., Liu Q., Bai G., Wang W., Xiong L., Ren W., Marsicano G., and Zhang X. (2012) Acute cannabinoids impair working memory through astroglial CB1 receptor modulation of hippocampal LTD. Cell 148, 1039–1050 [DOI] [PubMed] [Google Scholar]
- 137.Han X., Chen M., Wang F., Windrem M., Wang S., Shanz S., Xu Q., Oberheim N. A., Bekar L., Betstadt S., Silva A. J., Takano T., Goldman S. A., and Nedergaard M. (2013) Forebrain engraftment by human glial progenitor cells enhances synaptic plasticity and learning in adult mice. Cell Stem Cell 12, 342–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Moore C. I., and Cao R. (2008) The hemo-neural hypothesis: on the role of blood flow in information processing. J. Neurophysiol. 99, 2035–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Suzuki A., Stern S. A., Bozdagi O., Huntley G. W., Walker R. H., Magistretti P. J., and Alberini C. M. (2011) Astrocyte-neuron lactate transport is required for long-term memory formation. Cell 144, 810–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.