Abstract
We aimed to globally discover serum biomarkers for diagnosis of gastric cancer (GC). GC serum autoantibodies were discovered and validated using serum samples from independent patient cohorts encompassing 1,401 participants divided into three groups, i.e. healthy, GC patients, and GC-related disease group. To discover biomarkers for GC, the human proteome microarray was first applied to screen specific autoantibodies in a total of 87 serum samples from GC patients and healthy controls. Potential biomarkers were identified via a statistical analysis protocol. Targeted protein microarrays with only the potential biomarkers were constructed and used to validate the candidate biomarkers using 914 samples. To provide further validation, the abundance of autoantibodies specific to the biomarker candidates was analyzed using enzyme-linked immunosorbent assays. Receiver operating characteristic curves were generated to evaluate the diagnostic accuracy of the serum biomarkers. Finally, the efficacy of prognosis efficacy of the final four biomarkers was evaluated by analyzing the clinical records. The final panel of biomarkers consisting of COPS2, CTSF, NT5E, and TERF1 provides high diagnostic power, with 95% sensitivity and 92% specificity to differentiate GC patients from healthy individuals. Prognosis analysis showed that the panel could also serve as independent predictors of the overall GC patient survival. The panel of four serum biomarkers (COPS2, CTSF, NT5E, and TERF1) could serve as a noninvasive diagnostic index for GC, and the combination of them could potentially be used as a predictor of the overall GC survival rate.
Gastric cancer (GC)1 is the second leading cause of cancer-related deaths. A total of 952,000 new GC cases (6.8% of the total of the new cancer case) and 723,000 deaths (8.8% of the total new cancer case) occurred in 2012 (1). The highest mortality rates have been reported in East Asia, including China, Japan, and Korea (2–4), and ∼60% of new GC cases and deaths worldwide occur in this region. As GC has a 5-year survival rate of less than 15%, accurate diagnosis and prognostic assessment of patients are essential for optimizing therapeutic strategies, predicting the outcome of treatment, extending the survival period of patients, and potentially healing to reduce cancer mortality (5).
A variety of serum antigen biomarkers has been used for GC diagnosis and prognosis, e.g. carcinoembryonic antigen, carbohydrate antibody 19-9 (CA19-9), carbohydrate antibody 72-4 (CA72-4), and carbohydrate antibody 50 (CA50); the protein levels of these antigens in serum are usually used as signatures indicating cancer risk. However, generally, these serum antigen biomarkers lack sufficient sensitivity and specificity (6–8).
Autoantibodies against proteins that result from abnormal gene expression and gene mutation in patients with various tumors represent another type of serum biomarker (9–12). The corresponding antigens of the autoantibodies are usually recognized as tumor-specific antigens or tumor-associated antigens (13–16). There is particular interest in these autoantibodies due to the potential diagnostic and prognostic applications of the autoantibodies and their corresponding antigens. Indeed, there is a need to identify novel autoantibody-based biomarkers to improve the sensitivity and specificity for the diagnosis of GC.
In this study, we used a human proteome microarray containing 16,368 proteins to discover and independently validate serum autoantibodies with potential for diagnosis and prognosis of GC in a set of 1,401 serum samples. The samples were from 537 GC patients, 550 healthy controls, and 314 individuals of GC-related diseases. Four autoantigen serum biomarkers, COP9 constitutive photomorphogenic homolog subunit 2 (COPS2), CTSF, ecto-5′-nucleotidase (NT5E), and telomeric repeat binding factor 1 (TERF1), were identified with a combined diagnostic sensitivity of 95% and specificity of 92%. Furthermore, our data suggested COPS2, CTSF, NT5E, and TERF1 could also serve as potential predictors for prognostic assessment.
EXPERIMENTAL PROCEDURES
Serum Sample Collection
The cohort was composed of 1,401 serum samples from 537 GC patients, 550 healthy volunteers, and 314 people with gastric ulcer (GU)/gastric polyp (GP)/chronic atrophic gastritis (Tables I and II and Fig. 3A) who had given informed consent and met the eligibility criteria (supplemental Table S1). GC samples were collected from multiple centers and annotated with detailed clinical information, including seven hospitals, i.e. Ruijin Hospital, the First Affiliated Hospital of Fujian Medical University, Shanghai East Hospital, Tongren Hospital, Shanghai Putuo Center Hospital, Shanghai Pudong Gongli Hospital, and the Shanghai Fifth People's Hospital, between August 2008 and June 2013 (supplemental Table S2). The sera were prepared according to standard protocol. Briefly, 5 ml of whole blood was collected from each individual and placed in a Vacutainer (BD Biosciences) without anti-coagulant. The whole blood was then left undistributed at room temperature for 30 min. After centrifugation at 2,000 × g for 10 min in a refrigerated centrifuge, the sera were then transferred as a 0.5-ml aliquot immediately to clean Eppendorf tubes and stored at −70 °C.
Table I. Characteristics of study participants in the discovery phase.
| Variable | GC count (n = 37) |
Healthy count (n = 50) |
pa | ||||
|---|---|---|---|---|---|---|---|
| No. | Mean | % | No. | Mean | % | ||
| Age, years | 0.40 | ||||||
| Mean | 63.2 | 65.3 | |||||
| S.D. | 13.3 | 11.8 | |||||
| Sex | 0.013 | ||||||
| Male | 26 | 70 | 21 | 42 | |||
| Female | 11 | 30 | 29 | 58 | |||
| TNM stage | |||||||
| I | 12 | 32 | |||||
| II | 8 | 22 | |||||
| III | 8 | 22 | |||||
| IV | 9 | 24 | |||||
a Fisher's test.
Table II. Characteristics of the study participants in the training/testing phase.
The abbreviations used are as follow: S.D., standard deviation; SCC, squamous cell carcinoma; SRCC, signet ring cell carcinoma; CEA, carcinoembryonic antigen; AFP, alpha feto protein; TNM, tumor node metastasis.
| Variable | No. | Mean | % |
|---|---|---|---|
| GC count | 300 | ||
| Age, years | |||
| Mean | 59.55 | ||
| S.D. | 11.43 | ||
| Sex | |||
| Male | 222 | 74 | |
| Female | 78 | 26 | |
| CEA | |||
| ≤5 ng/ml | 250 | 83 | |
| >5 ng/ml | 50 | 17 | |
| CA 12-5 | |||
| ≤35 units/ml | 282 | 94 | |
| >35 units/ml | 18 | 6 | |
| CA 19-9 | |||
| ≤37 units/ml | 256 | 85 | |
| >37 units/ml | 44 | 15 | |
| CA 72-4 | |||
| ≤6 units/ml | 237 | 79 | |
| >6 units/ml | 63 | 21 | |
| AFP | |||
| ≤30 units/ml | 295 | 98 | |
| >30 units/ml | 5 | 2 | |
| Tumor size | |||
| ≤3 cm | 156 | 52 | |
| >3 cm | 144 | 48 | |
| Differentiation | |||
| Poor | 144 | 48 | |
| Moderate | 81 | 27 | |
| High | 7 | 2 | |
| Others | 68 | 23 | |
| Infiltration depth (T) | |||
| T1+T2 | 126 | 42 | |
| T3+T4 | 174 | 58 | |
| Positive lymph nodes (N) | |||
| ≤6 | 227 | 76 | |
| >6 | 73 | 24 | |
| Metastasis (M) | |||
| M1 | 13 | 4% | |
| M0 | 293 | 98 | |
| TNM stage | |||
| I | 95 | 32 | |
| II | 61 | 20 | |
| III | 131 | 44 | |
| IV | 13 | 4 | |
| Pathological type | |||
| Adenocarcinoma | 231 | 77 | |
| SCC | 4 | 1 | |
| SRCC | 18 | 6 | |
| Others | 47 | 16 | |
| Patients' status | |||
| Survival | 67 | 22 | |
| Death | 233 | 78 | |
| Median survival time (months) | |||
| 27.22 | |||
| Healthy count | |||
| Age, years | 300 | ||
| Mean | 54.89 | ||
| S.D. | 10.45 | ||
| Sex | |||
| Male | 143 | 48 | |
| Female | 157 | 52 | |
| CEA | 160 | ||
| ≤5 ng/ml | 155 | 97 | |
| >5 ng/ml | 5 | 3 | |
| CA 12-5 | 90 | ||
| ≤5 units/ml | 90 | 100 | |
| >35 units/ml | 0 | 0 | |
| CA 19-9 | 88 | ||
| ≤37 units/ml | 87 | 99 | |
| >37 units/ml | 1 | 1 | |
| CA 72-4 | 80 | ||
| ≤6 units/ml | 71 | 89 | |
| >6 units/ml | 9 | 11 | |
| AFP | 151 | ||
| ≤0 units/ml | 151 | 100 | |
| >30 units/ml | 0 | 0 |
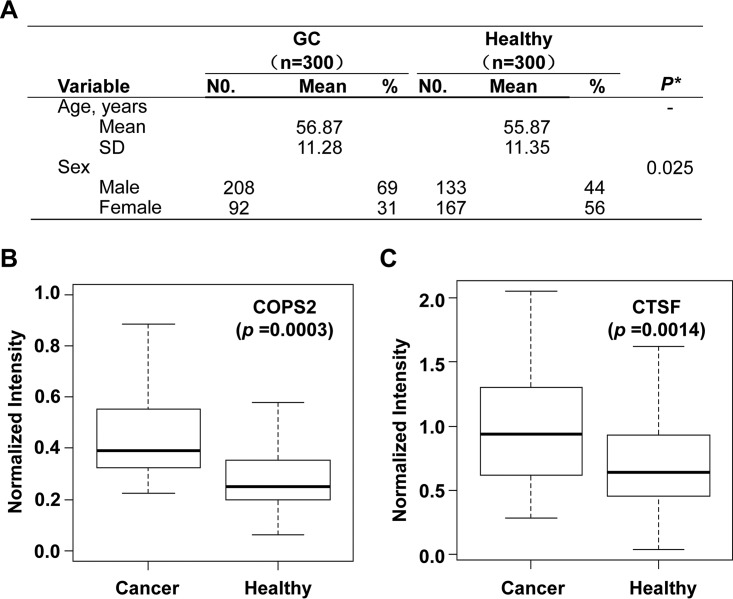
Fig. 3.
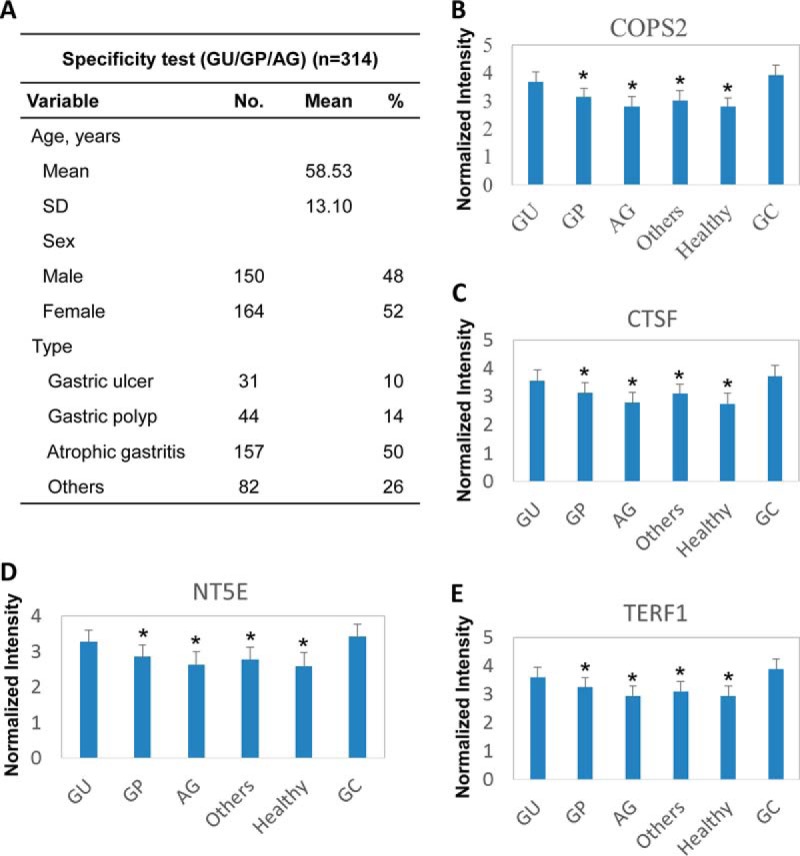
Set of four serum biomarkers, COPS2, CTSF, NT5E, and TERF1, were specific for gastric cancer but not for other gastric diseases. A, characteristics of study participants of GU/GP/chronic atrophic gastritis group. B–E, comparison of the GU/GP/CAG group, healthy people, and GC patient. All 314 cases were classified into the GU, GP, CAG, or other diseases group on the basis of clinical information. The signals of each of the four candidates are as follows: B, COPS2; C, CTSF; D, NT5E; and E, TERF1 reacting with the sera in each case are shown. (The y axis is log10 scale.) Asterisks indicate statistical difference as compared to the GC group (p < 0.05).
Protein Microarray Fabrication
A human proteome microarray was constructed as described previously (17). Briefly, the purified fusion proteins were printed onto protein slides (Full Moon BioSystems, Sunnyvale, CA), using a ChipWriter Pro 48-pin contact microarray (Bio-Rad). Each protein was spotted in duplicate. Control proteins, including histone H3, histone H4, BSA, and biotinylated BSA, were also spotted in duplicate. The targeted protein microarrays used for the training/testing phase were constructed following the same procedure as for the human proteome microarray, except that the proteins were printed onto three-dimensional protein slides (CapitalBio Inc., Beijing, China) with 12 identical subarrays per slide. Controls for the targeted protein microarrays included GST, anti-IgG, and elution buffer.
Serum Profiling on Proteome Microarray
Serum samples were diluted 1:100 in Tris-buffered saline containing 0.1% Tween 20 detergent (TBS-T). A total of 5 ml of the diluted serum sample was overlaid onto a protein microarray that was then incubated at room temperature for 1 h. The array was washed with TBS-T, and bound autoantibodies were detected by incubating with Alexa Fluor 532 goat anti-human IgG (H+L) (Jackson ImmunoResearch, West Grove, PA) diluted 1:1,000 in TBS-T at room temperature for 1 h. Arrays were washed with TBS-T and dried in a SlideWasher (CapitalBio Inc., Beijing, China) at room temperature. Microarrays were scanned in a GenePix 4200A (Molecular Devices, Sunnyvale, CA), and fluorescence data were analyzed with GenePix Pro 6.0 software (Molecular Devices). Targeted protein microarrays used in the training/testing phases were probed following the same procedure as that for the human proteome microarrays, except that 50 μl of diluted serum was loaded per subarray.
Protein Microarray Data Analysis
The median foreground and background intensities for each spot in the protein microarrays were obtained with GenePix Pro 6.0 software. The raw intensity for each spot was defined as the ratio of the foreground to the background median intensity. To remove the negative effects of nonspecific binding and spatial heterogeneity across the protein microarray, “normexp” (18), a background processing approach, was implemented in the R 2.15.1 package for background correction. To assess the quality of the protein microarrays, Array Quality Metrics in the Bioconductor R package (19) was employed to determine the quality and variability of the protein microarray experiment. Distance between Arrays and Boxplots functions were used to detect outlier microarrays, defined as those that failed both quality assessment methods using default settings in the Array Quality Metrics package. Outlier microarrays were excluded from further analysis.
Identifying Proteins Differentially Recognized by Serum Autoantibodies
After excluding outlier microarrays, the normalization was done twice during microarray data processing. The first round of normalization was performed by using the BackgroundCorrect package implemented in limma. The second round normalization was used to normalize between all the microarrays using Normalize between Arrays package in limma (20). Raw intensities were transformed to log2 values. Differentially expressed autoantibodies were identified by “limma” and “MASS” packages. p values were calculated for each protein using Fisher's test and were adjusted for multiple testing using the Benjamini and Hochberg approach to control for false discovery rates. Proteins with adjusted p values of no more than 0.05 were identified as candidates. The Daim package implemented in R 2.15.1 was used to generate receiver operating characteristic curves.
MASS package in R contains a powerful environment for the statistical and graphical analysis of data, which is also called the S environment. Many statistical methods, such as linear, nonlinear, and smooth regression models, tree-based methods, multivariate analysis, pattern recognition, survival analysis, time series, and spatial statistics, are included in the MASS package. Modern techniques such as robust methods, non-parametric smoothing, and bootstrapping can also be found in MASS package.
ELISA
EIA/RIA StripwellTM 96-well plates were obtained from Corning Glass. Plates were coated with 50 μg/ml antigens in PBS, 100 μl/well, and kept sealed with parafilm at 4 °C overnight. They were subsequently washed once with PBS-T, blocked for 1 h at room temperature with blocking buffer (0.2% bovine serum albumin (BSA), 0.1% gelatin, 0.02% thymerosal in PBS-T), and then washed again. Residual buffer was vacuum-aspirated from each well. Each serum sample was diluted 1:100 in blocking buffer, loaded into the wells, and incubated for 1 h at 37 °C, with duplicate wells being used for each serum sample. A 1:100 dilution was chosen because it gave the most reproducible results. The plates were then washed six times with PBS-T, and excess buffer was aspirated after the last wash. Incubation with anti-human IgG-peroxidase (Sangon Biotech, Shanghai, China) was performed for 1 h at 37 °C. After washing, the plates were developed using 100 μl/well TMB (Tetramethylbenzidine) substrate from Sigma, and the reaction was stopped with 50 μl/well of 1 m HCl. Optical density was read at 450 nm using a Behring EL311 ELISA microplate reader (Dade Behring Marburg Gmbh, Berlin, Germany). A typical four-parameter logistic nonlinear regression model was used for standard curve fitting for the ELISA, which was further used to estimate sample content (units/ml) from the absorbance measurement data. Outliers were detected using the Boxplots function in the Bioconductor package.
Clinical Data Analysis
The differences of the clinical characteristics in each groups were evaluated by Fisher's test or χ2 test. Survival time was calculated from the data of gastric cancer diagnosis to the date of death or the last follow up. The association between overall survival and protein expression levels was estimated by the Kaplan-Meier method and log-rank test.
RESULTS
Study Design
The whole study was composed of three phases as follows: discovery phase I, discovery phase II, and ELISA validation (Fig. 1). To ensure the reliability of the identified final serum autoantibody biomarkers, only serum samples with detailed clinical records and also meeting the eligibility criteria (supplemental Table S1) were collected. In each study phase, serum samples from GC patients with all four classical tumor stages (according to American Joint Committee on Cancer and Union for International Cancer Control 2010) were included. For global discovery and throughout the validation process, sera from 537 GC patients and 550 healthy volunteers were included for this study (supplemental Table S2). To avoid possible bias, these samples were collected from seven hospitals with similar cases for gastric cancer patients and healthy individuals from each hospital. To test the specificity of the identified serum autoantibody biomarker for GC diagnosis, sera from 314 people with other gastric diseases, e.g. GU, GP, CAG, and others, were also collected. In total, there were 1,401 samples included in this study, which is much larger than many other proteome microarray-based serum autoantibody biomarker studies, and thus a higher probability of identifying novel GC serum autoantibody biomarkers is expected. Because of the high cost of the human proteome microarray, it is neither economical nor efficient to carry out the whole study with the proteome microarray. In discovery phase II, the majority of samples probed on targeted protein microarray carries only the serum autoantibody biomarker candidates identified in discovery phase I. Using the targeted protein microarray, 12 samples could be assayed on a single microarray; the cost was significantly reduced, and the efficiency was highly improved. In the ELISA validation phase, the levels of autoantibodies specific to two candidates, i.e. COPS2 and CTSF, were assessed to use serum samples from 300 GC patients and 300 healthy controls using ELISA. This study was conducted with the approval of the ethics committee of Ruijin Hospital.
Fig. 1.
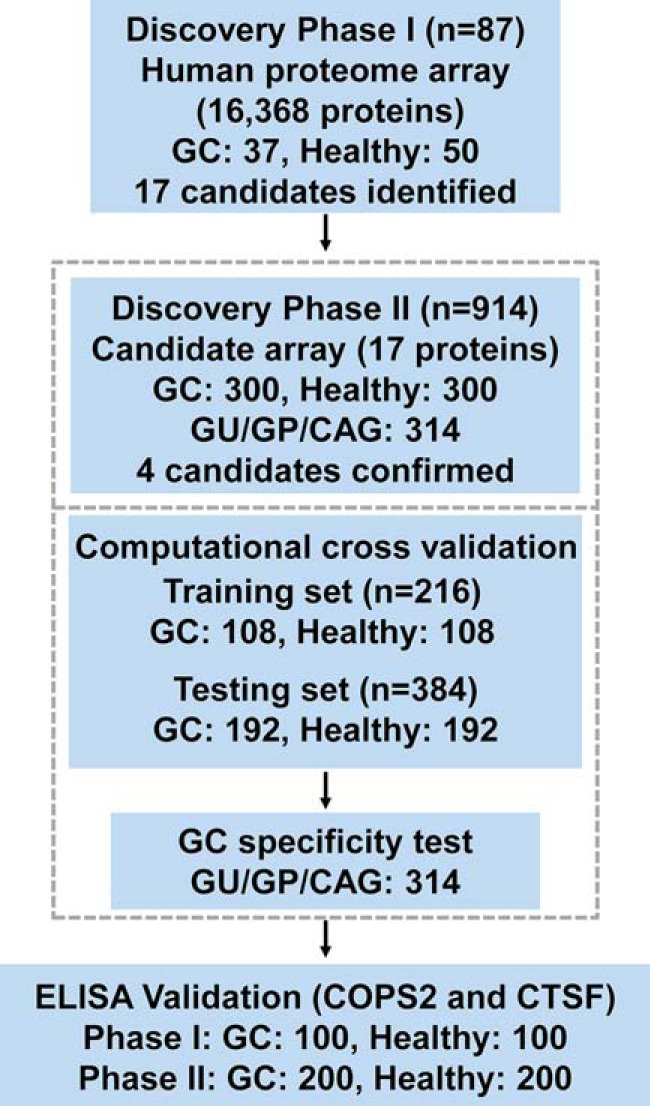
Study design. This study is composed of three major phases, i.e. discovery phase I, discovery phase II, and ELISA validation phase. For a better chance of discovering novel serum biomarkers, the human proteome microarray of 16,368 proteins was applied for comparing a small set of samples. A targeted protein microarray with the 17 candidates identified from discovery phase I was fabricated and applied for the discovery phase II. The final four candidates were then computationally validated by 100 times random sampling. The specificity of the final candidates was also tested by samples of other gastric diseases. Finally, the candidates were tested for the possibility of ELISA application. GU, gastric ulcer; GP, gastric polyp; CAG, chronic atrophic gastritis.
Serum Autoantibody Screening in Discovery Phase I
In discovery phase I, matched serum samples from 37 GC patients and 50 healthy controls were involved. Statistically, there is no bias of age and a slight difference of sex between the GC patient group and the healthy group (Table I). A microarray containing 16,368 human proteins (21) was initially used to screen for proteins that were differentially recognized by serum autoantibodies in the serum samples. The Bioconductor package limma (19) and MASS were applied to analyze the protein microarray data and to identify the differences in the response level of the serum autoantibodies between the GC patients and healthy controls. Seventeen proteins that were more significantly bound by the autoantibodies of the GC patient group than by those of the healthy group were identified using a p value of <0.01 and fold change > = 1.40 as cutoff (supplemental Table S3). The functions of these proteins were diverse and involved in a variety of biological pathways. This is the initial screening, and some of the candidates from this step may not be a true positive, such as OR5BU1, an olfactory receptor protein. As evidence, in GPMDB Global Proteome Machine Database and PeptideAtlas, the large number of olfactory receptor proteins reported from mass spectrometry studies falls to a very nearly null set upon reanalysis.
Set of Four Serum Autoantibody Biomarkers Were Determined in Discovery Phase II
The most reliable way of biomarker discovery is to test the candidates with a large cohort of samples, which have clear and sufficient clinical records. With this in mind, sera from 300 GC patients and 300 healthy controls with detailed clinical records were collected (Table II). To test these samples in an economical and efficient way, the 17 candidates from discovery phase I were purified and printed with a 12-well format to make the targeted protein microarray, and 12 samples could then be assayed on a single microarray. The Bioconductor package limma (19) and MASS were applied to analyze the protein microarray data and to identify the differences in the response level of the serum autoantibodies between the GC patients and healthy controls. Four proteins, COPS2, CTSF, NT5E, and TERF1, that were more significantly bound by the autoantibodies of the GC patient group than by those of the healthy group were determined. To test the four candidates, 100× computational cross-validations were conducted. For each cross-validation procedure, 108 healthy samples and 108 GC samples were randomly selected as the training phase. The rest of the samples were treated as the testing phase (192 healthy samples and 192 GC samples). The results of the computational cross-validation clearly showed that the four candidates alone could serve as potential autoantibody biomarkers for gastric cancer diagnosis based on the protein microarray format (Fig. 2, A–D). To further show the diagnosis accuracy of the four candidates, receiver operating characteristic analyses were also performed; an AUC of ∼0.9 was obtained for an individual candidate (Fig. 2, E–H) and as a combination (supplemental Fig. S1A). A classifier was also generated based on the best model (supplemental Fig. S1B). Surprisingly, the AUCs of known GC antigen biomarkers, such as CA12-5, CA19-9, CA72-4, and carcinoembryonic antigen, were all in the range of 0.5–0.6 (supplemental Fig. S2), which was significantly lower than that of the four candidates we identified on protein microarray.
Fig. 2.
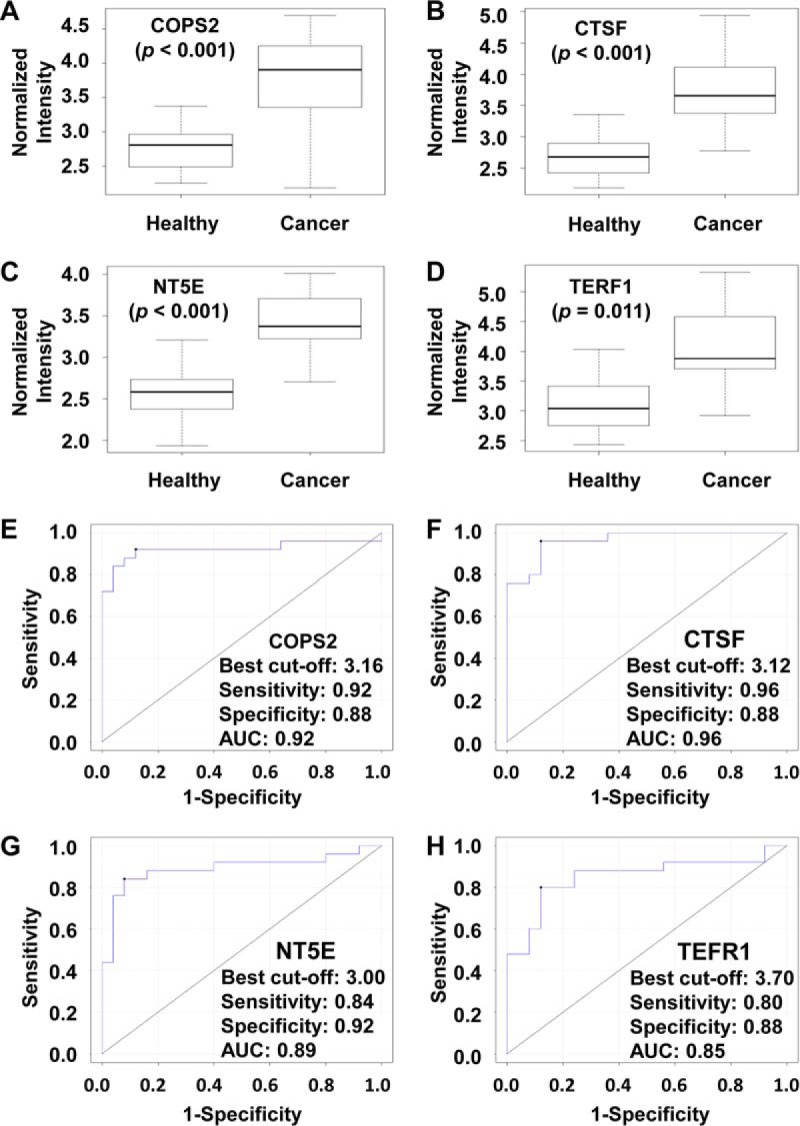
Four candidates of GC serum biomarker were confirmed in discovery phase II. Reactivity of the four serum biomarkers with sera of healthy people and GC patients on protein microarray is as follows: A, COPS2; B, CTSF; C, NT5E; and D, TERF1. (The y axis is log10 scale.) Receiver operating characteristic curve analysis of the four serum biomarkers for gastric cancer diagnosis is as follows: E, COPS2; F, CTSF; G, NT5E; and H, TERF1. The data presented here are based the 300 GC samples and 300 healthy people from discovery phase II.
It is possible that the final four autoantibody biomarkers may also react with the sera from other gastric diseases. To rule out this possibility, an additional set of sera from 314 people diagnosed with GU, GP, CAG, and other gastric diseases was also included in discovery phase II to test the GC specificity of the autoantibody biomarker candidates (Fig. 3A). For all four candidates, i.e. COPS2, CTSF, NT5E, and TERF1, the reactions to GC are significantly higher than that to GP, CAG, other gastric diseases, except the GU group, and healthy people. Additionally, there is no significant difference among healthy people and those with GP, CAG, and other gastric diseases (Fig. 3, B–E). These results indicate that all four autoantibody biomarker candidates are generally specific to GC, and they have potential serum autoantibody biomarkers for gastric cancer diagnostics. Clinically, gastric ulcer is a major precancerous condition of gastric cancer, and because the reactions of the four candidates to GU and GC sera are similar, an interesting possibility is that the four serum autoantibody biomarkers may potentially be applied for GC early diagnostics.
Association between Levels of Serum Autoantibodies and Patient Survival
We also tested the ability of the four autoantibody biomarker candidates for predicting GC survival in 300 GC patients. The low serum autoantibody levels against COPS2, CTSF, NT5E, and TERF1 were associated with unfavorable survival. As shown in Table III and supplemental Fig. S3, significant differences were observed for both median survival time and survival rate. Interestingly, patients with high autoantibody levels of COPS2, CTSF, NT5E, and TERF1 had a lower probability of cancer death and live longer. One plausible explanation is that CTSF and COPS2 are tumor-associated antigen/tumor-specific antigens for gastric cancer and also the possible drivers of progression, invasion, and metastasis. The immune system generates autoantibodies to block these antigens thus protects the patients from the harmful effects of gastric cancer. The autoantibody levels among individuals may be proportional to the robustness of their immune system against gastric cancer; thus, a higher level of autoantibody may indicate better protection and eventually longer survival.
Table III. Prognosis possibility of the final four candidates.
The correlation of the reactivities of the final four autoantibodies with the survival data of GC patients with follow-up records is shown. The median survival time (MST) was estimated based on the Kaplan-Meier estimates, and the p values were calculated using log-rank test.
| Candidate | Patients (n = 300) | Deaths (n = 233) | MST (months) | Survival rate | Log-rank p value |
|---|---|---|---|---|---|
| COPS2 | 0.01 | ||||
| Low, ≤3.16 | 112 | 95 | 15.93 | 15.18% | |
| High, >3.16 | 188 | 138 | 34.46 | 26.60% | |
| CTSF | 0.02 | ||||
| Low, ≤3.12 | 126 | 105 | 16.76 | 16.67% | |
| High, >3.12 | 174 | 128 | 34.61 | 26.44% | |
| NT5E | 0.01 | ||||
| Low, ≤3.00 | 114 | 99 | 16.21 | 13.16% | |
| High, >3.00 | 186 | 134 | 34.67 | 27.96% | |
| TERF1 | 0.01 | ||||
| Low, ≤3.70 | 121 | 102 | 16.83 | 15.70% | |
| High, >3.70 | 179 | 131 | 34.75 | 26.82% |
Validation of the Four Candidates Using ELISA and Tissue Microarray
ELISA is commonly used in clinical settings, and to test the probability of using the newly identified autoantibody biomarkers for GC diagnostics, we developed an ELISA with COPS2 and CTSF. These two candidates were chosen because they had the highest AUCs (Fig. 2) and could be easily purified. The sera for ELISA validation included samples from a set of 300 GC patients and 300 healthy controls; specifically, sera from 100 GC patients and healthy controls were randomly selected from the sample set used for microarray analysis (ELISA validation phase I); additional sera from 200 GC patients and healthy controls were freshly collected (ELISA validation phase II) (Fig. 4A). The 96-well plates were coated with COPS2 or CTSF and incubated with diluted serum samples, and readout of signals was with HRP-conjugated anti-human IgG. Results showed that for ELISA validation phase I and II, the GC group had significantly higher COPS2 and CTSF signals than that of the healthy control group (supplemental Fig. S4 and Fig. 4, B and C). As expected, the results of ELISA validation phase I are consistent with the same set of samples on the microarray. The correlation coefficiency r2 was 0.87 for COPS2 and 0.85 for CTSF, respectively. To examine the potential clinical relevance of the observed relationship between serum autoantibody levels against COPS2 and CTSF, and the protein levels in tissues, we employed a tissue microarray specific for gastric cancer containing 31 different paired samples (gastric cancer and healthy). The TMAs (Tissue microarrys) were incubated with antibodies for COPS2 and CTSF. As shown in supplemental Fig. S5, interestingly, lower levels of COPS2 and CTSF were observed in the gastric cancer group. To test whether the potential biomarkers are gastric cancer-specific, a tissue microarray of digestive tract tumor containing 30 samples was applied. There are six types of tumors, five samples for each, i.e. gastric cancer, esophagus cancer, colon cancer, rectal cancer, liver cancer, and pancreatic cancer. The results clearly demonstrated that CTSF is specific for gastric cancer, whereas COPS2 is not that specific (supplemental Fig. S6).
Fig. 4.
Two candidates of GC serum biomarker, i.e. COPS2 and CTSF, were tested by ELISA. A, characteristics of the study participants (300 GC sera and 300 healthy sera) in the ELISA validation phase. Statistical analysis showed that there is no bias for both age and gender between the GC group and healthy control. B and C, reactivity of the two serum biomarkers to the independently collected cohort of sera (200 healthy people and 200 GC patients) by ELISA. B, COPS2 and C, CTSF. (The y axis is log10 scale.)
DISCUSSION
Our study shows that COPS2, CTSF, NT5E, and TERF1 are recognized by GC-specific serum autoantibodies and that these four proteins have potential as biomarkers for GC diagnosis. Both the individual biomarkers and the combination of the four biomarkers have significantly higher sensitivity and specificity than that of routinely used serum autoantigen biomarkers such as carcinoembryonic antigen, CA19-9, CA72-4 and CA50. In addition to their potential for diagnosis, we found that these four autoantibody biomarkers, i.e. COPS2, CTSF, NT5E, and TERF1, are also associated with the overall survival of GC patients and could serve as a noninvasive predictor of GC prognosis.
In discovery phase II, the efficacy of the four candidates was successfully validated by microarray experiment and computational cross-validation with 100 times random sampling; these validations confirmed the reliability of the four serum autoantibody biomarkers that we identified. Because these serum autoantibody biomarkers are noninvasive and easy to detect in clinical settings, they may could also be used to investigate the drug response and chemosensitivity of GC patients by observing changes in expression levels and profiles before and after treatment.
According to the data that we obtained, two forms of test might be used for the clinical application of the identified autoantibody biomarkers, i.e. ELISA and microarray. However, the performance of the four autoantibody biomarkers was not as significant as that of protein microarray in the current form of ELISA; this could be improved when developing automatic ELISAs in future studies.
Our study is unique compared with studies on the discovery of circulating serum autoantibodies for GC diagnosis and prognosis (22–26), for the following reasons. First, we screened serum autoantibodies using a protein microarray, which included 16,368 human proteins, enabling us to have a better chance of globally identifying potential diagnostic and prognostic biomarkers. Second, a large cohort with 1,401 samples was tested for this study, which covers most of the stages of GC tumorigenesis. It is well known that the pathogenesis of GC is heterogeneous and that multiple mechanisms of tumorigenesis (such as tumor suppressor genes, oncogenes, viral effects, and angiogenesis) may be involved. It is well known that many types of cancer, including GC are heterogeneous, and some of them have clearly defined subgroups and some do not. It will be interesting to see the differences between/among cancer subgroups for biomarker purposes. However, as a first step, the aim of this study was set to identify serum biomarkers for gastric cancer in general. According to the significance and availability of the clinical resource, it will be interesting to try to identify subgroup-specific serum biomarkers for gastric cancer in future studies. Nonetheless, we hypothesized that similar to other types of cancer, most of GC cases fall into three stages as follows: healthy, gastritis, and GC. Because of its long development process, changes in protein expression may occur during any of the stages (gastritis or GC I, II, IV, and IV) before GC is clinically/pathophysiologically manifested. Thus, we included GC patients at four TNM stages of disease development in our study, and we also included 314 samples of other GC diseases during the training/testing phase. This design may give us an overall picture of the associated immune responses of GC progression and also strengthen the reliability of the identified biomarkers through cross-validations. In addition, our panel of four candidate autoantibody biomarkers was validated using a large and independent cohort from five medical centers.
All four autoantibody biomarkers identified in this study were related to key signaling pathways and tumorigenesis. COPS2 is a transcriptional co-repressor and was originally identified in a yeast two-hybrid screen as a thyroid hormone receptor-interacting protein that binds in a ligand-dependent manner (27) and participates in various signaling pathways. COPS2 interacts with a subset of nuclear hormone receptors such as the ecdysone receptor DAX-1 and thyroid hormone receptors (28). The COPS complex is composed of eight subunits, is localized in the cytoplasm and nucleus, and possesses an associated unidentified kinase activity that specifically phosphorylates transcriptional regulators such as p53, P105, and IκBα (29). These facts indicate that the COPS complex participates in various signal transduction pathways and in cell cycle progression. Cathepsins are cysteine proteases from the papain family that are responsible for protein breakdown in lysosomes (30). Human CTSF is a member of the cathepsin family and is composed of cathepsins B, C, H, K, L, O, S, V, W, and X (31). CTSF has been shown to be ubiquitously expressed in several tissues with higher levels of expression in skeletal muscle and the testis. CTSF transcripts have also been found in HeLa cells, suggesting that this enzyme could be involved in degradative processes during tumor progression (32). NT5E, known as CD73, is a glycosylphosphatidylinositol-linked 70-kDa cell surface enzyme found in most tissues (33, 34). CD73, originally identified as a lymphocyte differentiation antigen, is expressed in many cell types, including subsets of lymphocytes (35) and endothelial and epithelial cells (36). Recently, it has been shown that biological actions of CD73 are a consequence (at least in large part) of the regulated enzymatic phosphohydrolytic activity of extracellular nucleotides. This ectoenzymatic cascade, in tandem with CD39 (ecto-ATPase), generates adenosine from the ATP/AMP often released from damaged or inflamed target cells into the extracellular environment (37, 38) and mediates immune suppression (39, 40). Interestingly, CD73 is highly expressed in numerous human solid tumors (41–46), and its overexpression and elevated activity are associated with tumor invasiveness and metastasis (47, 48) and with shorter patient survival time (49), indicating that CD73 is closely involved in cancer progression. Telomeric repeat binding factor 1 (TERF1) is a double-stranded telomere DNA-binding protein (50). It forms a homodimer and nucleates protein complexes called shelterin at the telomeric TTAGGG repeats (51). As the DNA polymerase-dependent replication machinery cannot replicate the very ends of linear DNA, telomeres gradually shorten after every replication cycle. Critically shortened telomeres cannot protect the chromosome ends and thus trigger a DNA damage response, resulting in replicative senescence (52). Most immortal cells escape from replicative senescence by activating telomerase (53–55). Thus, this mechanism may be involved in tumor growth.
This study has some limitations. First, there is no overlap between the autoantibodies identified here and those from other related studies (56–58). This may due to the complex heterogeneity of GC. The proteome microarray-based strategy has the power of global and unbiased screening. It is quite different from the strategies/technologies used for the previous GC biomarker studies. It is possible that there are more biomarkers to be discovered, and each strategy could only reveal part of them. One plausible explanation for the observation of no overlap is that there are more existing biomarkers, and by using a variety of different strategies, each study, including ours, could only reveal part of the existing biomarkers. However, the autoantibody biomarkers that we have identified could be combined with those from other studies, thus providing higher sensitivity and specificity for both diagnosis and prognosis. Second, it was difficult for us to identify the autoantibodies to tumor antigens bearing structural changes and post-translational modification aberrance, as all proteins on the protein microarray were homogeneously expressed from normal human coding genes. However, our strategy is global and unbiased. On the protein microarray used here, there are 16,368 affinity-purified human proteins, representing ∼80% of human genome-encoded proteins. Thus, using human protein microarrays greatly increased our chances of discovering novel biomarkers.
In summary, we have identified a panel of four serum autoantibody biomarkers by using a large number of participants. These autoantibody-based biomarkers could differentiate GC patients from healthy individuals with a high degree of sensitivity and specificity. Our study demonstrates that this panel of serum biomarkers has considerable clinical value for the diagnosis of GC, in prospective studies of cancer outcome, and in therapy response evaluations.
Supplementary Material
Acknowledgments
We thank Dr. Daniel Czajkowsky for critical reading; Dr. Joy Fleming for English editing; and Dr. Zhi Xie and Dr. Ying Dong Zhao for statistical analysis.
Footnotes
Author contributions: Z.S., B. Liu, and S.T. designed research; L.Y., J.W., J.L., H. Zhang, S.G., M.Y., B. Lan, Y.D., M.X., W.L., X.G., and C.Q. performed research; M.Y., Z.Z., B. Lan, Y.D., M.X., W.L., X.G., C.Q., H. Zhu, and B. Liu contributed new reagents or analytic tools; L.Y., J.W., J.L., H. Zhang, S.G., M.Y., Z.Z., B. Lan, M.X., H. Zhu, Z.S., B. Liu, and S.T. analyzed data; L.Y., J.W., B. Liu, and S.T. wrote the paper.
* This work was supported by the National High Technology Research and Development Program of China Grants 2012AA020103 and 2012AA020203, the National Natural Science Foundation of China Grants 31370813 and 91229106, the Science and Technology Commission of Shanghai Municipality Grants 12XD1403700 and 13ZR1425600, and the Key Projects in the National Science and Technology Pillar Program of China Grant 2014BAI09B03. The authors declare that they have no conflicts of interest with the contents of this article.
 This article contains supplemental Materials and Methods, Tables S1 to S3, and Figs. S1 to S6.
This article contains supplemental Materials and Methods, Tables S1 to S3, and Figs. S1 to S6.
1 The abbreviations used are:
- GC
- gastric cancer
- CTSF
- cathepsins F
- GP
- gastric polyp
- GU
- gastric ulcer
- CAG
- chronic atrophic gastritis
- AUC
- areas under the curve.
REFERENCES
- 1.Jemal A., Bray F., Center M. M., Ferlay J., Ward E., and Forman D. (2011) Global cancer statistics. CA Cancer J. Clin. 61, 69–90 [DOI] [PubMed] [Google Scholar]
- 2.Chen W. Q., Zhang S. W., Zou X. N., and Zhao P. (2011) Cancer incidence and mortality in China. Chin. J. Cancer Res. 23, 3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel R., Naishadham D., and Jemal A. (2013) Cancer statistics, 2013. CA Cancer J. Clin. 63, 11–30 [DOI] [PubMed] [Google Scholar]
- 4.Yang L., Parkin D. M., Ferlay J., Li L., and Chen Y. (2005) Estimates of cancer incidence in China for 2000 and projections for 2005. Cancer Epidemiol. Biomarkers Prev. 14, 243–250 [PubMed] [Google Scholar]
- 5.Kang K. J., Kim K. M., Min B. H., Lee J. H., and Kim J. J. (2011) Endoscopic submucosal dissection of early gastric cancer. Gut Liver 5, 418–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emoto S., Ishigami H., Yamashita H., Yamaguchi H., Kaisaki S., and Kitayama J. (2012) Clinical significance of CA125 and CA72-4 in gastric cancer with peritoneal dissemination. Gastric Cancer 15, 154–161 [DOI] [PubMed] [Google Scholar]
- 7.Li Y., Yang Y., Lu M., and Shen L. (2011) Predictive value of serum CEA, CA19-9 and CA72.4 in early diagnosis of recurrence after radical resection of gastric cancer. Hepatogastroenterology 58, 2166–2170 [DOI] [PubMed] [Google Scholar]
- 8.Ucar E., Semerci E., Ustun H., Yetim T., Huzmeli C., and Gullu M. (2008) Prognostic value of preoperative CEA, CA 19-9, CA 72-4, and AFP levels in gastric cancer. Adv. Ther. 25, 1075–1084 [DOI] [PubMed] [Google Scholar]
- 9.Anderson K. S., Wong J., D'Souza G., Riemer A. B., Lorch J., Haddad R., Pai S. I., Longtine J., McClean M., LaBaer J., Kelsey K. T., and Posner M. (2011) Serum antibodies to the HPV16 proteome as biomarkers for head and neck cancer. Br. J. Cancer 104, 1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burford B., Gentry-Maharaj A., Graham R., Allen D., Pedersen J. W., Nudelman A. S., Blixt O., Fourkala E. O., Bueti D., Dawnay A., Ford J., Desai R., David L., Trinder P., Acres B., Schwientek T., Gammerman A., Reis C. A., Silva L., Osório H., Hallett R., Wandall H. H., Mandel U., Hollingsworth M. A., Jacobs I., Fentiman I., Clausen H., Taylor-Papadimitriou J., Menon U., and Burchell J. M. (2013) Autoantibodies to MUC1 glycopeptides cannot be used as a screening assay for early detection of breast, ovarian, lung or pancreatic cancer. Br. J. Cancer 108, 2045–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hudson M. E., Pozdnyakova I., Haines K., Mor G., and Snyder M. (2007) Identification of differentially expressed proteins in ovarian cancer using high-density protein microarrays. Proc. Natl. Acad. Sci. U.S.A. 104, 17494–17499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X., Yu J., Sreekumar A., Varambally S., Shen R., Giacherio D., Mehra R., Montie J. E., Pienta K. J., Sanda M. G., Kantoff P. W., Rubin M. A., Wei J. T., Ghosh D., and Chinnaiyan A. M. (2005) Autoantibody signatures in prostate cancer. N. Engl. J. Med. 353, 1224–1235 [DOI] [PubMed] [Google Scholar]
- 13.Chuang H. Y., Ren C. T., Chao C. A., Wu C. Y., Shivatare S. S., Cheng T. J., Wu C. Y., and Wong C. H. (2013) Synthesis and vaccine evaluation of the tumor-associated carbohydrate antigen RM2 from prostate cancer. J. Am. Chem. Soc. 135, 11140–11150 [DOI] [PubMed] [Google Scholar]
- 14.Dorvillius M., Garambois V., Pourquier D., Gutowski M., Rouanet P., Mani J. C., Pugnière M., Hynes N. E., and Pèlegrin A. (2002) Targeting of human breast cancer by a bispecific antibody directed against two tumour-associated antigens: ErbB-2 and carcinoembryonic antigen. Tumour Biol. 23, 337–347 [DOI] [PubMed] [Google Scholar]
- 15.Jaramillo A., Majumder K., Manna P. P., Fleming T. P., Doherty G., Dipersio J. F., and Mohanakumar T. (2002) Identification of HLA-A3-restricted CD8+ T cell epitopes derived from mammaglobin-A, a tumor-associated antigen of human breast cancer. Int. J. Cancer 102, 499–506 [DOI] [PubMed] [Google Scholar]
- 16.Li Y., Dong X., Yin Y., Su Y., Xu Q., Zhang Y., Pang X., Zhang Y., and Chen W. (2005) BJ-TSA-9, a novel human tumor-specific gene, has potential as a biomarker of lung cancer. Neoplasia 7, 1073–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong J. S., Jiang L., Albino E., Marrero J., Rho H. S., Hu J., Hu S., Vera C., Bayron-Poueymiroy D., Rivera-Pacheco Z. A., Ramos L., Torres-Castro C., Qian J., Bonaventura J., Boeke J. D., Yap W. Y., Pino I., Eichinger D. J., Zhu H., and Blackshaw S. (2012) Rapid identification of monospecific monoclonal antibodies using a human proteome microarray. Mol. Cell. Proteomics 11, O111.016253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ritchie M. E., Silver J., Oshlack A., Holmes M., Diyagama D., Holloway A., and Smyth G. K. (2007) A comparison of background correction methods for two-colour microarrays. Bioinformatics 23, 2700–2707 [DOI] [PubMed] [Google Scholar]
- 19.Kauffmann A., and Huber W. (2010) Microarray data quality control improves the detection of differentially expressed genes. Genomics 95, 138–142 [DOI] [PubMed] [Google Scholar]
- 20.Bolstad B. M., Irizarry R. A., Astrand M., and Speed T. P. (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 [DOI] [PubMed] [Google Scholar]
- 21.Chen Y., Yang L. N., Cheng L., Tu S., Guo S. J., Le H. Y., Xiong Q., Mo R., Li C. Y., Jeong J. S., Jiang L., Blackshaw S., Bi L. J., Zhu H., Tao S. C., and Ge F. (2013) Bcl2-associated athanogene 3 interactome analysis reveals a new role in modulating proteasome activity. Mol. Cell. Proteomics 12, 2804–2819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hao Y., Yu Y., Wang L., Yan M., Ji J., Qu Y., Zhang J., Liu B., and Zhu Z. (2008) IPO-38 is identified as a novel serum biomarker of gastric cancer based on clinical proteomics technology. J. Proteome Res. 7, 3668–3677 [DOI] [PubMed] [Google Scholar]
- 23.Jørgensen J. T., and Hersom M. (2012) HER2 as a prognostic marker in gastric cancer–a systematic analysis of data from the literature. J. Cancer 3, 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koskensalo S., Mrena J., Wiksten J. P., Nordling S., Kokkola A., Hagström J., and Haglund C. (2010) MMP-7 overexpression is an independent prognostic marker in gastric cancer. Tumour Biol. 31, 149–155 [DOI] [PubMed] [Google Scholar]
- 25.Mrena J., Wiksten J. P., Kokkola A., Nordling S., Ristimäki A., and Haglund C. (2010) COX-2 is associated with proliferation and apoptosis markers and serves as an independent prognostic factor in gastric cancer. Tumour Biol. 31, 1–7 [DOI] [PubMed] [Google Scholar]
- 26.Tseng C. W., Yang J. C., Chen C. N., Huang H. C., Chuang K. N., Lin C. C., Lai H. S., Lee P. H., Chang K. J., and Juan H. F. (2011) Identification of 14-3-3β in human gastric cancer cells and its potency as a diagnostic and prognostic biomarker. Proteomics 11, 2423–2439 [DOI] [PubMed] [Google Scholar]
- 27.Lee J. W., Choi H. S., Gyuris J., Brent R., and Moore D. D. (1995) Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol. Endocrinol. 9, 243–254 [DOI] [PubMed] [Google Scholar]
- 28.Altincicek B., Tenbaum S. P., Dressel U., Thormeyer D., Renkawitz R., and Baniahmad A. (2000) Interaction of the corepressor Alien with DAX-1 is abrogated by mutations of DAX-1 involved in adrenal hypoplasia congenita. J. Biol. Chem. 275, 7662–7667 [DOI] [PubMed] [Google Scholar]
- 29.Lykke-Andersen K., Schaefer L., Menon S., Deng X. W., Miller J. B., and Wei N. (2003) Disruption of the COP9 signalosome Csn2 subunit in mice causes deficient cell proliferation, accumulation of p53 and cyclin E, and early embryonic death. Mol. Cell. Biol. 23, 6790–6797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vazquez-Ortiz G., Pina-Sanchez P., Vazquez K., Duenas A., Taja L., Mendoza P., Garcia J. A., and Salcedo M. (2005) Overexpression of cathepsin F, matrix metalloproteinases 11 and 12 in cervical cancer. BMC Cancer 5, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wex T., Levy B., Wex H., and Brömme D. (2000) Human cathepsins W and F form a new subgroup of cathepsins that is evolutionary separated from the cathepsin B- and L-like cysteine proteases. Adv. Exp. Med. Biol. 477, 271–280 [DOI] [PubMed] [Google Scholar]
- 32.Santamaría I., Velasco G., Pendás A. M., Paz A., and López-Otín C. (1999) Molecular cloning and structural and functional characterization of human cathepsin F, a new cysteine proteinase of the papain family with a long propeptide domain. J. Biol. Chem. 274, 13800–13809 [DOI] [PubMed] [Google Scholar]
- 33.Colgan S. P., Eltzschig H. K., Eckle T., and Thompson L. F. (2006) Physiological roles for ecto-5′-nucleotidase (CD73). Purinergic Signal. 2, 351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Resta R., Yamashita Y., and Thompson L. F. (1998) Ecto-enzyme and signaling functions of lymphocyte CD73. Immunol. Rev. 161, 95–109 [DOI] [PubMed] [Google Scholar]
- 35.Yamashita Y., Hooker S. W., Jiang H., Laurent A. B., Resta R., Khare K., Coe A., Kincade P. W., and Thompson L. F. (1998) CD73 expression and fyn-dependent signaling on murine lymphocytes. Eur. J. Immunol. 28, 2981–2990 [DOI] [PubMed] [Google Scholar]
- 36.Strohmeier G. R., Lencer W. I., Patapoff T. W., Thompson L. F., Carlson S. L., Moe S. J., Carnes D. K., Mrsny R. J., and Madara J. L. (1997) Surface expression, polarization, and functional significance of CD73 in human intestinal epithelia. J. Clin. Invest. 99, 2588–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borsellino G., Kleinewietfeld M., Di Mitri D., Sternjak A., Diamantini A., Giometto R., Höpner S., Centonze D., Bernardi G., Dell'Acqua M. L., Rossini P. M., Battistini L., Rötzschke O., and Falk K. (2007) Expression of ectonucleotidase CD39 by Foxp3+ Treg cells: hydrolysis of extracellular ATP and immune suppression. Blood 110, 1225–1232 [DOI] [PubMed] [Google Scholar]
- 38.Dwyer K. M., Deaglio S., Gao W., Friedman D., Strom T. B., and Robson S. C. (2007) CD39 and control of cellular immune responses. Purinergic Signal. 3, 171–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deaglio S., Dwyer K. M., Gao W., Friedman D., Usheva A., Erat A., Chen J. F., Enjyoji K., Linden J., Oukka M., Kuchroo V. K., Strom T. B., and Robson S. C. (2007) Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J. Exp. Med. 204, 1257–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobie J. J., Shah P. R., Yang L., Rebhahn J. A., Fowell D. J., and Mosmann T. R. (2006) T regulatory and primed uncommitted CD4 T cells express CD73, which suppresses effector CD4 T cells by converting 5′-adenosine monophosphate to adenosine. J. Immunol. 177, 6780–6786 [DOI] [PubMed] [Google Scholar]
- 41.Canbolat O., Durak I., Cetin R., Kavutcu M., Demirci S., and Oztürk S. (1996) Activities of adenosine deaminase, 5′-nucleotidase, guanase, and cytidine deaminase enzymes in cancerous and non-cancerous human breast tissues. Breast Cancer Res. Treat. 37, 189–193 [DOI] [PubMed] [Google Scholar]
- 42.Eroglu A., Canbolat O., Demirci S., Kocaoglu H., Eryavuz Y., and Akgül H. (2000) Activities of adenosine deaminase and 5′-nucleotidase in cancerous and noncancerous human colorectal tissues. Med. Oncol. 17, 319–324 [DOI] [PubMed] [Google Scholar]
- 43.Kondo T., Nakazawa T., Murata S. I., and Katoh R. (2006) Expression of CD73 and its ecto-5′-nucleotidase activity are elevated in papillary thyroid carcinomas. Histopathology 48, 612–614 [DOI] [PubMed] [Google Scholar]
- 44.Sadej R., Spychala J., and Skladanowski A. C. (2006) Ecto-5′-nucleotidase (eN, CD73) is coexpressed with metastasis promoting antigens in human melanoma cells. Nucleosides Nucleotides Nucleic Acids 25, 1119–1123 [DOI] [PubMed] [Google Scholar]
- 45.Saitoh M., Nagai K., Nakagawa K., Yamamura T., Yamamoto S., and Nishizaki T. (2004) Adenosine induces apoptosis in the human gastric cancer cells via an intrinsic pathway relevant to activation of AMP-activated protein kinase. Biochem Pharmacol. 67, 2005–2011 [DOI] [PubMed] [Google Scholar]
- 46.Vannoni D., Bernini A., Carlucci F., Civitelli S., Di Pietro M. C., Leoncini R., Rosi F., Tabucchi A., Tanzini G., and Marinello E. (2004) Enzyme activities controlling adenosine levels in normal and neoplastic tissues. Med Oncol. 21, 187–195 [DOI] [PubMed] [Google Scholar]
- 47.Sadej R., Spychala J., and Skladanowski A. C. (2006) Expression of ecto-5′-nucleotidase (eN, CD73) in cell lines from various stages of human melanoma. Melanoma Res. 16, 213–222 [DOI] [PubMed] [Google Scholar]
- 48.Wang L., Zhou X., Zhou T., Ma D., Chen S., Zhi X., Yin L., Shao Z., Ou Z., and Zhou P. (2008) Ecto-5′-nucleotidase promotes invasion, migration and adhesion of human breast cancer cells. J. Cancer Res. Clin. Oncol. 134, 365–372 [DOI] [PubMed] [Google Scholar]
- 49.Spychala J., Lazarowski E., Ostapkowicz A., Ayscue L. H., Jin A., and Mitchell B. S. (2004) Role of estrogen receptor in the regulation of ecto-5′-nucleotidase and adenosine in breast cancer. Clin. Cancer Res. 10, 708–717 [DOI] [PubMed] [Google Scholar]
- 50.Salas T. R., Petruseva I., Lavrik O., Bourdoncle A., Mergny J. L., Favre A., and Saintomé C. (2006) Human replication protein A unfolds telomeric G-quadruplexes. Nucleic Acids Res. 34, 4857–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Lange T. (2005) Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19, 2100–2110 [DOI] [PubMed] [Google Scholar]
- 52.d'Adda di Fagagna F., Reaper P. M., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N. P., and Jackson S. P. (2003) A DNA damage checkpoint response in telomere-initiated senescence. Nature 426, 194–198 [DOI] [PubMed] [Google Scholar]
- 53.Bodnar A. G., Ouellette M., Frolkis M., Holt S. E., Chiu C. P., Morin G. B., Harley C. B., Shay J. W., Lichtsteiner S., and Wright W. E. (1998) Extension of life-span by introduction of telomerase into normal human cells. Science 279, 349–352 [DOI] [PubMed] [Google Scholar]
- 54.Kim N. W., Piatyszek M. A., Prowse K. R., Harley C. B., West M. D., Ho P. L., Coviello G. M., Wright W. E., Weinrich S. L., and Shay J. W. (1994) Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 55.Shay J. W., and Bacchetti S. (1997) A survey of telomerase activity in human cancer. Eur. J. Cancer 33, 787–791 [DOI] [PubMed] [Google Scholar]
- 56.Allgayer H., Babic R., Gruetzner K. U., Tarabichi A., Schildberg F. W., and Heiss M. M. (2000) c-erbB-2 is of independent prognostic relevance in gastric cancer and is associated with the expression of tumor-associated protease systems. J. Clin. Oncol. 18, 2201–2209 [DOI] [PubMed] [Google Scholar]
- 57.Chan A. O., Chu K. M., Lam S. K., Wong B. C., Kwok K. F., Law S., Ko S., Hui W. M., Yueng Y. H., and Wong J. (2003) Soluble E-cadherin is an independent pretherapeutic factor for long-term survival in gastric cancer. J. Clin. Oncol. 21, 2288–2293 [DOI] [PubMed] [Google Scholar]
- 58.Jüttner S., Wissmann C., Jöns T., Vieth M., Hertel J., Gretschel S., Schlag P. M., Kemmner W., and Höcker M. (2006) Vascular endothelial growth factor-D and its receptor VEGFR-3: two novel independent prognostic markers in gastric adenocarcinoma. J. Clin. Oncol. 24, 228–240 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.