Abstract
A robust method was developed and optimized for enrichment and quantitative analysis of posttranslational modifications (PTMs) in serum/plasma samples by combining immunoaffinity purification and LC-MS/MS without depletion of abundant proteins. The method was used to survey serum samples of patients with acute myeloid leukemia (AML), breast cancer (BC), and nonsmall cell lung cancer (NSCLC). Peptides were identified from serum samples containing phosphorylation, acetylation, lysine methylation, and arginine methylation. Of the PTMs identified, lysine acetylation (AcK) and arginine mono-methylation (Rme) were more prevalent than other PTMs. Label-free quantitative analysis of AcK and Rme peptides was performed for sera from AML, BC, and NSCLC patients. Several AcK and Rme sites showed distinct abundance distribution patterns across the three cancer types. The identification and quantification of posttranslationally modified peptides in serum samples reported here can be used for patient profiling and biomarker discovery research.
Biomarker identification is a key step for illustration of disease mechanisms, drug development, and diagnostics. Diagnostics research has focused on identifying biomarkers from viable biofluids, including serum/plasma, saliva, cerebrospinal fluid, and urine. Due to ease of collection and richness in proteins and metabolites, serum/plasma has been the preferred choice for diagnostic studies (1–3). Advancement of mass-spectrometry-based proteomic technologies has allowed identification and quantification of thousands of proteins in serum/plasma samples. Typically, these methods combine isotopic labeling, offline fractionation, and LC-MS/MS analysis. Facilitated by high-throughput proteomics analysis, researchers have collected vast amounts of comparative information about protein abundance in serum/plasma of patients of various types of diseases that accelerated the identification of potential biomarkers (4).
To date, the majority of serum/plasma proteomic work has been conducted to analyze total protein level abundance, with only a few studies to analyze posttranslational modifications (PTMs)1, usually glycosylation (5, 6). As one of the most important mechanisms for regulating protein function, PTMs, including phosphorylation, acetylation, ubiquitination, and methylation, have been identified and validated as critical for signaling transduction, protein degradation, and transcriptional regulation (7, 8). Currently, there exists very limited data about PTMs in serum/plasma beyond glycosylation. The abundant serum protein albumin has long been known to be acetylated by aspirin, and this reaction can occur in vitro without the presence of any acetyltransferase (9). Fibrinogen, another abundant serum protein, is also acetylated by aspirin both in vivo and in vitro (10, 11). These previous findings and the known importance of PTMs in cellular signaling provided the impetus for a large-scale survey of PTMs other than glycosylation by immunoaffinity enrichment of PTM-containing peptides.
One challenge for proteomic analysis of serum/plasma is the broad dynamic range of the serum/plasma proteome (12), including a high percentage of the total protein content of serum/plasma represented by only 12 proteins. This limitation can be partially overcome by immunodepletion of abundant proteins prior to enzymatic digestion (4, 13), however, generation of the large quantities of materials necessary for PTM enrichment with an immunodepletion workflow could be cost prohibitive. It was therefore of interest to develop a PTM enrichment workflow from serum/plasma without the need for depletion of the abundant proteins. This method allows PTM profiling from a reasonable volume of serum (∼250 μl for multiple PTM enrichment) followed by LC-MS/MS analysis. Among the PTMs surveyed, lysine acetylation (AcK) and arginine mono-methylation (Rme) were identified as the more prevalent PTMs in cancer patients' sera. These PTMs were profiled in sera from patients with acute myelogenous leukemia (AML), breast cancer (BC), and nonsmall cell lung cancer (NSCLC). At 1% FDR, we have identified 796 unique AcK sites and 808 unique Rme sites in the sera of 12 cancer patients.
The abundant serum protein human albumin was identified acetylated at 59 different sites, while other abundant proteins were also found to be acetylated, including A2M and serotransferrin. About 25% of the identified AcK sites (190 out of 796) were from the 12 most abundant serum proteins. In contrast, the Rme sites identified were from a more diverse complement of proteins, including transcriptional regulators and RNA processing proteins. Quantitative analysis identified a subset of peptides in each enrichment with differential abundance across the three cancer types surveyed. For example, the abundance of a Lys155-containing peptide from the complement component 3 protein was higher in the sera of NSCLC patients compared with AML and BC patients. Conversely, the abundance of an Arg1593 mono-methylated peptide from protein ARID1A was lower in the sera of NSCLC than the other two cancer types. Clustering of the quantitative data for the AcK and Rme enrichments revealed patterns of modification specific to cancer type as well as patient pathology. Together, these data demonstrate the utility of PTM profiling of human serum samples for disease characterization and the potential for biomarker discovery.
EXPERIMENTAL PROCEDURES
Cancer Patient Serum
Serum samples of four patients of AML, BC, and NSCLC were purchased from Proteogenex (Culver City, CA). Patient information and the concentration of total protein of each serum sample are provided in Supplemental Table S1.
Sample Preparation
Serum samples were processed using the PTMScan method as previously described (14). Equal volumes of serum (250 μl for individual sample) were mixed with urea lysis buffer (9 m sequanol-grade urea, 20 mm HEPES, pH 8.0, 1 mm β-glycerophosphate, 1 mm sodium vanadate, 2.5 mm sodium pyrophosphate) to a final concentration of 6 m urea. For the technical triplicate experiment, 150 μl serum from four nonsmall cell lung cancer patients were pooled together and split into three aliquots for independent processing. The samples were centrifuged at 16,000 × g for 15 min at 4 °C. Supernatants were collected and reduced with 4.5 mm DTT for 30 min at 55 °C. Reduced lysates were alkylated with iodoacetimide (0.095g per 5 ml H2O) for 15 min at room temperature in the dark. Samples were diluted 1:4 with 20 mm HEPES, pH 8.0, and digested overnight with 10 μg/ml trypsin-TPCK (Worthington, #LS003740, Lakewood, NJ). Digested peptide lysates were acidified with 1% TFA and peptides were desalted over 360 mg SEP PAK Classic C18 columns (Waters, #WAT051910, Milford, MA). Peptides were eluted with 40% acetonitrile in 0.1% TFA, dried under vacuum, and stored at −80 °C.
Immunoprecipitation
Enrichment of posttranslationally modified peptides was performed using the antibodies listed in Table I. following protocols described previously (14, 15). Briefly, saturating amounts of the indicated antibodies were bound to 30 μl packed Protein A agarose beads (Roche, Indianapolis, IN) overnight at 4 °C. Lyophilized serum peptides were resuspended in MOPS IAP buffer (50 mm MOPS, pH 7.2, 10 mm KH2PO4, 50 mm NaCl) and centrifuged 5 min at 10,000 × g. Supernatants were mixed with antibody bead slurries for 2 h at 4 °C. Beads were pelleted by centrifugation 30 s at 2,000 × g at 4 °C. Beads were washed three times with 1.5 ml IAP buffer containing 1% Nonidet P-40 and three times with 1 ml water (Burdick and Jackson, Morristown, NJ). Peptides were eluted from beads with 0.15% TFA (sequential elutions of 40 μl followed by 35 μl, 10 min each at room temperature). Eluted peptides were desalted over tips packed with Empore C18 (Sigma, St. Louis, MO) and eluted with 40% acetonitrile in 0.1% TFA. Eluted peptides were dried under vacuum and subject to a second in-solution trypsin digest using 250 ng of sequencing grade trypsin (Promega, Madison, WI) in 50 mm ammonium bicarbonate/5% acetonitrile for 2 h at 37 °C to minimize miscleavage and digest any antibody remaining in the sample. Samples were acidified with TFA and repurified over C18 tips as before.
Table I. Summary of posttranslationally modified peptides identified using various enrichments from pooled sera of cancer patients.
| Exp. # | Sample | Enrichment Method | PTM type | Raw file | Unique modified peptide |
|---|---|---|---|---|---|
| 1 | AML | IMAC-Fe3+ | pSTY | 23168 | 148 |
| 2 | BC | IMAC-Fe3+ | pSTY | 23169 | 184 |
| 3 | NSCLC | IMAC-Fe3+ | pSTY | 23170 | 145 |
| 4 | AML | pY motif antibody | pY | 23231 | 39 |
| 5 | BC | pY motif antibody | pY | 23232 | 39 |
| 6 | NSCLC | pY motif antibody | pY | 23233 | 44 |
| 7 | AML | pY → IMAC-Fe3+ | pY | 23234 | 19 |
| 8 | BC | pY → IMAC-Fe3+ | pY | 23235 | 26 |
| 9 | NSCLC | pY → IMAC-Fe3+ | pY | 23236 | 25 |
| 10 | AML | AcK motif antibody | AcK | 23171 | 355 |
| 11 | BC | AcK motif antibody | AcK | 23172 | 359 |
| 12 | NSCLC | AcK motif antibody | AcK | 23173 | 431 |
| 13 | AML | Rme motif antibody | Rme | 23238 | 206 |
| 14 | BC | Rme motif antibody | Rme | 23239 | 172 |
| 15 | NSCLC | Rme motif antibody | Rme | 23240 | 138 |
| 16 | AML | Pan-Kme antibody | Kme (mono, di, tri) | 23241 | 117 |
| 17 | BC | Pan-Kme antibody | Kme (mono, di, tri) | 23242 | 142 |
| 18 | NSCLC | Pan-Kme antibody | Kme (mono, di, tri) | 23243 | 148 |
IMAC
IMAC enrichment was performed as previously described (16). Nickel-agarose magnetic beads (Qiagen, Valencia, CA) were treated with EDTA to remove the nickel, washed three times with H2O, loaded with aqueous FeCl3 for 30 min, and washed. For phosphopeptide enrichment, 10 μl Fe3+-agarose slurry were added to peptide digested from 10 μl of serum in 1 ml 0.1% TFA/80% acetonitrile for 30 min at room temperature. Unbound peptides were removed by washing three times with 0.1% TFA/80% MeCN. Bound peptides were eluted with 2X 50 μl of 2.5% ammonia/50% acetonitrile solution for 5 min. The eluent was immediately acidified by 20% TFA and dried in a speed-vac. Samples were resuspended in 50 μl 0.15% TFA, desalted over C18 tips, and redried in a speed vac.
LC-MS/MS Analysis
Immunoprecipitated peptides were resuspended in 0.125% formic acid and separated on a reversed-phase C18 column (75 μm inner diameter × 10 cm) packed into a PicoTip emitter (∼8 μm inner diameter) with Magic C18 AQ (100Å × 5 μm, New Objective, Woburn, MA). Each sample was split, and analytical replicate injections were run to increase the number of identifications and provide metrics for analytical reproducibility of the method. A standard peptide mix (MassPREPTM Protein Digestion Standard Mix 1, Waters) was spiked in each sample vial in a total quantity of 100 fmol (33 fmol per injection) prior to LC-MS/MS analysis. For antibody enrichment, peptides from 120 μl serum were run per injection; for IMAC, peptides from 5 μl serum were run per injection. Replicate injections were run nonsequentially to reduce artificial changes in peptide abundance due to changes in instrument performance over time. One replicate of each sample was injected then the second replicate in reverse order. Peptides were eluted using a 120-min or 150-min linear gradient of acetonitrile in 0.125% formic acid delivered at 280 nl/min from 3% to 30% acetonitrile. Tandem mass spectra were collected in a data-dependent manner with an LTQ-Orbitrap ELITE mass spectrometer (Thermo Fisher Scientific, Waltham, MA) running XCalibur 2.0.7 SP1 using a top-20 MS/MS method, a dynamic repeat count of one, and a repeat duration of 30 s. The isolation window was set at 1.0 Da with a normalized collision energy of 35%. Real-time recalibration of mass error was performed using lock mass (17) with a singly charged polysiloxane ion m/z = 371.101237. The data associated with this manuscript including labeled MS2 spectra in Skyline library format may be downloaded from the ProteomeXchange Consortium via PRIDE with project accession numbers: PXD002931 [username: reviewer42095@ebi.ac.uk; password: wpPsK3wN] and PXD002932 [username: reviewer39066@ebi.ac.uk; password: qT4I4DrS].
MS/MS spectra were evaluated using SEQUEST and the Core platform from Harvard University (18–20). Files were searched against the NCBI homo sapiens FASTA database updated on June 27, 2011 containing 34,899 forward and 34,899 reverse sequences. A mass accuracy of ±5 ppm was used for precursor ions and 1 Da for product ions. Enzyme specificity was limited to trypsin, with at least one tryptic (K- or R-containing) terminus required per peptide and up to four miscleavages allowed. Cysteine carboxamidomethylation was specified as a static modification; oxidation of methionine residue and the appropriate PTM were allowed as variable modifications for each enrichment sample set. Reverse decoy databases were included for all searches to estimate false discovery rates, and filtered using a 1% FDR in the Linear Discriminant module of Core.
All quantitative results were generated using Progenesis V4.1 (Waters Cooperation) and Skyline Version 3.1 to extract the integrated peak area of the corresponding peptide assignments according to previously published protocols (21, 22). Extracted ion chromatograms for peptide ions that changed in abundance between samples were manually reviewed to ensure accurate quantitation in Skyline. Statistical analysis of the quantitative data was done using a two-tailed t test between two cancer groups. The maximum negative log-p value from three comparison pairs was used to indicate significance for abundance changes of a certain peptide between two cancer groups. False discovery rate for each binary comparison was further controlled by applying the Benjamini–Hochberg procedure. Heat maps of the quantitative data were generated and clustered in Spotfire DecisionSite (TIBCO Software AB) version 9.1.2.
Western Blotting
Equal volumes of serum samples were mixed with SDS-PAGE sample buffer (Cell Signaling Technology, Danvers, MA #7723,) and run on 4–20% gradient tris-glycine gels (Invitrogen Grand Island, NY). For pan-AcK and pan-Rme Western blots, serum was diluted 10-fold and 20 μl were loaded. For albumin, serum was diluted 10,000- fold and 20 μl were loaded. Proteins were transferred to nitrocellulose (Millipore, Bedford, MA) and blocked for 1 h in 5% nonfat dry milk (Sigma) in TBS. Primary antibodies (pan-AcK (#13420), pan-Rme (#8015/#8711), and albumin (#4929)) were incubated in 5% BSA in TBS plus 0.1% Tween-20 overnight at 4 °C. Membranes were washed three times with TBS plus 0.1% Tween-20, incubated with anti-rabbit secondary antibody (#5366) for 1 h at room temperature in 5% milk and TBS plus 0.1% Tween-20, washed three times with TBS plus 0.1% Tween-20, dried, and developed on the Odyssey near-infrared imaging system (LI-COR). All antibodies were from Cell Signaling Technology.
RESULTS
Enrichment Workflow for PTM Peptide Identification
To demonstrate an optimal workflow for analysis of posttranslational modifications in serum samples, we first tested enrichment using various PTM antibodies and IMAC-Fe3+ with pooled serum samples as outlined in Table I. Serum samples were directly processed for PTM enrichment without any prior depletion of abundant serum proteins. The enrichments performed included IMAC-Fe3+, phosphotyrosine enrichment, phosphotyrosine enrichment followed by IMAC-Fe3+, acetyl-lysine enrichment (AcK), arginine mono-methylation (Rme), and lysine pan-methylation. Of these enrichments, acetyl-lysine and mono-methyl-arginine showed the most promising results, with the highest number of peptides per sample (Table I). The number of identifications for the phospho-enrichment was low and failed to identify proteins/sites known to be important disease drivers or substrates for the cancer types profiled (data not shown).
PTM Profiling from Individual Patient Samples
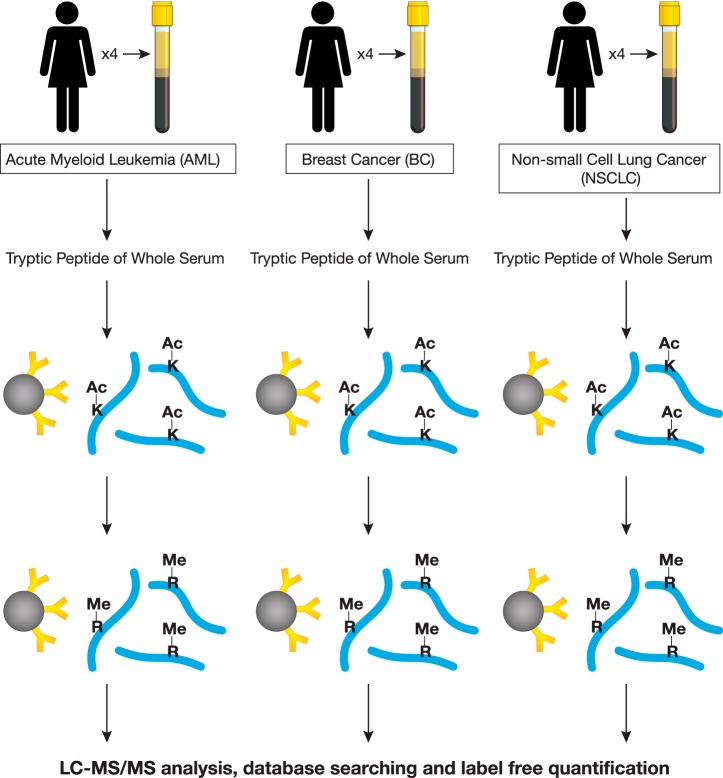
Having established the ability to profile PTMs directly from serum samples, we then applied the immunoaffinity enrichment method to profile lysine acetylation and arginine mono-methylation in patient sera of acute myeloid leukemia (AML), breast cancer (BC), and nonsmall cell lung cancer (NSCLC), respectively (n = 4 for each type of cancer, 12 samples total). As different PTMs were being profiled, PTM enrichment was performed sequentially on the same sample as previously described (23). To minimize interference from factors such as sex, age, and ethnic background, the study included only female patients of Caucasian background (Supplemental Table S1). The general workflow of the sequential enrichment is outlined in Fig. 1. Prior to immunoaffinity enrichment, Western blotting using pan-AcK and Rme motif antibodies were performed on the 12 patient samples (Supplemental Fig. S1). Most of the AcK signal in the Western blotting fell into a molecular weight range between 58–80 kDa, corresponding to human albumin (MW = 69kDa), with other lower intensity bands visible throughout the MW range. In the Rme blot, the strong signal from albumin was not observed, with other bands detected at various molecular weights.
Fig. 1.
General workflow of enrichment of AcK and Rme peptides from cancer patient serum samples. Equal volumes of serum samples (250 μl from each patient) were mixed with 9 m urea buffer and reduced, alkylated, digested with trypsin, desalted, lyophilized, and resuspended in MOPS IAP Buffer. Peptides were incubated with AcK and Rme antibody beads sequentially. Analytical replicates of enriched peptides were run in LC-MS/MS. Label-free quantification of peptide peak areas in the MS1 channel was performed to identify changes in abundance of AcK and Rme peptides across all samples.
Enrichment for AcK generated a range of 214 to 486 unique AcK peptides from each serum sample, while enrichment for Rme generated a range of 199 to 257 unique Rme peptides from each serum sample (Supplemental Tables S2 and S3, Details Tab). Many AcK and Rme sites identified were represented by multiple peptides due to methionine oxidation, miscleavage, and different charge states. Tables were made nonredundant by unique protein site (Supplemental Tables S2 and S3, Summary Tab) using the peptide with the largest number of MS/MS identifications (Count in Details Column, Summary Tab, Supplemental Table S2 and S3) as the best representative for a particular PTM site. In total, 796 and 808 unique sites were identified for AcK and Rme, respectively. Of these, 672 AcK sites and 619 Rme sites were previously unidentified and will be curated into the PhosphoSitePlus database as a public resource (24).
Classification of Proteins with Identified PTMs
This study identified 520 and 688 unique proteins from AcK and Rme enrichment, respectively (Supplemental Table S4). The top 10 protein classes represented in each enrichment are shown in the pie charts in Figs. 2A (AcK) and 2B (Rme). For AcK sites, the top five protein categories were secreted protein, receptor/channel/transporter/cell surface protein, adhesion/extracellular protein, chromatin/DNA-binding/DNA repair/DNA replication protein, and transcriptional regulator. Peptides from albumin were identified acetylated at a total of 59 unique AcK sites, which is consistent with the Western blotting signal shown in Supplemental Fig. S1A. High numbers of acetylation sites were also identified on other serum-abundant proteins, including alpha-2-macroglobulin and serotransferrin with a total number of 35 and 29 unique AcK sites identified, respectively. Overall, about 25% of all AcK sites identified (190 out of 796) were from the top 12 abundant serum proteins. Conversely, for Rme, only 4 out of 808 unique Rme sites from top 12 abundant serum proteins were identified. The top five protein categories for Rme were receptor/channel/transporter/cell surface protein, RNA processing, transcriptional regulator, adhesion/extracellular protein, and adaptor/scaffold. A large number of unique Rme sites was identified from various heterogeneous nuclear ribonucleoprotein (hnRNP) isoforms, most of which have been identified before in a previous study (14). The presence of these posttranslationally modified hnRNP peptides in serum/plasma has not been previously reported. The data for the two enrichments were largely complementary, with only 35 proteins identified in common (Supplemental Table S4, Supplemental Fig. S2). There was also a low degree of overlap between these results and a recent large-scale plasma proteome study using iTRAQ labeling and offline fractionation prior to LC-MS/MS analysis, which identified over 5300 proteins with high confidence (4), with 226 out of 520 proteins in common for AcK and 212 out of 688 for Rme (Supplemental Fig. S2).
Fig. 2.
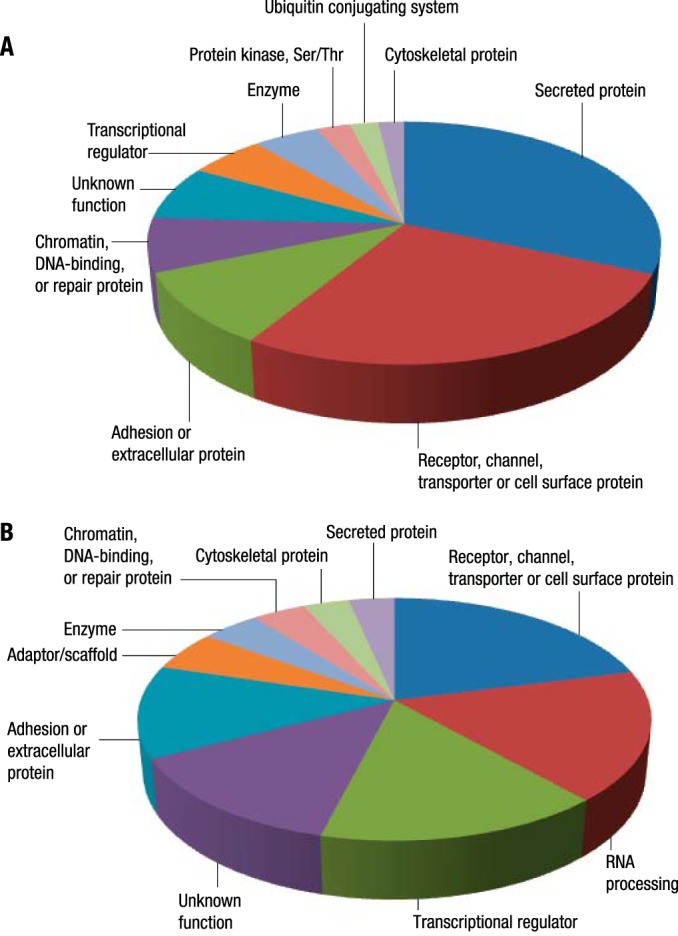
Functional classification of functions of proteins identified by AcK (A) and Rme enrichment (B). The size of each pie represents the number of unique modification sites identified in proteins belonging to each functional group. As most lysine acetylation sites were identified in serum-abundant proteins, secreted proteins including A2M, TF, HPX, HP, complement component 3, and transporter proteins including ALB are the most enriched functional groups. There were few Rme sites identified in serum-abundant proteins; instead, large numbers of RNA processing proteins and transcriptional regulators were identified methylated at arginine sites. Protein class information was retrieved from the PhosphoSite Plus database.
Reproducibility Assessment of the Method by Technical Triplicate Analysis
A pooled mixture of NSCLC patient serum was split into three aliquots and subject to parallel, independent trypsin digestion, sequential immuno-enrichment for AcK and Rme, and LC-MS/MS analysis. We have identified 555, 502, and 515 unique AcK peptides and 373, 357, and 377 unique Rme peptides from the three independent samples, respectively. A total of 778 unique AcK peptides and 564 unique Rme peptides were identified across the triplicate runs, of which, 361 (46%) AcK peptide identifications and 226 (40%) Rme peptide identifications were shared by all three samples (Supplemental Fig. S3). Of the unique peptides identified, 380 AcK and 356 Rme were quantified using the label-free quantification approach (Supplemental Tables S5 and S6). The distributions of %CV between technical triplicates for the two PTMs are shown in Supplemental Fig. S4. The median %CVs were 23 and 17% for lysine acetylation and arginine mono-methylation, respectively. About 78% of acetyl-lysine peptides and 94% of mono methyl-arginine peptides had %CVs lower than 40%, indicating good technical reproducibility of the method (Supplemental Fig. S4).
Quantitative Analysis of AcK and Rme Peptides across Three Cancer Types
A label-free quantification strategy was applied to analyze PTM site-specific abundance changes across the three cancer types profiled. Analytical replicates of each enriched peptide mixture were run to allow greater opportunity for identification of unique peptides and to provide data on analytical reproducibility of the method. To monitor the stability of the LC, ionization efficiency, and sensitivity, a standard peptide mix of HPLC-purified peptides from four proteins was added to each sample prior to LC-MS/MS. Sequences of the four standard proteins were added to the human database to allow identification of these peptides in each LC-MS/MS run. Standard peptides with retention times spanning the entire gradient range were chosen to monitor instrument performance. Standard peptide retention times and integrated MS1 peak areas across all 48 LC-MS/MS runs were consistent, indicating good performance of the instrument throughout the analysis. (Supplemental Fig. S5).
Both Skyline and Progenesis were used for the label-free quantification of posttranslationally modified peptides across all samples. Both programs perform retention time alignment across LC-MS/MS runs by either common MS2 identifications (Skyline) or common ion features (Progenesis) so that a narrow and accurate precursor window can be applied to each unique peptide or ion feature (21). This methodology allows determination of peptide peak intensities across all samples even if a particular peptide was not MS/MS identified in every sample run (14, 21, 25).
Other important quality control metrics for label-free quantification include coefficients of variation (%CV) between analytical replicates and chromatographic shape of the extracted ion chromatograms, both of which were monitored in Skyline. In total, we have quantified 612 and 621 unique sites for AcK and Rme, respectively (Summary Tab of Supplemental Tables S2 and S3). The median %CV was 14% for AcK out of 6983 analytical replicate measurements across all samples; and 15% for Rme out of 7200 analytical replicate measurements across all samples. Histograms plotting the distribution of %CVs for AcK and Rme data are shown in Supplemental Fig. S6.
The fold changes of each unique PTM site across three cancer types were calculated by comparing the average intensity of the chosen modified peptide across all four patients of each cancer type. Statistical analysis of each binary comparison was performed using a two-tailed t test (Summary Tab of Supplemental Tables S2 and S3). Those PTM sites with any fold change greater than 2.5 or less than –2.5 and a corresponding p value less than .01 were manually examined in Skyline and are indicated with bold intensity values in the Summary Tabs of Supplemental Tables S2 and S3.
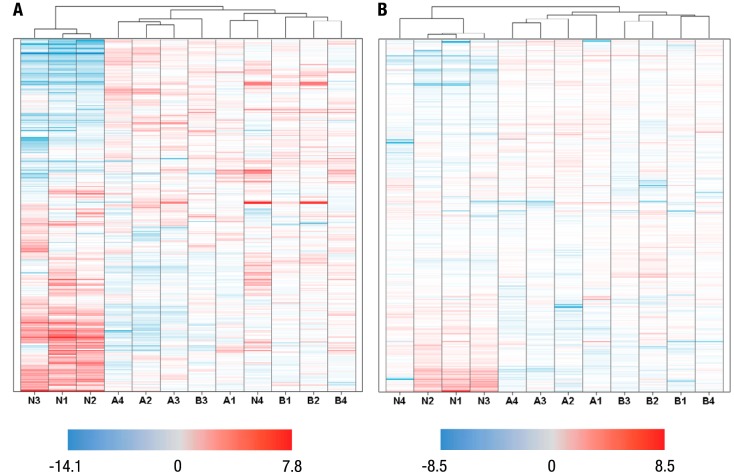
Peptide intensities in the 12 individual patient samples were compared with the median intensity across all samples to generate log2 ratios. Log2 ratios were used to generate heat maps in Spotfire and hierarchically clustered. For Rme data, samples from the same cancer type clustered together, with higher correlation between breast cancer and acute myeloid leukemia than with nonsmall cell lung cancer (Fig. 3B). For AcK data, not all patients of the same type of cancer were clustered in the same group. Specifically, patients #1, #2, and #3 of nonsmall cell lung cancer were clustered together, while patient #4 was an obvious outlier that exhibited a significantly different pattern of abundance for most unique AcK sites (Fig. 3A). Consistent with the clustering results of Rme data, the correlation between breast cancer group and acute myeloid leukemia is closer than those with nonsmall cell lung cancer.
Fig. 3.
Hierarchical clustering of quantitative data for AcK and Rme enrichments from 12 cancer patients' sera. (A) Each row in the heat map is a unique, label-free quantified AcK site. A: acute myeloid leukemia, AML; B: breast cancer, BC; N: nonsmall cell lung cancer, NSCLC. The relative intensity of the site in each patient's serum was represented by the log2 ratio of the site intensity in the individual patient divided by the median site intensity across all 12 patients. The range of log2 ratios is –14.1 (blue) and +7.8 (red). Patient #4 of NSCLC is an obvious outlier compared with the other three NSCLC patients and was diagnosed as squamous cell carcinoma, while NSCLC patients #1, 2, and 3 were diagnosed as adenocarcinoma. (B) Each row in the heat map is a unique, label-free quantified Rme site. The range of log2 ratios is –8.5 (blue) and +8.5 (red). Individual patients of the same cancer group were clustered together, and Rme sites in NSCLC patients showed a distinct intensity pattern compared with the other two cancer types.
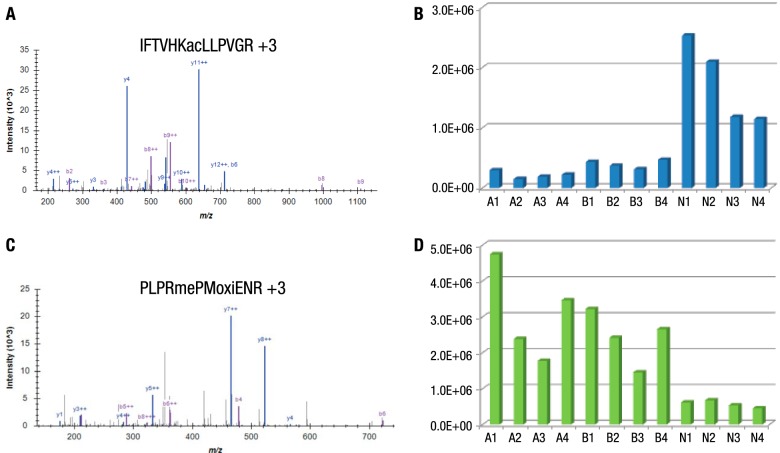
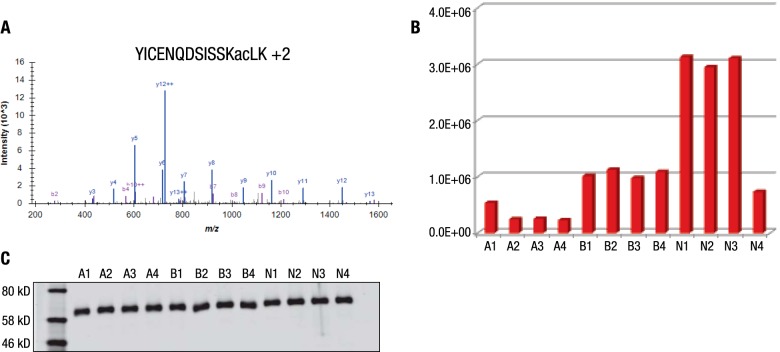
Specific examples of differential relative abundance between cancer types included a complement component 3 peptide acetylated at Lys155 (Fig. 4A). This peptide was 4.4- and 8.1-fold higher intensity in NSCLC samples than the average intensity in breast cancer and AML samples, respectively (Fig. 4B). An Arg1593-monomethylated peptide from the protein ARID1A was identified (Fig. 4C) and its abundance was 4.3- and 5.4-fold lower in NSCLC samples than the average intensity in breast cancer and AML samples, respectively (Fig. 4D). Some peptides showed an inconsistent intensity pattern even within the same cancer type, such as a peptide acetylated at Lys298 of albumin (Fig. 5A). This peptide was of higher abundance in NSCLC patients #1, #2, and #3 with 2.9- and 9.7-fold increase compared with the other two cancer types, whereas NSCLC patient #4 showed similar intensity to breast cancer and AML samples (Fig. 5B). Although there were differences in relative abundance for several acetylated peptides derived from albumin, the overall protein level of albumin was not changed (Fig. 5C). The histological diagnosis of NSCLC patient #4 (with 35 years of smoking history) was squamous cell carcinoma; while the histological diagnosis of #1, #2, and #3 patients (nonsmokers) of NSCLC was adenocarcinoma (Supplemental Table S1).
Fig. 4.
MS/MS spectra and intensity of Lys155 of complement component 3 (A and B) and Arg1593 of ARID1A (C and D). MS/MS spectra were annotated in Skyline. Site intensity is represented by the average integrated MS1 peak area of the modified peptide from two analytical replicates.
Fig. 5.
MS/MS spectra and intensity of Lys298 of Albumin (A and B). A number of unique AcK sites showed significantly different intensities in patient #4 of the NSCLC group (squamous cell carcinoma) compared with patients 1–3 (adenocarcinoma) such as Lys298 of albumin. The relative abundance of the Lys298-containing peptide is significantly higher in the BC group than the AML group, while the total protein abundance of human albumin showed no changes by Western blotting (C).
DISCUSSION
Extensive effort has been expended on method development for analyzing the serum/plasma proteome to identify potential biomarkers (1, 3, 26, 27). In the current study, we have developed a robust workflow combining PTM motif antibody enrichment and LC-MS/MS analysis for profiling different PTMs. The workflow has proven successful to analyze both plasma (data not shown) and serum samples for many commonly studied PTMs, including phosphorylation, acetylation, arginine methylation, and lysine methylation, with lysine acetylation and arginine mono-methylation yielding the highest number of unique peptide identifications.
Another important feature of the workflow is its compatibility with both label-free and isobaric-labeling quantification strategies. For this study, we employed label-free quantification based on MS1 precursor intensities for each identified PTM peptide (14, 22). The key factor for successful label-free quantification is the consistency of instrument performance during the entire data collection period. To validate the reproducibility of the method, we have performed a technical triplicate experiment using pooled NSCLC serum. At the MS2 identification level, 46% (361 out of 778 unique AcK peptides) and 40% (226 out of 564 unique Rme peptides) of peptides were identified across all three LC-MS/MS (Supplemental Fig. S3). The MS1 integrated peak areas of identified modified peptides showed a tight distribution of %CV across technical triplicate samples (Supplemental Fig. S4), with the median %CV for AcK and Rme peptides were 23 and 17%, respectively (Supplemental Tables S5 and S6). These quantitative data demonstrate the robustness of the method as well as the accuracy of label-free quantification for peptides with PTMs. These data are consistent with previous studies showing the reproducibility of label-free quantification, and label-free quantification followed by affinity purification has been broadly adopted for quantifying changes of various PTMs (14, 21, 28–33). Various software solutions, both academic and commercial, have been developed for label-free quantitative approaches, including MaxLFQ, Skyline, Ideal-Q, and OpenMS. These software packages have been extensively tested and validated in the literature (25, 34–36). Additionally, data from the spiked-in standard peptides showed reproducible retention time and integrated peak areas across all 48 LC-MS/MS runs that spanned over a week of continuous instrument operation (Supplemental Fig. S5). The accuracy of the quantitative data was also ensured by manual review of integrated peaks for those peptides with significant abundance changes across different cancer groups. The quantitative data from this study showed a distinct pattern for both AcK and Rme in which NSCLC patient samples were significantly different than AML and BC (Fig. 3). The hierarchical clustering based on the quantitative data also identified patient #4 of NSCLC as an outlier in the group for both modifications. Patient #4 was diagnosed as adenocarcinoma while the other three NSCLC patients were diagnosed as squamous cell carcinoma, raising the possibility that serum PTMs could also serve as biomarkers for histological classification of NSCLC. The small number of samples for each cancer type profiled in this study precludes definitive conclusions on this point but serves as an area worthy of further investigation. Therefore, it should be noted that this study is largely technical in nature, and further work on a larger cohort of samples would be necessary to more fully validate candidate disease biomarkers identified herein.
In order to achieve deeper proteome coverage of serum/plasma, recent studies have employed immunodepletion of abundant proteins followed by offline fractionation prior to LC-MS/MS analysis (4, 37). Combined with multiplex isotopic labeling, Keshishian et al. confidently identified over 5300 proteins from plasma samples of four patients (4). In the current study, we identified a total of 1173 unique proteins by combining the results of both AcK and Rme enrichments. Of these, there were 422 proteins overlapping between the two studies (Supplemental Table S4, Supplemental Fig. S2). It should be noted that two unique peptides were required for protein identification in the study by Keshishian et al., while in this study we reported all proteins with at least one unique modified peptide that passed 1% FDR filtering. We also compared our results with identified plasma proteins deposited in the Plasma Proteome Database (38). Roughly half of the proteins identified in our study (428 out of 1173) are absent from the repository database (Supplemental Table S4). We believe the complementarity of the results at the protein level is due to the specific enrichment by PTM motif antibodies that unveiled a large group of low abundance proteins, including transcriptional regulators, RNA processing proteins, and receptors. Interestingly, in the AcK enrichment samples, we identified several serum-abundant proteins, including albumin, APOA1, serotransferrin, and alpha-2-macroglobulin, and most were acetylated at multiple lysine sites. We have observed differential patterns of acetylation in NSCLC patients #1, #2, and #3 (histological diagnosis of adenocarcinoma) for these proteins compared with the NSCLC patient #4 (histological diagnosis of squamous cell carcinoma) (Fig. 3). The protein group showing higher abundance of acetylated peptides in NSCLC included albumin, with several lysine acetylation sites showing similar patterns as exemplified by Lys298 (Fig. 5B). The differential abundance of acetylated albumin peptides was in contrast to the total protein level of albumin, which showed minimal changes across all 12 cancer patients (Fig. 5C). As the most abundant protein in blood, albumin had previously been considered as an interfering protein rather than a potential biomarker when performing serum/plasma proteome profiling studies. However, the findings in this study of differential acetylation sites on albumin such as Lys160, Lys161, and Lys298 in different cancer types suggest that the potential exists for biomarker discovery even among abundant serum proteins.
Histone proteins, including H1, H3, and H4, were identified in the study heavily acetylated at a number of lysine sites. Hyper-acetylation of core histone tails is believed to neutralize the positive charge of these domains and weaken histone–DNA and nucleosome–nucleosome interactions. This weakening destabilizes chromatin structure and increases the accessibility of DNA to various DNA-binding proteins (39–41). Peptides from histone H3.3 acetylated at Lys15, Lys19 and H4 Lys6, Lys13, Lys17 showed significant abundance differences between cancer groups that may serve as indicators of the status of nucleosomes.
In contrast to the lysine acetylation results, in which many peptides were identified from abundant serum proteins, arginine mono-methylated peptides identified were distributed into diverse protein groups with few assignments to abundant serum proteins. The abundance differences of Rme sites were also generally less dramatic than AcK sites between the NSCLC group and the other two cancer groups (Fig. 3). Although there were not as many dramatic intensity changes of Rme sites across the three cancer groups, the four patients belonging to each cancer group clustered together, indicating more direct correlation between cancer type and arginine mono-methylation levels in serum than lysine acetylation levels. Consistent with the hierarchical clustering showing a closer relationship between AML and BC as cancer groups (Fig. 3), none of the p values derived from methyl-arginine quantitative data for site AML and BC samples passed Benjamini–Hochberg critical value analysis (Supplemental Table S3), which indicates similar levels of arginine mono-methylation levels between AML and BC. There were, however, significant differences in site abundance between NSCLC serum samples compared with AML or BC, including heterogeneous nuclear ribonucleoproteins, well-studied RNA processing proteins involved in posttranscriptional modification of pre-mRNA (42). Methylation of hnRNP family members containing the RGG motif promotes import into the nucleus (43). Therefore, changes in the levels of identified arginine methylation level in hnRNPs such as Arg291 of hnRNP A0, Arg206 of hnRNP A1, Arg203 of hnRNP A2/B1, and Arg272 of hnRNP D0 may help estimate their nucleo-cytoplasmic shuttling as well as posttranscriptional regulation in different cancers.
There have been few studies that detail posttranslational modification of serum/plasma proteins, so it was not clear whether the acetylation and arginine methylation identified from our study was regulated by well-known enzymatic catalysis or in an enzyme-independent manner. For example, we have identified several well-studied lysine acetylation sites on histones, including H3 and H4 (Supplemental Table S2). This finding is supported by the presence of several types of histone acetyltransferases as well as histones in the bloodstream even though they are nuclear proteins (44–46). Acetylation of albumin and hemoglobin by circulating aspirin has been previously shown in blood, providing one possible explanation for enzyme-independent lysine acetylation of serum-abundant proteins (47). The identification of most arginine methylation sites on low abundance proteins such as transcriptional regulators and RNA processing proteins in our study makes it less likely that there was a concentration-dependent, enzyme-independent reaction for methylation in blood. In fact, numerous protein arginine methyltransferases have been identified in plasma in previous studies (44, 48). Therefore, arginine methylation could occur either in intracellular compartments followed by release to the bloodstream or be catalyzed in the bloodstream directly.
In summary, we have completed a large-scale PTM profiling study in serum that identified many different types of PTMs in bloodstream. To our knowledge, this is the first study that systematically identified and quantified PTMs in serum. The workflow we have developed and optimized successfully provided accurate quantitative information for lysine acetylation and arginine mono-methylation in the serum samples of three cancer types. The successful identification of Rme modification of low abundance proteins demonstrated the specificity and sensitivity of PTM motif antibody based enrichment for serum samples. In addition, the identification of multiple lysine acetylation sites in abundant serum proteins such as albumin, some of which showed differential abundance patterns across cancer types, proved that immunodepletion of top abundant proteins in serum is unnecessary and potentially undesirable for serum PTM profiling studies. Together, these results provide novel identification data of posttranslational modifications of serum proteins and define a workflow that is suitable for large-scale quantitative proteomic studies of clinical serum/plasma samples for biomarker identification.
Supplementary Material
Footnotes
Author contributions: H.G. and J.C.S. designed the research; H.G., J.R., X.J., T.L., K.R., V.Y., and K.A.L. performed the research; H.G. contributed new reagents or analytic tools; H.G. analyzed data; and H.G. and M.P.S. wrote the paper.
 This article contains supplemental material Supplemental Tables S1-S6 and Supplemental Figs. S1-S6.
This article contains supplemental material Supplemental Tables S1-S6 and Supplemental Figs. S1-S6.
1 The abbreviations used are:
- PTM
- posttranslational modification
- AML
- acute myeloid leukemia
- BC
- breast cancer
- NSCLC
- nonsmall cell lung cancer
- AcK
- lysine acetylation
- Rme
- arginine mono-methylation
- IMAC
- immobilized metal ion affinity chromatography
- hnRNP
- heterogeneous nuclear ribonucleoprotein.
REFERENCES
- 1.Galasko D., and Golde T. E. (2013) Biomarkers for Alzheimer's disease in plasma, serum and blood—Conceptual and practical problems. Alzheimer's Res. Therapy 5, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn J. M., Sung H. J., Yoon Y. H., Kim B. G., Yang W. S., Lee C., Park H. M., Kim B. J., Kim B. G., Lee S. Y., An H. J., and Cho J. Y. (2014) Integrated glycoproteomics demonstrates fucosylated serum paraoxonase 1 alterations in small cell lung cancer. Mol. Cell. Proteomics 13, 30–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yotsukura S., and Mamitsuka H. (2015) Evaluation of serum-based cancer biomarkers: A brief review from a clinical and computational viewpoint. Crit. Rev. Oncol. Hematol. 93, 103–115 [DOI] [PubMed] [Google Scholar]
- 4.Keshishian H., Burgess M. W., Gillette M. A., Mertins P., Clauser K. R., Mani D. R., Kuhn E. W., Farrell L. A., Gerszten R. E., and Carr S. A. (2015) Multiplexed, quantitative workflow for sensitive biomarker discovery in plasma yields novel candidates for early myocardial injury. Mol. Cell. Proteomics 14, 2373–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berven F. S., Ahmad R., Clauser K. R., and Carr S. A. (2010) Optimizing performance of glycopeptide capture for plasma proteomics. J. Proteome Res. 9, 1706–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen S., Lu C., Gu H., Mehta A., Li J., Romano P. B., Horn D., Hooper D. C., Bazemore-Walker C. R., and Block T. (2012) Aleuria aurantia lectin (AAL)-reactive immunoglobulin G rapidly appears in sera of animals following antigen exposure. PloS One 7, e44422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prabakaran S., Lippens G., Steen H., and Gunawardena J. (2012) Post-translational modification: Nature's escape from genetic imprisonment and the basis for dynamic information encoding. Wiley Interdisciplinary Rev. Syst. Biol. Med. 4, 565–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang H., Lin S., Garcia B. A., and Zhao Y. (2015) Quantitative proteomic analysis of histone modifications. Chemical Rev. 115, 2376–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liyasova M. S., Schopfer L. M., and Lockridge O. (2010) Reaction of human albumin with aspirin in vitro: Mass spectrometric identification of acetylated lysines 199, 402, 519, and 545. Biochem. Pharmacol. 79, 784–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bjornsson T. D., Schneider D. E., and Berger H. Jr. (1989) Aspirin acetylates fibrinogen and enhances fibrinolysis. Fibrinolytic effect is independent of changes in plasminogen activator levels. J. Pharmacol. Exp. Therapeut. 250, 154–161 [PubMed] [Google Scholar]
- 11.Pinckard R. N., Hawkins D., and Farr R. S. (1968) In vitro acetylation of plasma proteins, enzymes and DNA by aspirin. Nature 219, 68–69 [DOI] [PubMed] [Google Scholar]
- 12.Mitchell P. (2010) Proteomics retrenches. Nature Biotech. 28, 665–670 [DOI] [PubMed] [Google Scholar]
- 13.Adkins J. N., Varnum S. M., Auberry K. J., Moore R. J., Angell N. H., Smith R. D., Springer D. L., and Pounds J. G. (2002) Toward a human blood serum proteome: Analysis by multidimensional separation coupled with mass spectrometry. Mol. Cell. Proteomics 1, 947–955 [DOI] [PubMed] [Google Scholar]
- 14.Guo A., Gu H., Zhou J., Mulhern D., Wang Y., Lee K. A., Yang V., Aguiar M., Kornhauser J., Jia X., Ren J., Beausoleil S. A., Silva J. C., Vemulapalli V., Bedford M. T., and Comb M. J. (2014) Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell. Proteomics 13, 372–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Svinkina T., Gu H., Silva J. C., Mertins P., Qiao J., Fereshetian S., Jaffe J. D., Kuhn E., Udeshi N. D., and Carr S. A. (2015) Deep, quantitative coverage of the lysine acetylome using novel anti-acetyl-lysine antibodies and an optimized proteomic workflow. Mol. Cell. Proteomics 14, 2429–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ficarro S. B., Zhang Y., Carrasco-Alfonso M. J., Garg B., Adelmant G., Webber J. T., Luckey C. J., and Marto J. A. (2011) Online nanoflow multidimensional fractionation for high efficiency phosphopeptide analysis. Mol. Cell. Proteomics 10, O111.011064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., and Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 18.Eng J. K., McCormack A. L., and Yates J. R. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectr. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 19.Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villén J., Haas W., Sowa M. E., and Gygi S. P. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villén J., Beausoleil S. A., Gerber S. A., and Gygi S. P. (2007) Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. U.S.A. 104, 1488–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schilling B., Rardin M. J., MacLean B. X., Zawadzka A. M., Frewen B. E., Cusack M. P., Sorensen D. J., Bereman M. S., Jing E., Wu C. C., Verdin E., Kahn C. R., Maccoss M. J., and Gibson B. W. (2012) Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: Application to protein acetylation and phosphorylation. Mol. Cell. Proteomics 11, 202–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gnad F., Young A., Zhou W., Lyle K., Ong C. C., Stokes M. P., Silva J. C., Belvin M., Friedman L. S., Koeppen H., Minden A., and Hoeflich K. P. (2013) Systems-wide analysis of K-Ras, Cdc42, and PAK4 signaling by quantitative phosphoproteomics. Mol. Cell. Proteomics 12, 2070–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mertins P., Qiao J. W., Patel J., Udeshi N. D., Clauser K. R., Mani D. R., Burgess M. W., Gillette M. A., Jaffe J. D., and Carr S. A. (2013) Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat. Methods 10, 634–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hornbeck P. V., Chabra I., Kornhauser J. M., Skrzypek E., and Zhang B. (2004) PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics 4, 1551–1561 [DOI] [PubMed] [Google Scholar]
- 25.Rotival M., Ko J. H., Srivastava P. K., Kerloc'h A., Montoya A., Mauro C., Faull P., Cutillas P. R., Petretto E., and Behmoaras J. (2015) Integrating phosphoproteome and transcriptome reveals new determinants of macrophage multinucleation. Mol. Cell. Proteomics 14, 484–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung L., Moore K., Phillips L., Boyle F. M., Marsh D. J., and Baxter R. C. (2014) Novel serum protein biomarker panel revealed by mass spectrometry and its prognostic value in breast cancer. Breast Cancer Res. 16, R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patz E. F. Jr., Campa M. J., Gottlin E. B., Kusmartseva I., Guan X. R., and Herndon J. E. 2nd (2007) Panel of serum biomarkers for the diagnosis of lung cancer. J. Clin. Oncol. 25, 5578–5583 [DOI] [PubMed] [Google Scholar]
- 28.Sharma K., D'Souza R. C., Tyanova S., Schaab C., Wiśniewski J. R., Cox J., and Mann M. (2014) Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 8, 1583–1594 [DOI] [PubMed] [Google Scholar]
- 29.Stokes M. P., Farnsworth C. L., Moritz A., Silva J. C., Jia X., Lee K. A., Guo A., Polakiewicz R. D., and Comb M. J. (2012) PTMScan direct: Identification and quantification of peptides from critical signaling proteins by immunoaffinity enrichment coupled with LC-MS/MS. Mol. Cell. Proteomics 11, 187–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sudhir P. R., Chen C. H., Pavana Kumari M., Wang M. J., Tsou C. C., Sung T. Y., Chen J. Y., and Chen C. H. (2012) Label-free quantitative proteomics and N-glycoproteomics analysis of KRAS-activated human bronchial epithelial cells. Mol. Cell. Proteomics 11, 901–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Graaf E. L., Giansanti P., Altelaar A. F., and Heck A. J. (2014) Single-step enrichment by Ti4+-IMAC and label-free quantitation enables in-depth monitoring of phosphorylation dynamics with high reproducibility and temporal resolution. Mol. Cell. Proteomics 13, 2426–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rardin M. J., Newman J. C., Held J. M., Cusack M. P., Sorensen D. J., Li B., Schilling B., Mooney S. D., Kahn C. R., Verdin E., and Gibson B. W. (2013) Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proc. Natl. Acad. Sci. U.S.A. 110, 6601–6606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hendriks I. A., D'Souza R. C., Yang B., Verlaan-de Vries M., Mann M., and Vertegaal A. C. (2014) Uncovering global SUMOylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 21, 927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., and Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsou C. C., Tsai C. F., Tsui Y. H., Sudhir P. R., Wang Y. T., Chen Y. J., Chen J. Y., Sung T. Y., and Hsu W. L. (2010) IDEAL-Q, an automated tool for label-free quantitation analysis using an efficient peptide alignment approach and spectral data validation. Mol. Cell. Proteomics 9, 131–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weisser H., Nahnsen S., Grossmann J., Nilse L., Quandt A., Brauer H., Sturm M., Kenar E., Kohlbacher O., Aebersold R., and Malmström L. (2013) An automated pipeline for high-throughput label-free quantitative proteomics. J. Proteome Res. 12, 1628–1644 [DOI] [PubMed] [Google Scholar]
- 37.Kimura A., Sogawa K., Satoh M., Kodera Y., Yokosuka O., Tomonaga T., and Nomura F. (2012) The application of a three-step serum proteome analysis for the discovery and identification of novel biomarkers of hepatocellular carcinoma. Int. J. Proteomics 2012, 623190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nanjappa V., Thomas J. K., Marimuthu A., Muthusamy B., Radhakrishnan A., Sharma R., Ahmad Khan A., Balakrishnan L., Sahasrabuddhe N. A., Kumar S., Jhaveri B. N., Sheth K. V., Kumar Khatana R., Shaw P. G., Srikanth S. M., Mathur P. P., Shankar S., Nagaraja D., Christopher R., Mathivanan S., Raju R., Sirdeshmukh R., Chatterjee A., Simpson R. J., Harsha H. C., Pandey A., and Prasad T. S. (2014) Plasma Proteome Database as a resource for proteomics research: 2014 update. Nucleic Acids Res. 42, D959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roth S. Y., Denu J. M., and Allis C. D. (2001) Histone acetyltransferases. Annu Rev. Biochem. 70, 81–120 [DOI] [PubMed] [Google Scholar]
- 40.Marmorstein R., and Roth S. Y. (2001) Histone acetyltransferases: Function, structure, and catalysis. Curr. Opin. Genetics Dev. 11, 155–161 [DOI] [PubMed] [Google Scholar]
- 41.Jaskelioff M., and Peterson C. L. (2003) Chromatin and transcription: Histones continue to make their marks. Nature Cell Biol. 5, 395–399 [DOI] [PubMed] [Google Scholar]
- 42.Han S. P., Tang Y. H., and Smith R. (2010) Functional diversity of the hnRNPs: Past, present and perspectives. Biochem. J. 430, 379–392 [DOI] [PubMed] [Google Scholar]
- 43.Nichols R. C., Wang X. W., Tang J., Hamilton B. J., High F. A., Herschman H. R., and Rigby W. F. (2000) The RGG domain in hnRNP A2 affects subcellular localization. Experiment. Cell Res. 256, 522–532 [DOI] [PubMed] [Google Scholar]
- 44.Liu X., Valentine S. J., Plasencia M. D., Trimpin S., Naylor S., and Clemmer D. E. (2007) Mapping the human plasma proteome by SCX-LC-IMS-MS. J. Am. Soc. Mass Spectr. 18, 1249–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bassani-Sternberg M., Barnea E., Beer I., Avivi I., Katz T., and Admon A. (2010) Soluble plasma HLA peptidome as a potential source for cancer biomarkers. Proc. Natl. Acad. Sci. U.S.A. 107, 18769–18776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeong S. K., Lee E. Y., Cho J. Y., Lee H. J., Jeong A. S., Cho S. Y., and Paik Y. K. (2010) Data management and functional annotation of the Korean reference plasma proteome. Proteomics 10, 1250–1255 [DOI] [PubMed] [Google Scholar]
- 47.Rendell M., Nierenberg J., Brannan C., Valentine J. L., Stephen P. M., Dodds S., Mercer P., Smith P. K., and Walder J. (1986) Inhibition of glycation of albumin and hemoglobin by acetylation in vitro and in vivo. J. Lab. Clin. Med. 108, 286–293 [PubMed] [Google Scholar]
- 48.Sennels L., Salek M., Lomas L., Boschetti E., Righetti P. G., and Rappsilber J. (2007) Proteomic analysis of human blood serum using peptide library beads. J. Proteome Res. 6, 4055–4062 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.