Abstract
AMPylation of mammalian small GTPases by bacterial virulence factors can be a key step in bacterial infection of host cells, and constitutes a potential drug target. This posttranslational modification also exists in eukaryotes, and AMP transferase activity was recently assigned to HYPE Filamentation induced by cyclic AMP domain containing protein (FICD) protein, which is conserved from Caenorhabditis elegans to humans. In contrast to bacterial AMP transferases, only a small number of HYPE substrates have been identified by immunoprecipitation and mass spectrometry approaches, and the full range of targets is yet to be determined in mammalian cells. We describe here the first example of global chemoproteomic screening and substrate validation for HYPE-mediated AMPylation in mammalian cell lysate. Through quantitative mass-spectrometry-based proteomics coupled with novel chemoproteomic tools providing MS/MS evidence of AMP modification, we identified a total of 25 AMPylated proteins, including the previously validated substrate endoplasmic reticulum (ER) chaperone BiP (HSPA5), and also novel substrates involved in pathways of gene expression, ATP biosynthesis, and maintenance of the cytoskeleton. This dataset represents the largest library of AMPylated human proteins reported to date and a foundation for substrate-specific investigations that can ultimately decipher the complex biological networks involved in eukaryotic AMPylation.
Covalent posttranslational modification (PTM) of hydroxyl-containing amino acids in proteins by adenosine monophosphate (AMP), called AMPylation or adenylylation, was first discovered almost a half century ago as a mechanism controlling the activity of bacterial glutamine synthetase (1). This unusual PTM was unknown in eukaryotes until it was identified in 2009 in the context of bacterial infection, when Yarbrough et al. reported AMPylation of host small GTPases by bacterial virulence factor Vibrio outer protein S (VopS) from Vibrio parahemeolyticus. In this context, AMPylation precludes interactions with downstream binding partners and causes actin cytoskeleton collapse leading to cell death (2). Since then, the field of AMPylation has grown substantially, with reports describing AMPylation activity of other bacterial effectors, like Immunoglobulin binding protein A (IbpA) in Histophilus somni (3) and Defects in Rab1 recruitment protein A (DrrA) in Legionella pneumophila (4). These new bacterial AMPylators share a common substrate class (small GTPases); however, they differed in the identity of their catalytic residues and architecture of their active sites. Accordingly, bacterial AMP transferases have been classified as either filamentation induced by cyclic AMP (FIC) or adenylyl transferase (AT)1 domain containing enzymes, with catalytic His or Asp residues, respectively.
Although adenylylation has been most extensively described in the context of bacterial infection, there is a growing interest in elucidating the scope of this PTM in a native eukaryotic context. Among the ca. 3000 FIC proteins identified so far by sequence alignment, only a single enzyme has been identified in eukaryotes: Huntingtin-associated protein E (HYPE), also known as FICD. HYPE is conserved from C. elegans to humans, and mRNA expression data suggest that it is present at low levels in all human tissues (3). Apart from the catalytic FIC domain, the protein consists of one transmembrane helix and two tetratricopeptide repeat motifs that point to localization at a membrane and amenability toward protein–protein interactions, respectively. We recently added to this picture by solving the first crystal structure of Homo sapiens HYPE (5), illustrating that the only human FIC is substantially different from its bacterial cousins (6, 7). HYPE was shown to form stable asymmetric dimers supported by the extended network of contacts exclusive to the FIC domains, while the tetratricopeptide repeat motifs have a more flexible arrangement and appear to be exposed for protein–protein interactions in the vicinity of the membrane. In addition, we confirmed the similarity of the active site architecture to other FIC proteins for which a crystal structure is available, with the catalytic loop comprising the invariant catalytic His363 (8), and further substantiated the role of a critical residue Glu234 in an inhibitory helix (9) that may be responsible for regulating HYPE enzymatic activity.
Various catalytic activities have been demonstrated for FIC proteins, including nucleotide (AMP, GMP, and UMP) transfer as well as phosphorylation and phosphocholination (10–13). We and others (3, 5, 14, 15) have demonstrated that HYPE can function in protein AMPylation, although the activity of the wild-type (WT) enzyme is very weak, consistent with active site obstruction by Glu234. It is hypothesized that this intramolecular inhibition can be relieved by specific but as yet unknown protein–protein interactions or by the removal of the conserved Glu. Indeed, the E234G mutation substantially boosts HYPE's activity as demonstrated by the elevated auto-AMPylation of HYPE itself (5, 9) and a few of its recently reported substrates, including the ER chaperone BiP in vivo (14, 15) and several histone proteins in vitro (16, 17). HYPE activity was initially implicated in visual neurotransmission in flies (18) and later in regulation of the unfolded protein response (UPR) in transfected cells, although there is limited consensus over the mechanism (14, 15). Most recently, it has been proposed that HYPE activity might have a role in regulation of gene expression; however, the mechanistic details remain to be elucidated (17).
AMPylation profiling is not a trivial task (19), and several strategies have emerged over the past few years ranging from labeling with radioactive ATP (2, 3) and immunoprecipitation with AMPylation-specific antibodies (20, 21) to mass spectrometry (MS) approaches focused on AMP fragmentation (22, 23). Although these methods contributed significantly to developments in the field, they also suffer from certain drawbacks, including low sensitivity, high background, limited quantitative power, and limited amenability to high-throughput (HT) substrate identification. In contrast, chemoproteomic strategies involving application of substrate analogues (substrate probes) equipped with small and inert chemical handles in combination with sensitive detection by MS can facilitate rapid visualization and/or robust enrichment of modified proteins and can provide superior performance in HT profiling of numerous challenging PTMs (24). AMPylation-specific substrate probes have been developed, and their robust performance was evaluated in vitro, albeit to date only in the context of bacterial effector-mediated AMPylation (25–27). We previously showed that a bioorthogonal substrate probe (26) is well tolerated in the active site of human HYPE and, moreover, that it has potential for chemoproteomic profiling of HYPE substrates in vitro when combined with ligation through copper-catalyzed azide alkyne cycloaddition (CuAAC) to a dedicated capture reagent decorated with a biotin affinity handle and carboxytetramethylrhodamine (TAMRA) fluorophore (5).
Herein, we present the first global AMPylation profile in a native eukaryotic context utilizing a bioorthogonal ATP analogue and chemoproteomic methodology. We first demonstrate efficient enrichment and fast visualization of potential HYPE substrates in cell lysates by in-gel fluorescence, followed by robust identification via shotgun proteomics on a QExactive mass spectrometer. Furthermore, we extensively validate candidate substrates via HYPE titration and ATP competition experiments with a quantitative MS-based readout, as well as Western blotting and direct MS/MS evidence for AMP modification. Finally, we analyze HYPE interaction partners in vivo, providing a link between our discoveries in lysates and a physiologically relevant context, delivering the first experimentally validated library of HYPE substrate proteins.
MATERIALS AND METHODS
General
Culture media and reagents for cell culture were obtained from Sigma Aldrich Ltd, Dundee Cell Products (Dundee, UK), and Gibco (Life Technologies Inc.). Yn-6-ATP was from Jena Bioscience (Jena, Germany) and Az-TB was synthesized as described (28). All CuAAC reagents (CuSO4, Tris (2-carboxyethyl phosphine) (TCEP), Tris (benzyltriazolylmethyl)amine (TBTA), buffer salts, ATP, DTT, and iodoacetamide were from Sigma Aldrich. EDTA-free complete protease inhibitor was obtained from Roche Diagnostics Ltd. Primary and secondary antibodies were from Abcam (Cambridge, UK) and Santa Cruz Biotechnology Inc. Absorbance in 96-well plates was measured using a SpectraMax M2/M2e Microplate Reader from Molecular Devices LLC. For proteomics, all buffers were filtered using a 0.2 μm filter. Low binding tubes (Protein LoBind tubes, Eppendorf, UK) were used to carry out the enrichment of proteins for MS-based proteomics. Sequencing grade Trypsin was obtained from Promega Corp. (Madison, WI).
Cloning and Protein Expression
Plasmid pcDNA-DEST40-HYPE-Bio, encoding HYPE WT-bio was prepared by cloning the biotinylation tag encoding the amino acid residues GLNDIFEAQKIEWHE at the COOH terminus of wild-type HYPE. This ORF was then cloned into the Gateway (Life Technologies) vector pcDNA-DEST40 and verified by sequencing. Plasmid pICC1394 encoding GFP-BirA was obtained as described in Mousnier et al. (29). Recombinant protein expression was performed as described previously (5). Briefly, HYPE constructs in a in-Fusion enabled, pET28a-based vector for expression of HIS6-SUMO-POI in E. coli (pOPINS) (OPPF, Oxford Protein Production Facility, UK) vector background were transformed into C41(DE3) (Lucigen Corp., Middleton, WI) and grown up in 2xYT medium at 37 °C. Cultures were cooled to 20 °C and subsequently induced with 0.1 mm isopropyl β-d-1-thiogalactopyranoside. Cultures were harvested after 16 h and frozen prior to purification (please see (5) for details).
Cell Culture and Lysis
HEK293 cells were grown in DMEM supplemented with 10% FBS in a humidified 10% CO2-containing atmosphere at 37 °C. For SILAC (30, 31) experiments, cells were grown as described above in R0K0 (light) and R10K8 (heavy) DMEM supplemented with dialyzed FBS, allowing >10 doublings to ensure efficient (>97%) incorporation of labeled amino acids. Cell lysis was performed on ice using AMPylation buffer (20 mm Hepes, pH 7.4, 100 mm NaCl, 5 mm MgCl2, 1% Triton X-100, 1 x EDTA-free complete protease inhibitor) and sheer stress. Lysates were kept on ice for 20 min and centrifuged at 17,000 × g for 20 min to remove insoluble material. Supernatants were collected and stored at −80 °C. Protein concentration was determined using the Bio-Rad DC Protein Assay.
In Vitro AMPylation, CuAAC and Enrichment
AMPylation in cell lysates (2 mg/ml) was carried out in AMPylation buffer supplemented with DTT (1 mm), Yn-6-ATP (100 μm) and ± recombinant AMPylators (1/10 w/w enzyme to total protein) for 3 h at 30 °C. AMPylation was stopped by protein precipitation (chloroform/methanol, 0.25:1, relative to the sample volume). Precipitates were isolated by centrifugation (17, 000 × g; 10 min), washed once with methanol (400 μl), and air dried (10 min). Dried pellets were then reconstituted (PBS, 0.4% SDS) at 2 mg/ml, and the CuAAC mixture was prepared by adding reagents in the following order and by vortex mixing between the addition of each reagent: Az-TB (final concentration 0.1 mm), CuSO4 (final concentration 1 mm), TCEP (final concentration 1 mm), and TBTA (final concentration 0.1 mm). Following the addition of the CuAAC mixture, the samples were vortexed (room temperature, 1 h), and the reaction was stopped by addition of EDTA (final concentration 10 mm). Subsequently, proteins were precipitated, isolated and reconstituted (1 mg/ml) as described above. The samples were then added to 15 μl (per 100 μg protein) of pre-washed (0.2% SDS in PBS (3 × 500 μl)) Dynabeads® MyOne™ Streptavidin C1 (Invitrogen Corp.) and gently vortex mixed for 90 min. The supernatant was removed and the beads were washed with 0.2% SDS in PBS (3 × 500 μl).
SDS-PAGE, in-gel Fluorescence and Western Blotting
30 μl of 2% SDS in PBS and 10 μl 4x SLB (Invitrogen) were added to the beads and 7 μl SLB were added to 20 μl of supernatant. The samples were then boiled (10 min), centrifuged (1,000 × g, 2 min) and loaded on a 12% SDS-PAGE gel (supernatant: 13 μl (∼10 μg of proteins); pull-down: 15 μl (∼40 μg of input proteins). Following electrophoresis (60 min, 180V), the gel was washed with MilliQ (3x), soaked in fixing solution (40% MeOH, 10% acetic acid, 50% water) for 5 min and washed with water (3x). In-gel fluorescence was detected using an Ettan DIGE Imager (GE Healthcare Uppsala, Sweden), and the protein loading was checked by Coomassie staining. For Western blotting, proteins were not fixed; instead, they were transferred onto PVDF membranes using an iBlot device (Invitrogen) according to manufacturer's instructions. After brief washing with TBS-T (1 x TBS, 0.1% Tween-20) membranes were blocked (5% milk, TBS-T, 1 h), washed with TBS-T (3x) and incubated with primary antibodies (5% milk, TBS-T, overnight, 4 °C) at supplier-recommended dilutions. Following washing (TBS-T, 3x), membranes were incubated with secondary antibodies (1:10,000, 5% milk, TBS-T, 1 h) and, after a final washing step, treated with Luminata Crescendo Western HRP substrate (Millipore Darmstadt, Germany) for chemiluminescence imaging using a LAS-3000 Imaging System (GE Healthcare). In case of biotin blots, membranes were blocked with 3% BSA and incubated with streptavidin-HRP in 0.3% BSA, TBS-T for 1 h.
Sample Preparation for MS-Based Proteomics
For SILAC-based identification of HYPE candidate substrates in cell lysates, AMPylation was carried out separately in 0.2 mg of light (no enzyme control) and 0.2 mg of heavy (various AMPylators) labeled HEK293 lysates as described above. After 3 h at 30 °C, the lysates were mixed in 1:1 ratio and proteins were immediately precipitated to quench the reaction. Samples were prepared in biochemical triplicates. For HYPE titration and ATP competition experiments, a “spike in” standard was prepared by performing AMPylation reaction with HYPE E234G for 3 h at 30 °C as described above in a heavy-labeled HEK293 lysate. This standard was then added in 1:2 ratio to light samples (0.2 mg each) prepared analogously and supplemented with decreasing amounts of HYPE E234G (1/10–1/1000 w/w enzyme to total protein) or increasing amounts of ATP (0–1000 μm) followed by immediate protein precipitation to quench the reaction. Those samples were prepared in biochemical duplicates.
CuAAC and Enrichment for MS-Based Proteomics
After protein reconstitution (2 mg/ml, 0.4%SDS in PBS), CuAAC was carried out as described above followed by protein precipitation and final reconstitution for substrate enrichment. NeutrAvidin agarose resin (Thermo Fisher Scientific Inc.) was washed with 0.2% SDS in PBS (3x). The samples were mixed with beads (25 μl slurry/sample), and the enrichment was carried out on a rotating wheel for 2 h at room temperature. Following the removal of supernatants, the beads were sequentially washed with 1% SDS in PBS (3x), 4 m urea in 50 mm AMBIC (2x), and 50 mm AMBIC (5x). Protein digestion was initiated upon addition of trypsin (ca. 1/1000 w/w protease to protein), and samples were incubated overnight at 37 °C. The samples were then briefly centrifuged, diluted twice (aqueous 0.1% TFA), and stage-tipped according to a published protocol (32). Elution from the sorbent (SDC-XC, from 3M Empore) with 70% acetonitrile in water was followed by speed-vac-assisted solvent removal, reconstitution of peptides in 0.5% TFA, 2% acetonitrile in water, and finally sample transfer into LC-MS sample vials.
Identification of HYPE Interactome In Vivo
HEK293 cells grown in R10K8 or R0K0 DMEM supplemented with biotin (4 μm) were reverse transfected with equal amounts of WT-bio and BirA or BirA alone, respectively, using TurboFect (Thermo Fisher Scientific) according to the manufacturer's instructions. After 18 h incubation, transfection media were replaced by a fresh aliquot, and cells incubated for additional 24 h. Next, cells were washed with PBS (2x) and lysed on ice using the lysis buffer (PBS 1x, 0.5% Triton X-100, 1 x EDTA-free complete protease inhibitor). Lysates were kept on ice for 20 min and carefully centrifuged at 5000 × g for 10 min only to remove insoluble material. Supernatants were collected and kept on ice while protein concentration was determined as described above. Heavy (WT-bio/BirA) and light (BirA) lysates (120 μg each) were used immediately to maintain native protein interactions. The samples were enriched separately on NeutrAvidin beads for 1 h at room temperature and washed carefully with the lysis buffer (4x) and 50 mm Ammonium bicarbonate (AMBIC) (4x). The beads were then mixed in 1:1 ratio, proteins were reduced (5 mm DTT, 30 min, 55 °C), alkylated (10 mm iodoacetamide, 30 min, in the dark), digested with trypsin, and processed for MS-based proteomics as described above. Samples were prepared in biological duplicates.
nLC-MS/MS and Data Analysis
The analysis was performed using an Acclaim PepMap RSLC column 50 cm × 75 μm inner diameter (Thermo Fisher Scientific) using a 2 h acetonitrile gradient in 0.1% aqueous formic acid at a flow rate of 250 nl/min. Easy nLC-1000 was coupled to a QExactive mass spectrometer via an easy-spray source (all Thermo Fisher Scientific). The QExactive was operated in data-dependent mode with survey scans acquired at a resolution of 75,000 at m/z 200 (transient time 256 ms). Up to 10 of the most abundant isotope patterns with charge +2 or higher from the survey scan were selected with an isolation window of 3.0 m/z and fragmented by HCD with normalized collision energy of 25. The maximum ion injection times for the survey scan and the MS/MS scans (acquired with a resolution of 17 500 at m/z 200) were 20 and 120 ms, respectively. The ion target value for MS was set to 106 and for MS/MS to 105, and the intensity threshold was set to 8.3 × 102. The data were processed with MaxQuant (33) (version 1.5.0.25), and the peptides were identified from the MS/MS spectra searched against human-referenced (with isoforms) proteome (UniProt (34), October 2014, 41,917 entries) using the Andromeda (35) search engine. SILAC-based experiments in MaxQuant were performed using the built-in quantification algorithm (33) with minimal ratio count = 2 and enabled “Match between runs” option (time window 0.7 min) and “Re-quantify” feature. Cysteine carbamidomethylation was selected as a fixed modification (protein–protein interaction experiments only) and methionine oxidation and acetylation of protein N terminus as variable modifications. For in silico digests of the reference proteome, the following peptide bond cleavages were allowed: arginine or lysine followed by any amino acid (a general setting referred to as Trypsin/P). Up to two missed cleavages were allowed. The false discovery rate was set to 0.01 for peptides, proteins, and sites. Other parameters were used as preset in the software (maximal mass error = 4.5 ppm and 20 ppm for precursor and product ions, respectively, minimum peptide length = 7, minimum razor + unique peptides = 1, minimum scores for unmodified and modified peptides = 0 and 40, respectively). “Unique and razor peptides” mode was selected to allow identification and quantification of proteins in groups (razor peptides are uniquely assigned to protein groups and not to individual proteins). Data were further analyzed using Microsoft Office Excel 2007 and Perseus (version 1.5.0.9) as described in the Results section and in Supplemental Tables S1-S3 and S6. Data are available via ProteomeXchange (identifier PXD002601).
Identification of AMPylated Peptides
AMPylation reaction was carried out in 0.3 mg of light lysate supplemented with E234G in the presence of Yn-6-ATP or ATP (control) as described above in biochemical triplicates. Instead of the standard capture reagent (Az-TB), a site ID reagent, Az-RTB (36), was applied for CuAAC under the above-mentioned conditions. Enriched proteome was digested with trypsin and analyzed by LC-MS/MS. Two injections per sample were performed and data from the two LC-MS/MS runs processed together with PEAKS7 suite (37). Samples were searched against the same UniProt Homo sapiens database that was used in MaxQuant analyses. Trypsin (specific, up to three missed cleavages allowed) was selected for database searches, and no enzyme was chosen in de novo searches (up to five candidates per spectrum reported). The maximal mass error was set to 10 ppm (de novo search) or 5 ppm (database search) for precursor ions and 0.01 Da for product ions. Methionine oxidation as well as the AMP-derived adduct (+624.1918 Da) to Ser, Thr, and Tyr were set as variable modifications. The maximal number of modifications per peptide was set as five. The false discovery rate was set to 0.01 for peptides, and a minimum of one unique peptide per protein was required.
Auto AMPylation of HYPE E234G (1 mg/ml) was carried out under standard conditions using ATP (1 mm) as the AMP donor. The protein was then precipitated, reconstituted in 50 mm AMBIC (0.5 mg/ml), and trypsinized before LC-MS/MS and data analysis with PEAKS7 as described above with the AMP adduct mass of +329.0525 Da. Data are available via ProteomeXchange (identifiers PXD002601 and PXD003053).
RESULTS
Screening HYPE Activity and Substrate Enrichment by In-Gel Fluorescence
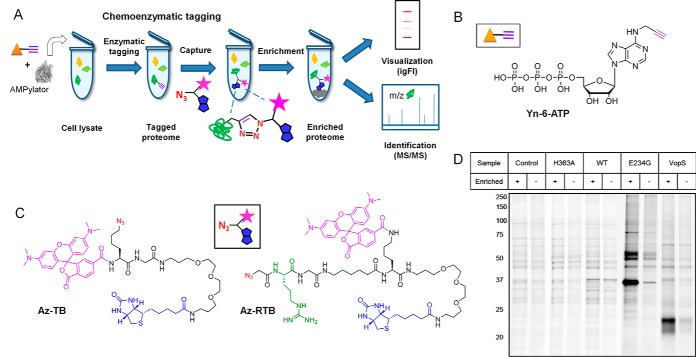
The process of chemoenzymatic tagging (Fig. 1A) relies on enzymatic transfer of a chemical handle, a so-called bioorthogonal reporter (usually an azide or an alkyne), from a synthetic ATP analogue substrate, Yn-6-ATP (Fig. 1B), onto substrate protein(s). This biotransformation is followed by a nonenzymatic but highly chemoselective reaction (CuAAC) to an appropriate ligation partner, or “capture reagent” (Fig. 1C), to introduce secondary labels; a TAMRA fluorophore and biotin affinity tag allow substrate detection and enrichment, respectively. Importantly, chemoenzymatic tagging introduces a covalent linkage between the chemical tag and the modified protein (Fig. 1A), thus allowing far more stringent washing following enrichment than is permitted by immunoprecipitation approaches, leading to background reduction. We previously demonstrated that HYPE activity can be efficiently visualized utilizing a chemoenzymatic strategy, with Yn-6-ATP and a capture reagent, Az-TB (5). To test if the methodology allows also for efficient enrichment of AMPylated substrates, we first identified the optimal AMPylation time for maximal labeling in lysate (3 h), and verified the probe specificity in our system through competition against ATP (Figs. S1A and S1B). We then performed AMPylation in HEK293 lysate either without AMP transferase (negative control) or with one of a range of HYPE constructs, alongside the well-characterized bacterial AMPylator VopS (positive control). As shown in Fig. 1D in the VopS-catalyzed reaction, we observed robust enrichment of potential AMPylation substrates clustered in the molecular weight range characteristic for small GTPases. In contrast, HYPE-catalyzed AMPylation resulted only in minimal enrichment for the WT and H363A proteins, consistent with the autoinhibited and catalytically inactive states of these HYPE variants, respectively. As in-gel fluorescence labeling patterns of no-enzyme control, WT and H363A HYPE are similar; the observed bands likely represent background labeling (proteins bound nonspecifically either to the enrichment beads or the ATP substrate analogue) and not potential substrate proteins. Upon relief of inhibition through E234G mutation, we observed a substantial increase in HYPE activity that translated to efficient enrichment of several protein bands as shown in Fig. 1D. Importantly, we did not observe any enrichment in the molecular weight range of small GTPases, which points to substantially different substrate specificities for bacterial and eukaryotic AMPylators.
Fig. 1.
Chemoenzymatic tagging strategy for AMPylome profiling. (A) Schematic representation of chemoenzymatic tagging of AMPylation substrates in vitro. (B) Structure of the bioorthogonally tagged substrate analogue, Yn-6-ATP. (C) Structures of capture reagents Az-TB and Az-RTB; Az - azide, T - TAMRA, B - biotin, R - arginine. (D) In-gel fluorescence imaging of enrichment efficiency for tested HYPE variants (WT, H363A, E234G) and bacterial AMPylator, VopS.
Identification of Candidate HYPE Substrates In Vitro
Efficient enrichment of substrate proteins is a crucial step in HT profiling of PTMs. Upon confirmation of enrichment efficiency, we implemented SILAC (30) shotgun proteomic experiments to identify HYPE AMPylation substrates in HEK293 cell lysate (Fig. S2). Cells cultured in heavy (R10K8) or light (R0K0) DMEM media were lysed as described in the Methods section. We then performed chemoenzymatic tagging with Yn-6-ATP in triplicate either in the presence of AMPylators (WT, E234G, VopS) or without enzyme (to account for potential nonspecificity) in heavy or light lysate, respectively. Lysates were mixed 1:1 and ligated to Az-TB by CuAAC, followed by enrichment of the tagged proteome by biotin-NeutrAvidin pull down, stringent washing, on-bead tryptic digestion, and analysis by shotgun proteomics on a nanoLC-QExactive mass spectrometer platform. RAW files were analyzed with MaxQuant software (33) as described in the Methods section, and heavy/light (H/L) ratios were generated for each protein. Quantification in at least two out of three replicates and all log2 H/L ratios >1 were required as threshold for protein assignments as potential AMPylation candidates. Applying these criteria, we were pleased to observe that all five protein IDs above this threshold in VopS-catalyzed AMPylation were indeed the expected substrates, i.e. Rho family smallGTPases (Table S1). Following this experimental validation, analogous thresholds were applied to HYPE-mediated AMPylations. In the case of E234G, we assigned 102 candidate substrates that matched these criteria from a total of 745 quantified proteins (Table S1). In the case of the WT protein, however, candidate substrates could not be assigned as all of the 616 quantified proteins were below the selected enrichment threshold (Table S1).
Candidate Substrate Validation through Quantitative Competition Experiments and Immunoblotting
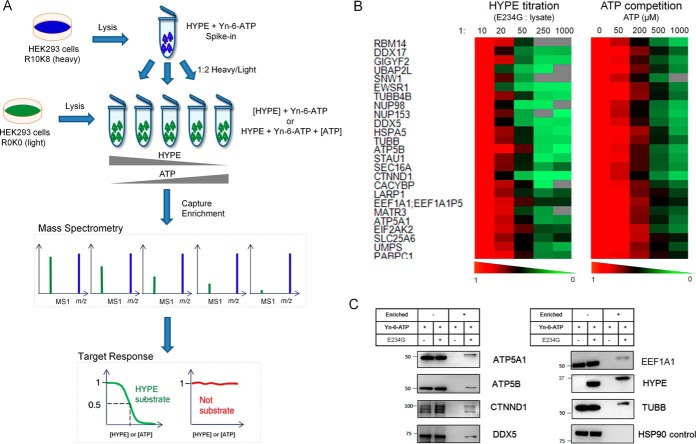
Although the design of the SILAC experiments above can select against false positive IDs originating from nonspecific interactions, it is important to note that HYPE-specific enrichment cannot exclude ATP-independent false discoveries as a result of tightly interacting substrate binding partners. To elevate the confidence of our identifications in terms of both HYPE and ATP specificity, we combined spike-in SILAC quantification (31) with HYPE titration and with ATP competition experiments (Fig. 2A). A spike-in standard, generated by performingAMPylation in the presence of HYPE E234G and Yn-6-ATP in heavy HEK293 lysate, was spiked in a 1:2 ratio into samples prepared analogously in the light lysate in the presence of either decreasing amounts of HYPE E234G or increasing amounts of ATP (five conditions in duplicate per experiment). Spiked samples were ligated to Az-TB, enriched, and subjected to digest and proteomic analysis, as described above. Quantification in 6 out of 10 samples (two replicates of five conditions) as well as a dose-dependent response >50% were set as specific thresholds. Application of these criteria to the 102 candidate substrates revealed robust response to HYPE titration for 34 proteins (Table S2), providing strong evidence for HYPE-specific enrichment, while 47 candidate substrates displayed high sensitivity toward competition with the natural substrate, ATP (Table S3). This double selection scenario accounting for both HYPE and ATP enrichment specificity delivered a pool of 25 high confidence substrates (Fig. 2B), including the previously reported ER chaperone BiP (HSPA5). Subsequently, several novel high-confidence substrates were cross-checked by a traditional immunoblotting approach, where protein-specific antibodies were applied to samples enriched in a HYPE-dependent manner (Fig. 2C).
Fig. 2.
Validation of AMPylation candidates. (A) Schematic diagram of the spike-in SILAC strategy used for substrate validation in terms of dose-dependent HYPE and ATP enrichment specificity. (B) Heat maps of quantitative MS-based response of 25 high-confidence AMPylation substrates to HYPE titration or ATP competition. Color coding represents normalized levels (five conditions normalized to 1:10 w/w enzyme to lysate or 0 μm ATP) of substrate AMPylation in response to a range of HYPE and ATP concentrations. Protein substrates are reported by gene names. (C) Western blot validation of selected high-confidence substrates.
Identification of AMPylation Sites
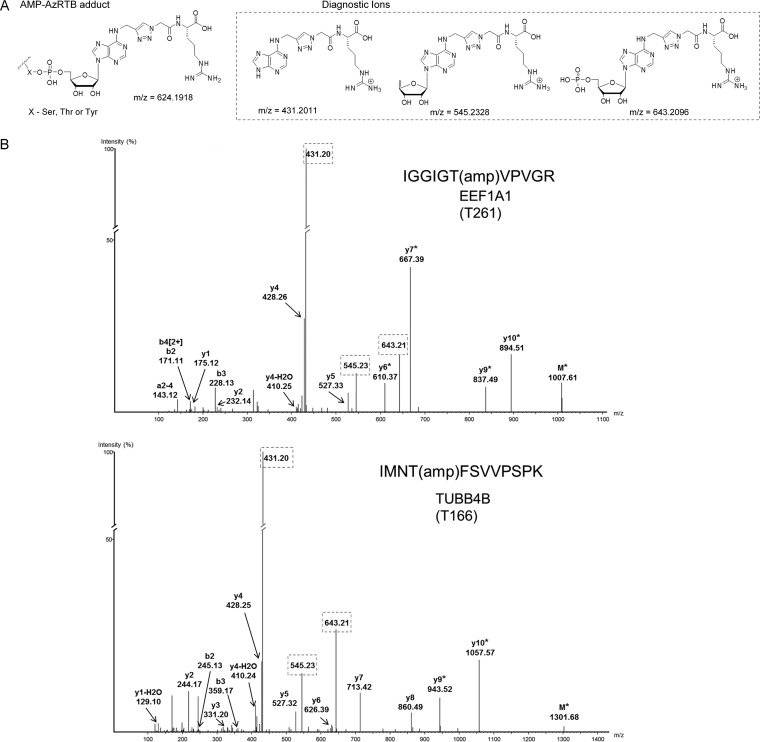
MS-based approaches for the identification of AMPylated peptides (22, 23) take advantage of characteristic fragmentation patterns of the AMP moiety that upon collision-induced dissociation give rise toseveral well-established diagnostic ions. Although the methodology has the potential to scan AMPylated peptides in complex protein mixtures, this feature has not been further explored. We decided to couple the confidence arising from the presence of AMP-characteristic ions with the HT feature of the chemoenzymatic approach and the site ID capture reagent, Az-RTB (Fig. 1C), previously developed in our laboratory for confident PTM site identification (36, 38). In contrast to Az-TB and owing to the presence of an Arg residue,Az-RTB enables facile release of modified peptides from NeutrAvidin beads during on-bead tryptic digestion (Fig. S3). AMPylation reactions were performed in HEK293 lysate supplemented with E234G and either Yn-6-ATP or ATP (control) in triplicate, followed by CuAAC ligation to Az-RTB, substrate enrichment, trypsinization, and shotgun proteomic analysis. RAW files were analyzed with the PEAKS7 suite (37) for the presence of an adduct derived from Yn-6-AMP coupled with AzRTB (Fig. 3A) on Ser, Thr, and Tyr residues, and spectra were manually inspected for their quality and the presence of diagnostic ions (Fig. 3A). In six samples analyzed in technical duplicate, we observed 115 peptide spectrum matches (Table S4) of which 30 did not contain at least two of three diagnostic ions and were therefore excluded. The remaining 85 peptide spectrum matches were found exclusively in Yn-6-ATP treated samples and matched 23 AMPylated peptides on 11 proteins. These 23 peptides were inspected further and divided into high and lower confidence annotations based on representation across triplicate samples (Table S4). Subsequent stringent manual examination of MS/MS fragmentation from the high-confidence group returned five AMPylation sites on four proteins (Fig. 3B and Fig. S4), including HYPE itself and proteins from the pool of 25 high-confidence substrates (HSPA5, EEF1A1, TUBB4B). We also identified AMPylated peptides for GLUD1 and TUBA1B (Table S4); however, due to the incomplete peak assignment of the obtained MS/MS spectra for these single peptide IDs (Fig. S5), they were not included in the high-confidence substrate pool. Az-RTB site ID methodology detected two high-confidence auto-AMPylation sites on HYPE (Thr168 and Thr183, Fig. S4), and these were further validated through AMPylation with isolated HYPE E234G and ATP in place of Yn-6-ATP since the absence of a complex cellular lysate removes the need for enrichment. The MS/MS searches were then executed in analogous manner as for Az-RTB but taking advantage of published masses for native AMP adduct formation and its diagnostic ions (22, 23). We observed a total of 12 auto-AMPylation sites (Table S5), including the two sites previously assigned using Az-RTB methodology (Fig. S6), thus validating Az-RTB as a robust tool for AMP site identification. Although we did not succeed in detection of AMPylated peptides for all the high-confidence substrates, our study provides a proof of principle and a starting point for optimization of AMPylated peptide profiling (e.g. using additional modes of ion fragmentation) and the first report of detection of AMPylated peptides from multiple proteins that have undergone AMPylation in cell lysates.
Fig. 3.
Identification of AMPylation sites. (A) Structure of AMP-AzRTB derived adduct on Ser, Thr, or Tyr residues of AMPylated proteins and respective diagnostic ions formed during MS/MS analysis. (B) Exemplary annotated MS/MS spectra of high-confidence assignments of AMPylated peptides/sites (see Fig. S4 for other examples). MS/MS spectra for a given peptide were selected based on the highest probability score (-10 lgP) assigned by PEAKS7. Diagnostic ions are framed. The asterisk (*) denotes loss of a fragment of diagnostic ion with delta mass of 642.2018.
WT HYPE Interaction Partners In Vivo
The experiments described above provide substantial and orthogonal evidence for substrate validation in vitro; nevertheless, whether these proteins are bona fide substrates in vivo remains a valid question. Due to poor cell penetrance and competition with endogenous ATP and nucleic acid biosynthetic pathways, Yn-6-ATP is unlikely to be an effective tool to investigate protein AMPylation in live cells; moreover, no alternative HT methodology exists to facilitate such experiments. To link our discovery in lysates with a more physiologically relevant environment, we sought to determine whether high-confidence substrates are also found as interacting partners of WT HYPE in vivo. We utilized BirA (39), a bacterial biotin ligase, and WT HYPE C-terminally tagged with a short BirA recognition sequence (WT-bio) in cotransfection experiments, allowing for selective HYPE biotinylation in vivo and subsequent enrichment together with its interacting partners. Accordingly, heavy HEK293 cells were cotransfected with WT-bio and BirA, whereas light cells were transfected with BirA only, to account for any unspecific biotinylation, in biological duplicate (Fig. S7). After mild cell lysis, biotinylated proteins were enriched on NeutrAvidin agarose beads separately in heavy and in light lysate, followed by beads mixing in 1:1 ratio, trypsin digest and shotgun proteomics. H/L ratios for each protein were then generated in MaxQuant, and quantification in both replicates with log2 H/L ratios > 1 was selected as the threshold for protein assignments. First, we observed >8-fold H/L enrichment for WT-bio (Table S6), which confirmed efficient cotransfection and tagging by BirA in vivo. Secondly, ca. fivefold enrichment was observed for the already reported (15) HYPE substrate and interaction partner BiP (HSPA5) (Table S6), validating this experimental approach. Furthermore, five high-confidence substrates reported herein, including ATP synthase subunits, tubulin chains, and elongation factor (Table S6), also passed the selection criteria, suggesting that they may indeed be novel bona fide HYPE substrates in vivo.
Functional Annotation of the Human AMPylome
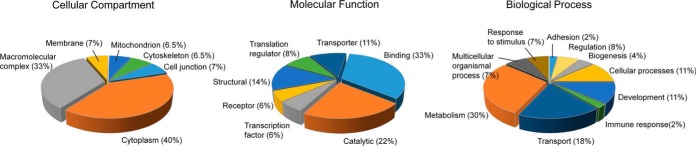
Following from the extensive validation of AMPylation substrates described above, we carried out gene ontology (40) annotation with The Database for Annotation, Visualization and Integrated Discovery v6.7 (41) (Table S7) and functional classification with Protein Analysis through Evolutionary Relationships v10.0 (42) (Fig. 4). Analysis of the cellular compartment domain (Fig. 4A) revealed propensity of AMPylation substrates for cytoplasmic localization as well as for macromolecular complex formation. Inspection of the molecular function domain (Fig. 4B) indicated prevalence of terms in the binding category with particular emphasis on protein, nucleotide, and RNA binding (Table S7). In addition, catalytic transporter as well as structural functions were also assigned. Within the biological process domain (Fig. 4C), we noted that AMPylated substrates show particular clustering within metabolic processes, including nucleotide metabolism and biosynthesis (Table S7), as well as transmembrane transport categories.
Fig. 4.
Functional classification of AMPylation substrates. (A) Distribution of cellular localization. (B) Distribution of molecular function. (C) Distribution of biological processes. Analysis was performed on a pool of 25 high-confidence substrates with Protein Analysis through Evolutionary Relationships classification tool.
DISCUSSION
AMPylation is a novel type of PTM in eukaryotes, with implications in essential cellular processes, including visual neurotransmission (18), heat shock and unfolded protein response (UPR) (14, 15), and potentially gene expression (17). Our understanding of this modification is, however, in its infancy since the number of known substrate proteins is strikingly small. Although several strategies have evolved to generate substrate identifications in the context of bacterial AMPylation (2, 26, 27), the scope of HYPE substrates has not yet been revealed. Profiling eukaryotic AMPylation is challenging due to the low level of HYPE expression and its autoinhibited state under physiological conditions. Moreover, triggers allowing for release of this intramolecular inhibition to yield catalytically active enzyme are currently unknown, as is the prevalence of de-AMPylating enzymatic activities in the cell. To overcome these obstacles, we performed chemoproteomic MS-based experiments in vitro utilizing a recombinant, activated form of HYPE (E234G mutant) and Yn-6-ATP substrate analogue (Fig. 1). This strategy allowed for efficient enrichment and robust identification of 102 AMPylation candidates (Table S1) that were subsequently subjected to extensive and stringent validation in terms of enrichment specificity (Fig. 2) as well as AMPylation site assignments (Fig. 3 and Fig. S4), resulting in a pool of 25 high-confidence HYPE substrates (Table S7), including the previously reported in vivo substrate BiP (HSPA5). This finding not only validates chemoproteomic methodologies as reliable tools for HYPE AMPylation profiling but also demonstrates the link between studies conducted in vitro and in vivo.
Apart from BiP, 24 identified proteins constitute novel substrates of eukaryotic AMPylation reported here for the first time. They represent mostly cytoplasmic proteins that can associate with diverse cellular organelles, e.g. ER, nucleus, and mitochondrion, and have a propensity to form larger complexes (Fig. 4A, Table S7). The variety of molecular functions and biological processes terms associated with the substrate pool (Figs. 4B and 4C, Table S7) significantly expands the previously reported repertoire of cellular phenomena potentially affected by AMPylation (14, 15, 18). For example, we identified several nuclear envelope proteins (NUP, CACYBP, MATR3) responsible for binding/transport of RNA and protein cargo across nuclear membranes and involved in up-regulation of DNA replication. In addition, we detected substrates (PABPC1, EWSR1, CTNND1, DDX5, DDX17) involved in regulation of transcription as well as several substrates (EEF1A, EIF2AK2, and LARP1) that participate in activation/regulation of translation. Together, this evidence strongly supports the previously suggested (17) involvement of eukaryotic AMPylation in pathways of gene expression and protein biosynthesis. Moreover, identification of substrates such as ATP synthase catalytic core subunits (ATP5B, ATP5A1) and further evidence for a direct interaction between WT HYPE and this large protein complex in vivo (Table S6) suggest that AMPylation may regulate ATP biosynthesis as well as transport across the mitochondrial membrane. This hypothesis is further corroborated by the identification of SLC25A6, which is involved in ATP/ADP exchange between mitochondria and the cytoplasm. Taken together, these results suggest a potential role of AMPylation in pathways involved in oxidative phosphorylation and cellular metabolism. Interestingly, it has been demonstrated that the UPR and mitochondrial metabolism are closely associated via so-called ER-mitochondrial coupling in the perinuclear space (43). This microtubule-dependent phenomenon promotes increase in mitochondrial respiration and ATP production during the onset of ER stress, providing favorable bioenergetics for chaperone expression and initiation of UPR. Identification of HYPE substrates with roles in UPR, ATP biosynthesis/transport, and gene expression as well as several members of the tubulin family (TUBB, TUBB4B) that are major constituents of microtubules provides interesting hypothesis that AMPylation by HYPE could be an important regulator of ER stress in eukaryotic cells. In addition, AMPylation of tubulins and potential involvement in microtubule biology could point to further implication of AMPylation in the maintenance of the cytoskeleton—an interesting perspective taking into account the fact that bacterial AMPylation of Rho GTPases also targets the cytoskeleton, with detrimental consequences for the infected cell (44). Our experimental evidence clearly excludes small GTPases as substrates of eukaryotic AMPylation in vitro, as supported by recent reports under physiological conditions (15) pointing to the conclusion that AMPylation is under stringent control within the eukaryotic cell.
Molecular consequences of protein AMPylation involve bulk and charge (–1) introduced by the AMP moiety and may affect not only the substrate itself (stability, activity, cofactor binding) but also its interaction landscape. Only recently, molecular consequences of HYPE-mediated AMPylation have been elucidated in the context of ER chaperone BiP (HSPA5), where modification with AMP in the vicinity of the ATPase domain was shown to enhance its activity and had no effect on binding of misfolded proteins (15). Interestingly, we detected another AMPylation site on HSPA5, i.e. Thr518 (Fig. S4, Table S4) within its substrate binding domain, and apart from potential implications in the regulation of substrate binding, this site is also targeted by phosphorylation (34). AMPylation thus has potential to (reversibly or irreversibly) mask this phosphosite, further substantiating a previously hypothesized (27) crosstalk between these classes of PTM. In addition to HSPA5, we confidently identified AMPylation sites on several other substrates (Table S4), including EEF1A1 and TUBB4B that were also identified herein as HYPE interaction partners in vivo (Table S6). AMPylation of EEF1A1 at Thr261 within domain 2, which is involved in binding to amino-acid-charged tRNA (45), supports a role of this PTM in the regulation of translation and gene expression as suggested above, while AMP modification of TUBB4B within the GTPase domain (Thr166) supports involvement of AMPylation in microtubule dynamics.
In contrast to bacterial AMPylation, which has received renewed interest in recent years, eukaryotic AMPylation and its cellular substrates have remained largely unexplored, precluding elucidation of the biological phenomena that may be regulated by this PTM. The results presented herein represent not only the first objective, multifaceted, and quantitative approach for AMPylation profiling but also dramatically extend the repertoire of HYPE substrates, delivering the first glimpse of the scope of the human AMPylome. Our confident identifications arising from extensive substrate validation by an array of state-of-the-art quantitative chemoproteomic techniques open new research avenues in the field of eukaryotic AMPylation and provide a road map for substrate-targeted investigations that will enable elucidation of specific consequences of AMPylation for substrate structure, function, and involvement in disease.
Supplementary Material
Acknowledgments
We would like to thank Gad Frankel's lab at Imperial College London for providing the BirA plasmid and recombinant VopS.
Footnotes
Author contributions: M.B. and E.W.T. designed the research; M.B. performed the research; R.A.S., T.D.B., and M.K. contributed new reagents or analytic tools; M.B., R.A.S., and E.W.T. analyzed data; M.B., R.A.S., T.D.B., M.K., and E.W.T. wrote the paper; and E.W.T. directed research.
* This work was supported by the European Union Marie Curie Intra European Fellowships to M.B. (PIEF-GA-2011–299740) and R.A.S. (PIEF-GA-2010-273868). MS proteomics data were deposited to the ProteomeXchange Consortium (46) via the PRIDE partner repository with dataset identifiers PXD002601 and PXD003053.
 This article contains supplemental material Tables S1-S7 and Figs. S1-S7.
This article contains supplemental material Tables S1-S7 and Figs. S1-S7.
1 The abbreviations used are:
- AT
- adenylyl transferase
- Az-TB
- azido-TAMRA-biotin
- Az-RTB
- azido-Arginine-TAMRA-biotin
- CuAAC
- copper-catalyzed azide-alkyne cycloaddition
- FIC
- filamentation induced by cyclic AMP
- HRP
- horseradish peroxidase
- HT
- high throughput
- PTM
- posttranslational modification
- SILAC
- stable isotope labeling by amino acids in cell culture.
REFERENCES
- 1.Shapiro B. M., and Stadtman E. R. (1968) 5′-adenylyl-O-tyrosine. The novel phosphodiester residue of adenylylated glutamine synthetase from Escherichia coli. J. Biol. Chem. 243, 3769–3771 [PubMed] [Google Scholar]
- 2.Yarbrough M. L., Li Y., Kinch L. N., Grishin N. V., Ball H. L., and Orth K. (2009) AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science 323, 269–272 [DOI] [PubMed] [Google Scholar]
- 3.Worby C. A., Mattoo S., Kruger R. P., Corbeil L. B., Koller A., Mendez J. C., Zekarias B., Lazar C., and Dixon J. E. (2009) The FIC domain: Regulation of cell signaling by adenylylation. Mol. Cell 34, 93–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Müller M. P., Peters H., Blumer J., Blankenfeldt W., Goody R. S., and Itzen A. (2010) The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329, 946–949 [DOI] [PubMed] [Google Scholar]
- 5.Bunney T. D., Cole A. R., Broncel M., Esposito D., Tate E. W., and Katan M. (2014) Crystal structure of the human, FIC-domain containing protein HYPE and implications for its functions. Structure 22, 1831–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luong P., Kinch L. N., Brautigam C. A., Grishin N. V., Tomchick D. R., and Orth K. (2010) Kinetic and structural insights into the mechanism of AMPylation by VopS FIC domain. J. Biol. Chem. 285, 20155–20163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao J., Worby C. A., Mattoo S., Sankaran B., and Dixon J. E. (2010) Structural basis of Fic-mediated adenylylation. Nat. Struct. Mol. Biol. 17, 1004–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Pino A., Zenkin N., and Loris R. (2014) The many faces of FIC: Structural and functional aspects of FIC enzymes. Trends Biochem. Sci. 39, 121–129 [DOI] [PubMed] [Google Scholar]
- 9.Engel P., Goepfert A., Stanger F. V., Harms A., Schmidt A., Schirmer T., and Dehio C. (2012) Adenylylation control by intra- or intermolecular active-site obstruction in FIC proteins. Nature 482, 107–110 [DOI] [PubMed] [Google Scholar]
- 10.Mattoo S., Durrant E., Chen M. J., Xiao J., Lazar C. S., Manning G., Dixon J. E., and Worby C. A. (2011) Comparative analysis of Histophilus somni immunoglobulin-binding protein A (IbpA) with other FIC domain-containing enzymes reveals differences in substrate and nucleotide specificities. J. Biol. Chem. 286, 32834–32842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng F., Yang F., Rong W., Wu X., Zhang J., Chen S., He C., and Zhou J. M. (2012) A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature 485, 114–118 [DOI] [PubMed] [Google Scholar]
- 12.Castro-Roa D., Garcia-Pino A., De Gieter S., van Nuland N. A., Loris R., and Zenkin N. (2013) The FIC protein Doc uses an inverted substrate to phosphorylate and inactivate EF-Tu. Nat. Chem. Biol. 9, 811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mukherjee S., Liu X., Arasaki K., McDonough J., Galán J. E., and Roy C. R. (2011) Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 477, 103–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ham H., Woolery A. R., Tracy C., Stenesen D., Krämer H., and Orth K. (2014) Unfolded protein response-regulated Drosophila FIC (dFic) protein reversibly AMPylates BiP chaperone during endoplasmic reticulum homeostasis. J. Biol. Chem. 289, 36059–36069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanyal A., Chen A. J., Nakayasu E. S., Lazar C. S., Zbornik E. A., Worby C. A., Koller A., and Mattoo S. (2015) A novel link between FIC (filamentation induced by cAMP)-mediated adenylylation/AMPylation and the unfolded protein response. J. Biol. Chem. 290, 8482–8499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewallen D. M., Sreelatha A., Dharmarajan V., Madoux F., Chase P., Griffin P. R., Orth K., Hodder P., and Thompson P. R. (2014) Inhibiting AMPylation: A novel screen to identify the first small molecule inhibitors of protein AMPylation. ACS Chem, Biol. 9, 433–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Truttmann M. C., Wu Q., Stiegeler S., Duarte J. N., Ingram J., and Ploegh H. L. (2015) HYPE-specific nanobodies as tools to modulate HYPE-mediated target AMPylation. J. Biol. Chem. 290, 9087–9100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rahman M., Ham H., Liu X., Sugiura Y., Orth K., and Krämer H. (2012) Visual neurotransmission in Drosophila requires expression of FIC in glial capitate projections. Nat. Neurosci. 15, 871–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hedberg C., and Itzen A. (2015) Molecular perspectives on protein adenylylation. ACS Chem. Biol. 10, 12–21 [DOI] [PubMed] [Google Scholar]
- 20.Smit C., Blümer J., Eerland M. F., Albers M. F., Müller M. P., Goody R. S., Itzen A., and Hedberg C. (2011) Efficient synthesis and applications of peptides containing adenylylated tyrosine residues. Angew Chem. Int. Ed. Engl. 50, 9200–9204 [DOI] [PubMed] [Google Scholar]
- 21.Hao Y. H., Chuang T., Ball H. L., Luong P., Li Y., Flores-Saaib R. D., and Orth K. (2011) Characterization of a rabbit polyclonal antibody against threonine-AMPylation. J. Biotechnol. 151, 251–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen T., Albers M., Hedberg C., and Sickmann A. (2013) Adenylylation, MS, and proteomics—Introducing a “new” modification to bottom-up proteomics. Proteomics 13, 955–963 [DOI] [PubMed] [Google Scholar]
- 23.Li Y., Al-Eryani R., Yarbrough M. L., Orth K., and Ball H. L. (2011) Characterization of AMPylation on threonine, serine, and tyrosine using mass spectrometry. J Am. Soc. Mass Spectrom. 22, 752–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tate E. W., Kalesh K. A., Lanyon-Hogg T., Storck E. M., and Thinon E. (2015) Global profiling of protein lipidation using chemical proteomic technologies. Curr. Opin. Chem. Biol. 24, 48–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewallen D. M., Steckler C. J., Knuckley B., Chalmers M. J., and Thompson P. R. (2012) Probing adenylation: Using a fluorescently labelled ATP probe to directly label and immunoprecipitate VopS substrates. Mol. Biosyst. 8, 1701–1706 [DOI] [PubMed] [Google Scholar]
- 26.Grammel M., Luong P., Orth K., and Hang H. C. (2011) A chemical reporter for protein AMPylation. J. Am. Chem. Soc. 133, 17103–17105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu X., Woolery A. R., Luong P., Hao Y. H., Grammel M., Westcott N., Park J., Wang J., Bian X., Demirkan G., Hang H. C., Orth K., and LaBaer J. (2014) Copper-catalyzed azide-alkyne cycloaddition (click chemistry)-based detection of global pathogen-host AMPylation on self-assembled human protein microarrays. Mol. Cell. Proteomics 13, 3164–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heal W. P., Wright M. H., Thinon E., and Tate E. W. (2012) Multifunctional protein labeling via enzymatic N-terminal tagging and elaboration by click chemistry. Nat. Protoc. 7, 105–117 [DOI] [PubMed] [Google Scholar]
- 29.Mousnier A., Schroeder G. N., Stoneham C. A., So E. C., Garnett J. A., Yu L., Matthews S. J., Choudhary J. S., Hartland E. L., and Frankel G. (2014) A new method to determine in vivo interactomes reveals binding of the Legionella pneumophila effector PieE to multiple rab GTPases. MBio 5, e01148–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., and Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 31.Geiger T., Wisniewski J. R., Cox J., Zanivan S., Kruger M., Ishihama Y., and Mann M. (2011) Use of stable isotope labeling by amino acids in cell culture as a spike-in standard in quantitative proteomics. Nat. Protoc. 6, 147–157 [DOI] [PubMed] [Google Scholar]
- 32.Rappsilber J., Ishihama Y., and Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 33.Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 34.UniProt C. (2015) UniProt: A hub for protein information. Nucleic Acids Res. 43, D204–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: A peptide search engine integrated into the MaxQuant environment. J Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 36.Broncel M., Serwa R. A., Ciepla P., Krause E., Dallman M. J., Magee A. I., and Tate E. W. (2015) Multifunctional reagents for quantitative proteome-wide analysis of protein modification in human cells and dynamic profiling of protein lipidation during vertebrate development. Angew Chem. Int. Ed. Engl. 54, 5948–5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J., Xin L., Shan B., Chen W., Xie M., Yuen D., Zhang W., Zhang Z., Lajoie G. A., and Ma B. (2012) PEAKS DB: De novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteomics 11, M111.010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thinon E., Serwa R. A., Broncel M., Brannigan J. A., Brassat U., Wright M. H., Heal W. P., Wilkinson A. J., Mann D. J., and Tate E. W. (2014) Global profiling of co- and post-translationally N-myristoylated proteomes in human cells. Nat. Commun. 5, 4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tenzer S., Moro A., Kuharev J., Francis A. C., Vidalino L., Provenzani A., and Macchi P. (2013) Proteome-wide characterization of the RNA-binding protein RALY-interactome using the in vivo-biotinylation-pulldown-quant (iBioPQ) approach. J. Proteome Res. 12, 2869–2884 [DOI] [PubMed] [Google Scholar]
- 40.Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., and Sherlock G. (2000) Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang da W., Sherman B. T., and Lempicki R. A. (2009) Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mi H., Muruganujan A., Casagrande J. T., and Thomas P. D. (2013) Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bravo R., Vicencio J. M., Parra V., Troncoso R., Munoz J. P., Bui M., Quiroga C., Rodriguez A. E., Verdejo H. E., Ferreira J., Iglewski M., Chiong M., Simmen T., Zorzano A., Hill J. A., Rothermel B. A., Szabadkai G., and Lavandero S. (2011) Increased ER-mitochondrial coupling promotes mitochondrial respiration and bioenergetics during early phases of ER stress. J. Cell Sci. 124, 2143–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woolery A. R., Yu X., LaBaer J., and Orth K. (2014) AMPylation of Rho GTPases subverts multiple host signaling processes. J. Biol. Chem. 289, 32977–32988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nissen P., Kjeldgaard M., Thirup S., Polekhina G., Reshetnikova L., Clark B. F., and Nyborg J. (1995) Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270, 1464–1472 [DOI] [PubMed] [Google Scholar]
- 46.Vizcaíno J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolomé S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.