Abstract
BRCA1-associated breast cancers are mostly basal-like high-grade ductal carcinomas that frequently overexpress epidermal growth factor receptor (EGFR). Aberrant EGFR expression is correlated with disease progression, resistance to radiation and chemotherapy, and poor clinical prognosis. While BRCA1 is involved in multiple cellular processes, its functional role in EGFR regulation remains enigmatic. Here, we report a previously unrecognized post-transcriptional mechanism by which BRCA1 regulates EGFR expression through the induction of miR-146a. We demonstrate that EGFR expression correlates negatively with BRCA1, while miR-146a levels increase with BRCA1. We show that BRCA1 binds to miR-146a promoter and activates transcription, which in turn attenuates EGFR expression. Knockdown of miR-146a in BRCA1-overexpressing cells negated this effect and suppressed its ability to inhibit proliferation and transformation. In archived triple negative breast cancer (TNBC) samples, we show a strong positive correlation between BRCA1 and miR-146a expression. We also show that low expression of miR-146a strongly predicts positive lymph node status and is associated with distinctively poor overall survival of patients. Together, these observations provide insight into a novel BRCA1→miR-146a→EGFR paradigm by which BRCA1 carries out an aspect of tumor suppressor function that is potentially amenable to therapeutic intervention.
Keywords: BRCA1, EGFR, miR-146a
INTRODUCTION
Mutations in the tumor suppressor BRCA1 (breast cancer 1, early onset) predispose women to an 80% lifetime risk of breast cancer and a 40% lifetime risk of ovarian cancer (1, 2). Breast cancer is a heterogeneous disease and gene expression profiling has demonstrated five major subtypes, including the basal-like (3) subtype that is enriched for tumors with genetic and/or epigenetic inactivation of BRCA1. BRCA1-associated tumors commonly display a triple-negative (TN) phenotype lacking expression of estrogen receptor (ER), progesterone receptor (PR) and the human epidermal growth factor receptor 2 (HER2). They are mostly basal-like high-grade ductal carcinomas, frequently overexpress epidermal growth factor receptor (EGFR/ErbB1/HER1) and basal cytokeratins (CK5, CK6, CK14 and CK17), and tend to be highly proliferative and invasive (4). A number of studies have implicated the basal cell as the cell of origin of BRCA1-associated basal-like breast cancers (BLBCs) while others identify them as luminal progenitor cells which could be targets for oncogenic events (5-7). About 30-40% of sporadic breast cancers either do not express or have significantly decreased levels of BRCA1 (8, 9) and show a strong histological resemblance to BRCA1-associated hereditary tumors (2, 10).
EGFR is a member of the human epidermal growth factor receptor (HER) family of transmembrane receptor tyrosine kinases that is widely expressed in different human tissues (11). While EGFR is essential for important cellular processes including proliferation, differentiation, and development, it can contribute to the development and growth of cancer when atypically expressed or activated (12). EGFR overexpression occurs in a range of solid tumors and is associated with tumor progression and metastasis, resistance to radiation and chemotherapy, and poor prognosis (13, 14). Low expression of EGFR is generally associated with good prognosis in cancers, and several studies have specifically demonstrated a negative correlation between EGFR status and relapse-free and overall survival from breast cancer (14-16). A significant association between increased EGFR expression and BRCA1 mutation/deficiency has been reported in several independent studies (17, 18), but whether there is a mechanistic basis for this consistent observation has thus far been undetermined.
BRCA1 is located on chromosome 17q21 and encodes a 1,863-amino-acid nuclear phosphoprotein with an N-terminal RING domain and C-terminal BRCT motifs. These highly conserved domains of BRCA1 are frequently affected by clinically important mutations resulting in genomic instability. BRCA1 exerts its tumor suppressor function primarily by interacting with multiple proteins through its RING domain and BRCT motifs to form large nuclear complexes that are critical for DNA repair and maintenance of genomic stability (19, 20). Other functions of BRCA1 that contribute to its tumor suppressor activity include mammary epithelial differentiation, transcriptional activation and repression, chromatin remodeling (5, 20-22).
MicroRNAs (miRNAs) are a class of evolutionarily conserved small noncoding RNAs approximately 21 nucleotides in length that regulate expression of more than one third of the genes in the human genome by inhibiting translation or by promoting mRNA degradation via sequence specific interaction with the 3'UTR of the cognate mRNA targets (23, 24). miRNAs function as oncogenes or tumor suppressors, and show characteristic expression signatures in different cancers predicting disease status and prognosis. They are implicated in stem and epithelial cell differentiation, epithelial-to-mesenchymal transition (EMT) and invasion and metastasis (25). A potential role for miRNAs in the regulation of EGFR in breast and other cancers has recently gained acceptance (26, 27).
EGFR upregulation is suggested to occur via multiple transcriptional and post-transcriptional mechanisms including gene amplification, C-terminal truncation, transcriptional activation and post-translational modifications (28). Yet, a mechanism that connects the loss of BRCA1 to the overexpression and activation of EGFR that contributes to BLBC pathogenesis has not been identified. In this regard, a unique mechanism involving miRNAs was investigated to reveal this critical missing link and to uncover novel therapeutic options for this deadly disease.
RESULTS
BRCA1 deficiency results in up-regulation of EGFR
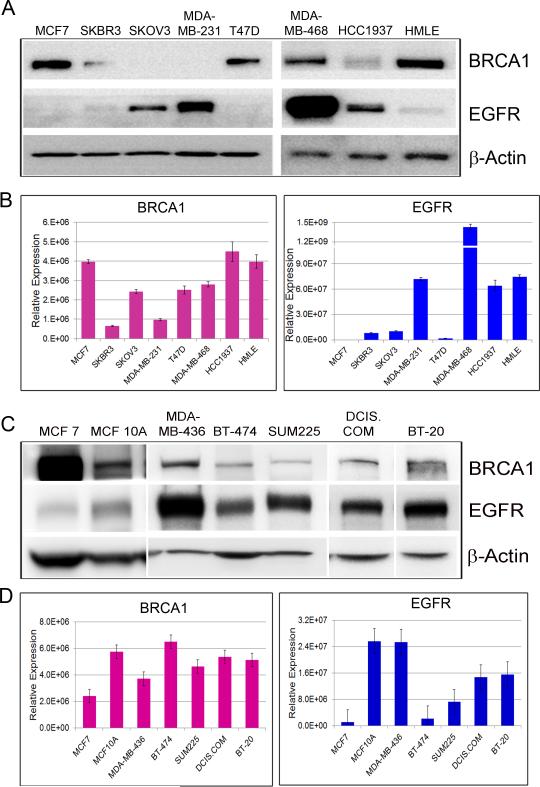
To investigate the relationship between EGFR expression and BRCA1 status, western blots for BRCA1 and EGFR were performed on extracts of breast and ovarian cancer cell lines representing different molecular subtypes, namely MCF7 and T47D (Luminal type, ER+/PR+/HER2−), BT-474 and SKBR3 (HER2 enriched, ER−/PR−/HER2+), MDA-MB-231, MDA-MB-436, MDA-MB-468 and BT-20 (Basal type, ER−/PR−/HER2−), MCF10DCIS.com and SUM-225 (ductal carcinoma in situ (DCIS) cell lines, basal like DCIS model and HER2 overexpressing DCIS model respectively), and SKOV3 (ovarian cancer cells, ER+/PR−/HER2+). In addition, we tested HCC1937, a basal breast cancer cell line that is homozygous for the BRCA1 5382insC mutation, HMLE, immortalized human mammary epithelial cells and MCF10A, immortalized non-transformed mammary epithelial cells. As shown in Fig.1A and C, high EGFR protein expression was associated with basal-type cell lines and low EGFR levels with luminal-type cell lines. HCC1937 cells that do not express functional BRCA1 display high EGFR protein expression (Fig.1A). HMLE and MCF10A cells on the other hand express considerable amounts of BRCA1 with minimal EGFR levels. An inverse correlation between BRCA1 and EGFR protein expression is observed in the cell lines tested. qRT-PCR of BRCA1 and EGFR (Fig.1B and D) revealed a somewhat similar pattern in most of the cell lines, further suggesting a negative relationship between BRCA1 and EGFR expression.
Figure 1. EGFR protein expression inversely correlates with BRCA1 expression in normal and breast cancer cell lines.
A&C. Western blots of BRCA1 and EGFR in breast and ovarian cancer cell lines. β-Actin was used as loading control. B&D. TaqMan qRT-PCR of BRCA1 and EGFR in breast and ovarian cancer cell lines. Data were normalized to 18SrRNA. (See also Figure S1)
Next, we analyzed HMLE cells infected with three different lentiviral clones of BRCA1 shRNA. Interestingly, depletion of BRCA1 resulted in a significant increase in EGFR protein expression, the increase reflecting the extent of BRCA1 knockdown (Fig.S1A). Densitometry of the immunoblots showed that EGFR levels increased as much as 600% upon BRCA1 depletion (Fig.S1B). qRT-PCR analysis revealed that BRCA1 knockdown elicited only marginal changes in EGFR mRNA expression (Fig.S1C) implicating a possible post-transcriptional mechanism for EGFR regulation. Similar results were observed in HeLa cells, further substantiating our findings (Fig.S1D &E).
BRCA1 regulates EGFR activation
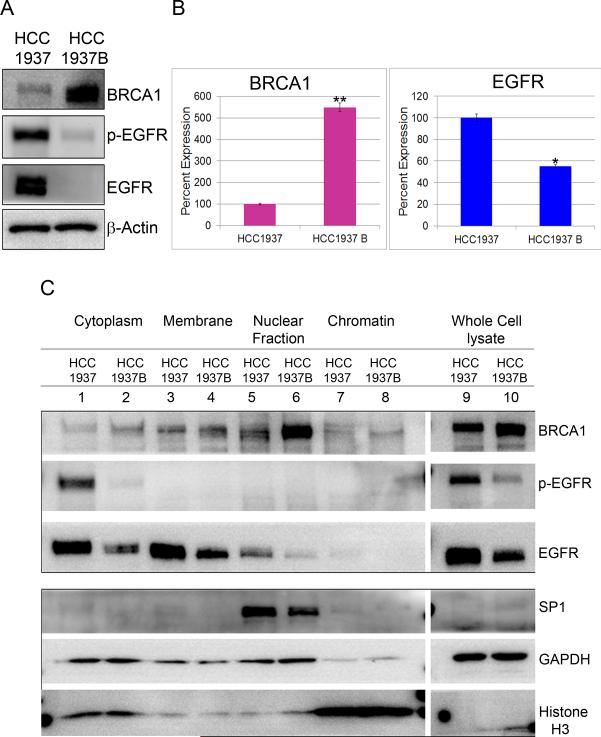
To investigate this further, we analyzed parental HCC1937 and HCC1937/WT BRCA1 (hereafter referred to as HCC1937B), a cell line in which BRCA1 has been restored by expression of full length BRCA1 cDNA. A significant decrease in total EGFR protein was observed when WT BRCA1 was re-expressed in the BRCA1-defective HCC1937 (Fig.2A & 2C-lanes 9, 10). In addition to overexpression of total EGFR, we also observed a noticeable increase in p-EGFR (Y-1068) in BRCA1-deficient cells. A moderate decrease in EGFR mRNA level was noticed in HCC1937B cells (Fig.2B). Subcellular fractionation of HCC1937 and HCC1937B cells showed accumulation of p-EGFR (Y-1068) in the cytoplasm of BRCA1 null cells (Fig 2C-lanes 1,2). We also observed a pronounced increase in total EGFR in the membrane and cytosol (Fig.2C-lanes 1-4), and a small amount in the nuclear fractions in BRCA1 null cells (Fig.2C-lanes 5,6). Together, the data confirm that augmented EGFR expression accompanying BRCA1 deficiency is also associated with activation of EGFR, as demonstrated by phosphorylation of specific tyrosine residue within the cytoplasmic domain.
Figure 2. BRCA1 re-expression in the BRCA1-defective breast cancer cell line decreases EGFR levels.
A. Western blots of BRCA1, p-EGFR and EGFR in extracts of HCC1937 and HCC1937B. β-Actin was used as loading control. B. TaqMan qRT-PCR of BRCA1 and EGFR in HCC1937 and HCC1937 WTBRCA1. Data were normalized to 18S rRNA. * p < 0.01, **p < 0.001 (Student's t test). C. BRCA1 deficiency results in increased activation of EGFR: Whole cell lysate and different subcellular fractions of HCC1937 and HCC1937B, cytoplasm, membrane, nuclear and chromatin were subjected to electrophoresis and probed for BRCA1, p-EGFR and EGFR. SP1, GAPDH and Histone H3 were used as cell fractionation markers for nuclear, cytoplasmic and chromatin fractions respectively.
We have shown an inverse correlation between BRCA1 and EGFR protein expression in multiple cell lines. Our data demonstrates that EGFR protein overexpression in BRCA1 deficiency far exceeds the change in its mRNA levels. Therefore we hypothesized that BRCA1 could regulate EGFR post-transcriptionally through a novel mechanism involving miRNAs. Using this rationale, we studied miRNAs with respect to BRCA1 and found that miR-146a plays a critical role in this process.
BRCA1 enhances miR-146 expression
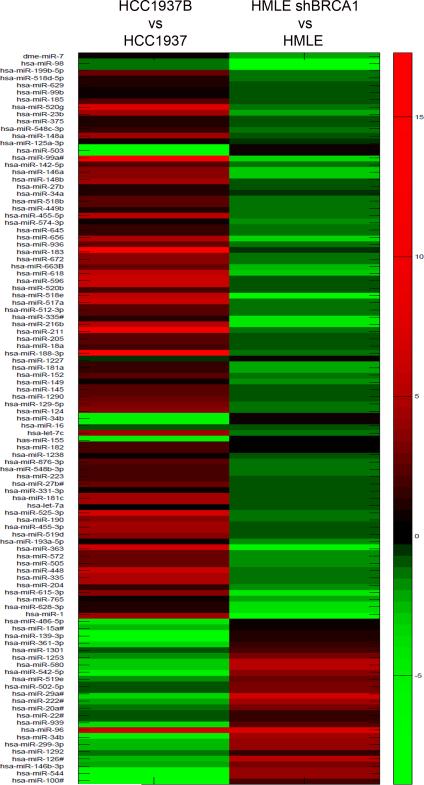
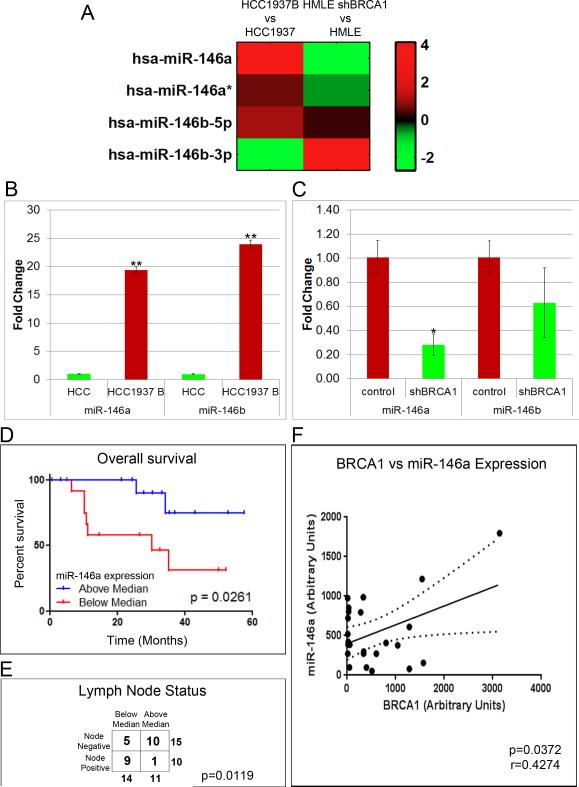
We performed miRNA profiling on HMLE and HCC1937, in which BRCA1 was depleted and overexpressed respectively. Analysis of array data revealed that distinct miRNAs were differentially regulated by BRCA1, exhibiting very high similarity regarding BRCA1 gain/loss in the two isogenic cell lines tested. The complete results of miRNA array are given in Fig.3. From the several miRNAs that showed BRCA1-dependent regulation that are also involved in EGFR signaling, we selected miR-146a based on its tumor suppressor function, its ability to target EGFR and its profound differential expression with respect to BRCA1 levels in both the cell lines. Further validation of our array data clearly illustrated significant regulation of the miR-146 family of miRNAs, with a profound increase in miR-146a in BRCA1 expressing cells (Fig.4A). Individual validation of miR-146a and miR-146b by qRT-PCR revealed that there was a 20-25 fold increase in both miR-146a and miR-146b in WT BRCA1 expressing HCC1937 (Fig.4B). In HMLE cells on the other hand, miR-146a was found to be down-regulated by about 80% in BRCA1 deficiency, while the effect was less significant for miR-146b (Fig.4C).
Figure 3. miRNA Array Comparative Heatmap in HMLE and HCC1937.
Heatmap showing miRNA expression profile in HCC1937B relative to HCC1937 and HMLE/shBRCA1 relative to HMLE. Red color represents upregulation and green color represents downregulation of miRNAs.
Figure 4. BRCA1 positively regulates the expression of tumor suppressor miRNAs, miR-146a and miR-146b.
A. Expression of miRNAs belonging to the miR-146 family in HCC1937 WTBRCA1 vs HCC1937 cells and HMLE shBRCA1 vs HMLE cells presented as heatmap. The log2 ratio of the relative fold change in the expression of the miRNA-146 family in the two cell lines is represented. B. Re-expression of BRCA1 increases miR-146a and miR-146b levels: TaqMan qRT-PCR for miR-146a and miR-146b in HCC1937 and HCC1937 WTBRCA1 cells. Data were normalized to U6 snRNA. C. Loss of BRCA1 decreases the expression of miR-146a and miR-146b levels: TaqMan qRT-PCR for miR-146a and miR-146b in control and BRCA1 depleted HMLE. Data were normalized to U6 snRNA. B&C. * p < 0.01, **p < 0.001 (Student's t test). D and E. Clinical evaluation of miR-146a: Kaplan-Meier overall survival curve, p=0.0261 (D) and lymph node status (E) with respect to miR-146a expression levels, p=0.0119. F. Scatter plot showing correlation of miR-146a levels with BRCA1 in TN samples.
To investigate if miR-146a expression could serve as an important prognostic biomarker in BRCA1-associated BLBC, we performed qRT-PCR analysis of 27 archived triple negative breast cancer (TNBC). From Kaplan-Meier survival analysis, we observed that low expression of miR-146a was associated with distinctively poor overall survival of patients (Fig.4D). Fisher's test shows a trend with miR-146a expression and lymph node status, with above median expression strongly predicting negative lymph node status (Fig.4E). miR-146a expression in the clinical samples was found to be closely correlated with BRCA1 expression levels (Fig.4F).
BRCA1 binds to miR-146a promoter and activates its transcription
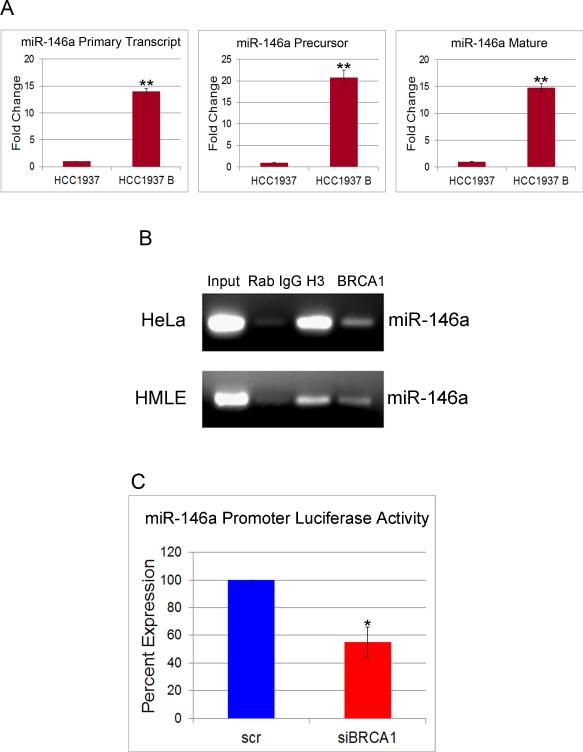
In order to understand the role of BRCA1 in the regulation of miR-146a biogenesis, we analyzed the levels of primary transcript, precursor and mature miR-146a levels in HCC1937 and HCC1937B cells by qRT-PCR. Our results indicated that in WT BRCA1 cells, the expression levels of all the three forms of miR-146a increased by about 15-20 fold compared to mutant BRCA1 cells (Fig.5A), suggesting a role of BRCA1 in miR-146a transcription and not biogenesis.
Figure 5. BRCA1 binds to the promoter and transcriptionally activates miR-146a.
A. TaqMan qRT-PCR for primary transcript, precursor and mature miR-146a in cultures of HCC1937 and HCC1937B cells. Data were normalized to U6 snRNA for precursor and mature and 18S rRNA for primary transcript. B. ChIP was performed using digested chromatin from HeLa and HMLE cells. IP was carried out with Rabbit IgG, anti-Histone H3 and anti-BRCA1. PCR was performed with miR-146a primer sets. Input lanes are shown. C. Promoter luciferase activity of a reporter construct carrying the miR-146a promoter upstream of the luciferase gene. Percent reduction in luciferase activity upon BRCA1 knockdown as compared to by scrambled control is shown. * p < 0.01, **p < 0.001 (Student's t test).
To investigate if BRCA1 augments transcription of miR-146a by binding to its putative promoters, we carried out a chromatin immunoprecipitation (ChIP) assays. Semi-quantitative PCR analysis of DNA, immunoprecipitated from HeLa and HMLE cells using the anti-BRCA1 C-terminal antibodies confirmed the recruitment of BRCA1 to miR-146a promoter (Fig.5B).
We performed luciferase reporter assays to confirm that miR-146a promoter is indeed activated by BRCA1. The promoter region for miR-146a is located between 17060 and 17550 bases upstream from it (29). Luciferase activity was measured after 48 hrs in HMLE cells co-transfected with pGL3-miR-146a-luc construct (containing the promoter region of miR-146a) along with either control siRNA or siRNA to BRCA1. Results showed that BRCA1 knockdown attenuated miR-146a promoter activity by about 45% compared to that of control (Fig 5C).
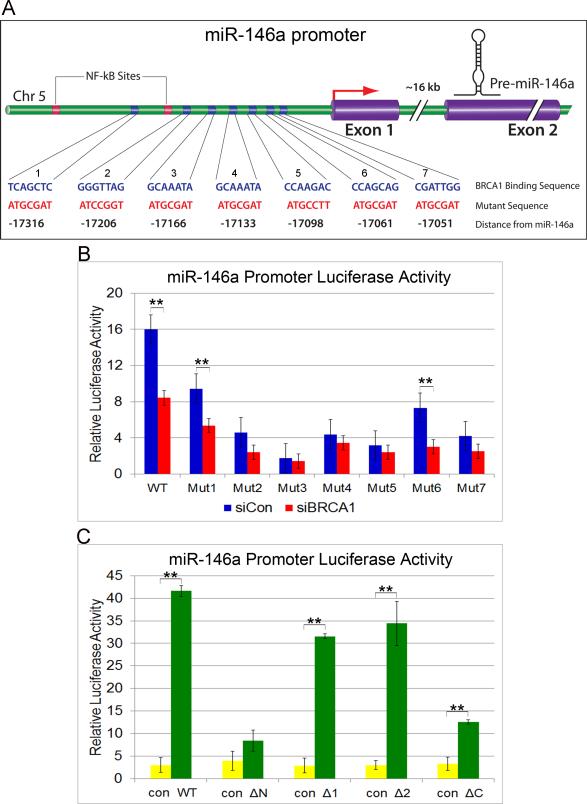
We further sought to identify the BRCA1 binding sites on the miR-146a promoter. We found 13 putative BRCA1 binding sites on miR-146a promoter (JASPAR database,(30)) (Fig.S2A). Of the 13, 8 matched Chip-Seq data (GEO Accession: GSM935377), with 5 of them showing frequency-matrix match over 0.80. To validate the interactions in vitro, we mutated seven of the eight target sequences individually (Fig.6A), transfected HMLE cells with luciferase constructs containing WT or miR-146a promoter mutants and measured luciferase activity following BRCA1 knockdown. All the seven mutants showed significant decrease in promoter activity compared to WT with mutants 3, 4 & 5 showing the most profound effect (Fig.6B). Mutations to BRCA1 binding sites 3, 4 or 5 led to the loss of BRCA1 sensitivity on the promoter as measured by luciferase activity, suggesting the importance of these binding sites in promoter activation by BRCA1 (Fig.6B).
Figure 6. BRCA1 binding regions on miR-146a promoter.
A. Schematic diagram showing the putative BRCA1 binding regions on miR-146a promoter. The seven binding sites are indicated in blue and the mutations generated in these regions are shown in red. Their relative distance from the mature miR-146a is represented below in black. B. Luciferase activity of WT and mutant miR-146a promoter in HMLE cells following BRCA1 silencing compared with scrambled control are shown. ** p < 0.001 (Student's t test). C. miR-146a luciferase activity in HMLE cells upon overexpression of WT BRCA1 and the different deletion mutants (WT: Full length, ΔΝ:Δ1−302 aa, Δ1:Δ305-770aa, Δ2: Δ775-1292aa, ΔC: Δ1527-1863aa) (See Fig.S2) compared to control virus are shown. Luciferase activity obtained with the WT and the different deletion mutants were normalized to the protein expression from Western blots. ** p < 0.001 (Student's t test). See also Fig.S2
To further identify the region of BRCA1 critical for binding to the miR-146a promoter, we infected HMLE cells with adenoviruses expressing HA-tagged WT and mutant BRCA1 (31) (Fig.S2B,S2C), then transfected the cells with WT miR-146a promoter luciferase construct and performed luciferase assay. Our results indicate that the N-terminal RING and the C-terminal BRCT domains of BRCA1 are important in binding to miR-146a promoter, as exogenous expression of protein lacking these domains did not have any significant effect on the miR-146a promoter luciferase activity compared to control (Fig.S2D &6C). Overexpression of WT and the 2 other deletion mutants on the other hand showed significant increase in luciferase activity. Our data corroborates earlier published reports on the importance of the RING and BRCT domains of BRCA1 in protein/protein and protein/DNA interactions (20).
miR-146a regulates EGFR expression
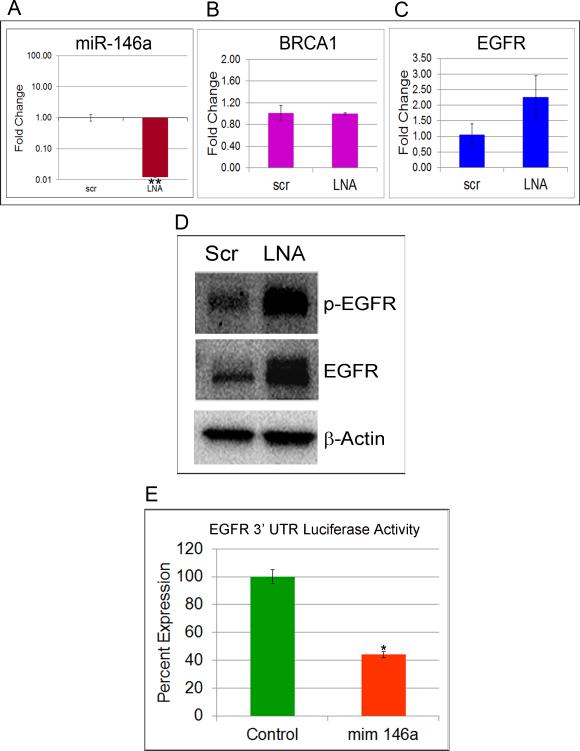
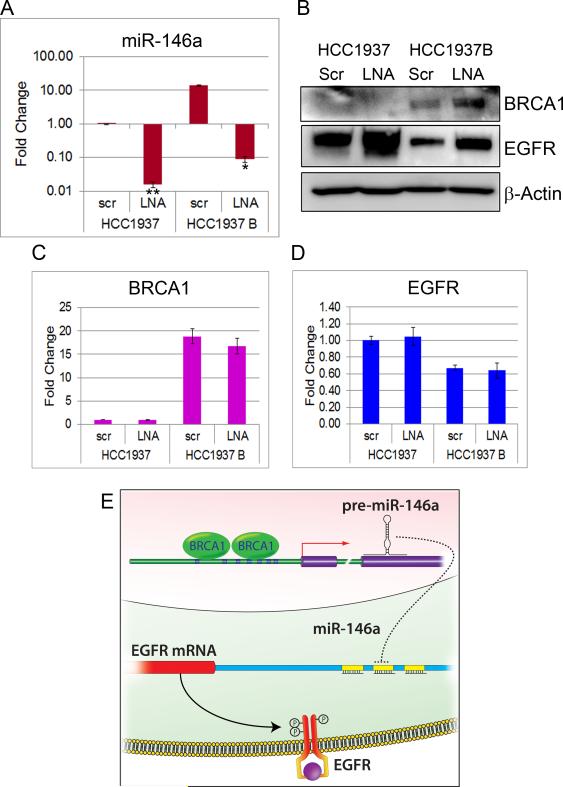
Both miR-146a and miR-146b have been shown to regulate EGFR in different cell types (26, 32, 33). To establish that miR-146a directly regulates the expression of EGFR in mammary epithelial cells, we transfected HMLE cells with miR-146a LNA or mimics. qRT-PCR of miR-146a after 48 hrs shows significant knockdown and overexpression upon transfection with LNA and mimic respectively (Fig.7A and S3A). BRCA1 and EGFR mRNA expression are shown in Fig.7B, 7C, S3B and S3C. Immunoblots revealed a significant increase in p-EGFR (Y-1068) and EGFR in HMLE upon miR-146a knockdown (Fig.7D) and a dramatic decrease upon its overexpression (Fig.S3D). Subcellular fractionation of LNA transfected HMLE revealed accumulation of EGFR in the membrane fraction (Fig.S3E) and also some expression in the nucleus similar to BRCA1-deficient cells. Similar results were observed in HeLa cells and MDA-MB231 (Fig.S3 F, G & H). These results collectively substantiate that miR-146a represses EGFR in mammary cells.
Figure 7. EGFR is a direct target of miR-146a.
A. TaqMan qRT-PCR for miR-146a in LNA-scrambled and LNA-miR-146a transfected HMLE. Data were normalized to U6 snRNA. * p < 0.001 (Student's t test). B. TaqMan qRT-PCR for BRCA1 and C. EGFR in LNA-scrambled and LNA-miR-146a transfected HMLE. Data were normalized to 18S rRNA. D. Western blots of p-EGFR and EGFR in cell lysates of HMLE transfected with scrambled and LNA-miR-146a. β-Actin was used as control. (See also Figure S3). F. Luciferase activity in HMLE cells transfected with miR-146a mimic and pEZX EGFR 3'UTR luciferase vector compared to control is shown. Values are mean ± SD of three independent experiments. * p < 0.01, **p < 0.001 (Student's t test). (See also Figure S3 and S4A).
Analysis of miR-146a binding to the 3'UTR of EGFR
In order to assess if miR-146a directly targets the 3' UTR of EGFR, we co-transfected HMLE and HeLA cells with a pEZX EGFR 3'UTR-luciferase-reporter construct and miR-146a mimic. We observed that the luciferase activity in cells transfected with miR-146a mimic decreased by 60% in HMLE (Fig.7E) and 30% in HeLa (FigS4A) cells compared to cells transfected with control mimic.
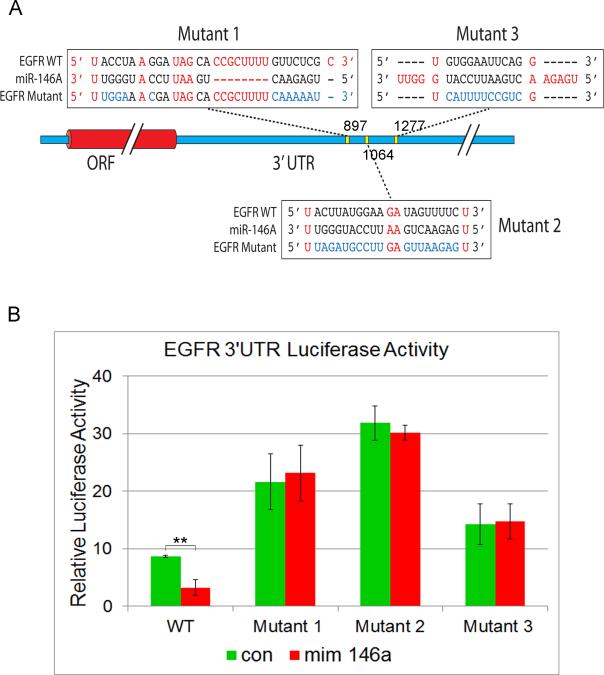
We further investigated miR-146a binding region to its specific target sequences within the 3'UTR of EGFR. We identified 21 high affinity (minimal free energy, MFE ≤ −20 kcal/mol) miR-146a targets in the 3'UTR of the EGFR gene with the strongest target located 1064 bp into the 3'UTR (Fig.S4B). In order to validate the miR-146a binding region on the 3'UTR of EGFR in vitro, we mutated three of the highest affinity target sites within the 3'UTR (Fig.8A, S4B). Co-transfection of miR-146a mimic with WT-EGFR 3'UTR luciferase construct resulted in significant inhibition of luciferase activity (Fig.8B). Mutagenesis of the miR-146a binding sites within the EGFR 3'UTR resulted in increase in luciferase activity and also completely abolished the ability of miR-146a to regulate EGFR 3'UTR luciferase expression. The result was more pronounced in mutant 2, located at 1064 bp which was also the strongest hit in our in silico analysis (Fig.S4B). Thus, we have demonstrated for the first time that EGFR is a direct target of miR-146a.
Figure 8. Analysis of miR-146a binding sites on EGFR 3'UTR.
A. Three high affinity miR-146a binding sites on the 3'UTR of EGFR used for validation with their positions. The expected base pairing and alignment of the seed region of miR-146a with EGFR 3'UTR are indicated in black and the sites of target mutagenesis are indicated in blue. B. Luciferase activity of WT and mutant EGFR 3'UTR in HMLE cells following miR-146a overexpression with miR-146a mimic compared to control mimic are shown. ** p < 0.001 (Student's t test). See also Fig.S4
miR-146a is downstream of BRCA1 and mediates the regulation of EGFR by BRCA1
We hypothesized that miR-146a is a key mediator of the effect of BRCA1 on EGFR expression. In order to test if knockdown of miR-146a can reverse the effect of BRCA1 on EGFR expression, we transfected HCC1937 and HCC1937B cells with LNA to miR-146a and evaluated the expression of miR-146a, BRCA1 and EGFR by qRT-PCR. There was a significant knockdown of miR-146a in both mutant and BRCA1-complemented HCC1937 cells upon LNA transfections (Fig.9A). Western blot analysis revealed that BRCA1 reconstitution in HCC1937 considerably decreased EGFR protein expression, consistent with our prior observations. Most importantly, knockdown of miR-146a in BRCA1-overexpressing HCC1937 cells was able to rescue EGFR protein expression (Fig. 9B). Results demonstrate that BRCA1 regulation of EGFR expression is dependent on miR-146a. The fact that miR-146a knockdown could rescue EGFR expression which is otherwise downregulated by BRCA1 suggests that miR-146a is critical to this function. There was no significant change in BRCA1 and EGFR mRNA levels upon LNA transfections (Fig.9C&D). We observed a small increase in BRCA1 protein expression upon LNA transfection in the HCC1937B (Fig. 9B) cell line. Similar results were noticed in mutant and BRCA1-complemented HCC1937 transfected with mimics (Fig.S5A-D). BRCA1 levels were found to decrease upon mimic transfection (Fig.10E) suggesting a feedback regulation. A model illustrating the post-transcriptional regulation of EGFR by BRCA1 is depicted in Fig.9E.
Figure 9. miR-146a mediates the regulation of EGFR by BRCA1.
A. qRT-PCR of miR-146a in LNA (scrambled and miR-146a) transfected HCC1937 and HCC1937 WT BRCA1. Data were normalized to U6 snRNA. * p < 0.01, **p < 0.001 (Student's t test). B. Western blots of EGFR in cell lysates of HCC1937 and HCC1937 WTBRCA1 transfected with scrambled and LNA-miR-146a. β-Actin was used as control. C&D. qRT-PCR of BRCA1 and EGFR in LNA (scrambled and miR-146a) transfected HCC1937 and HCC1937 WTBRCA1. Data were normalized to 18S rRNA. (See also Figure S4). E. Schematic showing post-transcriptional regulation of EGFR by BRCA1.
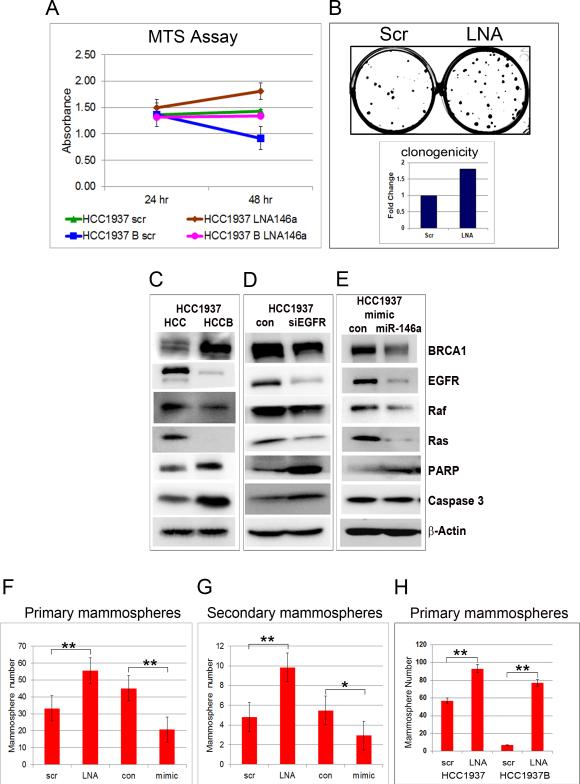
Figure 10. miR-146a loss results in increased proliferation in HCC1937 cells and clonogenic growth and mammosphere formation in HMLE cells.
A. Relative cell proliferation (as measured by the MTS Assay) of HCC1937 and HCC1937 WTBRCA1 cells transfected with scrambled and LNA miR-146a at 24 and 48 hrs after transfection. Results represent mean+/− SD of three independent experiments. B. Clonogenic images taken after 7-10 days of HMLE cells transfected with scrambled and LNA miR-146a are shown. Fold change is represented below the image. Data represent average of triplicates from three independent experiments. See also Fig.S6A,B,C. C,D&E. Western blots for BRCA1, EGFR, Raf, Ras, PARP and caspase 3 in HCC1937 cells after WTBRCA1 overexpression, EGFR knockdown and miR-146a mimic transfection. β-Actin was used as loading control. F&G. HMLE cells transfected with scrambled and LNA miR-146a and control and miR-146a mimic were cultured as primary and secondary mammospheres, and the number of spheres was counted per 1000 cells. Error bars denote +/− SD. ** p < 0.01, * p < 0.05 (Student's t test). (See also Figure S4). H, HCC1937 and HCC1937B cells transfected with scrambled and LNA miR-146a were cultured as primary mammospheres, and the number of spheres was counted per 1000 cells. Error bars denote +/− SD. ** p < 0.01, * p < 0.05 (Student's t test).
BRCA1 and miR-146a deficiency lead to hyperproliferation of cells and enhanced clonogenic growth
Additional functional studies were carried out to support the novel molecular mechanism by which BRCA1 and miR-146a regulate the initiation and development of BLBC by targeting EGFR and its downstream signaling pathway. BRCA1 overexpression resulted in significant reduction in cell proliferation (HCC1937 vs HCC1937B) (Fig.10A), corroborating previous reports that BRCA1-deficient cells proliferate faster than their BRCA1-proficient counterparts (20). miR-146a knockdown increased cell proliferation in both HCC1937 and HCC1937B cells. Most importantly, knockdown of miR-146a in HCC1937B cells significantly rescued cell growth by about 40% compared to control (Fig.10A) at 48 hours post-transfection. Cell cycle analysis showed a 20-30% increase in the percentage of cells in S phase in both HCC1937 and HCC1937B cells upon LNA miR-146a transfection (Fig.S6A & B). In clonogenic cell survival assay, knockdown of miR-146a significantly increased the number of surviving colonies by about two fold (Fig.10B), while its overexpression showed the opposite effect (Fig.S6C). Western blots performed after BRCA1 and miR-146a overexpression, and EGFR knockdown in HCC1937 show a strong decrease in EGFR, Raf-1 and Ras, and an increase in cleaved PARP and caspase-3 levels in HCC1937B compared to HCC1937 (Fig.10C), and in siEGFR transfected HCC1937 cells compared to control (Fig. 10D). Similar results were also observed in miR-146a mimic transfected cells compared to control with the exception of caspase-3 which did not show much difference (Fig.10E).
BRCA1 and miR-146a deficiency lead to enhanced mammosphere formation
It has been shown that signaling through EGFR is required for mammosphere formation and that breast cancer stem cells are EGFR positive (34). To understand the effect of miR-146a loss on anchorage independent growth, we selected the in vitro mammosphere culture system that enriches for putative stem/progenitor cells and that is capable of differentiation (35). Knockdown of miR-146a expanded both the primary and secondary mammospheres count by about two-fold, while overexpression diminished the count by 30-40% compared to control (Fig.10F&G). These experiments were also repeated using HCC1937 and HCC1937B cells. BRCA1 overexpression in the HCC1937 basal cell line profoundly decreased the mammosphere population, whereas knockdown of miR-146a in BRCA1 expressing cells offset its effect resulting in a 10-fold increase of mammospheres (Fig.10H).
DISCUSSION
Loss of functional BRCA1 and concomitant EGFR overexpression has been consistently observed in basal-like breast cancers by multiple groups (17, 18). Up to this point, this observation has always been a phenomenon indirectly-related-at-best, without a mechanistic explanation. In the current study, we investigated the relationship between EGFR expression and BRCA1 status and provide evidence that BRCA1 regulates EGFR expression post-transcriptionally. We demonstrate that BRCA1 binds to and transactivates the miR-146a promoter. We also show that the regulation of EGFR by BRCA1 is attenuated in the absence of miR-146a. Taken collectively, our results convincingly support a role for BRCA1 in the regulation of EGFR expression via miR-146a. This essential link in the mechanism, which connects the loss of functional BRCA1 to the overexpression of EGFR may provide novel options for preventive therapies in the future and may have functional consequences for the use of EGFR inhibitors in BRCA1-associated breast cancers.
Among TNBCs, the poor prognosis is strongly influenced by the presence of EGFR or basal cytokeratins (13, 14). Burga and colleagues have demonstrated that inhibition of BRCA1 leads to upregulation of EGFR and increased clonal growth in human mammary epithelial cells (hMEC) (36). Here, we show a direct negative correlation between BRCA1 and EGFR protein expression in breast cancer cell lines representing the different intrinsic tumor subtypes. In addition to the different cell lines where BRCA1 could either be mutated or epigenetically inactivated, we also show that BRCA1 knockdown in HMLE results in the upregulation of EGFR, while BRCA1 reconstitution in the BRCA1-mutant HCC1937 results in significant reduction in the expression of EGFR. Our results therefore demonstrate that in addition to BRCA1 mutational status, the overall expression levels of functional BRCA1 is very important in the regulation of EGFR.
Interestingly, the regulation of EGFR expression by BRCA1 appears to occur without major changes in its mRNA, suggesting a predominantly post-transcriptional mechanism, and our study reveals that this regulation occurs via miR-146a. miR-146a is a member of the miR-146 miRNA family consisting of two evolutionarily conserved miRNA genes, miR-146a and miR-146b that differ in their mature sequence only by 2 bases at the 3’ end. miR-146a was first discovered as an integral component of a negative feedback mechanism in the innate immune system. (29).
There was a 20-fold decrease or increase of miR-146a expression upon BRCA1 depletion or overexpression in HMLE and HCC1937, respectively. Our results are consistent with the Tanic et.al study (37) that reported up-regulation of miR-146a upon BRCA1 reconstitution in HCC1937. Garcia et al (38) have shown that miR-146a and miR-146b downregulate BRCA1 in TNBCs. In our experiments with LNA and mimic miR-146a transfections, we also observed an increase and decrease respectively of BRCA1 protein expression. The mechanism underlying this feedback regulation of BRCA1 and miR-146a that maintains their levels in a perfect homeostasis needs further investigation. Several epidemiological studies have evaluated the association between miR-146a polymorphisms, their capacity to bind to the 3'UTR of BRCA1, and breast cancer risk (39). Additional studies are required to assess the importance of this polymorphism in the potential feedback loop.
In a recent study, it has been reported that BRCA1 increases the expression of precursor and mature forms of cancer associated specific miRNAs by accelerating the microprocessor DROSHA-mediated processing of their primary transcript in vitro and in vivo (40). Our qRT-PCR analysis of the various stages of miR-146a biogenesis showed a uniform increase in expression levels of primary transcript, precursor and mature miR-146a with increase in BRCA1 suggesting that BRCA1 regulation of miR-146a is transcriptional and not through miRNA biogenesis.
ChIP analysis and promoter luciferase assays provide additional evidence that BRCA1 binds to the promoter of miR-146a and transcriptionally activates it. The role of BRCA1 as a transcriptional coregulator/corepressor is well known (21, 22, 41). Chang et al (42) have shown that BRCA1 plays a role in the epigenetic regulation of the oncogenic miR-155 via its interaction with the HDAC complex. Their study shows that BRCA1 associates with the miR-155 promoter and suppresses its transcription (42, 43). Evidence of transcriptional activation of any miRNA by BRCA1 has not been previously reported. Our data offers a new mechanism by which BRCA1 carries out its tumor suppressor function via a previously undescribed BRCA1→miR-146a→EGFR regulatory pathway. We have provided strong evidence that BRCA1 binds to specific regions on miR-146a promoter through its RING and BRCT domains and thus enhances its transcription.
It is widely reported that TRAF6 and IRAK 1 are bona fide targets of miR-146a (29), but recent studies have shown that it also targets mRNAs encoding other proteins, including EGFR (26, 32, 33, 44, 45). We demonstrate for the first time that miR-146a directly targets the 3'UTR of EGFR in breast cancer cells. We also provide evidence that EGFR downregulation following BRCA1 overexpression is dependent on the induction of miR-146a. Several independent studies have shown that miR-146a could inhibit tumor growth and metastasis in breast and other cancers (26, 45). Using genetically engineered mice, subsequent studies have demonstrated that miR-146a can function as a tumor suppressor gene. (46, 47) Our data showed that miR-146a levels in clinical samples strongly correlate with overall survival of TNBC patients, and also with expression levels of BRCA1. Further studies will determine if miR-146a can be used as a potential biomarker to identify the functional state of BRCA1 and EGFR in breast tumors.
Tight biologic control of EGFR pathway is necessary for proper downstream signaling. EGFR overexpression results in the activation of key downstream targets that induce hyperproliferation, resistance to apoptosis and enhanced cell migration and matrix remodeling (28). In our experiments with HCC1937 cells, we found that in addition to total EGFR, BRCA1-deficient cells exhibited increased autophosphorylation of EGFR at Y-1068 and increase in Raf-1 and Ras. Phosphorylation of specific tyrosine residues of EGFR within the cytoplasmic domain constitutes the initial activation process. Subsequent to this, the phosphotyrosine residues serve as docking sites for intracellular signaling molecules. Phosphotyrosine Y-1068 has been shown to mediate direct interaction of EGFR to Grb2 which in turn results in RAS/MAP kinase cascade activation (11). Although EGF signaling occurs through a variety of pathways, activation of the RAS/MAPK cascade has been associated with mammary epithelial cell growth and differentiation, as well as cancer development and progression (48). From gene expression profiling, Hoadley et al have shown that BLBCs preferentially use the EGFR-RAS-MEK pathway (49).
An interaction network was built to model the potential relationship of BRCA1, miR-146a and EGFR and their associated pathways. IPA analysis revealed their involvement in the MAPK signaling pathway (Fig.S7). MTS assay, clonogenicity assay and analysis of downstream targets of EGFR further confirm the interplay of BRCA1 and miR-146a in the regulation of RAS/MAPK signaling pathway in breast cancer cells, thereby contributing to tumor suppression.
In vitro studies on human breast luminal and basal cells have shown that basal cells express much higher levels of EGFR compared to their luminal counterpart (49). It has also been shown that EGFR is expressed in cancer stem cells (CSC) and hence could be involved in CSC renewal (50). We investigated the contribution of miR-146a to stem cell self-renewal. Malignant cells behave similarly to mammary stem and progenitor cells, proliferating in an undifferentiated state with a higher propensity for symmetric self-renewal. Our data showing an increase in mammosphere forming efficiency in miR-146a deficient cells suggests enhanced symmetric self-renewal capacity of HMLE due to increase in EGFR.
In this study, we report a post-transcriptional mechanism of EGFR regulation by BRCA1 mediated by miR-146a. Our data collectively provide an intriguing molecular mechanism for a role of BRCA1 in EGFR regulation. Our results suggest that the loss of miR-146a mediated EGFR regulation is a critical step in tumor pathogenesis in BRCA1 deficient breast cancer cells. Our results indicate that there exists an opportunity to prevent the development and progression of basal-like tumors from a state of cellular BRCA1 deficiency.
MATERIALS AND METHODS
RNA Isolation and Real-Time PCR
Total RNA inclusive of the small RNA fraction was extracted from cultured cells using RNeasy Mini kit (Qiagen), and from archived formalin fixed paraffin embedded (FFPE) tumor tissues, using the RecoverAll™ Total Nucleic Acid Isolation Kit (Ambion). Details of qRT-PCR are given in Supplemental Experimental Procedures.
Knockdown, Overexpression, Subcellular fractionation and Western blotting
Detailed protocols are given in Supplemental Experimental Procedures.
Dual Luciferase Reporter Assays
miR-146a promoter and EGFR 3'UTR luciferase activities were assayed using the Luciferase Assay System from Promega according to their instructions. The details are provided in Supplemental Experimental Procedures.
Chromatin Immunoprecipitation
ChIP assay was performed on HMLE and HeLa cells using SimpleChiP™ Enzymatic Chromatin IP Kit (Agarose Beads) from Cell Signaling Technology. The detailed protocol is given in Supplemental Experimental Procedures.
Mammosphere culture, MTS Assay, Clonogenicity and Cell Cycle analysis
The experiments were performed on HMLE, HCC1937 and HCC1937 WT BRCA1 cells. Please see Supplemental Experimental Procedures for detailed protocols.
Statistical Analyses
Unless otherwise noted, data are presented as mean ± SEM. Student's t test was used for assessing the significance of expression levels and comparisons, with p < 0.05 considered significant.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Vargheese Chennathukuzhi for critical review of the manuscript. The HA-tagged wild-type and deletion mutant adenoviruses were a kind gift from Jeffrey Parvin (The Ohio State University Medical Center). We thank Dr.David Baltimore (Caltech Institute of Technology) for the miR-146a promoter luciferase construct. We thank Dr.Ossama Tawfik, Tim Metcalf, Marsha Danley and the Biospecimen Shared Resources for assistance with clinical samples. We also thank Flow-Cytometry Shared Resource (supported by P30 CA168524 and P20 RR016443) of the University of Kansas Medical Center. This work was supported by the University of Kansas Cancer Center.
Footnotes
Disclosure of Potential Conflicts of Interest: None
REFERENCES
- 1.Futreal PA, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, Tavtigian S, et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994;266(5182):120–2. doi: 10.1126/science.7939630. Epub 1994/10/07. [DOI] [PubMed] [Google Scholar]
- 2.Shuen AY, Foulkes WD. Inherited mutations in breast cancer genes--risk and response. Journal of mammary gland biology and neoplasia. 2011;16(1):3–15. doi: 10.1007/s10911-011-9213-5. Epub 2011/04/05. [DOI] [PubMed] [Google Scholar]
- 3.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. doi: 10.1038/35021093. Epub 2000/08/30. [DOI] [PubMed] [Google Scholar]
- 4.Foulkes WD, Stefansson IM, Chappuis PO, Begin LR, Goffin JR, Wong N, et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. Journal of the National Cancer Institute. 2003;95(19):1482–5. doi: 10.1093/jnci/djg050. Epub 2003/10/02. [DOI] [PubMed] [Google Scholar]
- 5.Buckley NE, Mullan PB. BRCA1 - Conductor of the Breast Stem Cell Orchestra: The Role of BRCA1 in Mammary Gland Development and Identification of Cell of Origin of BRCA1 Mutant Breast Cancer. Stem cell reviews. 2012 doi: 10.1007/s12015-012-9354-y. Epub 2012/03/20. [DOI] [PubMed] [Google Scholar]
- 6.Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nature medicine. 2009;15(8):907–13. doi: 10.1038/nm.2000. Epub 2009/08/04. [DOI] [PubMed] [Google Scholar]
- 7.Molyneux G, Geyer FC, Magnay FA, McCarthy A, Kendrick H, Natrajan R, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7(3):403–17. doi: 10.1016/j.stem.2010.07.010. Epub 2010/09/02. [DOI] [PubMed] [Google Scholar]
- 8.Thompson ME, Jensen RA, Obermiller PS, Page DL, Holt JT. Decreased expression of BRCA1 accelerates growth and is often present during sporadic breast cancer progression. Nature genetics. 1995;9(4):444–50. doi: 10.1038/ng0495-444. Epub 1995/04/01. [DOI] [PubMed] [Google Scholar]
- 9.Holt JT, Thompson ME, Szabo C, Robinson-Benion C, Arteaga CL, King MC, et al. Growth retardation and tumour inhibition by BRCA1. Nature genetics. 1996;12(3):298–302. doi: 10.1038/ng0396-298. Epub 1996/03/01. [DOI] [PubMed] [Google Scholar]
- 10.Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer. 2004;4(10):814–9. doi: 10.1038/nrc1457. Epub 2004/10/29. [DOI] [PubMed] [Google Scholar]
- 11.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103(2):211–25. doi: 10.1016/s0092-8674(00)00114-8. Epub 2000/11/01. [DOI] [PubMed] [Google Scholar]
- 12.Yarden Y. The EGFR family and its ligands in human cancer. signalling mechanisms and therapeutic opportunities. Eur J Cancer. 2001;37(Suppl 4):S3–8. doi: 10.1016/s0959-8049(01)00230-1. Epub 2001/10/13. [DOI] [PubMed] [Google Scholar]
- 13.Rimawi MF, Shetty PB, Weiss HL, Schiff R, Osborne CK, Chamness GC, et al. Epidermal growth factor receptor expression in breast cancer association with biologic phenotype and clinical outcomes. Cancer. 2010;116(5):1234–42. doi: 10.1002/cncr.24816. Epub 2010/01/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sainsbury JR, Farndon JR, Needham GK, Malcolm AJ, Harris AL. Epidermal-growth-factor receptor status as predictor of early recurrence of and death from breast cancer. Lancet. 1987;1(8547):1398–402. doi: 10.1016/s0140-6736(87)90593-9. Epub 1987/06/20. [DOI] [PubMed] [Google Scholar]
- 15.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37(Suppl 4):S9–15. doi: 10.1016/s0959-8049(01)00231-3. Epub 2001/10/13. [DOI] [PubMed] [Google Scholar]
- 16.Viale G, Rotmensz N, Maisonneuve P, Bottiglieri L, Montagna E, Luini A, et al. Invasive ductal carcinoma of the breast with the “triple-negative” phenotype: prognostic implications of EGFR immunoreactivity. Breast cancer research and treatment. 2009;116(2):317–28. doi: 10.1007/s10549-008-0206-z. Epub 2008/10/08. [DOI] [PubMed] [Google Scholar]
- 17.Arnes JB, Begin LR, Stefansson I, Brunet JS, Nielsen TO, Foulkes WD, et al. Expression of epidermal growth factor receptor in relation to BRCA1 status, basal-like markers and prognosis in breast cancer. J Clin Pathol. 2009;62(2):139–46. doi: 10.1136/jcp.2008.056291. Epub 2008/08/07. [DOI] [PubMed] [Google Scholar]
- 18.Toyama T, Yamashita H, Kondo N, Okuda K, Takahashi S, Sasaki H, et al. Frequently increased epidermal growth factor receptor (EGFR) copy numbers and decreased BRCA1 mRNA expression in Japanese triple-negative breast cancers. BMC Cancer. 2008;8:309. doi: 10.1186/1471-2407-8-309. Epub 2008/10/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumaraswamy E, Shiekhattar R. Activation of BRCA1/BRCA2-associated helicase BACH1 is required for timely progression through S phase. Molecular and cellular biology. 2007;27(19):6733–41. doi: 10.1128/MCB.00961-07. Epub 2007/08/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huen MS, Sy SM, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol. 2010;11(2):138–48. doi: 10.1038/nrm2831. Epub 2009/12/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silver DP, Livingston DM. Mechanisms of BRCA1 Tumor Suppression. Cancer discovery. 2012;2(8):679–84. doi: 10.1158/2159-8290.CD-12-0221. Epub 2012/07/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorski JJ, James CR, Quinn JE, Stewart GE, Staunton KC, Buckley NE, et al. BRCA1 transcriptionally regulates genes associated with the basal-like phenotype in breast cancer. Breast cancer research and treatment. 2010;122(3):721–31. doi: 10.1007/s10549-009-0565-0. Epub 2009/11/03. [DOI] [PubMed] [Google Scholar]
- 23.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. Epub 2009/01/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436(7051):740–4. doi: 10.1038/nature03868. Epub 2005/06/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO molecular medicine. 2012;4(3):143–59. doi: 10.1002/emmm.201100209. Epub 2012/02/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, Welch DR. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer research. 2009;69(4):1279–83. doi: 10.1158/0008-5472.CAN-08-3559. Epub 2009/02/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uhlmann S, Mannsperger H, Zhang JD, Horvat EA, Schmidt C, Kublbeck M, et al. Global microRNA level regulation of EGFR-driven cell-cycle protein network in breast cancer. Molecular systems biology. 2012;8:570. doi: 10.1038/msb.2011.100. Epub 2012/02/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sebastian S, Settleman J, Reshkin SJ, Azzariti A, Bellizzi A, Paradiso A. The complexity of targeting EGFR signalling in cancer: from expression to turnover. Biochim Biophys Acta. 2006;1766(1):120–39. doi: 10.1016/j.bbcan.2006.06.001. Epub 2006/08/08. [DOI] [PubMed] [Google Scholar]
- 29.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(33):12481–6. doi: 10.1073/pnas.0605298103. Epub 2006/08/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic acids research. 2004;32(Database issue):D91–4. doi: 10.1093/nar/gkh012. Epub 2003/12/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiba N, Parvin JD. The BRCA1 and BARD1 association with the RNA polymerase II holoenzyme. Cancer research. 2002;62(15):4222–8. Epub 2002/08/03. [PubMed] [Google Scholar]
- 32.Xu B, Wang N, Wang X, Tong N, Shao N, Tao J, et al. MiR-146a suppresses tumor growth and progression by targeting EGFR pathway and in a p-ERK-dependent manner in castration-resistant prostate cancer. The Prostate. 2011 doi: 10.1002/pros.22466. Epub 2011/12/14. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Vandenboom TG, 2nd, Wang Z, Kong D, Ali S, Philip PA, et al. miR-146a suppresses invasion of pancreatic cancer cells. Cancer research. 2010;70(4):1486–95. doi: 10.1158/0008-5472.CAN-09-2792. Epub 2010/02/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Booth BW, Boulanger CA, Anderson LH, Jimenez-Rojo L, Brisken C, Smith GH. Amphiregulin mediates self-renewal in an immortal mammary epithelial cell line with stem cell characteristics. Experimental cell research. 2010;316(3):422–32. doi: 10.1016/j.yexcr.2009.11.006. Epub 2009/11/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17(10):1253–70. doi: 10.1101/gad.1061803. Epub 2003/05/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burga LN, Hu H, Juvekar A, Tung NM, Troyan SL, Hofstatter EW, et al. Loss of BRCA1 leads to an increase in epidermal growth factor receptor expression in mammary epithelial cells, and epidermal growth factor receptor inhibition prevents estrogen receptor-negative cancers in BRCA1-mutant mice. Breast cancer research : BCR. 2011;13(2):R30. doi: 10.1186/bcr2850. Epub 2011/03/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanic M, Zajac M, Gomez-Lopez G, Benitez J, Martinez-Delgado B. Integration of BRCA1-mediated miRNA and mRNA profiles reveals microRNA regulation of TRAF2 and NFkappaB pathway. Breast cancer research and treatment. 2011 doi: 10.1007/s10549-011-1905-4. Epub 2011/12/15. [DOI] [PubMed] [Google Scholar]
- 38.Garcia AI, Buisson M, Bertrand P, Rimokh R, Rouleau E, Lopez BS, et al. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO molecular medicine. 2011;3(5):279–90. doi: 10.1002/emmm.201100136. Epub 2011/04/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lian H, Wang L, Zhang J. Increased risk of breast cancer associated with CC genotype of Has-miR-146a Rs2910164 polymorphism in Europeans. PloS one. 2012;7(2):e31615. doi: 10.1371/journal.pone.0031615. Epub 2012/03/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawai S, Amano A. BRCA1 regulates microRNA biogenesis via the DROSHA microprocessor complex. The Journal of cell biology. 2012;197(2):201–8. doi: 10.1083/jcb.201110008. Epub 2012/04/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scully R, Anderson SF, Chao DM, Wei W, Ye L, Young RA, et al. BRCA1 is a component of the RNA polymerase II holoenzyme. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(11):5605–10. doi: 10.1073/pnas.94.11.5605. Epub 1997/05/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang S, Wang RH, Akagi K, Kim KA, Martin BK, Cavallone L, et al. Tumor suppressor BRCA1 epigenetically controls oncogenic microRNA-155. Nature medicine. 2011;17(10):1275–82. doi: 10.1038/nm.2459. Epub 2011/09/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang S, Sharan SK. BRCA1 and MicroRNAs: Emerging networks and potential therapeutic targets. Molecules and cells. 2012 doi: 10.1007/s10059-012-0118-y. Epub 2012/09/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mei J, Bachoo R, Zhang CL. MicroRNA-146a inhibits glioma development by targeting Notch1. Molecular and cellular biology. 2011;31(17):3584–92. doi: 10.1128/MCB.05821-11. Epub 2011/07/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhaumik D, Scott GK, Schokrpur S, Patil CK, Campisi J, Benz CC. Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene. 2008;27(42):5643–7. doi: 10.1038/onc.2008.171. Epub 2008/05/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. 2011;208(6):1189–201. doi: 10.1084/jem.20101823. Epub 2011/05/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao JL, Rao DS, Boldin MP, Taganov KD, O'Connell RM, Baltimore D. NF-kappaB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(22):9184–9. doi: 10.1073/pnas.1105398108. Epub 2011/05/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hynes NE, MacDonald G. ErbB receptors and signaling pathways in cancer. Current opinion in cell biology. 2009;21(2):177–84. doi: 10.1016/j.ceb.2008.12.010. Epub 2009/02/12. [DOI] [PubMed] [Google Scholar]
- 49.Hoadley KA, Weigman VJ, Fan C, Sawyer LR, He X, Troester MA, et al. EGFR associated expression profiles vary with breast tumor subtype. BMC genomics. 2007;8:258. doi: 10.1186/1471-2164-8-258. Epub 2007/08/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Farnie G, Clarke RB, Spence K, Pinnock N, Brennan K, Anderson NG, et al. Novel cell culture technique for primary ductal carcinoma in situ: role of Notch and epidermal growth factor receptor signaling pathways. Journal of the National Cancer Institute. 2007;99(8):616–27. doi: 10.1093/jnci/djk133. Epub 2007/04/19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.