Abstract
Natural chromosome ends resemble double-stranded DNA breaks, but they do not activate a damage response in healthy cells. Telomeres therefore have evolved to solve the ‘end-protection problem’ by inhibiting multiple DNA damage–response pathways. During the past decade, the view of telomeres has progressed from simple caps that hide chromosome ends to complex machineries that have an active role in organizing the genome. Here we focus on mammalian telomeres and summarize and interpret recent discoveries in detail, focusing on how repair pathways are inhibited, how resection and replication are controlled and how these mechanisms govern cell fate during senescence, crisis and transformation.
The end protection problem: inhibition of the DDR
Shelterin, the complex consisting of telomere repeat–binding factor (TRF) 1, TRF2, repressor activator protein 1 (Rap1), TRF1 interactor 2 (TIN2), TINT1–PTOP–PIP1 (TPP1) and protection of telomeres 1 (POT1), prevents the recognition of telomeres as sites of damage. Disruption of shelterin components leads to activation of the DNA-damage response (DDR), including phosphorylation of histone 2A family, member X (H2A.X) and ataxia telangiectasia mutated (ATM). Mediator of DNA damage checkpoint protein 1 (MDC1), RING-finger motif and FHA domain (RNF) 8 and RNF168, Nijmegen breakage syndrome 1 (NBS1) and p53 binding protein 1 (53BP1) are recruited to telomeres in discrete foci termed telomere dysfunction-induced foci (TIF). Eventually, fusion of chromosome ends and activation of p53 triggers cell-cycle arrest and senescence. The current state of knowledge of how shelterin and its accessory factors prevent DDR activation is considered in detail in the following section.
Inhibition of the ATM pathway
The shelterin component TRF2 is the main inhibitor of the ATM kinase pathway and classical non-homologous end joining (c-NHEJ) at telomeres1–4. Depletion of TRF2 or expression of TRF2ΔBΔM, a dominant-negative allele of human TRF2 (official symbol TERF2) that lacks both the C-terminal MYB-type DNA-binding domain and the N-terminal basic domain, induce strong DNA-damage signals at telomeres and extensive chromosomal fusions4 that are dependent on DNA-dependent protein kinase catalytic subunits (DNA-PKcs), ligase IV and the Ku70–Ku80 heterodimer5,6. The mechanism by which TRF2 inhibits both ATM activation and c-NHEJ is complex and highly redundant. TRF2 is essential in formation of the t loop7–9, a secondary structure that hides chromosome ends, thereby preventing ATM activation and blocking the loading of the Ku heterodimer (Fig. 1, steps 1 and 2). Whether t-loop DNA structures are sufficient to prevent the DDR and telomere fusions independently of TRF2 remains a matter of speculation because it is not yet possible to separate the roles of TRF2 in t-loop formation and direct inhibition of DDR. Moreover, t-loop structures are unlikely to protect telomeres throughout the cell cycle, because t loops require unfolding during telomere replication, thus suggesting the presence of additional mechanisms for effective DDR inhibition, such as a direct role of TRF2 in ATM inhibition.
Figure 1.
Repression of DNA-damage signaling pathways at telomeres. (a) ATM kinase pathway. (b) ATR kinase pathway. (c) Alternative NHEJ repair pathway. (d) Classical NHEJ repair pathway. (e) Homologous-recombination pathway. Steps of DNA damage–repair inhibition by shelterin or accessory factors are highlighted by red circles. (1) TRF2, potentially through its TRFH domain, and supported by Rap1, forms the t loop, a DNA structure that hides chromosome ends from the MRN–ATM factors7–9,18. (2) The t loop also prevents loading of the Ku heterodimer on chromosome ends. (3) TRF2iDDR inhibits the recruitment of RNF168 at telomeres10. (4) TRF2 interacts with the helix α5 domain of Ku70, thereby blocking its heterotetramerization13. (5) Rap1 has been proposed to directly block c-NHEJ17. (6) TPP1–POT1 prevents ATR activation, most probably by preventing binding of RPA on the overhang and on the ssTTAGGG at stalled replication forks20–23,25. (7) The Ku heterodimer, TRF2 and, more importantly, TPP1–POT1 redundantly repress alt-NHEJ at telomeres6,33,35. (8) The shelterin factors Rap1 and POT1 as well as Ku70–Ku80 are necessary to block HR between sister telomeres14,16,36,38. (9) The N-terminal basic domain of TRF2 blocks NBS1–XRCC3–mediated recombination events at the D loop that would lead to t-loop excision. Ku depletion induces t-loop excision as well39–41. (10) At deprotected telomeres, 53BP1, RIF1 and MAD2L2 protect chromosome ends from BRCA1–CtIP–EXO1–mediated 5′ resection59–63. LIG, ligase; Pol, polymerase; p, phospho-.
This direct role of TRF2 in the inhibition of the ATM pathway remained unclear until a recent study from Okamoto and colleagues, who used chimeric TRF2-TRF1 proteins to dissect the role of the domains of TRF2 in DDR inhibition10. TRF2 is composed of four distinct domains: the N-terminal basic domain; the TRFH domain, which is involved in homodimerization and binding to accessory factors; the hinge domain, which mediates interactions with Rap1 and TIN2; and the MYB domain, which confers specificity for TTAGGG repeats (Fig. 2). The study has revealed that the TRFH domain of TRF2 provides an essential but insufficient level of protection against early steps of ATM activation. The main protective property is provided by the so-called iDDR, a small region, located in the hinge domain of TRF2, that prevents RNF168 recruitment to telomeres (Fig. 1, step 3). Inhibition of RNF168 disrupts the subsequent recruitment of 53BP1, thus preventing the formation of telomere fusions. The authors suggest that iDDR acts through recruitment of the deubiquitination enzyme BRCA1–BRCA2–containing complex 3 (BRCC3), which prevents H2A polyubiquitination–dependent recruitment of RNF168 and ubiquitin protein ligase 5 (UBR5), an enzyme that mediates degradation of RNF168. Complete protection of telomeres therefore requires both the iDDR and the TRFH domains, because loss of either the TRFH or the hinge domain leads to a mild or strong telomere-fusion phenotype, respectively. 53BP1 has also been shown to increase the mobility of telomeres that lose TRF2, thereby facilitating c-NHEJ-based repair of distant break sites and dysfunctional telomeres, and adding another level of complexity11,12.
Figure 2.
Functions and interactions of TRF2 domains. TRF2 possesses four distinct domains, the N-terminal basic domain38,39,48, the TRFH domain10,44,46,96–100, the hinge domain10,14,93 and the MYB DNA-binding domain96,97. Human (H) or mouse (M) amino acid residues that are involved in protein-protein interactions or that disrupt interactions when mutated are specified in the third column.
Direct c-NHEJ repression
TRF2 also functions at a more downstream level, by directly blocking c-NHEJ (Fig. 1, step 4). The Ku70–Ku80 heterodimer, an essential initiator component of c-NHEJ, has a vital role in telomere maintenance and is recruited to telomeres through direct interactions with TRF1, TRF2 and Rap1 (ref. 13); however, the constitutive presence of Ku at telomeres is at odds with the need to prevent c-NHEJ. Ribes-Zamora and colleagues have recently discovered that TRF2 interacts with the α-helix 5 domain of Ku70, which mediates Ku70–Ku80 heterotetramerization and is required in DNA repair. By occupying this essential interaction site, TRF2 could block Ku activation by inhibiting heterotetramerization and thus prevent synapsis of chromosome ends even when Ku is present13.
TRF2 depletion is accompanied by loss of Rap1 at telomeres; therefore, some aspects of telomere protection that are potentially mediated by Rap1 may be masked by TRF2 depletion. Rap1 is dispensable in c-NHEJ inhibition, because loss of Rap1 does not cause telomere fusions or ATM activation14–16. However, a redundant role for Rap1 in the inhibition of c-NHEJ cannot be ruled out (Fig. 1, step 5). Indeed, tethering of Rap1 to telomeres has been shown to prevent TRF2ΔBΔM-induced fusions without inhibiting TIFs17, thus suggesting a potential direct role for Rap1 in NHEJ inhibition. Rap1 directly binds Ku70–Ku80 in vivo through an unidentified domain that is distinct from Ku70 α5 (ref. 13). Identification of this interaction domain would possibly unveil the role of Rap1 in this aspect of telomere protection.
Rap1 also supports TRF2 in preventing both ATM activation and c-NHEJ at telomeres. Electron microscopy analysis of t-loop formation has revealed that the Rap1–TRF2 complex, compared to TRF2 alone, is much more prone to bind telomeric DNA and to form t loops18. Rap1 binding to TRF2 also decreases electrostatic interactions between double-stranded DNA and the TRF2 basic domain, thereby decreasing nonspecific binding of TRF2 to DNA and increasing the specificity of TRF2 for telomeric DNA19.
The multiplicity of levels of c-NHEJ inhibition at telomeres, which include telomere structure and mobility and direct inhibition of ATM, Ku and 53BP1 by TRF2 and Rap1, reinforce the potential of c-NHEJ as a major threat to natural chromosome ends. In response, natural chromosome ends have evolved to effectively inhibit this repair pathway, thereby limiting chromosome end-to-end fusions and maintaining genome stability. However, c-NHEJ is not the only repair pathway that can act on natural chromosome ends.
Inhibition of ATR and suppression of alternative NHEJ
The ATM- and RAD3-related (ATR) kinase pathway is mainly activated by exposed single-stranded (ss) DNA, thus rendering the double-strand–to–single-strand transitions within telomeres a prime target. The TPP1–POT1 (POT1a in mice) heterodimer is the main inhibitor of the ATR pathway at telomeres, and the heterodimer is anchored to telomeres by TIN2 (refs. 20–22). Although the exact mechanism of how TPP1-POT1 inhibits ATR is not yet clear, it most probably acts through the exclusion of replication protein A (RPA) from the single-stranded overhang23 (Fig. 1, step 6). TPP1–POT1 also prevents ATR activation at stalled replication forks at telomeres, where it is recruited by TRF1–TIN2 (refs. 24,25) (Fig. 3a). Consequently, loss of TRF1 leads to ATR activation at stalled replication forks during S and G2 (ref. 24). Tethering TIN2–TPP1–POT1 to TRF2 rescues ATR activation without suppressing replication stress induced by TRF1 loss25. TPP1–POT1 may inhibit ATR by excluding RPA from the replicative G-rich single strand that accumulates at stalled replication forks23. Because TPP1–POT1 cannot protect the ssCCCTAA repeats, the lack of ATR activation may reflect the uncoupling of leading- and lagging-strand synthesis of the progressing replication forks upon replication stress at telomeres, which would prevent accumulation of ssCCCTAA repeats26.
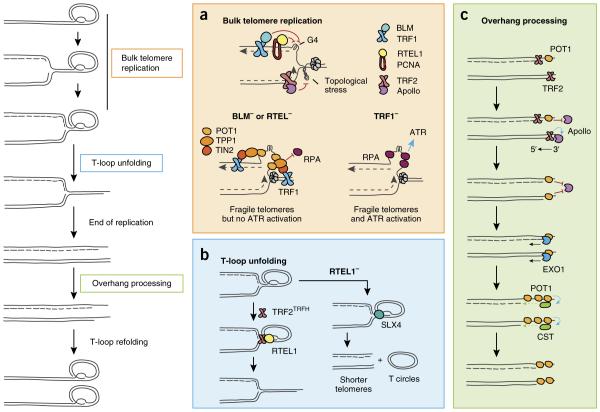
Figure 3.
Major DNA-repair factors involved in telomere maintenance during S phase. Replication initiates within subtelomeres and progresses until it encounters the D loop, which must be dismantled to allow replication to be completed. The overhangs are processed on both sister chromatids, and the t loop is refolded42. (a) TRF1 and proliferating cell nuclear antigen (PCNA) recruit BLM and RTEL1, both of which unfold G-quadruplex structures on the lagging strand25,94, while Apollo recruitment by TRF2 prevents accumulation of topological stress ahead of the fork95. Without BLM or RTEL1, the replication fork is blocked, thus leading to G-rich ssDNA accumulation. A bridge consisting of TRF1, TIN2 and TPP1 anchors POT1 on this ssDNA, preventing RPA loading and ATR activation24,25. (b) Once the bulk telomere is replicated, TRF2TRFH recruits RTEL1, which unfolds the t loop. In the absence of RTEL1, t loops are excised by SLX4, thus leading to telomere shortening and t-circle accumulation44,45. (c) Unidirectional replication results in blunt ends at the daughter chromatid formed by leading-strand replication, whereas lagging-strand replication leaves a short overhang that is bound by POT1. After termination of replication, TRF2 recruits Apollo to the leading-strand chromatid, and Apollo resects the 5′ end and generates a short overhang. Binding of POT1 blocks further resection by Apollo. A second round of overhang processing occurs during late S-G2, when EXO1 further resects the 5′ ends of both sister chromatids. Finally, the CST complex is recruited to the extended overhangs to fill in the lagging strand and reduce overhang length52–55.
Alternative NHEJ (alt-NHEJ) is a backup pathway for double-strand break (DSB) repair that depends on meiotic recombination 11 homolog (MRE11) and CtIP for end resection, whereas poly(ADP-ribose) polymerase 1 (Parp1), DNA ligase III and DNA polymerase θ are involved in the end-joining process27–32. Alt-NHEJ is activated at telomeres as a consequence of two distinct types of dysfunction. Deprotection of the ssDNA overhang upon TPP1–POT1 depletion is sufficient to induce low levels of telomere fusion by alt-NHEJ, which is in that case controlled by ATR33 (Fig. 1, 6). Loss of TRF2, in contrast, does not lead to alt-NHEJ, because the Ku heterodimer inhibits this repair pathway both at telomeres and at DSBs6,33–35. Co-depletion of Ku and TRF2 is therefore required to induce alt-NHEJ–dependent telomere fusions36 (Fig. 1, step 7). However, full activation of alt-NHEJ depends on suppression of both TRF2 and TPP1–POT1 as well as suppression of the Ku heterodimer or 53BP1 (refs. 33,35). Finally, RNF8 has an indirect role in alt-NHEJ inhibition at telomeres, because it polyubiquitinates and thereby stabilizes TPP1 at telomeres37.
The elaborate mechanisms that have evolved to block alt-NHEJ emphasize the potential threat that this pathway poses to telomeres and the complexity of efficient chromosome end–protection strategies in mammalian cells.
Inhibition of homologous recombination
Because telomeres are comprised of many kilobases of identical repeats at every chromosome end, they are excellent substrates for homologous recombination (HR). However, recombination between sister chromatids (telomeric sister-chromatid exchange (T-SCE)) or between distinct chromosome ends is toxic because it perturbs telomere-length homeostasis and can lead to chromosome fusions. The Ku70–Ku80 heterodimer, besides inhibiting alt-NHEJ, also prevents HR at deprotected telomeres. Suppression of Ku in mouse cells lacking either Rap1 or POT1 leads to high rates of T-SCEs, a byproduct of HR at telomeres14,36,38 (Fig. 1, step 8). However, these HR events are independent of classic DNA-damage signaling, because depletion of Rap1 in mouse cells lacking Ku70 occurs without the induction of TIFs14, thus indicating that the DDR machinery does not recognize such recombinogenic telomeres. In human cells, expression of a TRF2 allele mutated in the Rap1-binding domain (RBM) is sufficient to induce T-SCEs without Ku depletion16. It is however unlikely that Ku has no role in preventing HR at human telomeres, because another study has demonstrated that depletion of Rap1 alone does not lead to T-SCEs15. Induction of T-SCEs by the TRF2ΔRBM allele therefore raises the possibility that this domain also plays a part in the recruitment of Ku to telomeres.
Inhibition of HR is also essential in preventing t-loop excision by X-ray repair cross-complementing protein (XRCC3) and NBS1 (Fig. 1, step 9). Expression of the TRF2ΔB allele, which lacks the N-terminal basic domain, induces loss of the telomeric leading strand and XRCC3-dependent t-circle and C-overhang accumulation, which are believed to arise from t-loop excision39,40. Deletion of Ku86 in human cells leads to dramatic telomere loss and t-circle accumulation that rapidly causes cell death, although in that case it is not known whether t-loop excision is mediated by XRCC3 (ref. 41).
Inhibition of HR at telomeres is complex and redundant, and involves shelterin factors as well as Ku70–Ku80, a heterodimer that actually takes part in c-NHEJ, thus highlighting that many DDR factors also have major roles in telomere protection. This is most evident in the pathways that process telomeres after replication and protect them from excessive exonucleolytic attack.
DDR factors involved in telomere function and maintenance
It has been puzzling why telomeres, which must prevent activation of a DDR throughout most of the cell cycle, attract a variety of proteins with direct roles in the detection and repair of DNA. Recent findings have suggested that these factors are required for telomere processing, telomere replication and establishment of telomere protection.
T-loop unfolding and formation
Whereas t-loop structures protect telomeres against the ATM cascade and NHEJ, specific DDR factors are required for t-loop disassembly during replication and loop formation after S-phase42 as well as t-loop excision during trimming of very long telomeres43 (Fig. 3).
During S phase, TRF2 directly recruits the essential helicase regulator of telomere length 1 (RTEL1) through the interaction of TRF2TRFH and RTEL1C4C4 domains, thus facilitating unfolding of the t loop and enabling replication-fork progression. RTEL1–mediated t-loop disassembly has also been proposed to suppress t-loop formation between sister telomeres in trans. Failure to dismantle t loops during replication leads to excision of the persistent loops, thus resulting in t-circle production and rapid telomere shortening44,45 (Fig. 3b). In human and mouse cells, persistent t-loop excision is mediated by structure-specific endonuclease subunit homolog 4 (SLX4), a molecular scaffold protein that interacts with the DNA-repair endonucleases MUS81–EME1 (crossover-junction endonuclease), the xeroderma pigmentosum F (XPF)–excision repair cross-complementation group 1 (ERCC1) complex and SLX1, and is also involved in interstrand cross-link repair (ICL).
Unlike mouse SLX4, human SLX4 contains a sequence resembling the Apollo TRF2-binding motif, which drives TRF2TRFH-dependent permanent recruitment of the SLX4–MUS81–XPF–SLX1 complex to telomeres46–48. Using the SLX4–SLX1 complex, TRF2 has been proposed to control telomere-length homeostasis by regulating the trimming mechanism, in which very long telomeres are shortened by t-loop excision. TRF2TRFH-dependent recruitment of SLX4 would induce t-loop excision at very long telomeres, whereas the basic domain of TRF2 would inhibit SLX4 and therefore stabilize t loops at normal telomeres43,47,49. Of note, although mouse SLX4 is involved in persistent t-loop excision upon RTEL1 depletion, it does not bear the TRF2-binding motif and does not localize to mouse telomeres46. Mouse SLX4 is therefore not likely to be involved in telomere trimming, thus potentially explaining the very long telomeres tolerated in inbred mouse strains.
After replication and processing, t loops must be reformed, and TRF2 is required for this process in vitro and in vivo8,9. In vitro, TRF2 is sufficient to catalyze the invasion reaction of the single-stranded overhang into duplex TTAGGG repeats, thereby forming the D loop, which stabilizes the t loop. The Rad51 recombinase, which promotes HR by catalyzing D-loop formation, is inhibited by TRF2 but stimulated by TRF1 (ref. 50). The basic domain of TRF2 also inhibits resolvase activities at telomeres, thereby promoting telomere replication and stabilizing t loops48,49. Rad51 is also recruited to telomeres by BRCA2 during G2, when it has been suggested to play a role in facilitating telomere replication51, thus again demonstrating the complex interaction of the HR machinery with telomeres during and after replication.
Overhang processing
The shelterin complex and several DSB-repair factors act together to create and control the single-stranded telomeric overhangs that are required to form the t loop and to inhibit ATM52. Replication of telomeres is unidirectional, because it is primed by an origin located in the subtelomeric region24. As a consequence, lagging- and leading-strand synthesis replicate the G-rich and C-rich strands, respectively. During lagging-strand replication, the most distal RNA primer randomly anneals within 100 nucleotides from the chromosome end, thereby directly creating a single-stranded overhang at the ends of the replication product53. On the opposite (leading) strand, replication produces blunt ends that require 5′-to-3′ end resection to form an overhang. This is achieved by the exonuclease Apollo, which is recruited to telomeres by TRF2 and generates the leading-strand overhang immediately after replication (Fig. 3c). Depletion of Apollo induces chromatid-type fusions between leading-strand telomeres, and a splice variant of Apollo unable to bind TRF2 is one of the factors that can induce Hoyeraal-Hreidarsson telomere disorder syndrome, thus reinforcing the crucial role of Apollo-mediated overhang processing in telomere protection52,54–56. POT1b binding to the overhang inhibits Apollo-mediated resection at the lagging strand as well as at the newly formed leading-strand overhang. During late S-G2, both sister chromatids engage in a second round of overhang processing, in which exonuclease 1 (EXO1) nucleolytic digestion is buffered by CTC–STEN1–TEN1 (CST) complex–mediated fill-in synthesis55,57 (Fig. 3c). Whereas overhang formation is essential in chromosome-end protection, extensive resection must be inhibited because it would otherwise lead to a dramatic acceleration of telomere erosion. Protection against exonucleolytic digestion, which can be mediated by EXO1, Bloom syndrome helicase (BLM), CtIP and probably other factors, is therefore redundantly achieved by several shelterin factors as well as 53BP1 (ref. 35).
Regulation of telomere fusions
The fusion of critically short or deprotected telomeres is dependent on end resection, telomeric transcription, chromatin, and the cell cycle.
Control of resection
Competition between resection and protection of broken DNA ends is a determining event in pathway choice in DSB repair. On the one hand, BRCA1-mediated resection of DSBs by CtIP and EXO1, an initial step of homologous recombination, inhibits c-NHEJ58. On the other hand, recruitment of 53BP1, Rap1 interacting factor (RIF1) and MAD2 mitotic arrest deficient-like 2 (MAD2L2; also known as REV7) blocks end resection, thereby promoting c-NHEJ and preventing HR (Fig. 1, step 10). During this process, RIF1 is directly recruited by the N-terminal phospho-SQ/TQ domain of 53BP1, whereas MAD2L2 acts downstream of these factors and inhibits resection. At dysfunctional telomeres, HR is inhibited by Ku, yet the 53BP1–RIF1–MAD2L2 complex is required to prevent hyperresection of the C-rich strand. Deletion of TRF2 along with 53BP1, RIF1 or MAD2L2 leads to extensive 5′-end resection and suppresses telomere fusions, thus suggesting a crucial role of 53BP1–RIF1–MAD2L2 in end joining of dysfunctional telomeres59–63.
Telomeric chromatin and TERRA
Emerging evidence has suggested that chromatin state influences DSB repair64. Accordingly, the ability of deprotected telomeres to successfully carry out repair by NHEJ also partly depends on chromatin factors. Ring1b, a protein involved in heterochromatin maintenance, is associated with deprotected telomeres and stimulates NHEJ65. Depletion of Ring1b leads to reduced telomere fusions without affecting DNA damage–signaling capacities of chromosome ends65. The long noncoding RNA telomeric repeat–containing RNA (TERRA) has a major role in the control of telomeric chromatin structure, because it recruits SUV39H1, thus leading to an increase in trimethylated histone H3 K9, which is indicative of heterochromatin66–68. A global decrease in this trimethylation leads to reduced fusions at deprotected mouse and human telomeres65,68. Increased levels of TERRA at deprotected telomeres also lead to recruitment of lysine-specific histone demethylase 1 (LSD1), which stimulates nucleolytic processing of G overhangs by MRE11 (ref. 69). TERRA transcription is inhibited by the TRF2TRFH domain, thus adding another layer of NHEJ control by TRF2 (ref. 68).
Cell-cycle regulation
The dynamics of signaling from deprotected telomeres and their fusion are cell-cycle dependent. Unprotected telomeres fuse primarily in G1, thus suggesting active inhibition of NHEJ in other cell-cycle phases; however, how NHEJ is inhibited in a cell cycle–specific manner is clear neither at telomeres nor at intrachromosomal regions. Inhibition of specific repair pathways during specific cell-cycle phases represents a powerful way to control repair-pathway choice, as demonstrated by the finding that during mitosis RNF8 and 53BP1 are inhibited from binding to chromatin, owing to their phosphorylation. Restoration of RNF8 and 53BP1 binding during mitosis leads to sister-telomere fusions and, as a consequence, to dicentric chromosomes and genome instability, thus emphasizing the importance of cell-cycle control of repair pathways at telomeres and nontelomeric regions70. Cell cycle–dependent telomere shortening and deprotection has now been recognized to have major roles in controlling proliferative boundaries and cell fate during cellular aging and in prevention of tumorigenesis.
Control of proliferative boundaries and cellular fate
As discussed above, shelterin components have distinct roles in regulating damage signaling and end-joining pathways at telomeres. Experimental suppression of shelterin components has been highly informative in dissecting the mechanisms of telomere protection. Whereas POT1 mutations have recently been identified in melanomas, gliomas and chronic lymphocytic leukemia71–74, complete deletions of shelterin genes are not frequently observed in tumors. Partial or complete loss of telomere function in vivo is more likely to occur spontaneously as a consequence of extensive erosion, when telomeres become too short to be efficiently protected by shelterin. In humans, this tumor-suppressive mechanism is progressive with cell division and controls proliferative boundaries.
Entry into replicative senescence
In somatic human cells, which repress the expression of telomerase, telomeres shorten with every round of replication until they become critically short, and cells terminally differentiate in senescence (Fig. 4). Telomere shortening is a consequence of progressive erosion due to the end-replication problem, to processing and to rapid, stochastic shortening events caused by replication-fork collapse, t-loop excision or oxidative stress. As a consequence, telomere length is heterogeneous in presenescent cells, in which very short but also very long telomeres are present75. Entry into senescence is driven by the inability of the bulk of these short telomeres to fully protect the chromosome ends from DNA-damage signaling and the resulting production of TIFs. The number of TIFs increases with cellular aging and telomere shortening, but cells keep dividing until they accumulate at least five dysfunctional telomeres, which represent a critical damage threshold that leads to p53 activation and senescence entry76,77 (Fig. 4). At this point, telomeres are too short to be fully functional and are therefore unable to form the protective t-loop structure, but they retain enough TRF2 to prevent fusions. This model of an intermediate telomere state has been illustrated by work in our laboratory demonstrating that progressive suppression of TRF2 by short hairpin RNAs of varying efficiencies leads to TIF accumulation, ATM activation and p53-dependent cell-cycle arrest in G1, but it does not trigger fusions until TRF2 is completely removed78,79.
Figure 4.
Replicative senescence and crisis are two proliferative barriers controlled by telomere deprotection. Young fibroblasts contain long, fully protected telomeres (blue), which erode over the course of cellular division. Some telomeres become dysfunctional and enter the intermediate state of deprotection (orange), which is characterized by activation of the DNA-damage response, yet retain enough protective shelterin to inhibit fusions78,79. Telomere shortening and deprotection increase with population doublings until five or more dysfunctional telomeres are encountered, a damage threshold that activates p53 and leads to entry into replicative senescence77. Upon loss of p53 and retinoblastoma protein (Rb), cells bypass senescence and continue to grow. Telomeres continue to shorten until they become too short to retain any protective properties. Uncapped telomeres (red) fuse, thus leading to prolonged mitotic arrest that amplifies telomere deprotection and causes cell death in crisis89,90.
Fused chromosomes, which are unlikely to be separated properly during mitosis, lead to random chromosome-break events that cause genome instability. Entry into senescence occurs while telomeres still retain the capacity to inhibit fusions, thus representing a protective mechanism against genome instability. Furthermore, the DNA-damage response triggered by damaged telomeres, unlike that triggered by DSBs, does not activate Chk2 phosphorylation and the G2-M checkpoint. As a consequence, TIFs occurring during the final replication cycle before senescence are transmitted through mitosis, thus ensuring that senescent cells stop growth in the following cell cycle in a stable G1 arrest with a diploid genome78.
Although the number of TIFs correlates with bulk telomere shortening, TIFs in individual cells do not necessarily localize to the shortest telomeres75 (Fig. 4). This surprising observation raises the possibility that other types of damage, such as oxidative stress, may accumulate at telomeres over time, thus leading to deprotection of long chromosome ends80,81. Another potential explanation is that telomere shortening slightly and gradually diminishes the efficiency of t-loop formation. According to this model, the shortest telomeres have a higher probability of becoming dysfunctional, but they are not systematically deprotected. Given that there are 92 telomeres in a diploid cell, it is exceedingly unlikely that five telomeres will become dysfunctional when the bulk telomere length is high, but it is quite likely when the bulk length of telomeres is short. In this regard, it would be of great interest to determine the critical telomere length for efficient t-loop folding in vivo and to develop mathematical models to predict telomere deprotection.
Induction of crisis and cell fate
When the tumor suppressors p53 and retinoblastoma protein (Rb) are mutated, cells bypass senescence and continue to divide until crisis, a second proliferative boundary during which almost all of the cells in the population die (Fig. 4). Bypass of senescence is accompanied by further telomere shortening, until some telomeres become so short that they can no longer bind shelterin proteins and undergo fusion with other telomeric loci or with fragile nontelomeric loci. Such events eventually can lead to tumorigenesis, as first demonstrated by deletion of the telomerase template RNA in mice82. The telomere fusions observed in crisis cells and in human tumors display characteristic events of deletion and microhomology and are partially independent of DNA-PKcs and thus occur through both c- and alt-NHEJ83–88.
Breakage-fusion-bridge cycles initiated by telomere fusions have until now been considered to be a cause for the vast cell death that occurs during crisis. Although this model could contribute to crisis-driven genome instability, our laboratory has recently demonstrated that cell death is a consequence of a telomere-deprotection pathway initiated by telomere fusions. After human fibroblasts bypass senescence, fusion of a few very short telomeres causes mitotic arrest, most probably because of failure to segregate fused chromosomes. This prolonged mitotic arrest in turn induces further telomere deprotection through AuroraB-dependent removal of TRF2. The consequence is an amplification of the DNA-damage response, which leads to cell death during mitotic arrest or in the following G1 phase89,90.
Genome instability and transformation
Widespread telomere-induced cell death is the second barrier against tumor formation in cells that escape the first barrier of senescence. Some rare cells, however, manage to escape crisis by activating a telomere-maintenance mechanism through telomerase reactivation or alternative pathways. In these exceptional cases, telomere shortening and dysfunction pathways become protumorigenic and permit the accumulation of genome instability. This has been demonstrated in p53-negative models of constitutive telomere dysfunction, in which genome reduplication and tetraploidy in cells result in the emergence of tumors with subtetraploid karyotypes91,92. High levels of telomere fusions are also associated with poor prognosis in people with breast cancer93. Additionally, the specific pathway responsible for telomere fusion during crisis may account for the cell’s capacity to escape from crisis, because DNA ligase III, but not ligase IV, is essential in escaping crisis, thus highlighting differential roles for alt-NHEJ and c-NHEJ in the fusion of critically short telomeres before crisis87.
Conclusions
Less than two decades ago, TRF2 was discovered as a major protection factor at telomeres. At that time, the field could only dream of the exciting discoveries to come that would reveal the intricate and complex interactions of shelterin with various pathways of DNA-damage detection and repair. Many of these findings were made possible by advances in mouse genetics and the development of modern human and mouse genome-editing techniques, thus suggesting that more exciting discoveries are yet to be made. Telomeres, which were originally thought to be simple chromosome end caps, now must be regarded as delicate elements that use an interplay of protein and RNA complexes with DNA structure, thereby not only regulating numerous enzymatic pathways in cells but also taking advantage of these pathways to control cellular fate, including stem-cell maintenance, differentiation, senescence, crisis and transformation. The next exciting chapter will be to understand the role of telomeres in the regulation of genome-wide processes such as chromatin organization, chromosome structure and chromosomal territories during cellular and organismal aging as well as transformation.
ACKNOWLEDGMENTS
N.A. is supported by the Human Frontier Science Program (LT000284/2013). J.K. is supported by a Salk Institute Cancer Center Core Grant (P30CA014195), the US National Institutes of Health (R01GM087476 and R01CA174942), the Donald and Darlene Shiley Chair, the Highland Street Foundation, the Fritz B. Burns Foundation, the Emerald Foundation and the Glenn Center for Research on Aging.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 2.Karlseder J. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- 3.Smogorzewska A, Karlseder J, Holtgreve-Grez H, Jauch A, de Lange T. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr. Biol. 2002;12:1635–1644. doi: 10.1016/s0960-9822(02)01179-x. [DOI] [PubMed] [Google Scholar]
- 4.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 5.Smogorzewska A, de Lange T. Different telomere damage signaling pathways in human and mouse cells. EMBO J. 2002;21:4338–4348. doi: 10.1093/emboj/cdf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh S, Wang Y, Zimbric J, Hendrickson EA. Human LIGIV is synthetically lethal with the loss of Rad54B-dependent recombination and is required for certain chromosome fusion events induced by telomere dysfunction. Nucleic Acids Res. 2013;41:1734–1749. doi: 10.1093/nar/gks1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffith JD, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 8.Stansel RM, de Lange T, Griffith JD. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 2001;20:5532–5540. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doksani Y, Wu JY, de Lange T, Zhuang X. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell. 2013;155:345–356. doi: 10.1016/j.cell.2013.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okamoto K, et al. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature. 2013;494:502–505. doi: 10.1038/nature11873. This publication highlights the exact mechanisms of ATM-pathway suppression by TRF2, identifying the iDDR region in TRF2 as being inhibitory downstream of ATM, at the level of RNF168 suppression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–528. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, et al. Rapid telomere motions in live human cells analyzed by highly time-resolved microscopy. Epigenetics Chromatin. 2008;1:4. doi: 10.1186/1756-8935-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ribes-Zamora A, Indiviglio SM, Mihalek I, Williams CL, Bertuch AA. TRF2 interaction with Ku heterotetramerization interface gives insight into c-NHEJ prevention at human telomeres. Cell Reports. 2013;5:194–206. doi: 10.1016/j.celrep.2013.08.040. This manuscript explains the presence of Ku at protected telomeres and how c-NHEJ is suppressed despite the presence of Ku. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science. 2010;327:1657–1661. doi: 10.1126/science.1185100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kabir S, Hockemeyer D, de Lange T. TALEN gene knockouts reveal no requirement for the conserved human shelterin protein Rap1 in telomere protection and length regulation. Cell Reports. 2014;9:1273–1280. doi: 10.1016/j.celrep.2014.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, et al. A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms. Nat. Struct. Mol. Biol. 2011;18:213–221. doi: 10.1038/nsmb.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarthy J, Bae NS, Scrafford J, Baumann P. Human RAP1 inhibits non-homologous end joining at telomeres. EMBO J. 2009;28:3390–3399. doi: 10.1038/emboj.2009.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arat NÖ, Griffith JD. Human Rap1 interacts directly with telomeric DNA and regulates TRF2 localization at the telomere. J. Biol. Chem. 2012;287:41583–41594. doi: 10.1074/jbc.M112.415984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janoušková E, et al. Human Rap1 modulates TRF2 attraction to telomeric DNA. Nucleic Acids Res. 2015;43:2691–2700. doi: 10.1093/nar/gkv097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 21.Hockemeyer D, Palm W, Wang RC, Couto SS, de Lange T. Engineered telomere degradation models dyskeratosis congenita. Genes Dev. 2008;22:1773–1785. doi: 10.1101/gad.1679208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takai KK, Kibe T, Donigian JR, Frescas D, de Lange T. Telomere protection by TPP1/POT1 requires tethering to TIN2. Mol. Cell. 2011;44:647–659. doi: 10.1016/j.molcel.2011.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong Y, de Lange TA. Shld1-controlled POT1a provides support for repression of ATR signaling at telomeres through RPA exclusion. Mol. Cell. 2010;40:377–387. doi: 10.1016/j.molcel.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sfeir A, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zimmermann M, Kibe T, Kabir S, de Lange T. TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling. Genes Dev. 2014;28:2477–2491. doi: 10.1101/gad.251611.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnoult N, Saintome C, Ourliac-Garnier I, Riou JF, Londono-Vallejo A. Human POT1 is required for efficient telomere C-rich strand replication in the absence of WRN. Genes Dev. 2009;23:2915–2924. doi: 10.1101/gad.544009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang M, et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh S, et al. DNA ligase III and DNA ligase IV carry out genetically distinct forms of end joining in human somatic cells. DNA Repair (Amst.) 2014;21:97–110. doi: 10.1016/j.dnarep.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mateos-Gomez PA, et al. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature. 2015;518:254–257. doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ceccaldi R, et al. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature. 2015;518:258–262. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kent T, Chandramouly G, McDevitt SM, Ozdemir AY, Pomerantz RT. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase θ. Nat. Struct. Mol. Biol. 2015;22:230–237. doi: 10.1038/nsmb.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yousefzadeh MJ, et al. Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet. 2014;10:e1004654. doi: 10.1371/journal.pgen.1004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rai R, et al. The function of classical and alternative non-homologous end-joining pathways in the fusion of dysfunctional telomeres. EMBO J. 2010;29:2598–2610. doi: 10.1038/emboj.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frit P, Barboule N, Yuan Y, Gomez D, Calsou P. Alternative end-joining pathway(s): bricolage at DNA breaks. DNA Repair (Amst.) 2014;17:81–97. doi: 10.1016/j.dnarep.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 35.Sfeir A, de Lange T. Removal of shelterin reveals the telomere end-protection problem. Science. 2012;336:593–597. doi: 10.1126/science.1218498. This publication reports the complete removal of shelterin from telomeres, thereby establishing that the chromosome end-protection problem is specified by six DNA-damage and DNA-repair pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Celli GB, Denchi EL, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat. Cell Biol. 2006;8:885–890. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- 37.Rai R, et al. The E3 ubiquitin ligase Rnf8 stabilizes Tpp1 to promote telomere end protection. Nat. Struct. Mol. Biol. 2011;18:1400–1407. doi: 10.1038/nsmb.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palm W, Hockemeyer D, Kibe T, de Lange T. Functional dissection of human and mouse POT1 proteins. Mol. Cell. Biol. 2009;29:471–482. doi: 10.1128/MCB.01352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oganesian L, Karlseder J. 5′ C-rich telomeric overhangs are an outcome of rapid telomere truncation events. DNA Repair (Amst.) 2013;12:238–245. doi: 10.1016/j.dnarep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Ghosh G, Hendrickson EA. Ku86 represses lethal telomere deletion events in human somatic cells. Proc. Natl. Acad. Sci. USA. 2009;106:12430–12435. doi: 10.1073/pnas.0903362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 43.Pickett HA, Cesare AJ, Johnston RL, Neumann AA, Reddel RR. Control of telomere length by a trimming mechanism that involves generation of t-circles. EMBO J. 2009;28:799–809. doi: 10.1038/emboj.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vannier J-B, Pavicic-Kaltenbrunner V, Petalcorin MIR, Ding H, Boulton SJ. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149:795–806. doi: 10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 45.Sarek G, Vannier JB, Panier S, Petrini JH, Boulton SJ. TRF2 recruits RTEL1 to telomeres in S phase to promote t-loop unwinding. Mol. Cell. 2015;57:622–635. doi: 10.1016/j.molcel.2014.12.024. This publication elaborates on how RTEL1 is recruited to telomeres by TRF2 in a cell cycle–specific manner, explaining how catastrophic t-loop processing is avoided in S phase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson JS, et al. Localization-dependent and -independent roles of SLX4 in regulating telomeres. Cell Reports. 2013;4:853–860. doi: 10.1016/j.celrep.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wan B, et al. SLX4 assembles a telomere maintenance toolkit by bridging multiple endonucleases with telomeres. Cell Reports. 2013;4:861–869. doi: 10.1016/j.celrep.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarkar J, et al. SLX4 contributes to telomere preservation and regulated processing of telomeric joint molecule intermediates. Nucleic Acids Res. 2015;43:5912–5923. doi: 10.1093/nar/gkv522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saint-Léger A, et al. The basic N-terminal domain of TRF2 limits recombination endonuclease action at human telomeres. Cell Cycle. 2014;13:2469–2474. doi: 10.4161/cc.29422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bower BD, Griffith JD. TRF1 and TRF2 differentially modulate Rad51-mediated telomeric and nontelomeric displacement loop formation in vitro. Biochemistry. 2014;53:5485–5495. doi: 10.1021/bi5006249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Badie S, et al. BRCA2 acts as a RAD51 loader to facilitate telomere replication and capping. Nat. Struct. Mol. Biol. 2010;17:1461–1469. doi: 10.1038/nsmb.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu P, van Overbeek M, Rooney S, de Lange T. Apollo contributes to G overhang maintenance and protects leading-end telomeres. Mol. Cell. 2010;39:606–617. doi: 10.1016/j.molcel.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chow TT, Zhao Y, Mak SS, Shay JW, Wright WE. Early and late steps in telomere overhang processing in normal human cells: the position of the final RNA primer drives telomere shortening. Genes Dev. 2012;26:1167–1178. doi: 10.1101/gad.187211.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lam YC, et al. SNMIB/Apollo protects leading-strand telomeres against NHEJ-mediated repair. EMBO J. 2010;29:2230–2241. doi: 10.1038/emboj.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu P, Takai H, de Lange T. Telomeric 3′ overhangs derive from resection by Exo1 and Apollo and fill-in by POT1b-associated CST. Cell. 2012;150:39–52. doi: 10.1016/j.cell.2012.05.026. This publication analyzes the steps of overhang generation after telomere replication, elaborating on the balance between resection and polymerization of the lagging-strand product. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Touzot F, et al. Function of Apollo (SNM1B) at telomere highlighted by a splice variant identified in a patient with Hoyeraal-Hreidarsson syndrome. Proc. Natl. Acad. Sci. USA. 2010;107:10097–10102. doi: 10.1073/pnas.0914918107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dai X, et al. Molecular steps of G-overhang generation at human telomeres and its function in chromosome end protection. EMBO J. 2010;29:2788–2801. doi: 10.1038/emboj.2010.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 59.Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science. 2013;339:700–704. doi: 10.1126/science.1231573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chapman JR, et al. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell. 2013;49:858–871. doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Di Virgilio M, et al. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013;339:711–715. doi: 10.1126/science.1230624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boersma V, et al. MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5′ end resection. Nature. 2015;521:537–540. doi: 10.1038/nature14216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu G, et al. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature. 2015;521:541–544. doi: 10.1038/nature14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clouaire T, Legube G. DNA double strand break repair pathway choice: a chromatin based decision? Nucleus. 2015;6:107–113. doi: 10.1080/19491034.2015.1010946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bartocci C, et al. Isolation of chromatin from dysfunctional telomeres reveals an important role for Ring1b in NHEJ-mediated chromosome fusions. Cell Reports. 2014;7:1320–1332. doi: 10.1016/j.celrep.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arnoult N, Van Beneden A, Decottignies A. Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1α. Nat. Struct. Mol. Biol. 2012;19:948–956. doi: 10.1038/nsmb.2364. [DOI] [PubMed] [Google Scholar]
- 68.Porro A, et al. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat. Commun. 2014;5:5379. doi: 10.1038/ncomms6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Porro A, Feuerhahn S, Lingner J. TERRA-reinforced association of LSD1 with MRE11 promotes processing of uncapped telomeres. Cell Reports. 2014;6:765–776. doi: 10.1016/j.celrep.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 70.Orthwein A, et al. Mitosis inhibits DNA double-strand break repair to guard against telomere fusions. Science. 2014;344:189–193. doi: 10.1126/science.1248024. This publication highlights the importance of excluding 53BP1 and RNF8 from chromatin during mitosis, because tethering of these factors to DNA causes telomere fusions. [DOI] [PubMed] [Google Scholar]
- 71.Ramsay AJ, et al. POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat. Genet. 2013;45:526–530. doi: 10.1038/ng.2584. [DOI] [PubMed] [Google Scholar]
- 72.Bainbridge MN, et al. Germline mutations in shelterin complex genes are associated with familial glioma. J. Natl. Cancer Inst. 2015;107:384. doi: 10.1093/jnci/dju384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Robles-Espinoza CD, et al. POT1 loss-of-function variants predispose to familial melanoma. Nat. Genet. 2014;46:478–481. doi: 10.1038/ng.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shi J, et al. Rare missense variants in POT1 predispose to familial cutaneous malignant melanoma. Nat. Genet. 2014;46:482–486. doi: 10.1038/ng.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baird DM, Rowson J, Wynford-Thomas D, Kipling D. Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat. Genet. 2003;33:203–207. doi: 10.1038/ng1084. [DOI] [PubMed] [Google Scholar]
- 76.Cesare AJ, et al. Spontaneous occurrence of telomeric DNA damage response in the absence of chromosome fusions. Nat. Struct. Mol. Biol. 2009;16:1244–1251. doi: 10.1038/nsmb.1725. [DOI] [PubMed] [Google Scholar]
- 77.Kaul Z, Cesare AJ, Huschtscha LI, Neumann AA, Reddel RR. Five dysfunctional telomeres predict onset of senescence in human cells. EMBO Rep. 2012;13:52–59. doi: 10.1038/embor.2011.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cesare AJ, Hayashi MT, Crabbe L, Karlseder J. The telomere deprotection response is functionally distinct from the genomic DNA damage response. Mol. Cell. 2013;51:141–155. doi: 10.1016/j.molcel.2013.06.006. Here the authors demonstrate that partially deprotected telomeres do not activate the G2 DDR, explaining why senescent cells arrest in G1 with a diploid genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cesare AJ, Karlseder J. A three-state model of telomere control over human proliferative boundaries. Curr. Opin. Cell Biol. 2012;24:731–738. doi: 10.1016/j.ceb.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Opresko PL, Fan J, Danzy S, Wilson DM, III, Bohr VA. Oxidative damage in telomeric DNA disrupts recognition by TRF1 and TRF2. Nucleic Acids Res. 2005;33:1230–1239. doi: 10.1093/nar/gki273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hewitt G, et al. Telomeres are favoured targets of a persistent DNA damage response in ageing and stress-induced senescence. Nat. Commun. 2012;3:708. doi: 10.1038/ncomms1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Artandi SE, et al. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 83.Lin TT, et al. Telomere dysfunction and fusion during the progression of chronic lymphocytic leukemia: evidence for a telomere crisis. Blood. 2010;116:1899–1907. doi: 10.1182/blood-2010-02-272104. [DOI] [PubMed] [Google Scholar]
- 84.Roger L, et al. Extensive telomere erosion in the initiation of colorectal adenomas and its association with chromosomal instability. J. Natl. Cancer Inst. 2013;105:1202–1211. doi: 10.1093/jnci/djt191. [DOI] [PubMed] [Google Scholar]
- 85.Capper R, et al. The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev. 2007;21:2495–2508. doi: 10.1101/gad.439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Letsolo BT, Rowson J, Baird DM. Fusion of short telomeres in human cells is characterized by extensive deletion and microhomology, and can result in complex rearrangements. Nucleic Acids Res. 2010;38:1841–1852. doi: 10.1093/nar/gkp1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jones RE, et al. Escape from telomere-driven crisis is DNA ligase III dependent. Cell Reports. 2014;8:1063–1076. doi: 10.1016/j.celrep.2014.07.007. In this publication, the authors analyze the requirements for escape from telomere-driven crisis and find that suppression of ligase III, but not of ligase IV, is necessary. [DOI] [PubMed] [Google Scholar]
- 88.Maser RS, et al. DNA-dependent protein kinase catalytic subunit is not required for dysfunctional telomere fusion and checkpoint response in the telomerase-deficient mouse. Mol. Cell. Biol. 2007;27:2253–2265. doi: 10.1128/MCB.01354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hayashi MT, Cesare AJ, Fitzpatrick JA, Lazzerini-Denchi E, Karlseder J. A telomere-dependent DNA damage checkpoint induced by prolonged mitotic arrest. Nat. Struct. Mol. Biol. 2012;19:387–394. doi: 10.1038/nsmb.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hayashi MT, Cesare AJ, Rivera T, Karlseder J. Cell death during crisis is mediated by mitotic telomere deprotection. Nature. 2015;522:492–496. doi: 10.1038/nature14513. In this publication, the authors discover that few telomere fusions in precrisis cells lead to mitotic arrest. During mitotic arrest, telomere dysfunction is amplified, thus causing cell death in crisis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Davoli T, de Lange T. Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell. 2012;21:765–776. doi: 10.1016/j.ccr.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Davoli T, Denchi EL, de Lange T. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell. 2010;141:81–93. doi: 10.1016/j.cell.2010.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Simpson K, et al. Telomere fusion threshold identifies a poor prognostic subset of breast cancer patients. Mol. Oncol. 2015;9:1186–1193. doi: 10.1016/j.molonc.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vannier JB, et al. RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science. 2013;342:239–242. doi: 10.1126/science.1241779. [DOI] [PubMed] [Google Scholar]
- 95.Ye J, et al. TRF2 and apollo cooperate with topoisomerase 2alpha to protect human telomeres from replicative damage. Cell. 2010;142:230–242. doi: 10.1016/j.cell.2010.05.032. [DOI] [PubMed] [Google Scholar]
- 96.Broccoli D, Smogorzewska A, Chong L, de Lange T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 1997;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- 97.Bilaud T, et al. Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 1997;17:236–239. doi: 10.1038/ng1097-236. [DOI] [PubMed] [Google Scholar]
- 98.Chen Y, et al. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science. 2008;319:1092–1096. doi: 10.1126/science.1151804. [DOI] [PubMed] [Google Scholar]
- 99.Lenain C, et al. The Apollo 5′ exonuclease functions together with TRF2 to protect telomeres from DNA repair. Curr. Biol. 2006;16:1303–1310. doi: 10.1016/j.cub.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 100.van Overbeek M, de Lange T. Apollo, an Artemis-related nuclease, interacts with TRF2 and protects human telomeres in S phase. Curr. Biol. 2006;16:1295–1302. doi: 10.1016/j.cub.2006.05.022. [DOI] [PubMed] [Google Scholar]