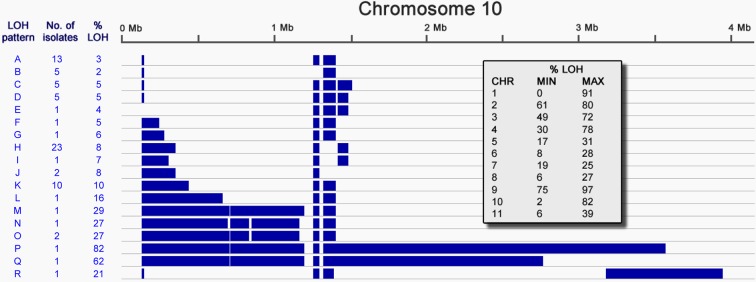
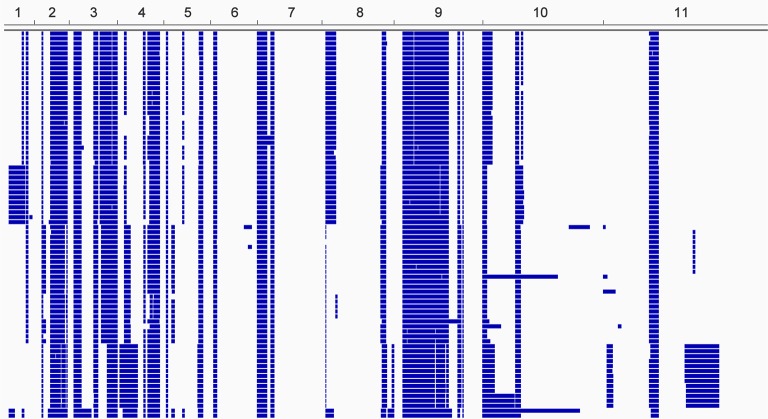
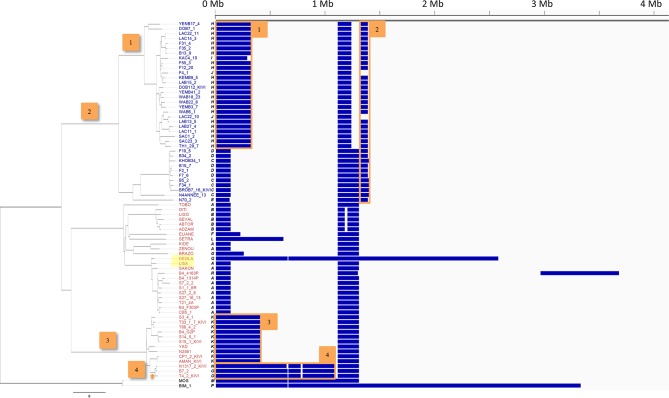
Figure 2. Loss of heterozygosity on chromosome 10.
Loss of heterozygosity regions (blue) spanning Chromosome 10 show 18 different patterns (A-R). The number of isolates possessing each pattern and the percentage of the chromosome affected are indicated. The table (inset) shows the extent of LOH (min and max) for each chromosome as a percentage of chromosome length.