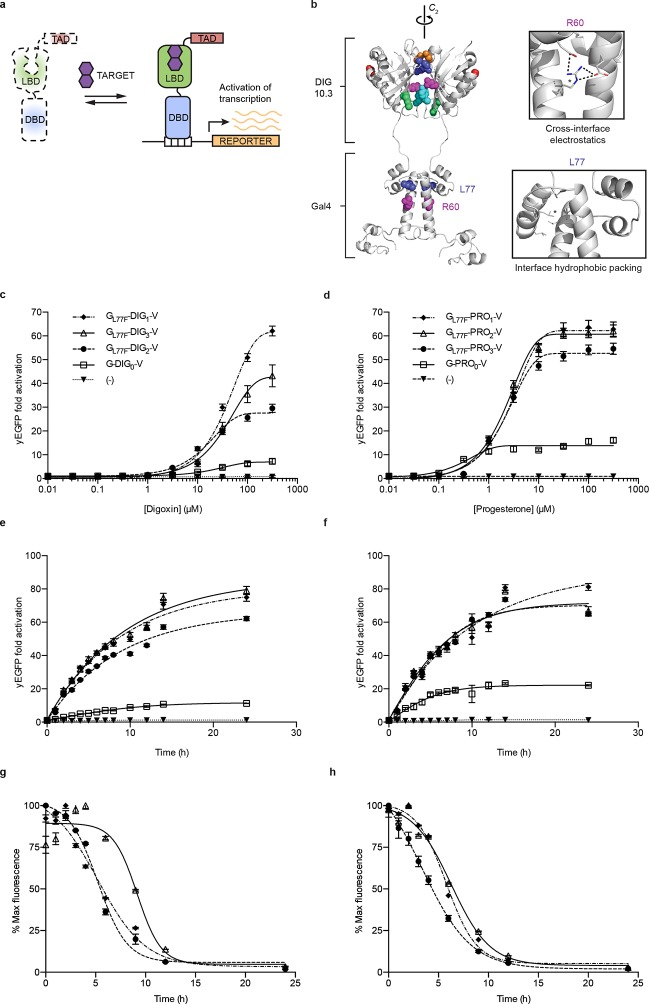
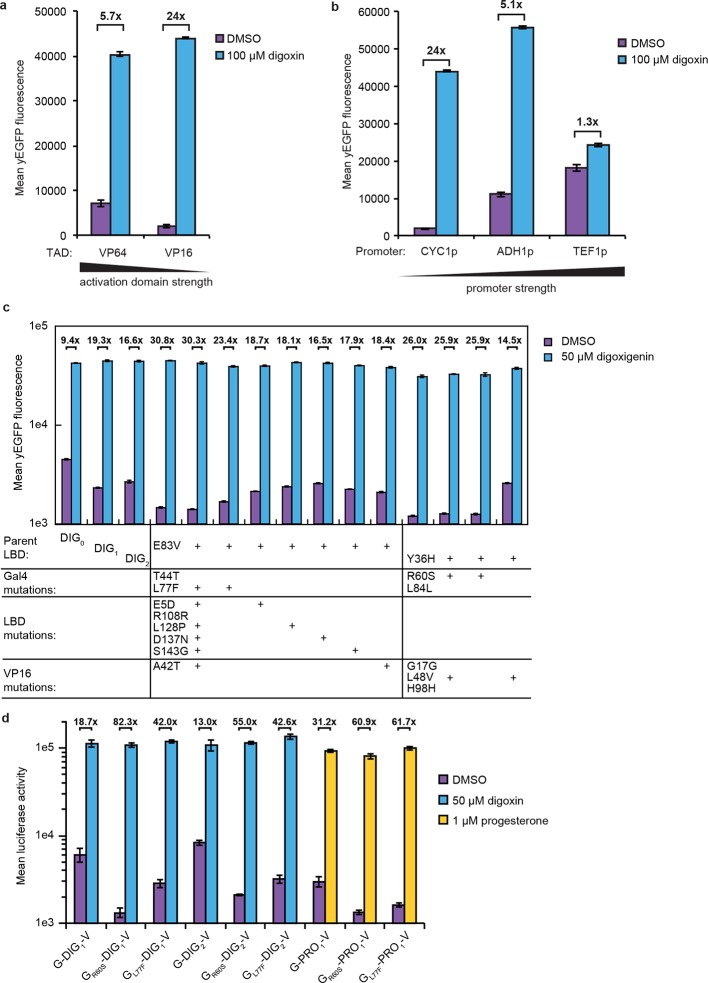
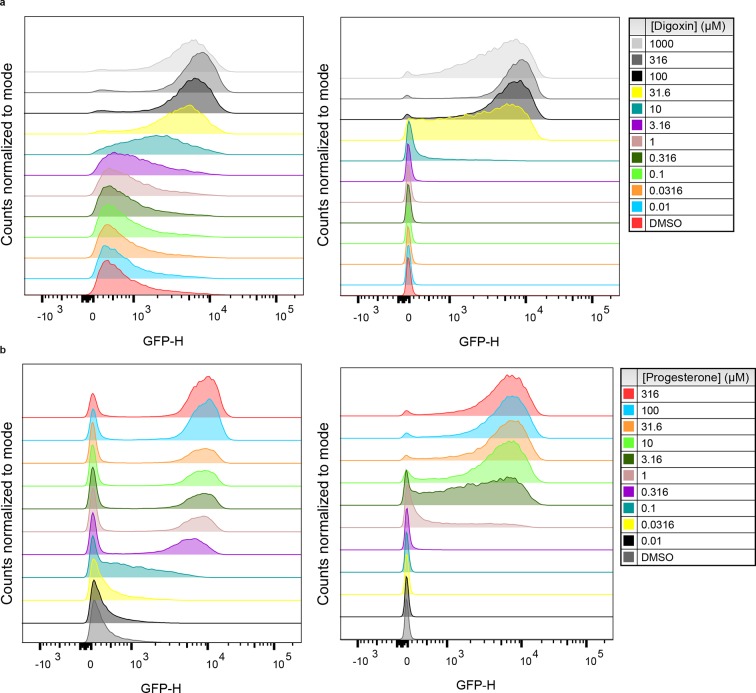
Figure 2. Ligand-dependent transcriptional activation.
(a) TF-biosensor construction from a conditionally destabilized LBD, a DNA-binding domain and a transcriptional activation (TAD) domain. (b) Positions of conditionally destabilizing mutations of Gal4 mapped to a computational model of Gal4-DIG0 homodimer. Residues are shown as colored spheres and key interactions are highlighted in insets. The TAD is not shown. (c) Concentration dependence of response to digoxin for digoxin TF-biosensors driving yEGFP expression. (d) Concentration dependence of response to progesterone for progesterone TF-biosensors driving yEGFP expression. (e) Time dependence of response to 250 μM digoxin for digoxin TF-biosensors. (f) Time dependence of response to 50 μM progesterone for progesterone TF-biosensors. (g) Time-dependent response to withdrawal of 250 μM digoxin for digoxin TF-biosensors. (h) Time-dependent response to withdrawal of 50 μM progesterone for progesterone TF-biosensors. In c-f, (-) indicates cells lacking biosensor plasmids and error bars represent s.e.m. of three biological replicates. Marker symbols in e and g are the same as in c. Marker symbols in f and h are the same as in d.