Abstract
Objective
Hydroxyurea has proven laboratory and clinical therapeutic benefits for sickle cell anemia (SCA) and other diseases, yet many questions remain regarding its in vivo pharmacokinetic and pharmacodynamic profiles. Previous reports suggest that hydroxyurea passively diffuses across cells, but its observed rapid absorption and distribution are more consistent with facilitated or active transport. We investigated the potential role of solute carrier (SLC) transporters in cellular uptake and accumulation of hydroxyurea.
Materials and Methods
Passive diffusion of hydroxyurea across cell membranes was determined using the parallel artificial membrane permeability assay. SLC transporter screens were conducted using in vitro intracellular drug accumulation and transcellular transport assays in cell lines and oocytes overexpressing SLC transporters. Gene expression of SLC transporters was measured by real-time PCR in human tissues and cell lines.
Results
Hydroxyurea had minimal diffusion across a lipid bilayer but was a substrate for 5 different SLC transporters belonging to the OCTN and OATP families of transporters and urea transporters A and B. Further characterization of hydroxyurea transport revealed that cellular uptake by OATP1B3 is time and temperature dependent and inhibited by known substrates of OATP1B3. Urea transporters A and B are expressed differentially in human tissues and erythroid cells, and transport hydroxyurea bidirectionally via facilitated diffusion.
Conclusions
These studies provide new insight into drug transport proteins that may be involved in the in vivo absorption, cellular distribution, and elimination of hydroxyurea. Elucidation of hydroxyurea transcellular movement should improve our understanding of its pharmacokinetics and pharmacodynamics, and may help explain some of the inter-patient drug variability observed in patients with SCA.
INTRODUCTION
Hydroxyurea was initially developed therapeutically in the 1960’s as an antineoplastic agent[1], but several decades later was found to have potential benefit for patients with sickle cell anemia (SCA)[2,3]. Subsequent studies demonstrated the laboratory and clinical efficacy of hydroxyurea for this disorder, primarily by increasing fetal hemoglobin (HbF)[4–7]. The drug received US FDA-approval in 1998 and European EMA-approval in 2007 for the treatment of SCA. Hydroxyurea acts as a potent ribonucleotide reductase inhibitor[8] that diminishes intracellular ribonucleotide pools and thereby affects cell proliferation and survival[9]. HbF induction is believed to involve guanylyl cyclase activation with altered erythroid kinetics[10], although other potential mechanisms of drug action may also exist[11].
Although hydroxyurea has clear therapeutic benefit for SCA and other diseases, many questions remain regarding its pharmacokinetic and pharmacodynamic profiles. Early pharmacokinetic studies of hydroxyurea in mice identified wide organ distribution including the liver, kidney, bladder, and intestine with a substantial portion excreted unchanged in the urine[12]. Additional studies documented hydroxyurea penetration to the CSF and brain[13]. In humans, hydroxyurea is rapidly absorbed orally with a Tmax of 1.22 hours and nearly complete bioavailability[14]. However, considerable variability exists for hydroxyurea pharmacokinetic parameters among individual patients[4,14–16], as well as pharmacodynamic parameters such as HbF level and maximum tolerated dose [7,11,17]. This observed variability could reflect differences in the transcellular transport of hydroxyurea, which is poorly understood. Hydroxyurea has been reported to enter cells via passive diffusion[18–20], but as a polar and hydrophilic molecule, a slow rate of passive diffusion across cell membranes would be predicted[21]. Instead, the rapid absorption and distribution profile of hydroxyurea is more consistent with a facilitated transport mechanism.
The solute carrier (SLC) gene superfamily encodes a variety of membrane transport proteins with significant roles in pharmacokinetic and pharmacodynamic profiles of many drugs, by influencing their absorption, metabolism, distribution, or elimination[22,23]. SLC transporters are widely distributed, but renal and hepatic transporter expression likely has the largest impact on drug pharmacology[24]. SLC-family proteins such as organic anion transporters (OATs), organic cation transporters (OCTs), organic cation/carnitine transporters (OCTNs) and organic anion transporting polypeptides (OATPs) are collectively called “drug transporters” because of their ability to transport xenobiotics in addition to ions, peptides and proteins of variable size, shape, and charge[25]. Urea transporters (UT) also belong to the SLC superfamily; UT proteins transport endogenous urea across cell membranes and are critical for urea recycling, urine concentration, and kidney function[26]. Limited studies suggest urea analogs including hydroxyurea can be substrates for UT[27,28].
The present study was designed to identify and characterize SLC transporters that may be involved in the cellular uptake and transcellular movement of hydroxyurea. Our in vitro uptake and transport assays identify hydroxyurea as a substrate for specific SLC drug transporters, including the urea transporters. These studies provide new insight into transport proteins that may be involved in vivo in the absorption, cellular distribution, and elimination of hydroxyurea. Elucidation of hydroxyurea transcellular movements should improve our understanding of its pharmacokinetics and pharmacodynamics, and may help explain inter-patient drug variability in SCA.
METHODS
Parallel artificial membrane permeability assay (PAMPA)
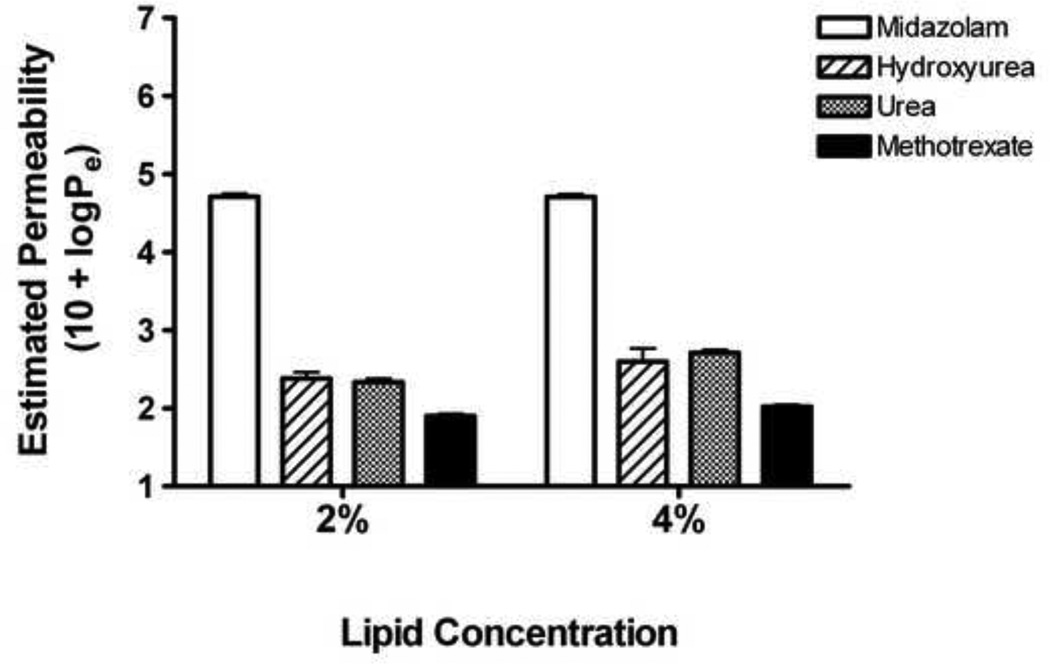
Passive diffusion of hydroxyurea across an artificial lipid membrane was investigated using the PAMPA (Millipore, Billerica, MA). For these experiments, PVDF filter membranes in a MultiScreen-IP PAMPA 96-well filter plate were coated with lipid layer consisting of 2% or 4% synthetic phospholipid blend (Avanti Polar Lipids, Alabaster, AL) mixed in dodecane organic solvent (Sigma, St Louis, MO). The following drugs and concentrations were placed in individual wells: 0.1–1.0µM 3H-midazolam (Moravek Biochemicals, Brea, CA), 50–500µM hydroxyurea (Sigma, St Louis, MO) spiked with 14C-hydroxyurea (American Radiolabeled Chemicals, St Louis, MO), 50–500µM urea (Fisher Scientific, Fairlawn, NJ) with 14C-urea (American Radiolabeled Chemicals), or 50–500µM methotrexate (Sigma) with 3H-methotrexate (Moravek Biochemicals). All drugs were dissolved in phosphate buffered solution (PBS) containing 5% DMSO. Filter plates were placed inside a 96-well PTFE acceptor plate (Millipore) filled with PBS/DMSO. After 16-hours incubation at room temperature, radioactivity was measured from the acceptor compartment to quantify drug diffusing across the artificial lipid membrane. The estimated permeability rate (log Pe) was calculated using the modified equation: log Pe= log {C−ln(1−DPMacceptor/DPMdrug eq)} as described[29]. Results are reported as 10 + log Pe so that higher values represent increased permeability across the lipid bilayer.
Cell lines and oocytes
The cell lines 3T3, Caco-2, HepG2, HEK293, and LLC-PK1 were maintained in DMEM supplemented with 10% FBS and 1% antibiotic/antimycotic (Invitrogen, Carlsbad, CA) at 37°C under 5% CO2. HEK293 cells that are stably transfected to overexpress transporters hOAT1, hOAT2, and hOAT3 (Dr. Yuichi Sugiyama, University of Tokyo); hOCT1, hOCT2, and hOCT3 (Dr. Heinz Bonisch, University of Bonn); hOCTN1 and hOCTN2 (Dr. Akira Tsuji, Kanazawa University) were used to evaluate in vitro hydroxyurea uptake. Transporter-overexpressing cell lines and pcDNA vector-transfected cells were maintained in DMEM with 10% FBS and G418 sulfate (400–800 µg/mL) at 37°C under 5% CO2. UTA or UTB-overexpressing cells were created by stably transfecting LLC-PK1 or HEK293 cells with UTA or UTB cDNA fragments spliced from the TrueClone plasmid (OriGene Technologies, Rockville, MD) and cloned into pIRES2-EGFP vector (BD Biosciences, Franklin Lakes, NJ) to create pIRES-UTA, and pIRES-UTB plasmids. After plasmid transfection using lipofectamine (Invitrogen), GFP-expressing cells containing pIRES-UTA or pIRES-UTB were sorted and maintained in DMEM supplemented with 10% FBS and G418. UTA and UTB overexpression was confirmed by real-time PCR using TaqMan® probes specific for UTA (Hs00998199_M1) or UTB (Hs00415414_M1) (Applied Biosystems, Carlsbad, CA). Oocytes from Xenopus laevis injected with cRNA to overexpress hOATP1A2, hOATP1B1, hOATP1B3 and water-injected control oocytes (BD Biosciences) were maintained in ND96 buffer at RT. Functional overexpression of transporters in oocytes was confirmed by uptake studies performed by the manufacturer prior to shipping each order.
Drug accumulation in human cell lines
3T3, Caco-2, HepG2, HEK 293, LLC-pK1, and HEK293 cell lines overexpressing transporter proteins were used to evaluate intracellular accumulation of hydroxyurea or prototypical drug substrates using uptake assays. Functional overexpression of each transporter was confirmed in all cell lines by performing uptake experiments using a prototypical substrate specific for each transporter. The substrates used were as follows: p-aminohippuric acid (3µM) for hOAT1–2 cells, estrone-3-sulfate (2µM) for OAT 3 cells, tetraethylammonium (10µM) for OCT 1–3 cells, and Carnitine (0.01 µM) for OCTN 1–2 cells. Cells were seeded in 6-well plates at 106 cells/well; at 95% confluence, DMEM containing radioactive drug was added. After incubation for a specified time (5–90 minutes) at 37°C, cells were washed 3 times with cold PBS, trypsinized, and lysed in 1.0 M NaOH. Total protein concentration of the cell lysate was measured (Bicinchoninic Acid Protein Assay kit, Thermo Fisher Scientific, Rockford, IL), and radioactivity was measured using an LS6500 scintillation counter (Beckman Coulter). Intracellular drug accumulation was determined by normalizing lysate 14C to total protein.
Hydroxyurea accumulation in oocytes
Hydroxyurea accumulation was measured in oocytes injected with OATP1A2, OATP1B1, OATP1B3 transporter cRNA or water. Oocytes were incubated in Transportocyte Sodium Buffer (pH 7.4; BD Biosciences) containing 50µM 14C-Hydroxyurea and washed 4 times in cold buffer; individual oocytes were lysed in 10% SDS and counted. For inhibition studies, 1mM of naringin (Sigma, St Louis, MO) or rifampin (Sigma, St Louis, MO) dissolved in 1% DMSO solution or 1mM methotrexate (Sigma, St Loius, MO) dissolved in water was added to hydroxyurea uptake medium. 1% DMSO solution was used as vehicle control. Inhibition is expressed as a percent of the OATP1B3-mediated hydroxyurea accumulation in the absence of all inhibitors.
Transcellular transport assay
LLC-PK1 cells transfected with pIRES2-UTA (LLC-UTA), pIRES2-UTB (LLC-UTB), or pIRES2 empty vector (LLC-pIRES) were used to evaluate transcellular movement of hydroxyurea. Cells were seeded at 2×106 cells/well on a 6-well transwell plate with 3.0µm pore size insert (Corning, Lowell, MA) in DMEM culture media supplemented 10% FBS and 400 ug/ml G418. After 3 days, media on the apical side was replaced and media on the basal side was removed. Plates were observed 15–30 min later for leakage of medium into the basal compartment. Upon confirming that no leaks were present, fresh DMEM media containing 14C-hydroxyurea was added to the appropriate compartment and the experiment was initiated. After incubation, radioactivity in media aliquots from the opposite compartment was measured. For inhibition studies, 1mM dimethylurea (Sigma, St Louis, MO) was added to the uptake medium containing hydroxyurea. The apparent Permeability coefficient (Papp) was calculated as described[30] and reported in cm/sec.
Isolation of human reticulocytes
CD71+ reticulocytes were isolated from children with SCA or from healthy normal adult volunteers. Children with SCA on hydroxyurea therapy were enrolled in the Hydroxyurea Study of Long-term Effects clinical protocol (HUSTLE, ClinicalTrials.gov NCT00305175). The HUSTLE protocol was approved by the Institutional Review Board and all patient families gave their written consent. As part of this protocol, whole blood was collected from each patient and centrifuged to remove platelets present in the plasma supernatant. Each sample was then depleted of CD45+ cells and enriched for CD71+cells by autoMACS magnetic cell sorter (Miltenyi Biotec, Auburn, CA). Purity of isolated CD71+ reticuloctyes was confirmed to be greater than 90% by flow cytometry analysis using fluorescently labeled antibodies to the cell surface antigens CD235a (glycophorin A), CD71 (transferrin receptor), and CD45 (LCA) (BD Biosciences, San Diego, CA).
Real-time PCR
Transporter mRNA expression was measured in cells and human tissues by real-time PCR using Taqman Gene Expression Primer and Probes for OATP1B1, OATP1B3, UTA and UTB (Applied Biosystems, Carlsbad, CA). Total RNA was isolated from 3T3, HepG2, Caco-2, and K562 cell lines or from human CD71+ cells using miRNeasy isolation kit (Qiagen, Valencia, CA). For each sample, the number of transcripts for the specific transporter was calculated using a standard curve generated from DNA plasmids containing coding sequences for the transporter. The transcript levels were normalized to the endogenous control gene β-actin. UTA and UTB mRNA expression was measured in human tissue cDNA (Origene Technologies, Rockville, MD) and human beta glucuronidase (GUSB) was included as an endogenous control. Relative mRNA expression levels in the human tissue samples were determined by comparative CT analysis that normalizes transcript levels and calculates UTA and UTB expression compared to a selected reference tissue known to express the transporters (kidney and bone marrow, respectively).
Statistics
Differences in hydroxyurea accumulation in cell lines were analyzed by two-tailed t-test statistical analysis, calculated using combined data from at least 3 independent experiments. Repeated-measure t-tests were used to determine differences in oocyte time course experiments. Changes in hydroxyurea accumulation in oocytes with OATP1B3 inhibitors were determined by one-way analysis of variance followed by Dunnett’s adjustment for multiple comparisons. For UT transwell time course experiments, differences between transporter-expressing cells and vector controls were analyzed by one-way repeated measures analysis of variance followed by Bonferroni’s adjustment for multiple comparisons. Transporter mRNA levels in cell lines were compared by one way analysis of variance followed by Bonferroni’s test.
RESULTS
PAMPA experiments
Hydroxyurea permeability across the artificial lipid bilayer was compared to drugs with known mechanisms of transcellular movement: urea and methotrexate (SLC-mediated facilitated transport) and midazolam (passive diffusion). The estimated permeability rate (log Pe + 10) across a 2% lipid bilayer for hydroxyurea (2.385) was lower than midazolam (4.710) and much closer to urea (2.335) and methotrexate (1.902), indicating that hydroxyurea does not readily diffuse across simple lipid membranes (Figure 1). Similar trends were seen for drug diffusion across 4% lipid bilayer where 10 + logPe values for midazolam, hydroxyurea, urea, and methotrexate were 4.707, 2.597, 2.712, and 2.022 respectively.
Figure 1. Limited permeability of hydroxyurea across lipid bilayers.
Hydroxyurea movement across synthetic lipid bilayers (2% or 4%) was compared to midazolam (passive diffusion) and to urea and methotrexate (SLC-mediated facilitated transport). Hydroxyurea had limited permeability across the bilayers, similar to urea and methotrexate but unlike midazolam, findings that are inconsistent with previous predictions of hydroxyurea movement exclusively by passive diffusion.
Screening of SLC transporters
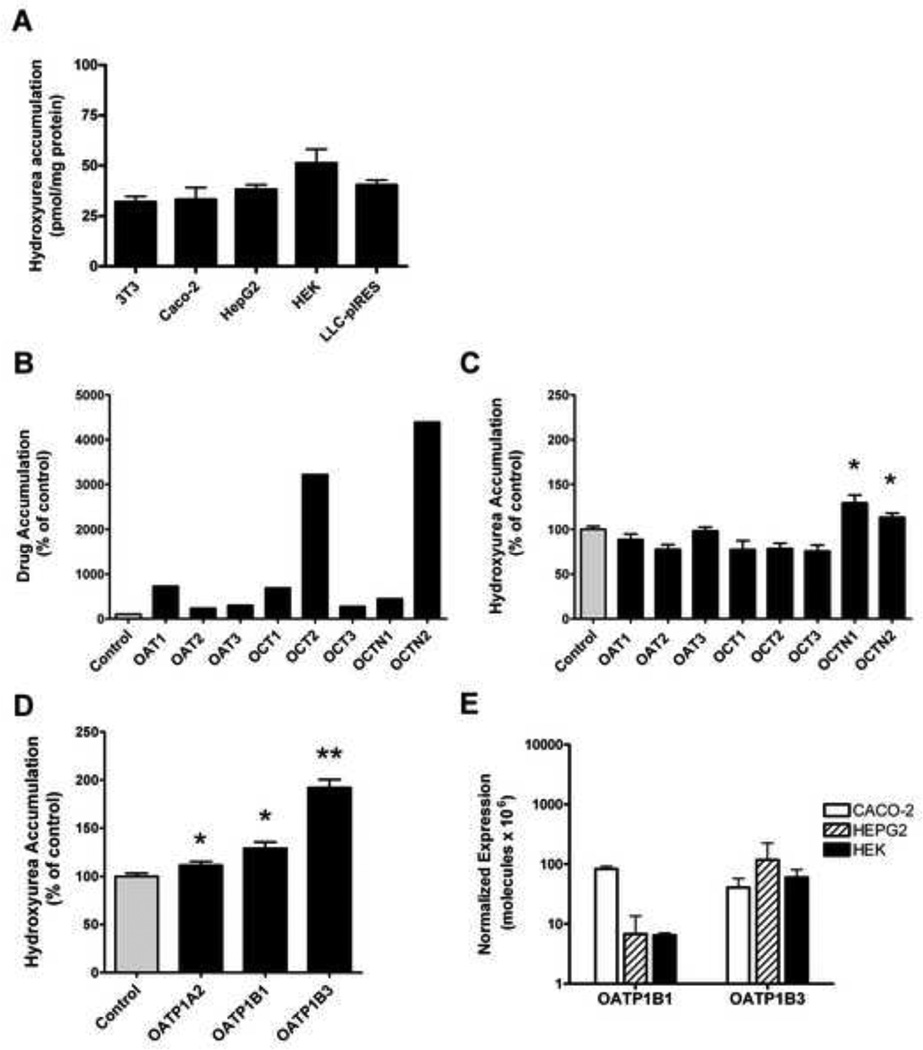
Hydroxyurea uptake and accumulation were analyzed in various cell lines and in transfected HEK cell lines overexpressing exogenous SLC transporters. Hydroxyurea uptake into cell lines without exogenous transporter overexpression was analyzed in fibroblasts 3T3, human colo-rectal Caco-2, liver HepG2, and kidney HEK cell lines and in the porcine kidney LLC-PK1 cell line. Hydroxyurea accumulation after 30 minutes was not different in any of these cell lines despite their various tissue origins (Figure 2A). In uptake studies with SLC transfected cells, accumulation of a previously identified prototypical drug ranged from 235% to over 4000% of control, which demonstrates a functional overexpression of each SLC transporter in the respective HEK cell lines (Figure 2B).
Figure 2. Uptake studies demonstrate that hydroxyurea is a specific substrate for some SLC transporters.
(A) Hydroxyurea accumulation in 3T3, Caco-2, HepG2, HEK, and LLC-PK1 cell lines is similar following 30 minute incubation period with 50µM hydroxyurea. (B–D) Movement of drug substrates by organic anion (OAT), organic cation (OCT), and organic cation/carnitine (OCTN), and organic anion transporting polypeptide (OATP) families of SLC transporters was determined by measuring accumulation of hydroxyurea in transporter overexpressing HEK293 cells and ooctyes. Drug substrate accumulation within cells or oocytes is expressed as a percentage of control cells or oocytes (100%, gray bar) from each experiment. An increase in accumulation of the prototypical drug substrates: p-aminohippuric acid (3µM) in OAT1 and OAT2 cells, estrone-3-sulfate (2µM) in OAT3 cells, tetraethylammonium (10µM) in OCT1, OCT2, and OCT3 cells; and Carnitine (0.01µM) in OCTN1 and OCTN2 cells shows the functional over expression of each transporter (B). Hydroxyurea accumulation is significantly increased in OCTN1 and OCTN2 cells (C). Over expression of OATP transporters significantly increase hydroxyurea accumulation in oocytes (D). Gene expression of endogenous OATP1B1 and OATP1B3 measured by real-time PCR in cell lines, is extremely low and similar in CACO-2, HepG2, and HEK cells (E). Each result represents the mean ± standard error of 3 or more independent experiments, each consisting of 3–10 measurements per cell line or oocyte (*P<.04; **P<.001 compared to control).
The SLC overexpressing cell lines were screened to determine if the SLC transporters influence hydroxyurea uptake. Significant increases in hydroxyurea uptake were observed only in cells overexpressing human OCTN1 (130%, p=.0031) or OCTN2 (113%, p=0.024), compared to water-injected controls (Figure 2C). Oocytes that overexpress human OATP transporters were also screened for their ability to transport hydroxyurea. Hydroxyurea accumulation in oocytes overexpressing OATP1B3 exceeded 190% of control (p<.001) and oocytes overexpressing OATP1A2 or OATP1B1 displayed small but statistically significant increases of 12–30% in hydroxyurea accumulation compared to controls (p<0.04, Figure 2D). Endogenous OATP1B1 and 1B3 mRNA expression was evaluated in Caco-2, HepG2 and HEK cell lines. Overall, OATP1B1 and 1B3 transporter expression in all three cell lines was extremely low, with OATP1B1 expression in HepG2 and HEK being nearly absent. OATP1B3 expression was similar in all three cell types (Figure 2E). Though transporter affinity for hydroxyurea cannot be directly compared from one SLC transporter to another, these studies indicate that OCTN1, OCTN2, OATP1A2, OATP1B1, and OATP1B3 all have the ability to transport hydroxyurea and influence intracellular accumulation.
Hydroxyurea transport by organic anion transporting polypeptide 1B3 (OATP1B3)
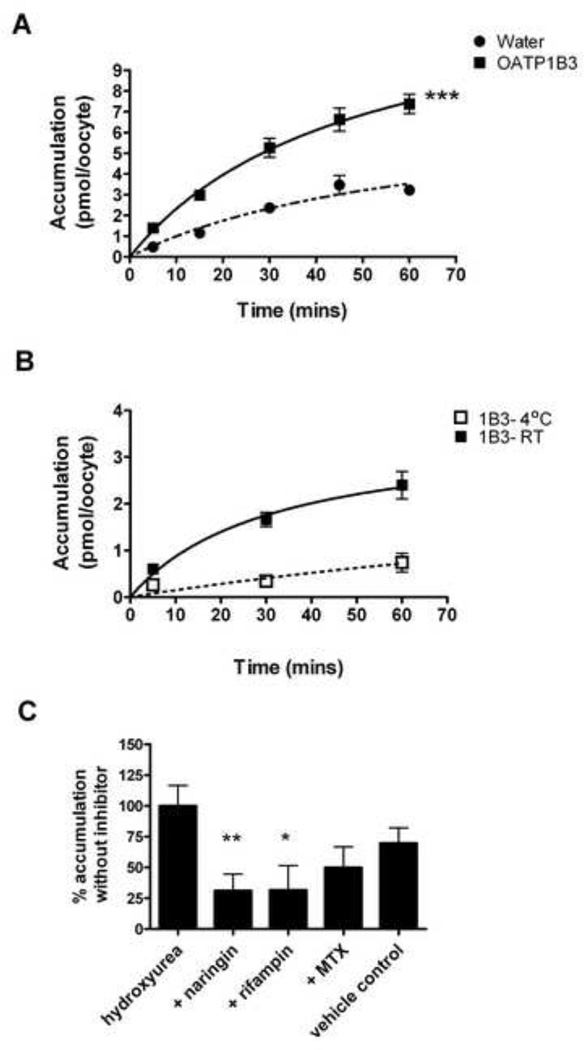
Because OATP1B3 overexpression was associated with significantly increased hydroxyurea uptake, additional experiments were performed to characterize this SLC-mediated drug transport further. Hydroxyurea accumulation in OATP1B3 oocytes was significantly higher compared to water-injected controls over a time course from 5 to 60 minutes (p=0.009) (Figure 3A). To confirm active uptake of hydroxyurea by OATP1B3, the assay temperature was lowered to inhibit transporter function. At 4°C, OATP1B3-mediated hydroxyurea accumulation decreased substantially showing a temperaturedependent inhibition of hydroxyurea transport at each time point (Figure 3B). With incubation at 4°C, accumulation was lowered to 30–40% of accumulation that occurs during incubation at room temperature. At room temperature, hydroxyurea accumulation becomes saturated over time whereas accumulation remains linear at 4°C. The time and temperature dependence of OATP1B3-mediated hydroxyurea transport indicates active transport mechanisms may be involved. To show specificity of hydroxyurea for OATP1B3 transporters, known substrates were then used as competitive inhibitors of OATP1B3-mediated hydroxyurea accumulation. Hydroxyurea accumulation in OATP1B3 oocytes was significantly reduced by co-incubation of 1mM naringin or 1mM rifampin to 30% of hydroxyurea accumulation without inhibitor. However, methotrexate co-incubation in the absence of DMSO lowered hydroxyurea accumulation by 50% (Figure 3C). There was a small additive impact of DMSO to naringin and rifampin inhibition which is noted by the decrease of hydroxyurea accumulation to 70% in vehicle control, but this decrease was not significantly different from that seen in either the absence or presence of inhibitor. The residual uptake of hydroxyurea into oocytes after inhibition by cold temperature or competitive inhibitors was most likely due to incomplete inhibition of the OATP1B3 and presence of other endogenous transporters, especially at the early time points.
Figure 3. Active transport of hydroxyurea is mediated by OATP1B3.
(A) Hydroxyurea uptake over time in OATP1B3 oocytes is significantly higher than uptake by controls (***P= .009). (B) OATP1B3 mediated accumulation of hydroxyurea at 4°C is reduced and remains linear over a time course compared to accumulation at room temperature that approaches saturation. (C) OATP1B3 mediated transport of hydroxyurea by is inhibited by co-incubation with naringin and other known OATP1B3 substrates. 1% DMSO solution was used as vehicle control. Each symbol or bar represents the mean ±SE of 3 independent experiments with at least 5 replicates per experimental point. **P<.01; *P<.05 compared to hydroxyurea alone
Hydroxyurea transport by Urea Transporters (UT)
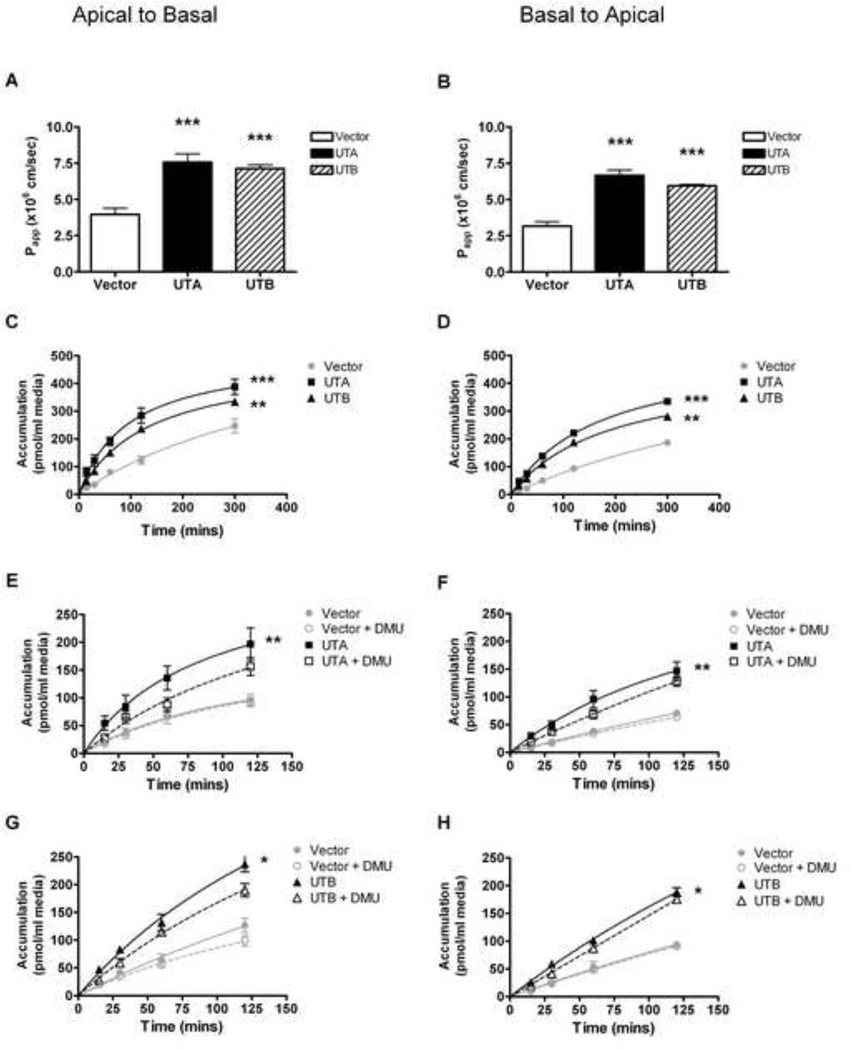
Hydroxyurea transport was examined in LLC-PK1 cells overexpressing UTA or UTB, in both the apical-basal and basal-apical directions. Hydroxyurea transport was direction independent and the apparent permeability coefficient was approximately 2-fold higher in LLC-UTA and LLC-UTB cells compared to vector control (p<.001, Figure4A and 4B). Vector control cells had measurable levels of hydroxyurea transport, which is most likely due to endogenous transporters on cell membrane. Over time, transcellular transport by LLC-UTA and LLC-UTB cells was more than doubled, and was significantly higher than that of vector control cells (p<.01, Figure4C and 4D). In the presence of the competitive inhibitor dimethylurea (DMU), however, hydroxyurea transport was marginally inhibited and was not significantly different from vector control(Figure 4E–H).
Figure 4. Urea Transporters mediate hydroxyurea transcellular movement.
(A–B) the apparent permeability coefficients of hydroxyurea transport from the apical to basal (A-to-B) direction and the basal to apical (B-to-A) direction is significantly higher for LLC-UTA cells (black bar) and LLC-UTB cells (striped bar) compared to LLC-pIRES control cells (white bar). (C–D) hydroxyurea transcellular transport over time in either direction is significantly higher for LLC-UTA cells (squares) and LLC-UTB cells (triangles) than for or LLC-pIRES cells (circles). (E–F) hydroxyurea transport measured in the presence of a competitive substrate inhibitor dimethylurea (DMU) is diminished in LLC-UTA cells (open squares) for both the A-to-B and B-to-A directions. (G–H) hydroxyurea transport with DMU is slightly diminished in LLC-UTB cells (open triangles) for both directions. Transport in control cells (open circle) is unaltered by DMU. All results represent mean ± SE of 2 independent experiments with 3 replicates of each experimental point. *** P<.001; **P<.01; * P<.05 compared to control without DMU.
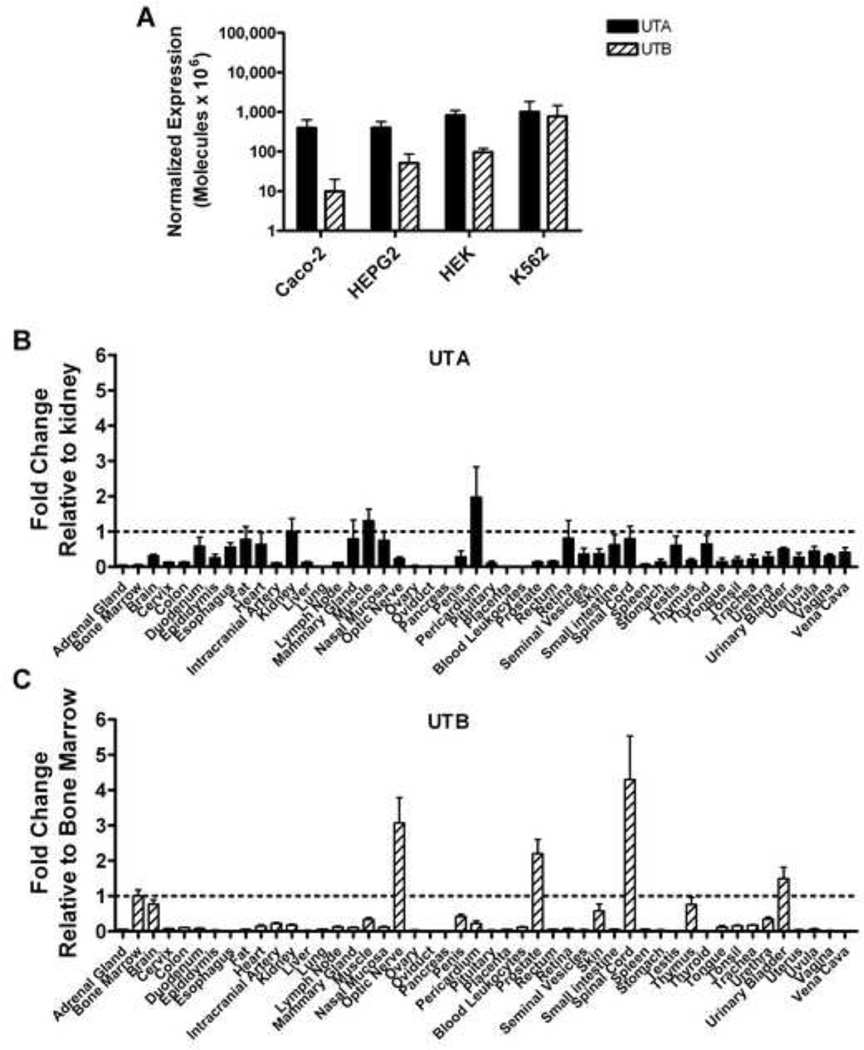
Urea transporter mRNA expression in human cells and tissues
UTA and UTB expression was measured in the erythroid cell line K562 and compared to expression in nonerythroid cell lines. UTA expression was similar in the cell lines including K562. Unlike UTA, UTB expression in cell lines showed more variability with K562 cells expressing the most though not significantly different (Figure 5A). Additionally, Real-time PCR analysis was performed to identify human tissues expressing UT transcripts that could be involved in hydroxyurea distribution. The mRNA expression was tissue specific with considerable variation: the highest expression of UTA mRNA was observed in pericardium, muscle, and kidney (Figure 5B) while UTB expression was highest in nervous system tissue along with prostate, urinary bladder, and bone marrow (Figure 5C). Interestingly, when expression of OATP1B1, OATP1B3, UTA, and UTB were examined in human CD71+ reticulocytes by real-time PCR, only UTB mRNA transcripts were present at a detectable level in both controls and patients with sickle cell anemia (Table 1).
Figure 5. Urea Transporter mRNA is differentially expressed in erythroid cells and in many human tissues.
(A) UTA (solid bars) and UTB (hatched bars) mRNA expression in Caco-2, HepG2, HEK, and K562 cell lines are quantified by real-time PCR and normalized to b-actin. (B–C) Real-time PCR analysis of cDNA from human tissues reveals wide expression for UTA (B) and UTB (C) as calculated by comparative CT analysis, in which UTA or UTB transcript levels are normalized to the endogenous control GUSB then compared to the reference tissues kidney (UTA) or bone marrow (UTB). Results represent the mean (±SE).
Table 1.
Mean of transporter expression in CD71+ cells. HEK cell lines that over express each of the transporters (UTA, UTB, 1B1, and 1B3) were used as positive controls and HEK-pIRES cell line was used as a negative control. Values were normalized to β-actin and expressed as number of molecules × 105. For experiments with CD71+ cells, results are the average from 6 healthy volunteers (CD71+ cells) and 9 patients with sickle cell disease (CD71+ cells-SCD).
| Cell Type | UTA | UTB | OATP1B1 | OATP1B3 |
|---|---|---|---|---|
| CD71+ | 6 | 6208 | 0 | 0 |
| CD71+ SCD | 9 | 9938 | 0 | 0 |
| HEK-UTA | 44547 | 3 | 0 | 3 |
| HEK-UTB | 1849 | 3773 | 0 | 0 |
| HEK-1B1 | 2 | 0 | 228 | 0 |
| HEK-1B3 | 2 | 0 | 0 | 2211 |
| HEK-IRES | 5 | 0 | 0 | 0 |
DISCUSSION
Like many other drugs, hydroxyurea has traditionally been thought to pass through cell membranes via passive diffusion. By comparison, urea also was once thought to utilize passive diffusion until specific urea transporters were identified and kinetic studies documented facilitated diffusion mechanisms[31,32]. Our in vitro studies demonstrate clearly that hydroxyurea transport across an artificial lipid bilayer by passive diffusion is limited (Figure 1), and further show that hydroxyurea is a substrate for certain SLC transporters (Figures 2 and 3). Although these studies do not allow for the direct comparison of the SLC transporters, they specifically identify hydroxyurea as a substrate for organic anion transporting polypeptide 1A2, 1B1, and 1B3 (OATP1A2; OATP1B1; OATP1B3), urea transporters A and B (UTA; UTB), organic cation/carnitine transporter 1 and 2 (OCTN1; OCTN2). In the case of the UTA and UTB transporters, hydroxyurea movement was bidirectional and significantly inhibited by competitive substrates (Figure 4). The variable transport capabilities and diverse locations of these physiologically important SLC proteins suggest that hydroxyurea transport mechanisms in vivo may have a clinically significant influence on the hydroxyurea pharmacokinetic or pharmacodynamic profiles, and could potentially help explain part of the observed inter-patient variability.
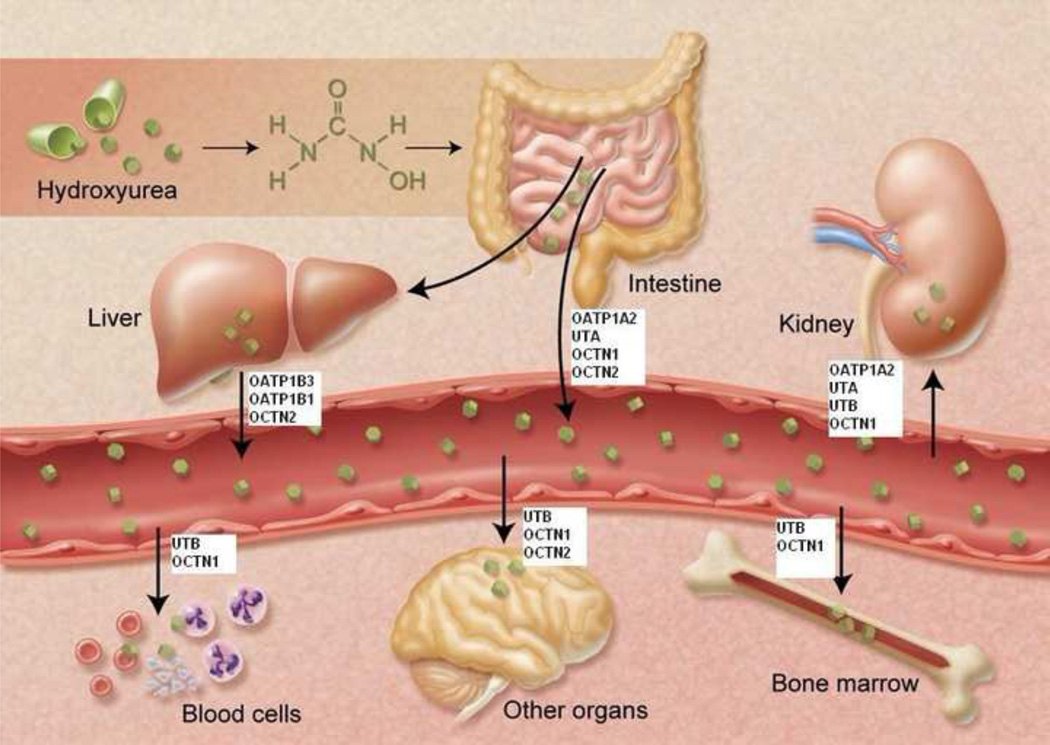
OCTN 1, OCTN2, and OATP1A2 have all been localized to the intestine and could, therefore, be involved in the oral absorption of hydroxyurea. Our current analysis also confirms UTA mRNA expression within the duodenum (Figure 5A). The localization of these potential hydroxyurea transporters to the early segments of the gut may account for its rapid but variable absorption[16]. Once in circulation, hydroxyurea is distributed to various organs and fluids, including the brain and cerebral spinal fluid as well as the mammary glands and breast milk[13,33]. Our studies show that UTA or UTB mRNA is expressed at relatively high levels in some of these same tissues (Figure 5). Further, OCTN1 and OCTN2 is also found in brain and bone marrow and in the lungs[34]. Thus, these transporters may influence drug distribution. Transporters located on bone marrow and peripheral blood cells could potentially directly influence hydroxyurea pharmacodynamic parameters such as efficacy (HbF induction) and toxicity (myelosuppression) that occurs with hydroxyurea treatment. Specific expression of UTB on early erythroid cells (Table 1) could be especially relevant to treatment in sickle cell anemia. Hepatic expression of OATP1B3, OATP1B1, and OCTN2 transporters may further impact hydroxyurea pharmacokinetic parameters by influencing the metabolism of hydroxyurea with subsequent elimination of the drug. Finally, because hydroxyurea is primarily eliminated through the kidney[12], the renal SLC transporters OATP1A2, UTA, UTB, and OCTN1 may facilitate drug elimination. Figure 6 summarizes the potential drug transporters affecting hydroxyurea in vivo, and illustrates the complexity of these drug interactions affecting movement of hydroxyurea between various body compartments.
Figure 6.
Hydroxyurea movement in vivo among various organs and compartments is influenced by numerous physiological SLC drug transporters that carry the drug to and from specific tissues.
Our kinetics experiments, which focused on OATP1B3 and urea transporters as examples, demonstrate two different models of carrier-mediated hydroxyurea transport: facilitated diffusion and active transport. The transport of hydroxyurea by UTA or UTB appears to occur through facilitated diffusion; in our transcellular transport assays, both the rate and total accumulation of hydroxyurea across the cell membrane were significantly increased by both UTA and UTB (Figure 4). These results are consistent with a model of facilitated diffusion of hydroxyurea by a channel protein. The recent elucidation of the crystalline structure of the bacterial UT reveals a channel motif and it is likely that human UT is also a channel because of conserved sequence homology[35]. Additionally, the movement of hydroxyurea from apical to basal did not differ greatly from basal to apical movement (Figure 4A and 4B). The lack of directionality in UT-mediated hydroxyurea transport is most likely explained by bidirectional flow of hydroxyurea through the UTA or UTB transporters, combined with the near-ubiquitous expression of UTA or UTB on both the basolateral and apical membranes of the polarized cells. Regardless of the direction, the presence of the transporter increased the rate and overall transcellular movement of hydroxyurea, as indicated by the rapid time course and over 2-fold increase in the apparent permeability (Figure 4). Urea transporters are most widely studied in the kidney for their role in maintaining the interstitial osmotic gradients allowing water conservation, but urea transporters are also located in other tissues and cells including testis, bone marrow, erythrocytes, and endothelial cells[36,37]. The physiological roles of urea transporters in these tissues and others identified by our real-time PCR experiments (Figure 5) are currently unclear. Similarly, the role of specific isoforms in hydroxyurea transport remains to be determined.
Conversely, OATP1B3 mediated uptake of hydroxyurea appears to act through active transport mechanisms. Encoded by the SLCO1B3 gene, OATP1B3 is a liver-specific transporter located on the basolateral membrane of hepatocytes and functions primarily as an uptake transporter carrying substrates from the portal blood system into the liver[38]. In oocytes, OATP1B3-mediated transport of hydroxyurea was time and temperature dependent suggesting active transport(Figure 3). Though we demonstrated that hydroxyurea movement across cell membranes is mediated by SLC transporters, we cannot exclude some additional transmembrane transport via passive diffusion. When OATP1B3 transporters were inhibited by cold temperatures, low levels of hydroxyurea accumulated in ooctyes following a linear trend which is consistent with models of passive diffusion. However, the rate and amount of hydroxyurea intracellular accumulation greatly increased with the presence of the SLC transporter.
In summary, our data demonstrate for the first time that in vitro transporter-mediated cellular entry of hydroxyurea occurs unequivocally, and the likely in vivo correlates of these observations have important implications regarding inter-individual variability in hydroxyurea pharmacokinetic and pharmacodynamic profiles. Further investigation into the mechanisms by which hydroxyurea is absorbed, distributed, and cleared will broaden our understanding of its therapeutic efficacy and toxicity. If hydroxyurea passage across cell membranes is mediated by SLC transport proteins in vivo, then the presence and function of these transporters located in key tissues have the potential to influence important pharmacokinetic and pharmacodynamic parameters. Alterations in transporter function by factors such as genetic polymorphisms or interactions with food and other drugs could further account for the observed variability in hydroxyurea efficacy and toxicity for patients with sickle cell anemia.
ACKNOWLEDGMENTS
Dr. Walker is the Lemuel Diggs Endowed Postdoctoral Fellow in Sickle Cell Disease. This work was supported by grants R01 HL090941 (REW) and the American Lebanese Syrian Associated Charities (ALSAC). We would like to thank Thad Howard, Shirley Steward, and Jonathan Flanagan for insightful discussions and laboratory assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST DISCLOSURE
No financial interest/relationships with financial interest relating to the topic of this article have been declared.
REFERENCES
- 1.Stevens M. Hydroxyurea: an overview. J Biol Regul Homeost Agents. 1999:13172–13175. [PubMed] [Google Scholar]
- 2.Platt OS. Hydroxyurea for the treatment of sickle cell anemia. N Engl J Med. 2008;358(13):1362–1369. doi: 10.1056/NEJMct0708272. [DOI] [PubMed] [Google Scholar]
- 3.Dover GJ, Humphries RK, Moore JG, et al. Hydroxyurea induction of hemoglobin F production in sickle cell disease: Relationship between cytotoxicity and F cell production. Blood. 1986;67(3):735–738. [PubMed] [Google Scholar]
- 4.Charache S, Dover GJ, Moore RD, et al. Hydroxyurea: Effects on hemoglobin F production in patients with sickle cell anemia [see comments] Blood. 1992;79(10):2555–2565. [PubMed] [Google Scholar]
- 5.Charache S, Terrin ML, Moore RD, et al. Effect of hydroxyurea on the frequency of painful crises in sickle cell anemia. N Engl J Med. 1995;332(20):1317–1322. doi: 10.1056/NEJM199505183322001. [DOI] [PubMed] [Google Scholar]
- 6.Kinney TR, Helms RW, O'Branski EE, et al. Safety of hydroxyurea in children with sickle cell anemia: Results of the hug-kids study, a phase I/II trial. Blood. 1999;94(5):1550–1554. [PubMed] [Google Scholar]
- 7.Zimmerman SA, Schultz WH, Davis JS, et al. Sustained long-term hematologic efficacy of hydroxyurea at maximum tolerated dose in children with sickle cell disease. Blood. 2004;103(6):2039–2045. doi: 10.1182/blood-2003-07-2475. [DOI] [PubMed] [Google Scholar]
- 8.Elford HL. Effect of hydroxyurea on ribonucleotide reductase. Biochem Biophys Res Commun. 1968;33(1):129–135. doi: 10.1016/0006-291x(68)90266-0. [DOI] [PubMed] [Google Scholar]
- 9.Snyder R. The role of deoxynucleoside triphosphate pools in the inhibition of DNA-excision repair and replication in human cells by hydroxyurea. Mutat Res. 1984;131:163–172. doi: 10.1016/0167-8817(84)90057-9. [DOI] [PubMed] [Google Scholar]
- 10.Cokic VP, Smith RD, Beleslin-Cokic BB, et al. Hydroxyurea induces fetal hemoglobin by the nitric oxide-dependent activation of soluble guanylyl cyclase. J Clin Invest. 2003;111(2):231–239. doi: 10.1172/JCI16672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ware RE. How I use hydroxyurea to treat young patients with sickle cell anemia. Blood. 2010;115(26):5300–5311. doi: 10.1182/blood-2009-04-146852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adamson RH, Ague SL, Hess SM, Davidson JD. The distribution, excretion, and metabolism of hydroxyurea-C14. J Pharmacol Exp Ther. 1965;150(2):322–327. [PubMed] [Google Scholar]
- 13.Gwilt PR, Manouilov KK, McNabb J, Swindells SS. Pharmacokinetics of hydroxyurea in plasma and cerebrospinal fluid of hiv-1-infected patients. J Clin Pharmacol. 2003;43(9):1003–1007. doi: 10.1177/0091270003256144. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez GI, Kuhn JG, Weiss GR, et al. A bioavailability and pharmacokinetic study of oral and intravenous hydroxyurea. Blood. 1998;91(5):1533–1541. [PubMed] [Google Scholar]
- 15.de Montalembert M, Bachir D, Hulin A, et al. Pharmacokinetics of hydroxyurea 1,000 mg coated breakable tablets and 500 mg capsules in pediatric and adult patients with sickle cell disease. Haematologica. 2006;91(12):1685–1688. [PubMed] [Google Scholar]
- 16.Ware RE, He J, Mortier NA, et al. Distinct phenotypes of hydroxyurea absorption among children with sickle cell anemia [abstract] Blood. 2008;112:709a. [Google Scholar]
- 17.Ware RE, Eggleston B, Redding-Lallinger R, et al. Predictors of fetal hemoglobin response in children with sickle cell anemia receiving hydroxyurea therapy. Blood. 2002;99(1):10–14. doi: 10.1182/blood.v99.1.10. [DOI] [PubMed] [Google Scholar]
- 18.Tagger AY, Boux J, Wright JA. Hydroxy [14C]urea uptake by normal and transformed human cells: evidence for a mechanism of passive diffusion. Biochem Cell Biol. 1987;65:925–929. doi: 10.1139/o87-120. [DOI] [PubMed] [Google Scholar]
- 19.Evered D, Selhi H. Transport characteristics of two carcinostatic compounds, hydroxyurea and hadacidin, with rat small intestine [abstract] Biochem J. 1972;126:26. doi: 10.1042/bj1260026pa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morgan JS, Creasey DC, Wright JA. Evidence that the antitumor agent hydroxyurea enters mammalian cells by a diffusion mechanism. Biochem Biophys Res Commun. 1986;134(3):1254–1259. doi: 10.1016/0006-291x(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 21.Finkelstein A. Water and nonelectrolyte permeability of lipid bilayer membranes. J Gen Physiol. 1976;68(2):127–135. doi: 10.1085/jgp.68.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oostendorp RL, Beijnen JH, Schellens JHM. The biological and clinical role of drug transporters at the intestinal barrier. Cancer Treat Rev. 2009;35(2):137–147. doi: 10.1016/j.ctrv.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Kitamura S, Maeda K, Sugiyama Y. Recent progresses in the experimental methods and evaluation strategies of transporter functions for the prediction of the pharmacokinetics in humans. Naunyn Schmiedebergs Arch Pharmacol. 2008;377(4):617–628. doi: 10.1007/s00210-008-0312-9. [DOI] [PubMed] [Google Scholar]
- 24.Grover A, Benet L. Effects of drug transporters on volume of distribution. AAPS J. 2009;11(2):250–261. doi: 10.1208/s12248-009-9102-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerb R. Implications of genetic polymorphisms in drug transporters for pharmacotherapy. Cancer Lett. 2006;234(1):4–33. doi: 10.1016/j.canlet.2005.06.051. [DOI] [PubMed] [Google Scholar]
- 26.Shayakul C, Hediger MA. The SLC14 gene family of urea transporters. Pflugers Archiv. 2004;447(5):603–609. doi: 10.1007/s00424-003-1124-x. [DOI] [PubMed] [Google Scholar]
- 27.Zhao D, Sonawane ND, Levin MH, Yang B. Comparative transport efficiencies of urea analogues through urea transporter UT-B. Biochim Biophys Acta. 2007;1768(7):1815–1821. doi: 10.1016/j.bbamem.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 28.Stewart GS, O'Brien JH, Smith CP. Ubiquitination regulates the plasma membrane expression of renal UT-A urea transporters. Am J Physiol Cell Physiol. 2008;295:C121–C129. doi: 10.1152/ajpcell.00444.2007. [DOI] [PubMed] [Google Scholar]
- 29.Wohnsland F, Faller B. High-throughput permeability pH profile and highthroughput alkane/water log p with artificial membranes. J Med Chem. 2001;44(6):923–930. doi: 10.1021/jm001020e. [DOI] [PubMed] [Google Scholar]
- 30.Xia CQ, Liu N, Miwa GT, Gan L-S. Interactions of cyclosporin a with breast cancer resistance protein. Drug Metab Dispos. 2007;35(4):576–582. doi: 10.1124/dmd.106.011866. [DOI] [PubMed] [Google Scholar]
- 31.Brahm J. Urea permeability of human red cells. J Gen Physiol. 1983;82(1):1–23. doi: 10.1085/jgp.82.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.You G, Smith CP, Kanai Y, Lee W-S, Stelzner M, Hediger MA. Cloning and characterization of the vasopressin-regulated urea transporter. Nature. 1993;365(6449):844–847. doi: 10.1038/365844a0. [DOI] [PubMed] [Google Scholar]
- 33.Sylvester RK, Lobell M, Teresi ME, Brundage D, Dubowy R. Excretion of hydroxyurea into milk. Cancer. 1987;60(9):2177–2178. doi: 10.1002/1097-0142(19871101)60:9<2177::aid-cncr2820600911>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 34.Koepsell H, Lips K, Volk C. Polyspecific organic cation transporters: structure, function, physiological roles, and biopharmaceutical implications. Pharm Res. 2007;24(7):1227–1251. doi: 10.1007/s11095-007-9254-z. [DOI] [PubMed] [Google Scholar]
- 35.Levin EJ, Quick M, Zhou M. Crystal structure of a bacterial homologue of the kidney urea transporter. Nature. 2009;462:757–762. doi: 10.1038/nature08558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsukaguchi H, Shayakul C, Berger UV, Tokui T, Brown D, Hediger A. Cloning and characterization of the urea transporter UT3: localization in rat kidney and testis. J Clin Invest. 1997;99(7):1506–1515. doi: 10.1172/JCI119313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner L, Klein JD, Sands JM, Baylis C. Urea transporters are distributed in endothelial cells and mediate inhibition of l-arginine transport. Am J Physiol Renal Physiol. 2002;283(3):F578–F582. doi: 10.1152/ajprenal.00355.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalliokoski A, Niemi M. Impact of OATP transporters on pharmacokinetics. Br J Pharmacol. 2009;158(3):693–705. doi: 10.1111/j.1476-5381.2009.00430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]