Abstract
Plants are an incredibly rich source of compounds that activate the Nrf2 transcription factor, leading to upregulation of a battery of cytoprotective genes. This perspective surveys established and proposed molecular mechanisms of Nrf2 activation by phytochemicals with a special emphasis on a common chemical property of Nrf2 activators: the ability as “soft” electrophiles to modify cellular thiols, either directly or as oxidized biotransformants. In addition, the role of reactive oxygen/nitrogen species as secondary messengers in Nrf2 activation is discussed. While the uniquely reactive C151 of Keap1, an Nrf2 repressor protein, is highlighted as a key target of cytoprotective phytochemicals, also reviewed are other stress-responsive proteins, including kinases, which play non-redundant roles in the activation of Nrf2 by plant-derived agents. Finally, the perspective presents two key factors accounting for the enhanced therapeutic windows of effective phytochemical activators of the Keap1–Nrf2 axis: enhanced selectivity toward sensor cysteines and reversibility of addition to thiolate molecules.
7.1 Introduction
Numerous phytochemicals have shown great promise for prevention and treatment of various human diseases. For example, ClinicalTrials.gov lists 25 different intervention trials investigating the effects of standardized preparations of broccoli sprouts. These trials include the amelioration of symptoms of diseases as diverse as autism1, cystic fibrosis, influenza, asthma, and chronic obstructive pulmonary disease. The trials also encompass studies on the prevention of breast, lung, and prostate cancers, carcinogenesis from aflatoxin exposure, and cardiovascular disease. The purpose of this Perspective is to i) review a key molecular mechanism of phytochemicals in the prevention and amelioration of diseases: the activation of the transcription factor NF-E2-related factor 2 (Nrf2), ii) outline common chemical features of potent Nrf2 activators, and iii) provide perspectives for harnessing these features for more effective disease prevention or treatment.
7.2 Upregulation of Cytoprotective Genes by Nrf2
Oxidative stress and associated inflammation contribute to progression of many chronic degenerative diseases in humans [1, 2]. Exposure to oxidant and electrophilic agents from air, water, food, and other environmental sources has also been implicated in a large (70–90%) component of cancer and cardiovascular disease risks [3]. The Nrf2 transcription factor has emerged as a key player in protecting cells against various intrinsic and extrinsic assaults. Nrf2 regulates more than 600 genes, including over one hundred that encode cytoprotective proteins [4], named for their ability to protect cells against oxidative stress, reactive electrophilic species, and other types of stress (reviewed in [5]). In brief, these proteins include antioxidant enzymes, NADPH regeneration enzymes, glutathione biosynthesis enzymes, heat shock proteins, enzymes that facilitate the elimination of xenobiotic toxicants such as detoxification enzymes and drug-efflux pumps, as well as subunits of the 26S proteasome. Collectively, these Nrf2-regulated genes share at least one copy of the antioxidant response element (ARE, ) in their promoter region [4]. Nrf2 binds to the ARE as a heterodimer with one of several small Maf transcription factors, leading to upregulation of gene transcription.
Upregulation of this battery of cytoprotective proteins through Nrf2 activation is a critical component of an organism's ability to cope with intrinsic and extrinsic stress factors, including inflammation, reactive oxygen/nitrogen species (ROS/RNS), shear stressors such as endothelial stressors, and environmental toxins. Studies on Nrf2−/− mice, which have low and largely non-inducible levels of many cytoprotective proteins [4], serve as striking and comprehensive examples of the importance of these proteins in maintaining health and preventing disease. Nrf2-deficient mice are prone to develop disorders that are caused by ROS and inflammation, including macular degeneration [6], neurodegeneration in a murine model of Parkinson's disease [7], cardiac disorders [8, 9], and chemically-induced tumorigenesis [10-12]. Furthermore, Nrf2−/− mice are more susceptible to damage of the blood–brain barrier following brain injury [13], formation of carcinogen–DNA adducts in the lung exposed to diesel exhaust [14], acute pulmonary injury induced by butylated hydroxytoluene [15], and hepatic damage by acetaminophen [16]. In addition, they are deficient in their intrinsic capacity for skin wound healing [17]. Since loss of Nrf2 increases pathological cell and tissue damage in response to intrinsic and extrinsic factors, it is believed that upregulation of Nrf2 serves both cytoprotective and preventive roles in diverse pathophysiological situations.
In support of the importance of Nrf2 in preventing human diseases, inherited DNA polymorphisms that reduce the abundance of Nrf2 are associated with various pathologies, including chronic gastrisis, ulcerative colitis, skin pathologies such as skin vitiligo, and adult respiratory distress syndrome (reviewed in [18]). In fact, a number of clinical trials that assess the effects of broccoli sprouts include direct measurements of Nrf2 activation. For example, Nrf2 levels, along with markers of oxidative stress, will be assessed after administration of macerated broccoli sprouts in both healthy volunteers and those with cystic fibrosis2, in nasal epithelial cells obtained by curettage, as well as in alveolar macrophages and bronchial epithelial cells of patients with chronic obstructive pulmonary disease3.
7.3 Phytochemical Activation of Nrf2
7.3.1 Overview
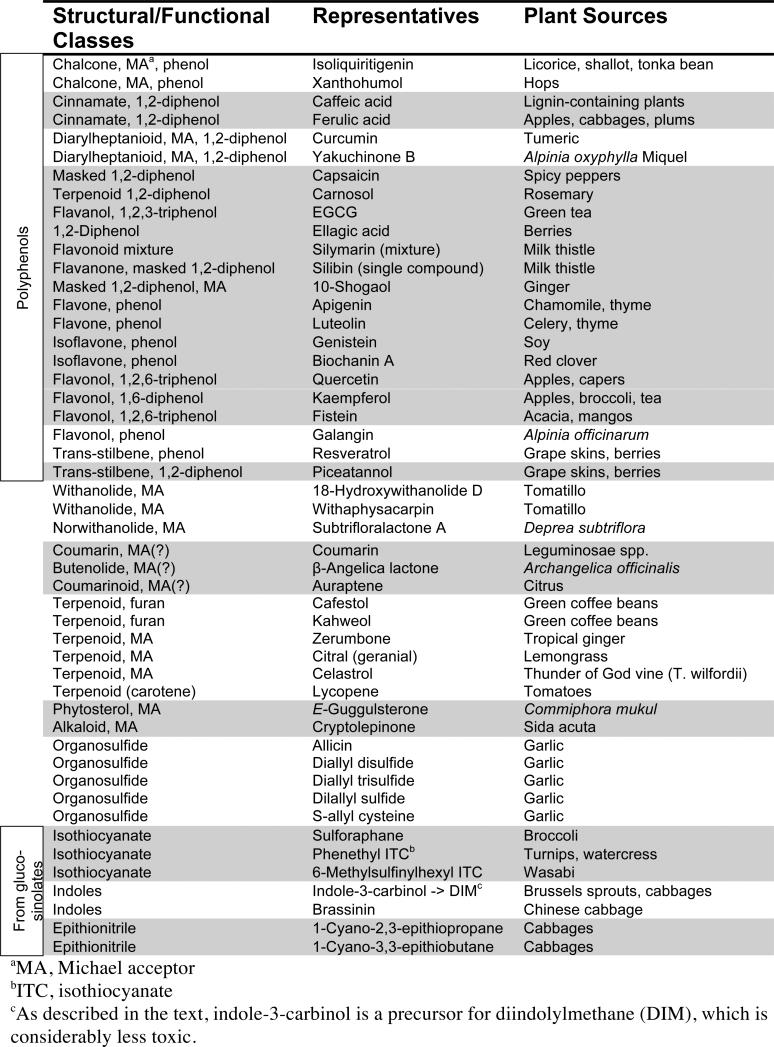
Plants have been an incredibly rich source for the identification of compounds that activate cytoprotective genes. Development of a simple microtiter-plate based assay [19] to assess induction of the cytoprotective enzyme NAD(P)H: quinone oxidoreductase 1 (QR1) in mouse Hepa1c1c7 cells has greatly facilitated the ability to screen for and identify many cytoprotective phytochemicals. For example, a collective effort of colleagues at the University of Illinois at Chicago and Purdue University has identified 66 compounds from 18 plant species that are active in the QR1 assay [20]. Representative phytochemicals that have been shown to activate cytoprotective genes are listed in Table 1.
Table 1.
Structures of Phytochemicals that Upregulate Cytoprotective Enyzmes through Nrf2
Several plant families important for human diets are particularly rich in ARE inducers. For example, many organosulfur activators have been isolated from garlic and onion, edible members of Allium family. The Cruciferous family of vegetables, including broccoli, cabbage, Brussels sprouts, horseradish, mustard and watercress, produces a particularly large and functionally diverse number of potent ARE inducers (Table 1). These plants contain glucosinolates, the thioglucoside conjugates of the ARE-activating species. Altogether, over 120 glucosinolates have been identified from various plants [21]. These are enzymatically converted to the ARE-inducing forms either by myrosinase, a thioglucosidase that is localized in a separated cellular compartment and is released upon maceration or chewing, or by intestinal microflora after ingestion [22]. Three general classes of ARE inducers produced by myrosinase-catalyzed hydrolysis of glucosinolates are isothiocyanates, indoles and epithionitriles (Table 1). The enzymatic hydrolysis mechanisms involved in their release are reviewed elsewhere [23, 24]. Finally, phenolic compounds, another important class of Nrf2-activating agents, have been isolated from diverse plant families, including grapes (Vitaceae) and teas (Theaseae), which are rich sources of flavonoids and related catechins.
Importantly, Nrf2−/− mice experiments highlight the key role of Nrf2 in mediating the cytoprotective effects of the phytochemicals discussed above. Thus, sulforaphane, one of the active components of broccoli sprout extracts has been reported to inhibit skin [12] and forestomach [10] carcinogenesis in wild-type (wt) mice, but its ability to do so is significantly attenuated in Nrf2−/− animals. In addition, sulforaphane was able to protect the blood-brain barrier post-injury only in wt Nrf2 mice [13]. Similarly, resveratrol, a flavonoid-like molecule produced by many plants, protected against high-fat-diet-induced oxidative stress in aortas of wt but not Nrf2−/− mice [9].
7.3.2 The Chemistry Required for Phytochemicals to Activate Nrf2
7.3.2.1 Reactivity with Thiolates
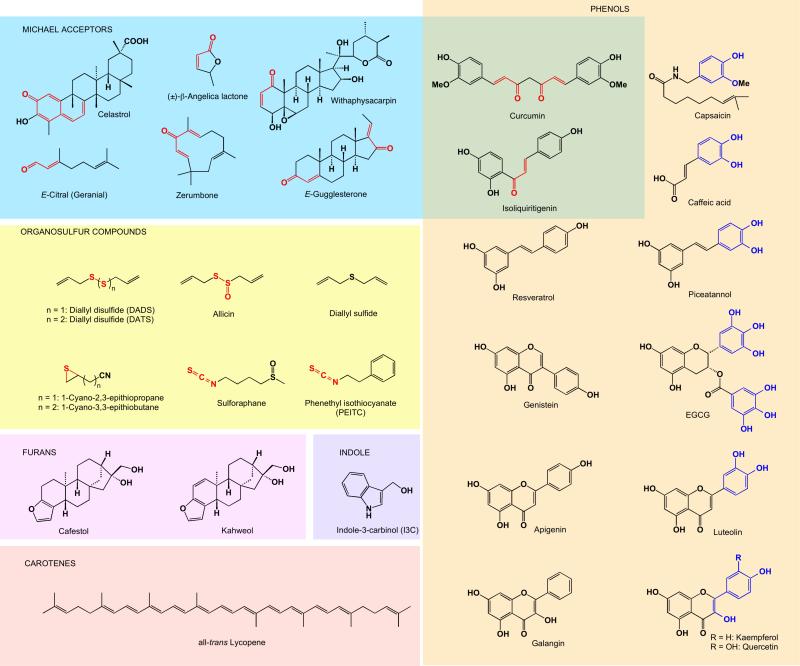
Despite the high level of overall structural diversity among ARE inducers, many cytoprotective phytochemicals from different plant sources share common thiol-reactive chemical motifs (collectively shown in Figure 7.1 as red moieties). For example, the presence of an α,β-unsaturated carbonyl group, a potential Michael acceptor, is a particularly common feature in ARE inducers, including withanolides (e.g. withaphysacarpin), chalcones (e.g., isoliquritigenin), butenolides (e.g. β-angelica lactone), oxidized terpenoids (e.g. zerumbone, E-gugglesterone, and citrals), and curcuminoids (e.g. curcumin). Other inducer classes are similarly electrophilic epithionitriles (e.g. 1-cyano-2,3-epithiopropane), isothiocyanates (e.g. sulforaphane), and organopolysulfides (e.g. allicin). Talalay and colleagues in 1988 first recognized that the diverse structures share the ability to react with thiolate groups [25]. They hypothesized that gene induction takes place by virtue of an intracellular sensor that contains one or more reactive cysteine residues, modification of which by inducing agents would lead to target gene activation. This seminal hypothesis is supported by numerous studies that followed, which directly linked the presence of particular functional groups to Nrf2–ARE induction, as shown with zerumbone [26], chalcones [27], flavonoids [28], and withanolides [20, 29]. Furthermore, a strong correlation has been identified between inducer potencies and chemical reactivities toward thiolates [30].
Fig. 7.1.
Structures of phytochemicals arranged by the chemistry that leads to Nrf2 activation. All compounds shown activate Nrf2. Thiol-reactive chemical motifs are shown in red, and “additive” distribution of electron-donating groups, which can be oxidized to quinoids, are shown in blue.
Finally, in 1999 a key repressor of Nrf2 was discovered, the Keap1 protein, which was found to possess an unusually large number of cysteines (25 and 27 in the mouse and human proteins, respectively) [31]. As described in detail in Section 7.3.2.2, a subset of these cysteines, C151 in particular, has been found to be important for Nrf2 activation by phytochemicals.
7.3.2.2 Phytochemicals that Do Not Have the Ability to React with Thiolates
While the critical feature of an Nrf2 activator appears to be the ability to react with thiolates, a large number of phytochemicals, including phenols, monosulfides, furans, and indoles, would need to acquire this property through metabolic and/or chemical processing. Studies on biotransformations of many such molecules are surprisingly limited, despite the current interest in health benefits of phytochemicals. Herein, we summarize the available experimental evidence from literature implicating mechanisms of converting phytochemicals to thiol-reactive species. We also discuss two classes for which transformation to a thiolate-reactive species is considerably more difficult to ascertain, carotenoids and 1,3-polyphenols.
One relatively well-characterized chemical class of phytochemicals that require electrophilic conversion for inducer activity are electron-rich phenolic compounds featuring “additive” distribution of electron-donating groups (Figure 7.1, blue structural fragments). These are compounds that contain phenolic hydroxyl groups in a conjugated system with an even number of carbons separating them. Representative examples of this class include 1,2-diphenols (e.g., epigallocatechin gallate (EGCG), caffeic acid, and piceatannol) and vinylogous 1,6-diphenols, with an alkene spacer acting as the “electron conduit” (e.g., quercetin and kaempferol). The 1,2- 1,4-, and 1,6-diphenols, but not the mono- or 1,3-dihydroxy variants, can be readily oxidized in vivo by a variety of mechanisms (Figure 7.2) to the corresponding quinoids and will be referred to herein as quinoid-forming phenols. In fact, such phenols have been associated with induction of carcinogen-detoxifying enzymes by one of the earliest observations in the field [32]. The resulting Michael acceptors can then readily react with cysteine thiolates (Figure 7.2). For example, the well-characterized flavonoid quercetin is expected to be a very weak electrophile intrinsically due to the electron-donating effect of the 3-hydroxyl group and both aromatic rings (see Figure 7.3.A for numbering scheme). However, in human blood plasma, quercetin is readily oxidized to a significantly more electrophilic quinone methide [33] (Figure 7.3.A). Importantly, the oxidized species is much more reactive toward thiolates, and its conjugation products have been detected with glutathione (GSH), N-acetylcysteine (NAC), cysteine [34, 35], and protein cysteine residues [36]. Similarly, EGCG contains several aromatic 1,2-dihydroxy units, and thus can form quinones that react with isolated and protein thiolates, as demonstrated in both biochemical experiments and cells [37, 38]. EGCG has also been found in mouse urine as the S-cysteinyl–EGCG conjugate after a high oral dose [39]. Furthermore, the oxidative conversion of phenolic compounds to Michael acceptors has been shown to correlate strongly with cytoprotective enzyme induction in studies evaluating tert-butylhydroquinone (tBHQ) [40] and EGCG analogs [38], as well as a broad series of phenols [41]. As further evidence that the oxidation of phenols is a prerequisite to ARE induction, Cu2+ or other oxidized transition metal cations in the media strongly stimulated the ARE induction potential of para- and ortho-hydroquinones [42]. These metal ions act as catalysts in the oxidation of phenols to Michael reaction acceptors (Figure 7.2) under aerobic conditions. Importantly, transition metal salts had no effect on inducer activity of the corresponding quinones or sulforaphane [43].
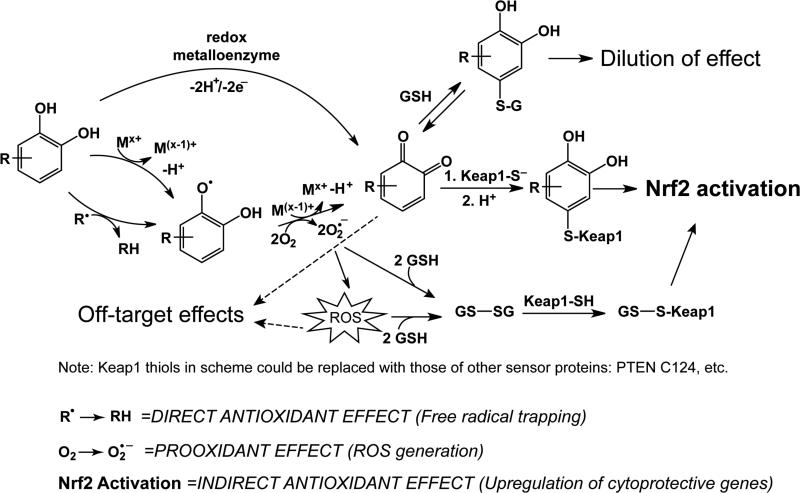
Fig. 7.2.
The three roles a quinoid-forming polyphenol (represented by the R-catechol) can play: prooxidant, direct antioxidant, and indirect antioxidant. As a direct antioxidant, in the presence of a high concentration of free radical species, a polyphenol can trap the radical, forming a relatively stable radical species. As a prooxidant, in the presence of catalytic amounts of a transition metal, a polyphenol can promote the formation of superoxide and other ROS, enroute to formation of a Michael acceptor. An alternate path to oxidation of the polyphenol is catalyzed by a metalloenzyme and occurs without the production of ROS. Once the quinoid group is formed, the Michael acceptor group can react with a thiolate molecule. There is evidence that a quinone reacts with a key Keap1 sensor cysteine, C151, leading to Nrf2 activation, as described in the text. Upon activation, Nrf2 upregulates a battery of antioxidant enzymes and other cytoprotective enzymes, known as the indirect antioxidant effect. Reaction of the quinoid with GSH and subsequent elimination from the cell will lead to dilution of the effect. Alternative mechanisms of Nrf2 activation by radicals or ROS not depicted are oxidation of sensor cysteines, or formation of disulfides among sensor cysteines.
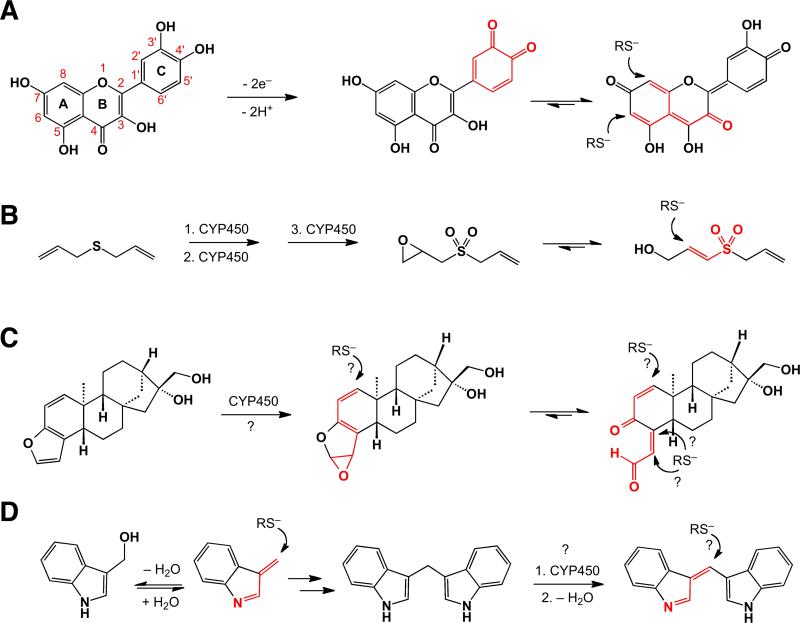
Fig. 7.3.
Biotransformations implicated in conversions of quercetin (A), DAS (B), coffee triterpenoids (kahweol shown here) (C), and I3C and DIM (D) into thiol-reactive conjugated electrophiles: quinoids (quercetin), sulfone (DAS), epoxide or γ-ketoenal (kahweol) and indolenines (I3C & DIM). The established or proposed reactive groups are highlighted in red.
The organosulfur compounds from garlic and onions were shown by the Wattenberg group in 1988 to have interesting structural requirements for inducer activity [44]. Garlic organopolysulfides and derivative thiosulfinates, such as allicin (Figure 7.1), were able to induce the cytoprotective enzyme glutathione S-transferase, as might be expected from their ability to modify cellular thiols. However, the monosulfide diallylsulfide (DAS) is also an inducer, and the mechanism by which it activates Nrf2 is yet to be delineated. In addition, the diallyl forms of the di- and tri-sulfides were much more potent than the propyl version. In surmising why the diallyl sufides might have higher potency, it is interesting to note that cytochrome P450 2E1 (CYP2E1) is implicated in sequential conversion of the sulfide to the corresponding sulfoxide (DASO) and sulfone (DASO2) derivatives [45]. Further oxidation of the sulfone metabolite generated an electrophile that was shown to act as a suicide inhibitor of CYP2E1, as well as other unidentified cytochromes implicated in bioactivation of various cytotoxins [45]. While the exact mechanism of this inhibition remains to be established, the irreversible nature of the antagonism implies generation of a reactive intermediate capable of covalent modification of the enzyme involving, in all likelihood, an active site cysteine [46]. In addition, LC–MS/MS analysis of bile fluids from rats treated with DAS, DASO and DASO2 identified several GSH conjugates, implicating epoxidation of the allylic group as an important metabolic activation step for all three compounds [47]. Unlike the epoxides of DAS and DASO, the DASO2-derived epoxide provides a unique entry into a sulfone-activated Michael acceptor containing a sulfonylprop-2-en-1-ol group (see Figure 7.3.B for the proposed mechanism). This so-called “soft” electrophile, with highly distributed charge density is much more likely to react with a thiolate, a similarly polarizable “soft” nucleophile, rather than the “hard” epoxide (see Section 7.3.3 for a brief discussion of hard and soft reagents, and other effects affecting electrophile chemoselectivity). Therefore, oxidation of the sulfide, in combination with epoxidation of the allylic group, could be responsible for the inactivation of CYP2E1 and, possibly, induction of Nrf2 by modifying sensor cysteines.
Furan-containing compounds, such as the diterpenes cafestol and kahweol, are known to act as the principal Nrf2 activators in coffee despite lacking a thiol-reactive functional group [48, 49]. Although the exact mechanism of induction is yet to be established, recent mass spectrometric study of bile fluids from mice injected with cafestol was interpreted by the authors to suggest that epoxidation is the key step in converting coffee furans into thiol-reactive species [50]. However, the study did not distinguish between direct addition to the epoxide and addition at a remote double bond conjugated to the epoxide or Michael addition to a γ-ketoenal, arising from ring-opening reaction of the oxidized furan (depicted in Figure 7.3.C). The distinction is important because furan epoxides and corresponding dicarbonyl derivatives have long been associated with severe cytotoxicity of furan-containing compounds [51], due to both high reactivity and tendency to react with oxygen and nitrogen nucleophiles, in addition to thiolates. This, however, is inconsistent with the generally cytoprotective properties of the coffee furans. The particular structural environment of electrophiles derived from these furans may account for the observed shift in the balance of toxic and protective effects (Figure 7.3.C). Thus, kahweol epoxide presents a doubly conjugated epoxide (epoxydiene) requiring a soft thiolate nucleophile to attack the soft electrophilic center next to a highly sterically congested quaternary carbon. The corresponding ketoenal is also deactivated toward non-specific additions by both cross-conjugation of the dienone functionality and β,β-dialkyl substitution of the unsaturated aldehyde [52]. Alternatively, participation of less reactive metabolites, such as conjugated lactones, for example, which have been detected as metabolic derivatives of furan-containing compounds [53], may account for the low toxicity of these phytochemicals. Further studies will be required for understanding the unique properties of these furans.
Indole-3-carbinol (I3C), a glucosinolate breakdown product, and its digestive product 3,3’-diindolylmethane (DIM) are ARE inducers [54], and have many other cancer chemopreventive effects [55]. However, while DIM has been described as an effective inducer of the ARE with little associated long-term toxicity [56], I3C has a weak level of Nrf2 activation and is associated with a complex interplay of pro- and anticarcinogen effects (for a review see [56]). Although no specific mechanism accounting for the ARE induction ability of DIM has been established, reaction schemes for producing electrophilic species capable of covalent modification of thiols could be inferred from known metabolic and chemical transformations of I3C, DIM, and 3-methylindole (3MI), a byproduct of tryptophan metabolism by intestinal microflora [57]. In the case of the latter compound, a highly electrophilic thiolate-reactive derivative 3-methyleneindolenine, an α,β-unsaturated imine, is produced via cytochrome P450-mediated dehydrogenation [58]. This species is postulated to be responsible for the cytotoxicity of this substituted indole [58, 59]. The reactivity of the α,β-unsaturated imine toward nucleophiles is promoted by a concomitant rearomatization of the indole nucleus, which makes it a rather non-discriminating and thus toxic agent, capable of modifying a variety of functionalities found in a cellular environment. 3-Methyleneindolenine also serves as an intermediate in the dehydration–conjugate addition–retro-aldol cascade in the acid-catalyzed conversion of I3C to DIM [60] (Figure 7.3.D). Therefore the low level of Nrf2 activation and pro- and anticarcinogen effects associated with I3C could be explained by the formation of both the toxic electrophile and DIM, respectively, in the course of the same chemical process. On the basis of established biotransformations of DIM [61], it is tempting to propose that an oxidation event similar to that seen for 3MI can lead to the extended conjugation-stabilized Michael acceptor, 3-(3-indolylmethylene)-indolenine (Figure 7.3.D). We must note that no thiol conjugates of DIM metabolites have been isolated, perhaps, due to reversibility of such additions to the conjugation-stabilized electrophile (see Section 7.3.2.4 for further discussion of reversibility importance). Significantly, a sulfate-conjugated hydration product of the proposed species has been isolated as one of the major biotransformants from cultured cancer cells [61]. The Michael acceptor produced by both I3C and 3MI may be the principal agent responsible for associated toxicities. Unlike 3MI, however, biotransformation of I3C can also lead to DIM, a more stable indole derivative [62] that is unlikely to produce a non-discriminating electrophile. This example underscores that the level of reactivity of phytochemical-derived electrophiles could be the key determinant in a sensitive balance of cytotoxic and cytoprotective effects.
Carotenoids, lycopene in particular, have been shown to activate the ARE via Nrf2 [63]. The carotenes are unsaturated hydrocarbons and thus contain no thiol-reactive species. However, the authors point out that the ethanolic extract of the lycopene preparation, containing unidentified hydrophilic lycopene derivatives, activated the ARE with a similar potency as lycopene. Therefore, it seems likely that oxidation of the polyene, leading to formation of an electrophilic species (e.g. citral [64]), is a prerequisite for Nrf2 activation.
In addition, there are a handful of phenols (non-quinoid forming) that have no electrophilic moieties, or facile non-enzymatic autoxidative paths to obtaining these moieties, which have been shown to activate cytoprotective enzymes, such as resveratrol [65] and galangin [28]. The Nrf2-dependence of ARE activation by resveratrol has been particularly well-established in studies in Nrf2−/− mice [9] and normal human small airway epithelial cells (SAEC) [66]. Resveratrol and other related phenols can be hydroxylated to quinoid-forming species by a member of the cytochrome P450 (CYP) superfamily of monooxygenases, such as pro-carcinogen activating CYP1A1, CYP1A2, and CYP1B1 [67, 68]. These enzymes are transcriptionally controlled by the aryl hydrocarbon receptor (AhR). So-called bifunctional ARE inducers, such as β-naphthoflavone (β-NF), activate Nrf2 by first binding to and activating AhR, which in turn leads to upregulation of CYP enzymes [69]. However, resveratrol is a known inhibitor of AhR [70, 71], and therefore in uninduced normal cells resveratrol may not be hydroxylated by CYPs prior to Nrf2 activation. Resveratrol activation of QR1 through the estrogen receptor β (ERβ) in breast cancer cells has been explored as another mechanistic possibility. Various studies support a model in which binding of phytoestrogens, including resveratrol, to ERβ causes ERβ to bind and activate the QR1 ARE [72-75]. However, this mechanism appears to be restricted to cancer cells overexpressing ERβ [75]. A third mechanism has been suggested based on the observation that resveratrol binds to and inhibits quinone reductase 2 (QR2) with low nanomolar affinity [76]. QR2's function is not clearly elucidated, but it is known to catalyze the reduction of quinones, among several other classes of electron-deficient compounds. The authors suggested that resveratrol may activate Nrf2 by inhibiting QR2, resulting in the accumulation of endogenous quinones that can then induce electrophilic stress by modifying cellular thiols [76]. A similar indirect induction mechanism could apply to other phytochemical inhibitors of QR2 that inhibit this reductase at physiologically relevant concentrations, including quercetin, kampferol, apigenin, or genistein [76, 77]. This hypothesis warrants further investigation. Finally, a fourth mechanism accounting for activation of Nrf2 by non-quinoid-forming phenols by means of kinase activation is explored for genistein in Section 7.3.4.3.
7.3.2.3 Role of ROS in Nrf2 Activation by Phenolic Compounds
One important area of consideration is to what extent ROS are involved in the activation of Nrf2 by phytochemical inducers. Phenolic ARE inducers can play multiple roles in redox status of a cell (Figure 7.2). They are able to act as both direct antioxidants, scavenging free radicals, and, if converted to thiolate-reactive electrophiles, as indirect antioxidants by upregulating antioxidant genes [78]. Depending on the experimental conditions, they can also act as pro-oxidants [79]. As shown in Figure 7.2, the non-enzymatic autoxidation of a polyphenol can lead to ROS formation. This generation of ROS may play a role in Nrf2 activation, as well as have potential off-target effects such as toxicity. The ability of ROS to upregulate Nrf2 is well-established, for example by treatment of cells with H2O2 [80]. One mechanism by which ROS may activate Nrf2 is depicted in Figure 7.2. In this scenario, generation of superoxide and other ROS can lead to oxidation of GSH to GSSG [81]. GSSG then could modify sensor thiolates such as Keap1 cysteines [82, 83], thereby activating Nrf2 (see also section 7.3.4.1). In addition, sensor thiolates can be directly oxidized by ROS (reviewed in [84] and [85]).
There are indications that ROS mediate signaling for some ARE inducers. For example, the generation of ROS in cells [86, 87] and cell media [88] by EGCG has been well established. Importantly, ROS-scavengers NAC, GSH, superoxide dismutase (SOD), and catalase all inhibited the induction of heme oxygenase 1 (HO-1) by EGCG, as shown in bovine aortic endothelial cells (BAECs) [89]. Thus, EGCG-induced ROS, rather than an EGCG-derived electrophile, mediated EGCG-induced HO-1 expression under these conditions. A role for ROS as a secondary messenger in Nrf2 activation, specifically H2O2, has also been shown for a different ARE inducer class, dithiolethiones, in Hepa 1c1c7 cells [80]. In addition, a role for ROS in Nrf2 activation by 21 flavonoids (including fisetin, kaempferol, and quercetin) was explored [90]. A high level of correlation was observed between the flavonoids’ ability to activate the ARE and their computed energy levels of the highest occupied molecular orbitals (EHOMO), representing the tendency of the flavonoid to donate electrons in redox processes, for example. Thus, more oxidizable flavonoids possessing less negative EHOMO values were generally more potent inducers of ARE-mediated gene expression. Therefore ROS may be important secondary messengers for flavonoids.
Interestingly, we note that there are three outliers in the flavonoid correlation analysis [90] with much greater abilities to activate the ARE than predicted by their EHOMO values (and hence their tendency to donate electrons in the formation of ROS). This boost in induction potency could be associated with a particular set of structural features. Thus, only these three flavonoids (quercetin, morin, myricetin) out of 21 tested can be predicted to form extended conjugation-stabilized quinone methides involving the C ring, as products of a two-electron oxidation sequence (Figure 7.3.A). We hypothesize that while generation of ROS by flavonoids may contribute to activation of Nrf2, the ability to form a conjugation-stabilized electrophile, which can react with thiolates such as quinoid, produces a much greater extent of ARE activation due to its ability to react directly with sensor thiolates, such as Keap1 cysteines. Importantly, the quinone methide derived from quercetin has a remarkably high stabilization effect of the extended conjugation, as shown by its ability to form reversible thiol adducts [34]. Finally, in support of this hypothesis, out of a series of five structurally similar flavonoids, only the quinoid-forming ones (kaempferol, quercetin and luteolin) were able to react with a thiol (GSH) to form mono- and bis-GSH conjugates, without forming radicals and ROS [91, 92]. In contrast, the other two flavonoids (apigenin and naringenin), which are not able to form conjugation-stabilized electrophiles, generated radicals and ROS and could not form conjugates with GSH.
In considering whether generation of ROS is a secondary messenger in Nrf2 activation by flavonoids and other phytochemicals, it is important to note that the two-electron oxidation of phenols needs not necessarily lead to ROS formation. If a polyphenol can be recognized as a substrate of an oxidizing metalloenzyme, it can be converted to the Michael acceptor without production of ROS (Figure 7.2). To our knowledge, this theory remains to be tested. Dosage is likely critical as to whether useful or harmful levels of ROS are produced by treatment with ARE inducers [93]. Much work remains to determine the role(s) that ROS play in Nrf2 activation by phytochemicals, including whether ROS generated by therapeutic and physiologically relevant concentrations of phytochemicals have deleterious off-target effects, or are relatively harmless and perhaps relevant for participating in numerous signaling mechanisms, which are beyond the scope of this Perspective (reviewed in [94], [95], and [85]). The dosage amount is likely very important, as illustrated through the example of the synthetic oleanane triterpenoid CDDO-Im. While at low concentrations (≤ 100 nM) CDDO-Im is a potent Nrf2 inducer with undetectable adverse effects, above 300 nM ROS-mediated toxicity is observed [96]).
7.3.3 Basis of Phytochemicals as Therapeutic, Rather than Toxic, Agents
The cytoprotective roles of phytochemicals discussed thus far could be considered surprising, given that most if not all were produced for biodefense against insects, bacterial parasites and other animals, including humans, consuming plant parts. The questions arises then as to how these molecules have a cumulatively cytoprotective effect. One critical factor is dosage, as at high concentrations most of these molecules have been shown to display at least some level of toxicity. At low concentrations then, the residual “toxicity” maintains cells in a state of adaptive stress, providing them tools for counteracting a variety of adverse conditions [97]. The ability of a phytochemical to induce such a state selectively plays a critical role in the cytotoxicity/cytoprotection balance. This balance appears to be rather sensitive to subtle changes in a chemical structure. Two considerations that appear to be significant are selectivity for stress-sensing cysteines and reversibility of thiol modification.
There are various factors that contribute to preference of an electrophile for stress-sensing cysteines. First, cytoprotective compounds (or their bioactivated derivatives) contain “soft” electrophilic centers, which generally favor an attack by a corresponding soft nucleophile, best represented in a cellular environment by a thiolate anion. In phytochemicals, soft electrophilicity is associated with a carbon center conjugated through a network of π-bonds to a reactive functional group, such as a carbonyl (Figure 7.3.A), sulfone (Figure 7.3.B), epoxide (Figure 7.3.C), or imine (Figure 7.3.D), or with an electrophilic center activated directly by large polarizable atoms (e.g. epithionitriles, isothiocyanates, etc.). The molecular orbital effects at the root of these preferences are important but are beyond the scope of this perspective (See [98] for relevant discussion). Soft electrophilicity, however, is not sufficient for shifting the therapeutic window toward cytoprotection, as highly reactive albeit soft agents will modify unintended thiols (i.e., hemoglobin, human serum albumin, glutathione, etc) [3] and even non-thiol targets (i.e., nucleic acids).
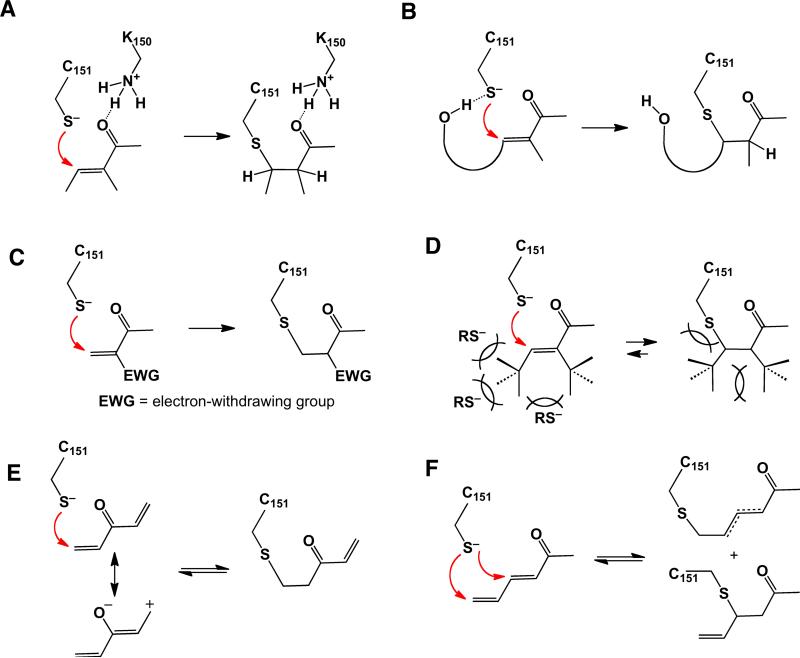
Second, stress-sensing cysteines are likely maintained by their immediate environment in a highly reactive state. This could include pKa reduction [99] and the presence of Brønsted acids to orient and activate the incoming electrophiles through hydrogen bonding [52] (See Figure 7.4.A). It is highly likely that one or several Keap1 cysteines are in fact presented in such an environment (see Section 7.3.4.1).
Fig 7.4.
Structural features of Michael acceptors affecting the rates of forward and reverse reactions with thiols. Keap1 C151 is shown as an example thiol. (A) Brønsted acid catalysis by a neighboring residue (Keap1 K150). (B) Neighboring group (proximity) catalysis through hydrogen bonding with a hydroxyl adjacent to the β-carbon. (C) Alkene polyactivation, favoring addition. (D) Steric congestion, preventing additions by less reactive nucleophiles via transition state crowding. Cross-conjugation (E) and extended conjugation (F) stabilize an electrophile by diluting the partial positive charges at the electrophilic centers and thereby reducing its reactivity and promoting reversibility.
Third, the structural context of presenting the electrophilic functionalities is likely to play an essential role in an electrophile's ability to react selectively. Thus, another contributing factor noted in enhancing reactivity with sulfur nucleophiles is the presence of a neighboring group, such as a hydroxyl functionality in a vicinity of a reactive β-carbon of cinnamates, chalcones, curcuminoids, bis(2-hydroxybenzylidene)-derivatives [30], and some withanolides [20]. A neighboring hydroxyl can guide an incoming thiolate anion via a transient charge-dipole interaction (hydrogen bond) resulting in a highly selective process that enhances the effectiveness of such agents (Figure 7.4.B).
In addition, a sterically hindered electrophile may exercise a superior degree of selectivity toward uniquely reactive thiolates (e.g., Keap1 cysteines). For example, both the high effectiveness and low toxicity profile of synthetic oleanane triterpenoids has been ascribed to a special combination of high intrinsic electrophilicity of the Michael acceptor activated by both keto and cyano groups (Figure 7.4.C) and sterics, specifically the presence of a Michael acceptor functionality directly adjacent to a highly congested quaternary carbon center [100] (Figure 7.4.D). Congestion in the vicinity of the reactive β-carbon reduces the reactivity of the Michael acceptor by crowding the transition state, where trigonal planar geometry is converted into a more sterically demanding tetrahedral state. This should allow only the most reactive thiolates to overcome the steric barrier and form covalent adducts. It must be noted, however, that mere reduction of reactivity without adjustment of other factors is expected to result in a situation where the enhanced selectivity and reduced toxicity come at the expense of potency. Thus, of two stereoisomers of guggulsterone, a terpenoid that lacks any known toxicity, only the E-form displays moderate ARE induction [101], due to, presumably, a poor accessibility by nucleophiles of the trisubstituted α,β-unsaturated ketone furnished with a quaternary center as the α-carbon substituent. The relatively low toxicity of coffee furans, as compared to other furans, can also be explained by steric factors, if the operative electrophile is a conjugated epoxydiene formed directly from kahweol (Figure 7.3.C) or after oxidative processing of cafestol.
The reversibility of addition, the fourth factor on our list, is emerging as a crucial characteristic of highly effective inducers with low associated toxicity [102]. Clearly, the entropically-favored reversal of electrophilic additions is expected to i) alleviate many potentially adverse outcomes associated with unintended alkylation events, ii) activate multiple stress sensors at a greatly reduced concentration, and iii) exhibit catalytic turnover (recycling) in maintaining the state of adaptive stress. ARE inducers must compete with prominent blood proteins—hemoglobin and serum albumin—and glutathione, among many other potential nucleophiles. Glutathione, the most available cellular thiol responsible for detoxification, can be conjugated with electrophiles in an uncatalyzed manner, or the conjugation can be catalyzed by glutathione S-transferases (GST) (reviewed in [103]). Once formed, if sufficiently stable, these conjugates are excreted from the cell by trans-membrane multidrug resistance-associated proteins (MRPs), leading to dilution of the ARE inducer and the signaling event (Figure 7.2). Thus, highly reversible ARE inducers may escape elimination from the cell by de-conjugating from GSH prior to being transported by an MRP, maintaining their cellular concentration more effectively than ARE inducers that form much more stable conjugates. For “reversible” ARE inducers, conjugation with glutathione has been hypothesized to be an intermediate step [104], leading via continuous de-conjugations to activation of less represented but highly reactive “sensory” thiolates, responsible for antioxidant effects (Figure 7.2).
For example, the reversible addition of sulforaphane to thiols has long been suspected to be responsible for pleiotropic nature of its cellular activity [78, 105]. In addition, the dramatically enhanced Nrf2-induction potential of CDDO and its variants over traditional phytochemical inducers may be due in large part to the facile reversibility of their thiol conjugates [100]. While a systematic analysis of chemical features responsible for the reversibility of thiol conjugations would benefit the field, certain generalizations about these features can be made herein. Thus, unusual stabilization of the electrophile through cross-conjugation (Figure 7.4.E) or extension of conjugation (Figure 7.4.F) could be invoked in a few cases documenting the favorability of addition reversal [34, 52, 100]. Both effects should lead to the enhanced delocalization of the partial positive charge induced by the associated carbonyl, reducing the electrophilicity of β- (δ-, etc.) carbon centers. For example, in celastrol, a quinone methide triterpene that has sub-micromolar potency for ARE activation, extended conjugation can account for the observed reversibility of its reaction with GSH [106]. Similarly, reversibility due to enhanced stabilization of the electrophile could explain the reduced toxicity and unique effectiveness of quercetin, when compared to structurally related flavonoids and flavonols in particular. Thus, the glutathionyl adducts of the quercetin-derived quinone methide have been shown to be reversible on the order of minutes to hours [34]. In addition, we suggest that reversibility of addition may play an important role in shifting the effectiveness-to-toxicity balance in yet-to-be established cases. For example, the divergent toxicity profiles of I3C and DIM (see section 7.3.2.1) could be explained by the reversibility of the DIM-derived thiol conjugate due to conjugation extension by a second aromatic group.
Alternatively, the destabilization of an adduct through steric congestion at positions adjacent to the nascent C–S bond may also facilitate elimination, as carbon hybridization changes from trigonal planar to tetrahedral. Thus, we hypothesize that the unusually low toxicity of CDDO [100] is likely the result of addition reversibility due to steric congestion in the corresponding adduct (Figure 7.4.D). In contrast, ARE inducers that do not form relatively reversible adducts, such as the EGCG o-quinone, for example, may be eliminated from the cell rather quickly through sequestration by GSH and subsequent elimination by an MRP. For these compounds, generation of superoxide and ROS as signaling intermediates may represent a more effective pathway for Nrf2 activation (Figure 7.2).
7.3.4 Mechanisms of Nrf2 Activation
7.3.4.1 Modification of Keap1 Cysteines: Identification of Key Sensors for ARE Inducers
A key mechanism by which phytochemicals activate Nrf2 is revealed by their common ability to react with cysteine residues. As mentioned in Section 7.3.2.1, the Nrf2-repressor protein Keap1 is particularly cysteine-rich. Keap1 directly binds to Nrf2 [31] and represses the transcription factor in at least two ways. First, Keap1 contains a Crm1-dependent nuclear export signal sequence that appears to be important for maintaining Nrf2 primarily in the cytoplasm under basal conditions [107-109]. In addition, and likely most important for phytochemical signaling, Keap1 serves as a bridge between Nrf2 and the Cul3-E3-ubiquitin ligase complex, leading to ubiquitination of Nrf2 and subsequent degradation by the 26S proteasome (reviewed in [110]). There are two separate sites in Nrf2 that bind to Keap1, termed the ETGE and DLG motifs [111]. Keap1 forms a homodimer through its N-terminal BTB domain, and it is the C-terminal Kelch domain of Keap1 that binds to the Nrf2 ETGE and DLG motifs. The seven lysines of Nrf2 that are targets for Cul3-mediated ubiquitination are located between the ETGE and DLG motifs [112]. Binding of the two Kelch domains of a homodimer of Keap1 to the ETGE and DLG sites of an Nrf2 protein molecule is believed to be critical for presentation of those lysines to an E2 ligase for ubiquitination [113]. Ubiquitination-directed degradation maintains the Nrf2 protein at a low level under normal conditions. Upon exposure to stress or compounds that modify cysteines, Nrf2 escapes Keap1 repression, and thereby is no longer ubiquitinated and degraded. This in turn leads to Nrf2 accumulation, resulting in activation of ARE-regulated genes.
Two primary methods have been used to illustrate which of the Keap1 cysteines (27 in human Keap1) are implicated in sensing ARE inducers: overexpression of Keap1 containing cysteine mutated to serine or alanine in mammalian cell lines or zebrafish, and mapping of cysteine modification in the purified protein by peptidic digestion and mass spectrometry. These complementary methods have revealed that a “cysteine code” may exist, in which particular types of ARE inducers react most readily with particular Keap1 cysteines. Cysteines 226 and 613 were shown to sense heavy metals [114], while C273 and C288 have been implicated in sensing alkenals, some cyclopentenone prostaglandins and nitro-fatty acids [114-116]. Keap1 C151 has been shown to be required for sensing many electrophilic ARE inducers [115, 117-121]. For example, a potent imidazole derivative of CDDO (CDDO-Im), tBHQ, a quinone-forming phenol, and sulforaphane are all highly dependent on C151 to upregulate the ARE [107, 115, 120]. Finally, the importance of Keap1 C151 in sensing ARE inducers is illustrated by the Keap1 C151S transgenic mouse [122]. This mouse is not only less responsive to tBHQ for upregulation of cytoprotective enzymes but also the basal levels of these enzymes are repressed [122]. Thus Keap1 C151 emerges as the key sensor for both endogenous agents as well as phytochemicals. The role of this cysteine as a sensor has been proposed to have evolved as a means of sensing nitric oxide [114]. Interestingly, mutation of C226 and C623 to serine has no effect on ARE activation by sulforaphane or tBHQ [114]. Therefore C151 is currently the only known Keap1 sensor cysteine for a phytochemical, sulforaphane, and a synthetic quinoid-forming polyphenol, tBHQ.
The role of Keap1 C151 as a principal sensor for phytochemicals is further supported by peptide mapping studies. Keap1 C151 is the only cysteine consistently and highly modified by all phytochemicals tested thus far: isoliquiritigenin, 10-shogaol, xanthohumol [123] and sulforaphane [124] (Figure 7.5). There have been discrepant results for modification of Keap1 cysteines, in particular C151 by sulforaphane. Initially, peptide-mapping studies of human Keap1 cysteines modified by sulforaphane found C151 to be one of the least readily modified cysteines (Figure 7.5)[125]. Based on the high dependence of sulforaphane ARE activation on C151, as well as the known labile, reversible nature of dithiocarbamates [126], i.e. the products of sulforaphane reaction with thiols, further experiments were conducted using a streamlined method to limit the reverse reaction after labeling Keap1 cysteines with sulforaphane [124]. Under these conditions, C151 emerged as one of the four most readily modified cysteines of Keap1. The adduction of Keap1 C151 by sulforaphane was also observed indirectly for Keap1 overexpressed in Cos1 cells using a biotin-switch technique, where C151 appeared to be modified to a greater extent than other cysteines [114]. The entire pattern of modification of Keap1 by sulforaphane generally shows large variability depending on the method used (Figure 7.5 and [124]), in particular as to whether C151 is detected as modified, illustrating the importance of considering the reversible nature of electrophile-cysteine adducts. Interestingly, several cysteines were detected as readily modified regardless of the method used, including C77, C226, C368, and C489 (Figure 7.5), indicating that certain dithiocarbamate-cysteine adducts are much more stable than others. The C151 dithiocarbamate adduct, on the other hand, is both one of the most reactive and reversible. Modeling of the BTB domain of Keap1 containing C151 [114, 120] depicts various residues nearby, including K131, R135, K150 and H154, that could participate both in lowering the pKa of C151 and providing acid/base catalysis for promoting both the forward (addition) and reverse (elimination) reactions. As a confirmation, a single mutation of K150 to threonine significantly reduces the response to C151-dependent ARE inducers [115].
Fig. 7.5.
Keap1 cysteines readily modified by phytochemicals, glutathione, and a quinone-forming polyphenol. Abbreviated name, chemical name and reference: XAN, xanthohumol [123]; ISO, isoliquiritigenin [123]; SHO, 10-shogaol [123]; SUL, sulforaphane [124, 125]; tBQ, tert-butylquinone [127]; and GSSG, oxidized glutathione [82]. Cysteines are ranked in order of most readily modified for all but tBQ, as determined by increasing concentrations of the electrophile, with the darkest circles being the most readily modified, and the empty circle indicating weakly modified or not modified cysteines. For tBQ, the cysteines are not ranked.
While the modification pattern of a quinoid-forming polyphenol phytochemicals has not yet been assessed, the oxidized form of tBHQ was found to react with Keap1 C151, shown directly for purified Keap1 [127] (Figure 7.5) and indirectly for Keap1 overexpressed in Cos1 cells [114]. Interestingly, C151 is not found to be readily modified by the oxidized form of GSH, GSSG [82] (Figure 7.5). Phytochemicals for which ROS production is a key means of activating Nrf2 (Section 7.3.2.3) likely activate Nrf2 by first generating GSSG (Figure 7.2). Thus, their Nrf2 activation would presumably be much less dependent on Keap1 C151.
While C151 is the cysteine most readily modified by phytochemicals based on current knowledge, other cysteines are sites of phytochemical addition in purified Keap1 protein (Figure 7.5). For example C319 is readily modified by all four phytochemicals shown in Figure 7.5 as well as oxidized glutathione. To our knowledge this cysteine has not yet been evaluated as a sensor for phytochemicals, and it has been shown not to be required to sense the soft cations, such as As3+ and Se4+, but there is an indication that it may play a role in sensing the Zn2+ ion [114].
Notably, none of the phytochemicals evaluated in Figure 7.5 modified C273 or C288 appreciably, although glutathione modified C288 to some extent. These cysteines are considered to be potential sensors, as mutation of these cysteines to serine renders Keap1 unable to repress Nrf2 [107, 122]. As these cysteines have been implicated in sensing more reactive Michael acceptors—alkenals, prostaglandins and nitro-fatty acids [114-116]—they may serve to detect primarily these endogenous signaling agents, rather than more stable phytochemical electrophiles. Keap1 C226 and C613, which sense heavy metal salts and other soft cations [114], are readily modified by phytochemicals, with isoliquiritigenin and sulforaphane targeting the former and xanthohumol, 10-shogaol, and to some extent, sulforaphane targeting the latter. However, as described above, mutation of C226 or C613 to serine had no effect on the ability of Keap1 to sense sulforaphane [114]. It is likely that the C151 residue present in these mutant proteins could be modified by sulforaphane, leading to a loss of Keap1 repression of Nrf2. Further work is required to ascertain whether modification of Keap1 cysteines other than C151 contributes to Nrf2 activation by phytochemicals.
7.3.4.2 Mechanism of Nrf2 activation upon modification of Keap1 Cysteines
The means by which Nrf2 escapes Keap1-Cul3-directed ubiquitination and degradation is still under active investigation. Disruption of the Keap1–Nrf2 interaction upon modification of Keap1 cysteines, allowing Nrf2 to escape Keap1, was originally proposed on the basis of experiments using recombinant proteins [126]. However, further investigations of a similar nature showed that the overall affinity of the Keap1-Nrf2 complex is maintained after modification of Keap1 at sensor cysteines, including C151 and C288, by ARE inducers including sulforaphane and isoliquiritigenin [128]. While a reduced affinity of Keap1 for Nrf2 after treatment of cells with tBHQ has also been observed by pull-down assays [129], other groups have reported that Keap1 and Nrf2 remain associated after treatment of cells with either tBHQ [107, 130], sulforaphane [107], a synthetic triterpenoid derivative, dh404 [131], or quercetin [132]. Pull-down assays are a useful but imprecise means of determining relative affinities of protein–protein interactions, making it difficult to ascertain whether the observed perturbation of the Keap1–Nrf2 complex occurs in response to inducers in a cellular context. It is noteworthy that preventing Nrf2 degradation is sufficient to promote Nrf2 nuclear accumulation and ARE activation [133]. It has been hypothesized that the affinity of the weaker Nrf2 DLG site is decreased after modification of Keap1 cysteines including C151, C273 and/or C288, leading to decreased ubiquitination, without affecting substantially the overall stability of the complex [122].
Significantly, the stability of the Keap1–Cul3 interaction, also essential for ubiquitination, is reduced upon treatment with ARE inducers as observed by pull-down assays [112, 134]. These studies show that the reduced affinity is dependent on Keap1 C151, as mutation of C151 to serine largely abolished the effect. Modification of the C151 residue has clearly been shown to disrupt Keap1-mediated ubiquitination of Nrf2, as mutation of C151 to tryptophan had the same effect as electrophile treatment of cells [120]. The Keap1 C151W mutant was largely unable to mediate Nrf2 ubiquitination, leading to stabilization of Nrf2 with concurrent activation of ARE-regulated genes, and had reduced affinity for Cul3, as assessed by the pull-down assay. Remarkably, Nrf2 binding to Keap1 in this assay was unaffected by the tryptophan mutation. While other Keap1 cysteines do very likely participate as well in Nrf2 activation, the results with Keap1 C151W in cells, as well as in a zebrafish model [115], show that modification of Keap1 C151 alone is sufficient for signaling for Nrf2 activation.
It has been proposed that the negative charge of the C151 thiolate anion is essential for Nrf2 turnover, and that modification of C151 by an electrophile neutralizes the negative charge, leading to the signaling event [114]. However, a series of 13 mutations at position 151 showed that the size of the residues was the key determinant, rather than hydrophobicity or charge, with the largest residue, tryptophan, showing the largest effect [120]. In particular, asparagine, with a relatively small partial molar volume (i.e. size) and no capacity to carry a negative charge, rendered the corresponding Keap1 mutant effective in suppressing Nrf2. Therefore, the size of the residue at position appears to be the major determining factor for Keap1's ability to repress Nrf2. Since all phytochemicals that were found to react with the C151 thiolate have partial molar volumes larger than that of tryptophan, they would all be large enough to trigger the signaling mechanism.
Alternatively, formation of disulfide bonds between Keap1 cysteines has been proposed to mediate loss of Nrf2 repression. Disulfide-linked Keap1 dimers have been observed in extracts of cells treated with the chalcone derivative bis(2-hydroxybenzylidene)acetone, which is a phenolic Michael acceptor [135]. In addition, treatment of cells with oxidizing agents (e.g., H2O2, nitric oxide, hypochlorous acid, or S-nitrosocysteine) leads to a Keap1 dimer linked through C151 [136]. Non-enzymatic oxidation of phenolic compounds will generate phenoxyl radicals of variable stability (Figure 7.2), and it has been proposed that these reactive intermediates could mediate the formation of thiyl radicals, promoting the formation of a disulfide bond or bonds between Keap1 cysteines [78]. Furthermore, both ROS and GSSG could also participate in Keap1 disulfide bond formation (Figure 7.2). In addition, a Keap1 dimer that was stable under reducing conditions was induced by treatment of cells with a quinoid-forming polyphenol, tBHQ [107]. Formation of this non-disulfide-linked dimer was dependent on C151. Radical-mediated hydrogen abstraction from surface tryptophans or tyrosines by subsequent cross-linking may account for the formation of the non-reducible dimer [137]. Further studies are required to determine on a structural/molecular level how direct modification of C151 and other Keap1 residues by ARE inducers or GSSG, or Keap1 dimer formation, lead to the loss of its ability to repress Nrf2.
7.3.4.3 Role of Protein Kinases in Nrf2 Activation by Phytochemicals
In addition to the direct Keap1-repression-inactivation mechanism discussed above, a number of kinases have been implicated in Nrf2 activation. The kinase pathways that currently appear to be the most important for Nrf2 activation by phytochemicals are the ones mediated by phosphatidylinositol 3-kinase (PI3K)/Akt and mitogen-activated protein kinases (MAPKs). The PI3K/Akt pathway mediates carnosol-induced expression of the cytoprotective enzyme HO-1 in rat pheochromocytoma PC12 cells [138]. In addition, EGCG, guggulsterone and piceatannol activate expression of cytoprotective enzymes including HO-1 in the human mammary epithelial (MCF10A) cells in a PI3K/Akt–dependent manner [101, 139, 140]. EGCG activation of Nrf2 in BAECs is also PI3K/Akt dependent [89]. The Nrf2 activation by CDDO-Im is dependent upon PI3K/Akt activity in ARPE-19 retinal epithelial cells [141].
The mechanism by which Nrf2 is regulated by PI3K/Akt has been investigated in some detail. The GSK-3β kinase, which is inactivated upon phosphorylation by Akt1, phosphorylates and negatively regulates Nrf2 via two distinct mechanisms [142, 143]. The phosphorylation of Nrf2 by GSK-3β leads to both cytoplasmic localization of Nrf2 [142], and Keap1-independent ubiquitination by the SCF/β-TrCP/Cul1 complex, leading to Nrf2 degradation [143]. A key cysteine residue, C124 in PTEN, a negative regulator of the PI3K/Akt axis [144], is directly modified by biotinylated CDDO in ARPE-19 cells [141], and the same cysteine is required for guggulsterone-induced Nrf2 accumulation in MCF10A cells [101]. Remaining to be demonstrated is whether the modification of PTEN C124 by electrophilic agents also inactivates GSK-3β, and this in turn leads to Nrf2 accumulation and Nrf2 nuclear localization.
While the inhibition of GSK-3β is implicated in the initial response to Nrf2 activators, the re-activation of GSK-3β has been proposed to occur several hours later, leading to eventual downregulation of the response by increasing Nrf2 nuclear export [145]. In this proposed mechanism, H2O2 treatment of cells activates GSK-3β, leading to subsequent activation of Fyn/Src kinases, which then phosphorylate Nrf2 Y568. Phosphorylation of Y568 enhances the interaction of Nrf2 with the Crm1 nuclear export protein. This mechanism has not yet been evaluated for phytochemical ARE inducers. However, these investigations were prompted by the observation that genistein, an ARE inducer, is a tyrosine kinase inhibitor. Whether inhibition of Nrf2 Y568 phosphorylation is in part or whole responsible for the ARE induction by genistein or other natural phenols is unknown.
The role of MAPKs in activation of Nrf2 is somewhat controversial. A large number of studies with various phytochemicals, including sulforaphane, phenethyl isothiocyanate (PEITC), curcumin, quercetin and EGCG, have implicated MAPKs in Nrf2 activation (reviewed in [146]). In order to assess the importance of this kinase family in Nrf2 signaling, MAPKs were overexpressed in HEK293T cells, and potential sites of MAPK-dependent phosphorylation on Nrf2 were identified by mass spectrometry. As a result, five phosphorylated residues were identified [146]. An Nrf2 mutant with all five sites mutated to alanine showed a moderate decrease in the transcriptional activity of Nrf2, concomitant with a slight reduction in its nuclear accumulation. However the stability of Nrf2 protein, which is primarily controlled by Keap1, was not affected. The general conclusion drawn from this work is that direct phosphorylation of Nrf2 by MAPKs has limited contribution to modulating Nrf2 activity [110, 146]. However, the Nrf2 protein with five mutated residues was only evaluated for blocked phosphorylation by overexpression of JNK2, rather than each MAPK relevant for Nrf2 activation [146]. Therefore, an additional site on Nrf2 beyond those five identified by mass spectrometry may in fact be a relevant target. Regardless of whether direct Nrf2 phosphorylation is the means by which MAPKs activate Nrf2, the sheer number of studies that implicate this pathway in Nrf2 activation by phytochemicals calls for further mechanistic investigation.
Activation of Nrf2 is also mediated by other kinases such as PERK [147, 148] and casein kinase 2 [149, 150]. However, to our knowledge, these pathways have not been evaluated for phytochemical activation of Nrf2. In addition, protein kinase C (PKC) isoforms are involved in the upregulation of cytoprotective genes by several phytochemicals. PKC-dependent ARE gene upregulation was observed for curcumin in HUH7 hepatoma cells and human monocytes [151, 152], piceatannol in BAECs [153], and epigallocatechin in human monocytic cells [154]. Nrf2 nuclear accumulation and ARE activation are also PKC-dependent for tBHQ in HepG2 and rat H4IIEC3 hepatoma cell lines [129, 155, 156], as well as nontumorigenic human keratinocytes (HaCaT cells) [157]. However, ARE activation by quercetin or kaempferol was not dependent on PKC in Hepa1c1c7 hepatoma cells [90], nor was diallyl trisulfide ARE activation dependent on PKC in HepG2 cells [158]. Further investigations are required to ascertain under what circumstances a phytochemical might activate Nrf2 through PKC isoforms.
7.4 Summary and Future Directions
Nrf2 is emerging as a master control of cytoprotective mechanisms important for defense against environmental challenges and stress factors. Phytochemical activation of Nrf2 promises to be an important mechanism in the prevention and amelioration of a wide variety of human diseases. In addition, Nrf2 is under investigation as a drug target [159], and there is much to be learned from investigations of phytochemical ARE inducers with regards to what makes an effective agent with minimal toxicity. A large effort has been put towards understanding the mechanisms by which Nrf2 is activated, although phytochemicals are still relatively understudied compared to other compounds [160].
In general, a common trait of agents that activate Nrf2 effectively is the ability to react with thiols either directly or upon bioactivation (spontaneous or enzyme-catalyzed). Alternative mechanisms of Nrf2 de-repression involving induction of oxidative stress, for example, have also been discussed. The electrophilic and/or oxidative nature of Nrf2 activators means that a balance between toxic and protective effects is necessary for safe administration of these agents. This balance is described in conventional drug development as a “therapeutic window” or effective dosage range. Within this range, beneficial effects are exhibited at optimal strength without being compromised by adverse processes. Our primary recommendation for future studies in phytochemical activation of Nrf2 is that, as with conventional drugs, efforts are put towards understanding how this dosage range is widened. This will enable identification or development of more effective agents for disease prevention and strategies for administering those agents.
Widening this dosage range does not appear to be a trivial exercise. The challenge lies in the fact that a strategy for increasing potency that merely relies on higher reactivity of these electrophiles is likely to enhance the extent of deleterious events. These may range from effect dilution through GSH conjugation followed by efflux to severe cytotoxicity associated with covalent modification of unintended targets. At the same time, reduction of toxicity through attenuation of electrophilicity would be expected to compromise potency. However, nature provides some important examples of how an effective therapeutic window can be achieved, which involves both partners in the modification reaction: sensor cysteines and naturally occurring cytoprotective agents.
The chemistry of the sensor cysteines plays an important role in ensuring advantageous biological effects of phytochemicals. The unique reactivity of soft thiolates of the sensor cysteines implicated in ARE induction, including Keap1 C151, ensures a high degree of “chemoselectivity” through the intrinsic preference for π-conjugated and thereby polarizable (soft) electrophilic centers, such as those found in effective phytochemical activators of Nrf2 (Figures 1 and Section 7.3.3). Chemoselectivity is further ensured by the high reactivity of the sensor cysteines implicated in ARE induction, including Keap1 C151. What remains to be explored is whether yet further preference toward particular sensor cysteines could be provided by the remaining structural features of the covalent modifiers, such as overall size or presence of particular functionalities that could interact favorably with the protein environment surrounding the thiol.
Certain phytochemical ARE inducers, either relatively potent ones or those with low or no observable toxicity, also provide clues as to features that widen the therapeutic window. Thus, preference for sensor thiolates can be enhanced by neighboring hydroxyl groups guiding a thiolate toward an attack on an electrophile. Alternatively, since sensor thiolates are so reactive, poor Michael acceptors could conjugate to them preferentially and, thereby, act as viable ARE inducers, while evading less reactive “off-target” nucleophiles. Examples of phytochemicals that are expected to be relatively unreactive Michael acceptors include those deactivated by alkene polysubstitution, bulky neighbors, and/or extended conjugation (Section 7.3.3). Remarkably, these deactivating factors are also expected to facilitate the reverse elimination process either by enhancing the stability of the electrophile, or by destabilizing the adduct. The reversibility of conjugates is emerging as an important feature for high potency and low toxicity. As discussed in Section 7.3.3, continuous de-conjugation may simultaneously contribute to both toxicity reduction due to instability of off-target conjugates and potency enhancement through efflux prevention. In addition, there is much to learn as to how modification of Keap1 cysteines leads to Nrf2 de-repression, for example, whether the size of modifiers influences the extent of the response, as suggested by a mutagenesis study [120].
Another factor important in the therapeutic window for phytochemicals is the ability to induce Nrf2 by a variety of pathways. Key sensor cysteines of phytochemical ARE inducers are being identified—Keap1 C151 and C124 of PTEN kinase—and other cysteines will likely emerge as important in sensing ARE inducers. Depending on the nature of the chemical and biological events associated with a given phytochemical in a given condition, different signaling “nodes” can be activated, all leading to Nrf2-ARE regulation. For example, the ultimate messenger molecule producing the Nrf2-dependent signal may not only be the phytochemical itself and/or its metabolite, but a concomitantly formed ROS/RNS or GSSG, as the GSH:GSSG ratio drops. These secondary signaling messengers target not only Keap1 and PTEN, but also numerous other kinases, including those involved in Nrf2 regulation. For example, various flavonoids including quercetin and EGCG can non-covalently associate with and modulate a number of protein kinases with high affinity (reviewed in [161]). There appears to be an intricate network of regulatory proteins that modulate Nrf2 activity, and multiple sensing steps can lead to the ability of Nrf2 upregulation to be tuned as is appropriate. Importantly, the presence of such a network implies that, if multiple nodes are activated by a single phytochemical, the convergence of the responses may lead to a more pronounced effect. This rationale also provides the basis for the unexpected effectiveness exhibited by mixtures of Nrf2 activators. Indeed, impressive synergism has been observed in a few studies conducted thus far, including for glucosinolate break-down products [54] and a commercially-available phytochemical supplement mixture [162]. Nrf2 activation, while clearly important, is certainly not the only mediator of the cytoprotective and disease-preventive attributes of phytochemicals (reviewed in [163]). Thus, phytochemicals that modulate a number of targets in disease prevention within their “therapeutic window” will likely be most effective in humans.
In summary, despite the multitude of cellular targets affected by phytochemicals, upregulation of Nrf2 is emerging as a key factor in their cytoprotective properties. And while it is unlikely that a single phytochemical or even a plant source will emerge as a magic bullet for disease prevention or amelioration, we expect that unraveling the chemical and biological aspects of their action may lead to unprecedented opportunities. These prospects could range from dietary/supplement recommendations to phytochemical “cocktails” specially formulated for synergistic effects and to nature-inspired synthetic molecules harnessing the most effective features of the plant-derived prototypes.
Acknowledgements
We thank Young-Joon Surh, Mark Hannink, Wendy A. Peer, Andrew D. Mesecar, and Albena Dinkova-Kostova for their thoughtful comments on this Perspective. We are grateful to National Institutes of Health for financial support through the R03 CA128095 grant.
Footnotes
Reference as: Eggler, A. L. and S. N. Savinov (2013). Chemical and Biological Mechanisms of Phytochemical Activation of NRF2 and Importance in Disease Prevention. 50 Years of Phytochemistry Research. D. R. Gang, Springer International Publishing. 43: 121-155.
ClinicalTrials.gov identifiers, in order as listed above. Amelioration studies: NCT01474993, NCT01315665, NCT01269723, NCT01183923, NCT00994604. Prevention studies: NCT00982319, NCT00255775, NCT00607932, NCT01437501, NCT00252018.
Clinicaltrials.gov identifier NCT01315665
Clinicaltrials.gov identifier NCT01335971
Contributor Information
Aimee L. Eggler, Department of Chemistry, Villanova University, 215a Mendel Science Hall, 800 Lancaster Avenue, Villanova, PA 19085
Sergey N. Savinov, Purdue University Center for Cancer Research, West Lafayette, Indiana, 47907
References
- 1.Pashkow FJ. Oxidative stress and inflammation in heart disease: do antioxidants have a role in treatment and/or prevention? Int J Inflam. 2011;2011:514623. doi: 10.4061/2011/514623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kasai H, Kawai K. Oxidative DNA damage: mechanisms and significance in health and disease. Antioxid Redox Signal. 2006;8:981–983. doi: 10.1089/ars.2006.8.981. [DOI] [PubMed] [Google Scholar]
- 3.Rappaport SM, Li H, Grigoryan H, Funk WE, Williams ER. Adductomics: Characterizing exposures to reactive electrophiles. Toxicol Lett. 2011 doi: 10.1016/j.toxlet.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malhotra D, Portales-Casamar E, Singh A, Srivastava S, Arenillas D, Happel C, Shyr C, Wakabayashi N, Kensler TW, Wasserman WW, Biswal S. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38:5718–5734. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayes JD, McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci. 2009;34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Zhao Z, Chen Y, Wang J, Sternberg P, Freeman ML, Grossniklaus HE, Cai J. Age-related retinopathy in NRF2-deficient mice. PLoS ONE. 2011;6:e19456. doi: 10.1371/journal.pone.0019456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen P-C, Vargas MR, Pani AK, Smeyne RJ, Johnson DA, Kan YW, Johnson JA. Nrf2-mediated neuroprotection in the MPTP mouse model of Parkinson's disease: Critical role for the astrocyte. Proc Natl Acad Sci USA. 2009;106:2933–2938. doi: 10.1073/pnas.0813361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sussan TE, Rangasamy T, Blake DJ, Malhotra D, El-Haddad H, Bedja D, Yates MS, Kombairaju P, Yamamoto M, Liby KT, Sporn MB, Gabrielson KL, Champion HC, Tuder RM, Kensler TW, Biswal S. Targeting Nrf2 with the triterpenoid CDDO- imidazolide attenuates cigarette smoke-induced emphysema and cardiac dysfunction in mice. Proc Natl Acad Sci USA. 2009;106:250–255. doi: 10.1073/pnas.0804333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ungvari Z, Bagi Z, Feher A, Recchia FA, Sonntag WE, Pearson K, de Cabo R, Csiszar A. Resveratrol confers endothelial protection via activation of the antioxidant transcription factor Nrf2. Am J Physiol Heart Circ Physiol. 2010;299:H18–24. doi: 10.1152/ajpheart.00260.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fahey JW, Haristoy X, Dolan PM, Kensler TW, Scholtus I, Stephenson KK, Talalay P, Lozniewski A. Sulforaphane inhibits extracellular, intracellular, and antibiotic-resistant strains of Helicobacter pylori and prevents benzo[α]pyrene-induced stomach tumors. Proc Natl Acad Sci USA. 2002;99:7610–7615. doi: 10.1073/pnas.112203099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Becks L, Prince M, Burson H, Christophe C, Broadway M, Itoh K, Yamamoto M, Mathis M, Orchard E, Shi R, McLarty J, Pruitt K, Zhang S, Kleiner-Hancock H. Aggressive mammary carcinoma progression in Nrf2 knockout mice treated with 7,12-dimethylbenz[a]anthracene. BMC Cancer. 2010;10:540. doi: 10.1186/1471-2407-10-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu C, Huang M-T, Shen G, Yuan X, Lin W, Khor TO, Conney AH, Kong A-NT. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2–related factor 2. Cancer Res. 2006;66:8293–8296. doi: 10.1158/0008-5472.CAN-06-0300. [DOI] [PubMed] [Google Scholar]
- 13.Zhao J, Moore AN, Redell JB, Dash PK. Enhancing expression of Nrf2-driven genes protects the blood brain barrier after brain injury. J Neurosci. 2007;27:10240–10248. doi: 10.1523/JNEUROSCI.1683-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aoki Y, Sato H, Nishimura N, Takahashi S, Itoh K, Yamamoto M. Accelerated DNA adduct formation in the lung of the Nrf2 knockout mouse exposed to diesel exhaust. Toxicol Appl Pharmacol. 2001;173:154–160. doi: 10.1006/taap.2001.9176. [DOI] [PubMed] [Google Scholar]
- 15.Chan K, Kan YW. Nrf2 is essential for protection against acute pulmonary injury in mice. Proc Natl Acad Sci USA. 1999;96:12731–12736. doi: 10.1073/pnas.96.22.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enomoto A, Itoh K, Nagayoshi E, Haruta J, Kimura T, O'Connor T, Harada T, Yamamoto M. High sensitivity of Nrf2 knockout mice to acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes. Toxicol Sci. 2001;59:169–177. doi: 10.1093/toxsci/59.1.169. [DOI] [PubMed] [Google Scholar]
- 17.Braun S, Hanselmann C, Gassmann MG, auf dem Keller U, Born-Berclaz C, Chan K, Kan YW, Werner S. Nrf2 transcription factor, a novel target of keratinocyte growth factor action which regulates gene expression and inflammation in the healing skin wound. Mol Cell Biol. 2002;22:5492–5505. doi: 10.1128/MCB.22.15.5492-5505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sykiotis GP, Bohmann D. Stress-activated cap'n'collar transcription factors in aging and human disease. Sci Signal. 2010;3:re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prochaska HJ, Santamaria AB, Talalay P. Rapid detection of inducers of enzymes that protect against carcinogens. Proc Natl Acad Sci USA. 1992;89:2394–2398. doi: 10.1073/pnas.89.6.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinghorn AD, Su BN, Jang DS, Chang LC, Lee D, Gu JQ, Carcache-Blanco EJ, Pawlus AD, Lee SK, Park EJ, Cuendet M, Gills JJ, Bhat K, Park HS, Mata-Greenwood E, Song LL, Jang M, Pezzuto JM. Natural inhibitors of carcinogenesis. Planta Med. 2004;70:691–705. doi: 10.1055/s-2004-827198. [DOI] [PubMed] [Google Scholar]
- 21.Fahey JW, Zalcmann AT, Talalay P. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry. 2001;56:5–51. doi: 10.1016/s0031-9422(00)00316-2. [DOI] [PubMed] [Google Scholar]
- 22.Hecht SS. Inhibition of carcinogenesis by isothiocyanates. Drug Metab Rev. 2000;32:395–411. doi: 10.1081/dmr-100102342. [DOI] [PubMed] [Google Scholar]
- 23.Burow M, Wittstock U. Regulation and function of specifier proteins in plants. Phytochemistry Reviews. 2009;8:87–99. [Google Scholar]
- 24.Agerbirk NN, De Vos M, Kim JH, Jander G. Indole glucosinolate breakdown and its biological effects. Phytochemistry Reviews. 2009;8:101–120. [Google Scholar]
- 25.Talalay P, De Long MJ, Prochaska HJ. Identification of a common chemical signal regulating the induction of enzymes that protect against chemical carcinogenesis. Proc Natl Acad Sci USA. 1988;85:8261–8265. doi: 10.1073/pnas.85.21.8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murakami A, Takahashi D, Kinoshita T, Koshimizu K, Kim HW, Yoshihiro A, Nakamura Y, Jiwajinda S, Terao J, Ohigashi H. Zerumbone, a Southeast Asian ginger sesquiterpene, markedly suppresses free radical generation, proinflammatory protein production, and cancer cell proliferation accompanied by apoptosis: the α,β-unsaturated carbonyl group is a prerequisite. Carcinogenesis. 2002;23:795–802. doi: 10.1093/carcin/23.5.795. [DOI] [PubMed] [Google Scholar]
- 27.Su BN, Jung Park E, Vigo JS, Graham JG, Cabieses F, Fong HH, Pezzuto JM, Kinghorn AD. Activity-guided isolation of the chemical constituents of Muntingia calabura using a quinone reductase induction assay. Phytochemistry. 2003;63:335–341. doi: 10.1016/s0031-9422(03)00112-2. [DOI] [PubMed] [Google Scholar]
- 28.Uda Y, Price KR, Williamson G, Rhodes MJ. Induction of the anticarcinogenic marker enzyme, quinone reductase, in murine hepatoma cells in vitro by flavonoids. Cancer Lett. 1997;120:213–216. doi: 10.1016/s0304-3835(97)00311-x. [DOI] [PubMed] [Google Scholar]
- 29.Su BN, Gu J-Q, Kang Y-H, Park E-J, Pezzuto JM, Kinghorn AD. Induction of the phase II enzyme, quinone reductase, by withanolides and norwithanolides from Solanaceous species. Mini-Reviews in Organic Chemistry. 2004;1:115–123. [Google Scholar]
- 30.Dinkova-Kostova AT, Massiah MA, Bozak RE, Hicks RJ, Talalay P. Potency of Michael reaction acceptors as inducers of enzymes that protect against carcinogenesis depends on their reactivity with sulfhydryl groups. Proc Natl Acad Sci USA. 2001;98:3404–3409. doi: 10.1073/pnas.051632198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prochaska HJ, Bregman HS, De Long MJ, Talalay P. Specificity of induction of cancer protective enzymes by analogues of tert-butyl-4-hydroxyanisole (BHA). Biochem Pharmacol. 1985;34:3909–3914. doi: 10.1016/0006-2952(85)90443-5. [DOI] [PubMed] [Google Scholar]
- 33.Jacobs H, Moalin M, Bast A, van der Vijgh WJF, Haenen GRMM. An essential difference between the flavonoids monoHER and quercetin in their interplay with the endogenous antioxidant network. PLoS ONE. 2010;5:e13880. doi: 10.1371/journal.pone.0013880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Awad HM, Boersma MG, Boeren S, van Bladeren PJ, Vervoort J, Rietjens IMCM. Quenching of quercetin quinone/quinone methides by different thiolate scavengers: stability and reversibility of conjugate formation. Chem Res Toxicol. 2003;16:822–831. doi: 10.1021/tx020079g. [DOI] [PubMed] [Google Scholar]
- 35.Galati G, Moridani MY, Chan TS, O'Brien PJ. Peroxidative metabolism of apigenin and naringenin versus luteolin and quercetin: glutathione oxidation and conjugation. Free Radic Biol Med. 2001;30:370–382. doi: 10.1016/s0891-5849(00)00481-0. [DOI] [PubMed] [Google Scholar]
- 36.van Zanden JJ, Ben Hamman O, van Iersel MLPS, Boeren S, Cnubben NHP, Lo Bello M, Vervoort J, van Bladeren PJ, Rietjens IMCM. Inhibition of human glutathione S-transferase P1-1 by the flavonoid quercetin. Chem Biol Interact. 2003;145:139–148. doi: 10.1016/s0009-2797(02)00250-8. [DOI] [PubMed] [Google Scholar]
- 37.Ishii T, Mori T, Tanaka T, Mizuno D, Yamaji R, Kumazawa S, Nakayama T, Akagawa M. Covalent modification of proteins by green tea polyphenol (−)-epigallocatechin-3-gallate through autoxidation. Free Radic Biol Med. 2008;45:1384–1394. doi: 10.1016/j.freeradbiomed.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 38.Muzolf-Panek M, Gliszczyńska-Świgło A, de Haan L, Aarts JMMJG, Szymusiak H, Vervoort JM, Tyrakowska B, Rietjens IMCM. Role of catechin quinones in the induction of EpRE-mediated gene expression. Chem Res Toxicol. 2008;21:2352–2360. doi: 10.1021/tx8001498. [DOI] [PubMed] [Google Scholar]
- 39.Sang S, Lambert JD, Hong J, Tian S, Lee M-J, Stark RE, Ho C-T, Yang CS. Synthesis and structure identification of thiol conjugates of (−)-epigallocatechin gallate and their urinary levels in mice. Chem Res Toxicol. 2005;18:1762–1769. doi: 10.1021/tx050151l. [DOI] [PubMed] [Google Scholar]
- 40.Nakamura Y, Kumagai T, Yoshida C, Naito Y, Miyamoto M, Ohigashi H, Osawa T, Uchida K. Pivotal role of electrophilicity in glutathione S-transferase induction by tert-butylhydroquinone. Biochemistry. 2003;42:4300–4309. doi: 10.1021/bi0340090. [DOI] [PubMed] [Google Scholar]
- 41.Bensasson RV, Zoete V, Dinkova-Kostova AT, Talalay P. Two-step mechanism of induction of the gene expression of a prototypic cancer-protective enzyme by diphenols. Chem Res Toxicol. 2008;21:805–812. doi: 10.1021/tx7002883. [DOI] [PubMed] [Google Scholar]
- 42.Wang XJ, Hayes JD, Higgins LG, Wolf CR, Dinkova-Kostova AT. Activation of the NRF2 signaling pathway by copper-mediated redox cycling of para- and ortho-hydroquinones. Chem Biol. 2010;17:75–85. doi: 10.1016/j.chembiol.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 43.Dinkova-Kostova AT, Wang XJ. Induction of the Keap1/Nrf2/ARE pathway by oxidizable diphenols. Chem Biol Interact. 2011;192:101–106. doi: 10.1016/j.cbi.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 44.Sparnins VL, Barany G, Wattenberg LW. Effects of organosulfur compounds from garlic and onions on benzo[a]pyrene-induced neoplasia and glutathione S-transferase activity in the mouse. Carcinogenesis. 1988;9:131–134. doi: 10.1093/carcin/9.1.131. [DOI] [PubMed] [Google Scholar]
- 45.Brady JF, Ishizaki H, Fukuto JM, Lin MC, Fadel A, Gapac JM, Yang CS. Inhibition of cytochrome P-450 2E1 by diallyl sulfide and its metabolites. Chem Res Toxicol. 1991;4:642–647. doi: 10.1021/tx00024a008. [DOI] [PubMed] [Google Scholar]
- 46.Gurtoo HL, Marinello AJ, Struck RF, Paul B, Dahms RP. Studies on the mechanism of denaturation of cytochrome P-450 by cyclophosphamide and its metabolites. J Biol Chem. 1981;256:11691–11701. [PubMed] [Google Scholar]
- 47.Jin L, Baillie TA. Metabolism of the chemoprotective agent diallyl sulfide to glutathione conjugates in rats. Chem Res Toxicol. 1997;10:318–327. doi: 10.1021/tx9601768. [DOI] [PubMed] [Google Scholar]
- 48.Cavin C, Bezencon C, Guignard G, Schilter B. Coffee diterpenes prevent benzo[a]pyrene genotoxicity in rat and human culture systems. Biochem Biophys Res Commun. 2003;306:488–495. doi: 10.1016/s0006-291x(03)00994-x. [DOI] [PubMed] [Google Scholar]
- 49.Hwang YP, Jeong HG. The coffee diterpene kahweol induces heme oxygenase-1 via the PI3K and p38/Nrf2 pathway to protect human dopaminergic neurons from 6-hydroxydopamine-derived oxidative stress. FEBS Lett. 2008;582:2655–2662. doi: 10.1016/j.febslet.2008.06.045. [DOI] [PubMed] [Google Scholar]
- 50.van Cruchten STJ, de Haan LHJ, Mulder PPJ, Kunne C, Boekschoten MV, Katan MB, Aarts JMMJG, Witkamp RF. The role of epoxidation and electrophile-responsive element-regulated gene transcription in the potentially beneficial and harmful effects of the coffee components cafestol and kahweol. The Journal of Nutritional Biochemistry. 2010;21:757–763. doi: 10.1016/j.jnutbio.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 51.Boyd MR. Toxicity mediated by reactive metabolites of furans. Adv Exp Med Biol. 1981;136(Pt B):865–879. [PubMed] [Google Scholar]
- 52.Avonto C, Taglialatela-Scafati O, Pollastro F, Minassi A, Di Marzo V, De Petrocellis L, Appendino G. An NMR Spectroscopic Method to Identify and Classify Thiol-Trapping Agents: Revival of Michael Acceptors for Drug Discovery? Angewandte Chemie International Edition. 2011;50:467–471. doi: 10.1002/anie.201005959. [DOI] [PubMed] [Google Scholar]
- 53.Khojasteh-Bakht SC, Chen W, Koenigs LL, Peter RM, Nelson SD. Metabolism of (R)-(+)-pulegone and (R)-(+)-menthofuran by human liver cytochrome P-450s: evidence for formation of a furan epoxide. Drug Metab Dispos. 1999;27:574–580. [PubMed] [Google Scholar]
- 54.Saw CL-L, Cintrón M, Wu T-Y, Guo Y, Huang Y, Jeong W-S, Kong A-NT. Pharmacodynamics of dietary phytochemical indoles I3C and DIM: Induction of Nrf2-mediated phase II drug metabolizing and antioxidant genes and synergism with isothiocyanates. Biopharm Drug Dispos. 2011;32:289–300. doi: 10.1002/bdd.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aggarwal BB, Ichikawa H. Molecular targets and anticancer potential of indole-3-carbinol and its derivatives. Cell Cycle. 2005;4:1201–1215. doi: 10.4161/cc.4.9.1993. [DOI] [PubMed] [Google Scholar]
- 56.Leibelt DA, Hedstrom OR, Fischer KA, Pereira CB, Williams DE. Evaluation of chronic dietary exposure to indole-3-carbinol and absorption-enhanced 3,3′-diindolylmethane in sprague-dawley rats. Toxicol Sci. 2003;74:10–21. doi: 10.1093/toxsci/kfg103. [DOI] [PubMed] [Google Scholar]
- 57.Fordtran JS, Scroggie WB, Polter DE. Colonic absorption of tryptophan metabolites in man. J Lab Clin Med. 1964;64:125–132. [PubMed] [Google Scholar]
- 58.Thornton-Manning J, Appleton ML, Gonzalez FJ, Yost GS. Metabolism of 3-methylindole by vaccinia-expressed P450 enzymes: correlation of 3-methyleneindolenine formation and protein-binding. J Pharmacol Exp Ther. 1996;276:21–29. [PubMed] [Google Scholar]
- 59.Regal KA, Laws GM, Yuan C, Yost GS, Skiles GL. Detection and characterization of DNA adducts of 3-methylindole. Chem Res Toxicol. 2001;14:1014–1024. doi: 10.1021/tx0100237. [DOI] [PubMed] [Google Scholar]
- 60.Grose KR, Bjeldanes LF. Oligomerization of indole-3-carbinol in aqueous acid. Chem Res Toxicol. 1992;5:188–193. doi: 10.1021/tx00026a007. [DOI] [PubMed] [Google Scholar]
- 61.Staub RE, Onisko B, Bjeldanes LF. Fate of 3,3′-diindolylmethane in cultured MCF-7 human breast cancer cells. Chem Res Toxicol. 2006;19:436–442. doi: 10.1021/tx050325z. [DOI] [PubMed] [Google Scholar]
- 62.Sepkovic DW, Bradlow HL, Bell M. Quantitative determination of 3,3′-diindolylmethane in urine of individuals receiving indole-3-carbinol. Nutr Cancer. 2001;41:57–63. doi: 10.1080/01635581.2001.9680612. [DOI] [PubMed] [Google Scholar]
- 63.Ben-Dor A, Steiner M, Gheber L, Danilenko M, Dubi N, Linnewiel K, Zick A, Sharoni Y, Levy J. Carotenoids activate the antioxidant response element transcription system. Mol Cancer Ther. 2005;4:177–186. [PubMed] [Google Scholar]
- 64.Lewinsohn E, Sitrit Y, Bar E, Azulay Y, Meir A, Zamir D, Tadmor Y. Carotenoid pigmentation affects the volatile composition of tomato and watermelon fruits, as revealed by comparative genetic analyses. J Agric Food Chem. 2005;53:3142–3148. doi: 10.1021/jf047927t. [DOI] [PubMed] [Google Scholar]
- 65.Kondratyuk TP, Park E-J, Marler LE, Ahn S, Yuan Y, Choi Y, Yu R, van Breemen RB, Sun B, Hoshino J, Cushman M, Jermihov KC, Mesecar AD, Grubbs CJ, Pezzuto JM. Resveratrol derivatives as promising chemopreventive agents with improved potency and selectivity. Mol Nutr Food Res. 2011;55:1249–1265. doi: 10.1002/mnfr.201100122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kode A, Rajendrasozhan S, Caito S, Yang S-R, Megson IL, Rahman I. Resveratrol induces glutathione synthesis by activation of Nrf2 and protects against cigarette smoke-mediated oxidative stress in human lung epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2008;294:L478–L488. doi: 10.1152/ajplung.00361.2007. [DOI] [PubMed] [Google Scholar]
- 67.Piver B, Fer M, Vitrac X, Merillon JM, Dreano Y, Berthou F, Lucas D. Involvement of cytochrome P450 1A2 in the biotransformation of trans-resveratrol in human liver microsomes. Biochem Pharmacol. 2004;68:773–782. doi: 10.1016/j.bcp.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 68.Potter GA, Patterson LH, Wanogho E, Perry PJ, Butler PC, Ijaz T, Ruparelia KC, Lamb JH, Farmer PB, Stanley LA, Burke MD. The cancer preventative agent resveratrol is converted to the anticancer agent piceatannol by the cytochrome P450 enzyme CYP1B1. Br J Cancer. 2002;86:774–778. doi: 10.1038/sj.bjc.6600197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rushmore TH, Kong AN. Pharmacogenomics, regulation and signaling pathways of phase I and II drug metabolizing enzymes. Curr Drug Metab. 2002;3:481–490. doi: 10.2174/1389200023337171. [DOI] [PubMed] [Google Scholar]
- 70.Casper RF, Quesne M, Rogers IM, Shirota T, Jolivet A, Milgrom E, Savouret J-F. Resveratrol has antagonist activity on the aryl hydrocarbon receptor: implications for prevention of dioxin toxicity. Mol Pharmacol. 1999;56:784–790. [PubMed] [Google Scholar]
- 71.Miao W, Hu L, Kandouz M, Batist G. Oltipraz is a bifunctional inducer activating both phase I and phase II drug-metabolizing enzymes via the xenobiotic responsive element. Mol Pharmacol. 2003;64:346–354. doi: 10.1124/mol.64.2.346. [DOI] [PubMed] [Google Scholar]
- 72.Montano MM, Katzenellenbogen BS. The quinone reductase gene: A unique estrogen receptor-regulated gene that is activated by antiestrogens. Proc Natl Acad Sci USA. 1997;94:2581–2586. doi: 10.1073/pnas.94.6.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Montano MM, Jaiswal AK, Katzenellenbogen BS. Transcriptional regulation of the human quinone reductase gene by antiestrogen-liganded estrogen receptor-alpha and estrogen receptor-beta. J Biol Chem. 1998;273:25443–25449. doi: 10.1074/jbc.273.39.25443. [DOI] [PubMed] [Google Scholar]
- 74.Montano MM, Deng H, Liu M, Sun X, Singal R. Transcriptional regulation by the estrogen receptor of antioxidative stress enzymes and its functional implications. Oncogene. 2004;23:2442–2453. doi: 10.1038/sj.onc.1207358. [DOI] [PubMed] [Google Scholar]
- 75.Bianco NR, Chaplin LJ, Montano MM. Differential induction of quinone reductase by phytoestrogens and protection against oestrogen-induced DNA damage. Biochem J. 2005;385:279–287. doi: 10.1042/BJ20040959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Buryanovskyy L, Fu Y, Boyd M, Ma Y, Hsieh T-c, Wu JM, Zhang Z. Crystal structure of quinone reductase 2 in complex with resveratrol. Biochemistry. 2004;43:11417–11426. doi: 10.1021/bi049162o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Boutin JA, Chatelain-Egger F, Vella F, Delagrange P, Ferry G. Quinone reductase 2 substrate specificity and inhibition pharmacology. Chem Biol Interact. 2005;151:213–228. doi: 10.1016/j.cbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 78.Dinkova-Kostova AT, Talalay P. Direct and indirect antioxidant properties of inducers of cytoprotective proteins. Mol Nutr Food Res. 2008;52:S128–S138. doi: 10.1002/mnfr.200700195. [DOI] [PubMed] [Google Scholar]
- 79.Nakamura Y, Miyoshi N. Electrophiles in Foods: The Current Status of Isothiocyanates and Their Chemical Biology. Biosci Biotechnol Biochem. 2010;74:242–255. doi: 10.1271/bbb.90731. [DOI] [PubMed] [Google Scholar]
- 80.Holland R, Navamal M, Velayutham M, Zweier JL, Kensler TW, Fishbein JC. Hydrogen peroxide is a second messenger in phase 2 enzyme induction by cancer chemopreventive dithiolethiones. Chem Res Toxicol. 2009;22:1427–1434. doi: 10.1021/tx900110n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Winterbourn CC. Superoxide as an intracellular radical sink. Free Radic Biol Med. 1993;14:85–90. doi: 10.1016/0891-5849(93)90512-s. [DOI] [PubMed] [Google Scholar]
- 82.Holland R, Hawkins AE, Eggler AL, Mesecar AD, Fabris D, Fishbein JC. Prospective type 1 and type 2 disulfides of Keap1 protein. Chem Res Toxicol. 2008;21:2051–2060. doi: 10.1021/tx800226m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mieyal JJ, Gallogly MM, Qanungo S, Sabens EA, Shelton MD. Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid Redox Signal. 2008;10:1941–1988. doi: 10.1089/ars.2008.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Na H-K, Surh Y-J. Transcriptional regulation via cysteine thiol modification: A novel molecular strategy for chemoprevention and cytoprotection. Mol Carcinog. 2006;45:368–380. doi: 10.1002/mc.20225. [DOI] [PubMed] [Google Scholar]
- 85.Janssen-Heininger YMW, Mossman BT, Heintz NH, Forman HJ, Kalyanaraman B, Finkel T, Stamler JS, Rhee SG, van der Vliet A. Redox-based regulation of signal transduction: Principles, pitfalls, and promises. Free Radic Biol Med. 2008;45:1–17. doi: 10.1016/j.freeradbiomed.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nakagawa H, Hasumi K, Woo J-T, Nagai K, Wachi M. Generation of hydrogen peroxide primarily contributes to the induction of Fe(II)-dependent apoptosis in Jurkat cells by (−)-epigallocatechin gallate. Carcinogenesis. 2004;25:1567–1574. doi: 10.1093/carcin/bgh168. [DOI] [PubMed] [Google Scholar]
- 87.Yang G-Y, Liao J, Li C, Chung J, Yurkow EJ, Ho C-T, Yang CS. Effect of black and green tea polyphenols on c-jun phosphorylation and H2O2 production in transformed and non-transformed human bronchial cell lines: possible mechanisms of cell growth inhibition and apoptosis induction. Carcinogenesis. 2000;21:2035–2039. doi: 10.1093/carcin/21.11.2035. [DOI] [PubMed] [Google Scholar]
- 88.Hou Z, Sang S, You H, Lee M-J, Hong J, Chin K-V, Yang CS. Mechanism of action of (−)-epigallocatechin-3-gallate: auto-oxidation-dependent inactivation of epidermal growth factor receptor and direct effects on growth inhibition in human esophageal cancer KYSE 150 cells. Cancer Res. 2005;65:8049–8056. doi: 10.1158/0008-5472.CAN-05-0480. [DOI] [PubMed] [Google Scholar]
- 89.Wu CC, Hsu MC, Hsieh CW, Lin JB, Lai PH, Wung BS. Upregulation of heme oxygenase-1 by Epigallocatechin-3-gallate via the phosphatidylinositol 3-kinase/Akt and ERK pathways. Life Sci. 2006;78:2889–2897. doi: 10.1016/j.lfs.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 90.Lee-Hilz YY, Boerboom A-MJF, Westphal AH, van Berkel WJH, Aarts JMMJG, Rietjens IMCM. Pro-oxidant activity of flavonoids induces EpRE-mediated gene expression. Chem Res Toxicol. 2006;19:1499–1505. doi: 10.1021/tx060157q. [DOI] [PubMed] [Google Scholar]
- 91.Galati G, Moridani MY, Chan TS, O'Brien PJ. Peroxidative metabolism of apigenin and naringenin versus luteolin and quercetin: glutathione oxidation and conjugation. Free Radic Biol Med. 2001;30:370–382. doi: 10.1016/s0891-5849(00)00481-0. [DOI] [PubMed] [Google Scholar]
- 92.Galati G, Chan T, Wu B, O'Brien PJ. Glutathione-dependent generation of reactive oxygen species by the peroxidase-catalyzed redox cycling of flavonoids. Chem Res Toxicol. 1999;12:521–525. doi: 10.1021/tx980271b. [DOI] [PubMed] [Google Scholar]
- 93.Liby KT, Yore MM, Sporn MB. Triterpenoids and rexinoids as multifunctional agents for the prevention and treatment of cancer. Nat Rev Cancer. 2007;7:357–369. doi: 10.1038/nrc2129. [DOI] [PubMed] [Google Scholar]
- 94.Forman HJ, Fukuto JM, Torres M. Redox signaling: thiol chemistry defines which reactive oxygen and nitrogen species can act as second messengers. Am J Physiol Cell Physiol. 2004;287:C246–256. doi: 10.1152/ajpcell.00516.2003. [DOI] [PubMed] [Google Scholar]
- 95.Magder S. Reactive oxygen species: toxic molecules or spark of life? Crit Care. 2006;10:208. doi: 10.1186/cc3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liby K, Hock T, Yore MM, Suh N, Place AE, Risingsong R, Williams CR, Royce DB, Honda T, Honda Y, Gribble GW, Hill-Kapturczak N, Agarwal A, Sporn MB. The synthetic triterpenoids, CDDO and CDDO-imidazolide, are potent inducers of heme oxygenase-1 and Nrf2/ARE signaling. Cancer Res. 2005;65:4789–4798. doi: 10.1158/0008-5472.CAN-04-4539. [DOI] [PubMed] [Google Scholar]
- 97.Calabrese EJ. Cancer biology and hormesis: human tumor cell lines commonly display hormetic (biphasic) dose responses. Crit Rev Toxicol. 2005;35:463–582. doi: 10.1080/10408440591034502. [DOI] [PubMed] [Google Scholar]
- 98.LoPachin RM, Barber DS, Gavin T. Molecular mechanisms of the conjugated alpha,beta-unsaturated carbonyl derivatives: relevance to neurotoxicity and neurodegenerative diseases. Toxicol Sci. 2008;104:235–249. doi: 10.1093/toxsci/kfm301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brandes N, Schmitt S, Jakob U. Thiol-based redox switches in eukaryotic proteins. Antioxid Redox Signal. 2009;11:997–1014. doi: 10.1089/ars.2008.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sporn MB, Liby KT, Yore MM, Fu L, Lopchuk JM, Gribble GW. New synthetic triterpenoids: potent agents for prevention and treatment of tissue injury caused by inflammatory and oxidative stress. J Nat Prod. 2011;74:537–545. doi: 10.1021/np100826q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Almazari I, Park J-M, Park S-A, Suh J-Y, Na H-K, Cha Y-N, Surh Y-J. Guggulsterone induces heme oxygenase-1 expression through activation of Nrf2 in human mammary epithelial cells: PTEN as a putative target. Carcinogenesis. 2012;33:368–376. doi: 10.1093/carcin/bgr259. [DOI] [PubMed] [Google Scholar]
- 102.Lin D, Saleh S, Liebler DC. Reversibility of covalent electrophile-protein adducts and chemical toxicity. Chem Res Toxicol. 2008;21:2361–2369. doi: 10.1021/tx800248x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 104.Talalay P, Fahey JW, Holtzclaw WD, Prestera T, Zhang Y. Chemoprotection against cancer by phase 2 enzyme induction. Toxicol Lett. 1995;82-83:173–179. doi: 10.1016/0378-4274(95)03553-2. [DOI] [PubMed] [Google Scholar]
- 105.Thornalley PJ. Isothiocyanates: mechanism of cancer chemopreventive action. Anticancer Drugs. 2002;13:331–338. doi: 10.1097/00001813-200204000-00001. [DOI] [PubMed] [Google Scholar]
- 106.Klaić L, Morimoto RI, Silverman RB. Celastrol analogues as inducers of the heat shock response. Design and synthesis of affinity probes for the identification of protein targets. ACS Chem Biol. 2012;7:928–937. doi: 10.1021/cb200539u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Karapetian RN, Evstafieva AG, Abaeva IS, Chichkova NV, Filonov GS, Rubtsov YP, Sukhacheva EA, Melnikov SV, Schneider U, Wanker EE, Vartapetian AB. Nuclear oncoprotein prothymosin alpha is a partner of Keap1: Implications for expression of oxidative stress-protecting genes. Mol Cell Biol. 2005;25:1089–1099. doi: 10.1128/MCB.25.3.1089-1099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Velichkova M, Hasson T. Keap1 regulates the oxidation-sensitive shuttling of Nrf2 into and out of the nucleus via a Crm1-dependent nuclear export mechanism. Mol Cell Biol. 2005;25:4501–4513. doi: 10.1128/MCB.25.11.4501-4513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hayes JD, McMahon M, Chowdhry S, Dinkova-Kostova AT. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid Redox Signal. 2010;13:1713–1748. doi: 10.1089/ars.2010.3221. [DOI] [PubMed] [Google Scholar]
- 111.Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol Cell Biol. 2006;26:2887–2900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang DD, Lo S-C, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. Dimerisation of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a 'tethering' mechanism: A two-site interaction model for the Nrf2-Keap1 complex. J Biol Chem. 2006;281:24756–24768. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
- 114.McMahon M, Lamont DJ, Beattie KA, Hayes JD. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc Natl Acad Sci USA. 2010;107:18838–18843. doi: 10.1073/pnas.1007387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kobayashi M, Li L, Iwamoto N, Nakajima-Takagi Y, Kaneko H, Nakayama Y, Eguchi M, Wada Y, Kumagai Y, Yamamoto M. The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol Cell Biol. 2009;29:493–502. doi: 10.1128/MCB.01080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kansanen E, Bonacci G, Schopfer FJ, Kuosmanen SM, Tong KI, Leinonen H, Woodcock SR, Yamamoto M, Carlberg C, Ylä-Herttuala S, Freeman BA, Levonen A-L. Electrophilic nitro-fatty acids activate NRF2 by a KEAP1 cysteine 151-independent mechanism. J Biol Chem. 2011;286:14019–14027. doi: 10.1074/jbc.M110.190710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sakurai T, Kanayama M, Shibata T, Itoh K, Kobayashi A, Yamamoto M, Uchida K. Ebselen, a seleno-organic antioxidant, as an electrophile. Chem Res Toxicol. 2006;19:1196–1204. doi: 10.1021/tx0601105. [DOI] [PubMed] [Google Scholar]
- 118.Ohnuma T, Nakayama S, Anan E, Nishiyama T, Ogura K, Hiratsuka A. Activation of the Nrf2/ARE pathway via S-alkylation of cysteine 151 in the chemopreventive agent-sensor Keap1 protein by falcarindiol, a conjugated diacetylene compound. Toxicol Appl Pharmacol. 2010;244:27–36. doi: 10.1016/j.taap.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 119.Satoh T, Okamoto S-i, Cui J, Watanabe Y, Furuta K, Suzuki M, Tohyama K, Lipton SA. From the Cover: Activation of the Keap1/Nrf2 pathway for neuroprotection by electrophillic phase II inducers. Proc Natl Acad Sci U S A. 2006;103:768–773. doi: 10.1073/pnas.0505723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Eggler AL, Small E, Hannink M, Mesecar AD. Cul3-mediated Nrf2 ubiquitination and antioxidant response element (ARE) activation are dependent on the partial molar volume at position 151 of Keap1. Biochem J. 2009;422:171–180. doi: 10.1042/BJ20090471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hur W, Sun Z, Jiang T, Mason DE, Peters EC, Zhang DD, Luesch H, Schultz PG, Gray NS. A small-molecule inducer of the antioxidant response element. Chem Biol. 2010;17:537–547. doi: 10.1016/j.chembiol.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 122.Yamamoto T, Suzuki T, Kobayashi A, Wakabayashi J, Maher J, Motohashi H, Yamamoto M. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol Cell Biol. 2008;28:2758–2770. doi: 10.1128/MCB.01704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Luo Y, Eggler AL, Liu D, Liu G, Mesecar AD, van Breemen RB. Sites of alkylation of human Keap1 by natural chemoprevention agents. J Am Soc Mass Spectrom. 2007;18:2226–2232. doi: 10.1016/j.jasms.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hu C, Eggler AL, Mesecar AD, van Breemen RB. Modification of Keap1 cysteine residues by sulforaphane. Chem Res Toxicol. 2011;24:515–521. doi: 10.1021/tx100389r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hong F, Freeman ML, Liebler DC. Identification of sensor cysteines in human Keap1 modified by the cancer chemopreventive agent sulforaphane. Chem Res Toxicol. 2005;18:1917–1926. doi: 10.1021/tx0502138. [DOI] [PubMed] [Google Scholar]
- 126.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci USA. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Abiko Y, Miura T, Phuc BH, Shinkai Y, Kumagai Y. Participation of covalent modification of Keap1 in the activation of Nrf2 by tert-butylbenzoquinone, an electrophilic metabolite of butylated hydroxyanisole. Toxicol Appl Pharmacol. 2011;255:32–39. doi: 10.1016/j.taap.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 128.Eggler AL, Liu G, Pezzuto JM, van Breemen RB, Mesecar AD. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc Natl Acad Sci U S A. 2005;102:10070–10075. doi: 10.1073/pnas.0502402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Niture SK, Jain AK, Jaiswal AK. Antioxidant-induced modification of INrf2 cysteine 151 and PKC-delta-mediated phosphorylation of Nrf2 serine 40 are both required for stabilization and nuclear translocation of Nrf2 and increased drug resistance. J Cell Sci. 2009;122:4452–4464. doi: 10.1242/jcs.058537. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 130.Kobayashi A, Kang M-I, Watai Y, Tong KI, Shibata T, Uchida K, Yamamoto M. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol Cell Biol. 2006;26:221–229. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ichikawa T, Li J, Meyer CJ, Janicki JS, Hannink M, Cui T. Dihydro-CDDO-trifluoroethyl amide (dh404), a novel Nrf2 activator, suppresses oxidative stress in cardiomyocytes. PLoS ONE. 2009;4:e8391. doi: 10.1371/journal.pone.0008391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tanigawa S, Fujii M, Hou D-X. Action of Nrf2 and Keap1 in ARE-mediated NQO1 expression by quercetin. Free Radic Biol Med. 2007;42:1690–1703. doi: 10.1016/j.freeradbiomed.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 133.Dreger H, Westphal K, Wilck N, Baumann G, Stangl V, Stangl K, Meiners S. Protection of vascular cells from oxidative stress by proteasome inhibition depends on Nrf2. Cardiovasc Res. 2010;85:395–403. doi: 10.1093/cvr/cvp279. [DOI] [PubMed] [Google Scholar]
- 134.Rachakonda G, Xiong Y, Sekhar KR, Stamer SL, Liebler DC, Freeman ML. Covalent modification at Cys151 dissociates the electrophile sensor Keap1 from the ubiquitin ligase CUL3. Chem Res Toxicol. 2008;21:705–710. doi: 10.1021/tx700302s. [DOI] [PubMed] [Google Scholar]
- 135.Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang MI, Kobayashi A, Yamamoto M, Kensler TW, Talalay P. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci USA. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fourquet S, Guerois R, Biard D, Toledano MB. Activation of NRF2 by nitrosative agents and H2O2 involves KEAP1 disulfide formation. J Biol Chem. 2010;285:8463–8471. doi: 10.1074/jbc.M109.051714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lund MN, Luxford C, Skibsted LH, Davies MJ. Oxidation of myosin by haem proteins generates myosin radicals and protein cross-links. Biochem J. 2008;410:565–574. doi: 10.1042/BJ20071107. [DOI] [PubMed] [Google Scholar]
- 138.Martin D, Rojo AI, Salinas M, Diaz R, Gallardo G, Alam J, De Galarreta CM, Cuadrado A. Regulation of heme oxygenase-1 expression through the phosphatidylinositol 3-kinase/Akt pathway and the Nrf2 transcription factor in response to the antioxidant phytochemical carnosol. J Biol Chem. 2004;279:8919–8929. doi: 10.1074/jbc.M309660200. [DOI] [PubMed] [Google Scholar]
- 139.Na H-K, Kim E-H, Jung J-H, Lee H-H, Hyun J-W, Surh Y-J. (−)-Epigallocatechin gallate induces Nrf2-mediated antioxidant enzyme expression via activation of PI3K and ERK in human mammary epithelial cells. Arch Biochem Biophys. 2008;476:171–177. doi: 10.1016/j.abb.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 140.Lee H-H, Park S-A, Almazari I, Kim E-H, Na H-K, Surh Y-J. Piceatannol induces heme oxygenase-1 expression in human mammary epithelial cells through activation of ARE-driven Nrf2 signaling. Arch Biochem Biophys. 2010;501:142–150. doi: 10.1016/j.abb.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 141.Pitha-Rowe I, Liby K, Royce D, Sporn M. Synthetic triterpenoids attenuate cytotoxic retinal injury: cross-talk between Nrf2 and PI3K/AKT signaling through inhibition of the lipid phosphatase PTEN. Invest Ophthalmol Vis Sci. 2009;50:5339–5347. doi: 10.1167/iovs.09-3648. [DOI] [PubMed] [Google Scholar]
- 142.Salazar M, Rojo AI, Velasco D, de Sagarra RM, Cuadrado A. Glycogen synthase kinase-3beta inhibits the xenobiotic and antioxidant cell response by direct phosphorylation and nuclear exclusion of the transcription factor Nrf2. J Biol Chem. 2006;281:14841–14851. doi: 10.1074/jbc.M513737200. [DOI] [PubMed] [Google Scholar]
- 143.Rada P, Rojo AI, Chowdhry S, McMahon M, Hayes JD, Cuadrado A. SCF/β-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol. 2011;31:1121–1133. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gericke A, Munson M, Ross AH. Regulation of the PTEN phosphatase. Gene. 2006;374:1–9. doi: 10.1016/j.gene.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 145.Jain AK, Jaiswal AK. GSK-3beta acts upstream of Fyn kinase in regulation of nuclear export and degradation of NF-E2 related factor 2. J Biol Chem. 2007;282:16502–16510. doi: 10.1074/jbc.M611336200. [DOI] [PubMed] [Google Scholar]
- 146.Sun Z, Huang Z, Zhang DD. Phosphorylation of Nrf2 at multiple sites by MAP kinases has a limited contribution in modulating the Nrf2-dependent antioxidant response. PLoS ONE. 2009;4:e6588. doi: 10.1371/journal.pone.0006588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cullinan SB, Zhang D, Hannink M, Arvisais E, Kaufman RJ, Diehl JA. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol Cell Biol. 2003;23:7198–7209. doi: 10.1128/MCB.23.20.7198-7209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cullinan SB, Diehl JA. Coordination of ER and oxidative stress signaling: The PERK/Nrf2 signaling pathway. The International Journal of Biochemistry & Cell Biology. 2006;38:317–332. doi: 10.1016/j.biocel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 149.Pi J, Bai Y, Reece JM, Williams J, Liu D, Freeman ML, Fahl WE, Shugar D, Liu J, Qu W, Collins S, Waalkes MP. Molecular mechanism of human Nrf2 activation and degradation: Role of sequential phosphorylation by protein kinase CK2. Free Radic Biol Med. 2007;42:1797–1806. doi: 10.1016/j.freeradbiomed.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Apopa PL, He X, Ma Q. Phosphorylation of Nrf2 in the transcription activation domain by casein kinase 2 (CK2) is critical for the nuclear translocation and transcription activation function of Nrf2 in IMR-32 neuroblastoma cells. J Biochem Mol Toxicol. 2008;22:63–76. doi: 10.1002/jbt.20212. [DOI] [PubMed] [Google Scholar]
- 151.McNally SJ, Harrison EM, Ross JA, Garden OJ, Wigmore SJ. Curcumin induces heme oxygenase 1 through generation of reactive oxygen species, p38 activation and phosphatase inhibition. Int J Mol Med. 2007;19:165–172. [PubMed] [Google Scholar]
- 152.Rushworth SA, Ogborne RM, Charalambos CA, O'Connell MA. Role of protein kinase C δ in curcumin-induced antioxidant response element-mediated gene expression in human monocytes. Biochem Biophys Res Commun. 2006;341:1007–1016. doi: 10.1016/j.bbrc.2006.01.065. [DOI] [PubMed] [Google Scholar]
- 153.Wung B-S, Hsu M-C, Wu C-C, Hsieh C-W. Piceatannol upregulates endothelial heme oxygenase-1 expression via novel protein kinase C and tyrosine kinase pathways. Pharmacol Res. 2006;53:113–122. doi: 10.1016/j.phrs.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 154.Ogborne RM, Rushworth SA, O'Connell MA. Epigallocatechin activates haem oxygenase-1 expression via protein kinase Cdelta and Nrf2. Biochem Biophys Res Commun. 2008;373:584–588. doi: 10.1016/j.bbrc.2008.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Numazawa S, Ishikawa M, Yoshida A, Tanaka S, Yoshida T. Atypical protein kinase C mediates activation of NF-E2-related factor 2 in response to oxidative stress. Am J Physiol Cell Physiol. 2003;285:C334–342. doi: 10.1152/ajpcell.00043.2003. [DOI] [PubMed] [Google Scholar]
- 156.Sherratt PJ, Huang HC, Nguyen T, Pickett CB. Role of protein phosphorylation in the regulation of NF-E2-related factor 2 activity. Methods Enzymol. 2004;378:286–301. doi: 10.1016/S0076-6879(04)78022-2. [DOI] [PubMed] [Google Scholar]
- 157.Zhu M, Zhang Y, Bowden GT. Involvement of mitogen-activated protein kinases and protein kinase C in regulation of antioxidant response element activity in human keratinocytes. Cancer Lett. 2006;244:220–228. doi: 10.1016/j.canlet.2005.12.023. [DOI] [PubMed] [Google Scholar]
- 158.Chen C, Pung D, Leong V, Hebbar V, Shen G, Nair S, Li W, Kong AN. Induction of detoxifying enzymes by garlic organosulfur compounds through transcription factor Nrf2: effect of chemical structure and stress signals. Free Radic Biol Med. 2004;37:1578–1590. doi: 10.1016/j.freeradbiomed.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 159.Kern JT, Hannink M, Hess JF. Disruption of the Keap1-containing ubiquitination complex as an antioxidant therapy. Curr Top Med Chem. 2007;7:972–978. doi: 10.2174/156802607780906825. [DOI] [PubMed] [Google Scholar]
- 160.Eggler AL, Gay KA, Mesecar AD. Molecular mechanisms of natural products in chemoprevention: induction of cytoprotective enzymes by Nrf2. Mol Nutr Food Res 52 Suppl. 2008;1:S84–94. doi: 10.1002/mnfr.200700249. [DOI] [PubMed] [Google Scholar]
- 161.Hou DX, Kumamoto T. Flavonoids as protein kinase inhibitors for cancer chemoprevention: direct binding and molecular modeling. Antioxid Redox Signal. 2010;13:691–719. doi: 10.1089/ars.2009.2816. [DOI] [PubMed] [Google Scholar]
- 162.Velmurugan K, Alam J, McCord JM, Pugazhenthi S. Synergistic induction of heme oxygenase-1 by the components of the antioxidant supplement Protandim. Free Radic Biol Med. 2009;46:430–440. doi: 10.1016/j.freeradbiomed.2008.10.050. [DOI] [PubMed] [Google Scholar]
- 163.Murakami A, Ohnishi K. Target molecules of food phytochemicals: Food science bound for the next dimension. Food Funct. 2012;3:462–476. doi: 10.1039/c2fo10274a. [DOI] [PubMed] [Google Scholar]