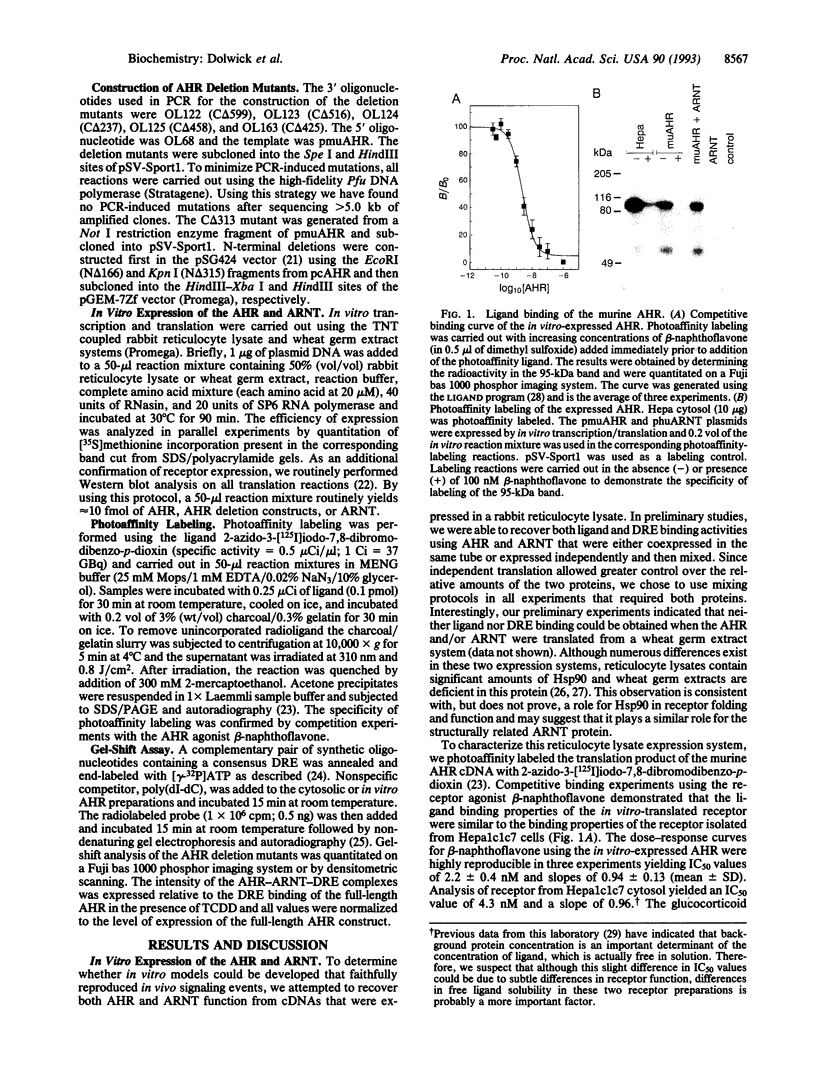
Abstract
The Ah receptor (AHR) is a basic helix-loop-helix protein that mediates the effects of 2,3,7,8-tetrachloro-dibenzo-p-dioxin. In this report, we describe a rabbit reticulocyte system that allows functional expression of both the AHR and its dimeric partner, the AHR nuclear translocator protein (ARNT). By using this in vitro system, we were able to reconstitute agonist binding to the AHR and agonist-induced AHR-ARNT recognition of a cognate DNA enhancer sequence. Expression of AHR deletion mutants revealed the location of N-terminal domains responsible for ligand and DNA recognition and C-terminal domains that play roles in agonist-induced DNA recognition.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blackwood E. M., Eisenman R. N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991 Mar 8;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Bradfield C. A., Kende A. S., Poland A. Kinetic and equilibrium studies of Ah receptor-ligand binding: use of [125I]2-iodo-7,8-dibromodibenzo-p-dioxin. Mol Pharmacol. 1988 Aug;34(2):229–237. [PubMed] [Google Scholar]
- Burbach K. M., Poland A., Bradfield C. A. Cloning of the Ah-receptor cDNA reveals a distinctive ligand-activated transcription factor. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):8185–8189. doi: 10.1073/pnas.89.17.8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalman F. C., Bresnick E. H., Patel P. D., Perdew G. H., Watson S. J., Jr, Pratt W. B. Direct evidence that the glucocorticoid receptor binds to hsp90 at or near the termination of receptor translation in vitro. J Biol Chem. 1989 Nov 25;264(33):19815–19821. [PubMed] [Google Scholar]
- Denis M., Gustafsson J. A. Translation of glucocorticoid receptor mRNA in vitro yields a nonactivated protein. J Biol Chem. 1989 Apr 15;264(11):6005–6008. [PubMed] [Google Scholar]
- Denison M. S., Fisher J. M., Whitlock J. P., Jr Protein-DNA interactions at recognition sites for the dioxin-Ah receptor complex. J Biol Chem. 1989 Oct 5;264(28):16478–16482. [PubMed] [Google Scholar]
- Denison M. S., Yao E. F. Characterization of the interaction of transformed rat hepatic cytosolic Ah receptor with a dioxin responsive transcriptional enhancer. Arch Biochem Biophys. 1991 Jan;284(1):158–166. doi: 10.1016/0003-9861(91)90278-q. [DOI] [PubMed] [Google Scholar]
- Durrin L. K., Jones P. B., Fisher J. M., Galeazzi D. R., Whitlock J. P., Jr 2,3,7,8-Tetrachlorodibenzo-p-dioxin receptors regulate transcription of the cytochrome P1-450 gene. J Cell Biochem. 1987 Oct;35(2):153–160. doi: 10.1002/jcb.240350208. [DOI] [PubMed] [Google Scholar]
- Elferink C. J., Gasiewicz T. A., Whitlock J. P., Jr Protein-DNA interactions at a dioxin-responsive enhancer. Evidence that the transformed Ah receptor is heteromeric. J Biol Chem. 1990 Nov 25;265(33):20708–20712. [PubMed] [Google Scholar]
- Ema M., Sogawa K., Watanabe N., Chujoh Y., Matsushita N., Gotoh O., Funae Y., Fujii-Kuriyama Y. cDNA cloning and structure of mouse putative Ah receptor. Biochem Biophys Res Commun. 1992 Apr 15;184(1):246–253. doi: 10.1016/0006-291x(92)91185-s. [DOI] [PubMed] [Google Scholar]
- Gielen J. E., Goujon F. M., Nebert D. W. Genetic regulation of aryl hydrocarbon hydroxylase induction. II. Simple Mendelian expression in mouse tissues in vivo. J Biol Chem. 1972 Feb 25;247(4):1125–1137. [PubMed] [Google Scholar]
- Godowski P. J., Picard D., Yamamoto K. R. Signal transduction and transcriptional regulation by glucocorticoid receptor-LexA fusion proteins. Science. 1988 Aug 12;241(4867):812–816. doi: 10.1126/science.3043662. [DOI] [PubMed] [Google Scholar]
- Hoffman E. C., Reyes H., Chu F. F., Sander F., Conley L. H., Brooks B. A., Hankinson O. Cloning of a factor required for activity of the Ah (dioxin) receptor. Science. 1991 May 17;252(5008):954–958. doi: 10.1126/science.1852076. [DOI] [PubMed] [Google Scholar]
- Huang Z. J., Edery I., Rosbash M. PAS is a dimerization domain common to Drosophila period and several transcription factors. Nature. 1993 Jul 15;364(6434):259–262. doi: 10.1038/364259a0. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987 Oct 26;15(20):8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Munson P. J., Rodbard D. Ligand: a versatile computerized approach for characterization of ligand-binding systems. Anal Biochem. 1980 Sep 1;107(1):220–239. doi: 10.1016/0003-2697(80)90515-1. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Nambu J. R., Lewis J. O., Wharton K. A., Jr, Crews S. T. The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell. 1991 Dec 20;67(6):1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- Nebert D. W., Gonzalez F. J. P450 genes: structure, evolution, and regulation. Annu Rev Biochem. 1987;56:945–993. doi: 10.1146/annurev.bi.56.070187.004501. [DOI] [PubMed] [Google Scholar]
- Neuhold L. A., Shirayoshi Y., Ozato K., Jones J. E., Nebert D. W. Regulation of mouse CYP1A1 gene expression by dioxin: requirement of two cis-acting elements during induction. Mol Cell Biol. 1989 Jun;9(6):2378–2386. doi: 10.1128/mcb.9.6.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdew G. H. Association of the Ah receptor with the 90-kDa heat shock protein. J Biol Chem. 1988 Sep 25;263(27):13802–13805. [PubMed] [Google Scholar]
- Poland A., Glover E. 2,3,7,8,-Tetrachlorodibenzo-p-dioxin: segregation of toxocity with the Ah locus. Mol Pharmacol. 1980 Jan;17(1):86–94. [PubMed] [Google Scholar]
- Poland A., Glover E., Bradfield C. A. Characterization of polyclonal antibodies to the Ah receptor prepared by immunization with a synthetic peptide hapten. Mol Pharmacol. 1991 Jan;39(1):20–26. [PubMed] [Google Scholar]
- Poland A., Glover E., Ebetino F. H., Kende A. S. Photoaffinity labeling of the Ah receptor. J Biol Chem. 1986 May 15;261(14):6352–6365. [PubMed] [Google Scholar]
- Poland A., Glover E., Kende A. S. Stereospecific, high affinity binding of 2,3,7,8-tetrachlorodibenzo-p-dioxin by hepatic cytosol. Evidence that the binding species is receptor for induction of aryl hydrocarbon hydroxylase. J Biol Chem. 1976 Aug 25;251(16):4936–4946. [PubMed] [Google Scholar]
- Poland A., Knutson J. C. 2,3,7,8-tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annu Rev Pharmacol Toxicol. 1982;22:517–554. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- Pongratz I., Mason G. G., Poellinger L. Dual roles of the 90-kDa heat shock protein hsp90 in modulating functional activities of the dioxin receptor. Evidence that the dioxin receptor functionally belongs to a subclass of nuclear receptors which require hsp90 both for ligand binding activity and repression of intrinsic DNA binding activity. J Biol Chem. 1992 Jul 5;267(19):13728–13734. [PubMed] [Google Scholar]
- Reyes H., Reisz-Porszasz S., Hankinson O. Identification of the Ah receptor nuclear translocator protein (Arnt) as a component of the DNA binding form of the Ah receptor. Science. 1992 May 22;256(5060):1193–1195. doi: 10.1126/science.256.5060.1193. [DOI] [PubMed] [Google Scholar]
- Sadowski I., Ptashne M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989 Sep 25;17(18):7539–7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telakowski-Hopkins C. A., King R. G., Pickett C. B. Glutathione S-transferase Ya subunit gene: identification of regulatory elements required for basal level and inducible expression. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1000–1004. doi: 10.1073/pnas.85.4.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner R., Tjian R. Leucine repeats and an adjacent DNA binding domain mediate the formation of functional cFos-cJun heterodimers. Science. 1989 Mar 31;243(4899):1689–1694. doi: 10.1126/science.2494701. [DOI] [PubMed] [Google Scholar]
- Van Doren K., Hanahan D., Gluzman Y. Infection of eucaryotic cells by helper-independent recombinant adenoviruses: early region 1 is not obligatory for integration of viral DNA. J Virol. 1984 May;50(2):606–614. doi: 10.1128/jvi.50.2.606-614.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Davis R., Tapscott S., Thayer M., Krause M., Benezra R., Blackwell T. K., Turner D., Rupp R., Hollenberg S. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991 Feb 15;251(4995):761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- Whitelaw M., Pongratz I., Wilhelmsson A., Gustafsson J. A., Poellinger L. Ligand-dependent recruitment of the Arnt coregulator determines DNA recognition by the dioxin receptor. Mol Cell Biol. 1993 Apr;13(4):2504–2514. doi: 10.1128/mcb.13.4.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]