Abstract
Human immunodeficiency virus type 1 (HIV-1) latency is a major barrier to a cure of AIDS. Latently infected cells harbor an integrated HIV-1 genome but are not actively producing HIV-1. Histone deacetylase (HDAC) inhibitors, such as vorinostat (SAHA), have been shown to reactivate latent HIV-1. AR-42, a modified HDAC inhibitor, has demonstrated efficacy against malignant melanoma, meningioma, and acute myeloid leukemia and is currently used in clinical trials for non-Hodgkin’s lymphoma and multiple myeloma. In this study, we evaluated the ability of AR-42 to reactivate HIV-1 in the two established CD4+ T-cell line models of HIV-1 latency. In HIV-1 chronically infected ACH-2 cells, AR-42-induced histone acetylation was more potent and robust than that of vorinostat. Although AR-42 and vorinostat were equipotent in their ability to reactivate HIV-1, AR-42-induced maximal HIV-1 reactivation was twofold greater than vorinostat in ACH-2 and J-Lat (clone 9.2) cells. These data provide rationale for assessing the efficacy of AR-42-mediated HIV-1 reactivation within primary CD4+ T-cells.
Keywords: HIV-1, histone deacetylase, HIV reactivation, kick and kill, AR-42
During primary infection, human immunodeficiency virus type 1 (HIV-1) infects permissive cells and converts its single-stranded RNA genome into a double-stranded DNA genome that integrates into the host-cell genome.1 A subset of the cells harboring integrated HIV-1, termed the latent reservoir, does not actively produce HIV-1 progeny and is thus refractory-to-current antiviral therapy.2,3 The posttranslational modifications of chromatin, such as histone deacetylation, cause chromatin condensation, which restricts RNA polymerase-mediated HIV-1 transcription and results in viral latency (reviewed in Siliciano and Greene).4 Previous reports have demonstrated the ability of histone deacetylase (HDAC) inhibitors, including vorinostat (also known as SAHA) and valproic acid, to reactivate latent HIV-1 through the reversal of chromatin condensation, although there have been inconsistent reports on the effectiveness of valproic acid.5,6 Clinical studies of vorinostat investigating the kick and kill strategy indicate consistent HIV-1 reactivation from cell lines and HIV-infected patients, but at high dosages.7,8 Additionally, recent studies with panobinostat and romidepsin in patients on suppressive antiretroviral therapy indicate the potential utility of more potent HDAC inhibitors.5
The histone deacetylation activity within chromatin indicates HDAC inhibitors as potentially valuable therapeutic agents for HIV-1 reactivation.9–11 Currently, the most potent HDAC inhibitors belong to the hydroxamic acid family.12 This class of HDAC inhibitors includes the US Food and Drug Administration-approved vorinostat and a novel compound AR-42.5,10 AR-42 is a novel anticancer drug candidate that inhibits deacetylation on both histone and nonhistone proteins.13,14 AR-42, a modified hydroxamic acid, was rationally designed with an aromatic linker and two Zn2+-binding motifs that bind a zinc cation in the catalytic domain of class I and II HDACs with an IC50 value of 30 nM.15
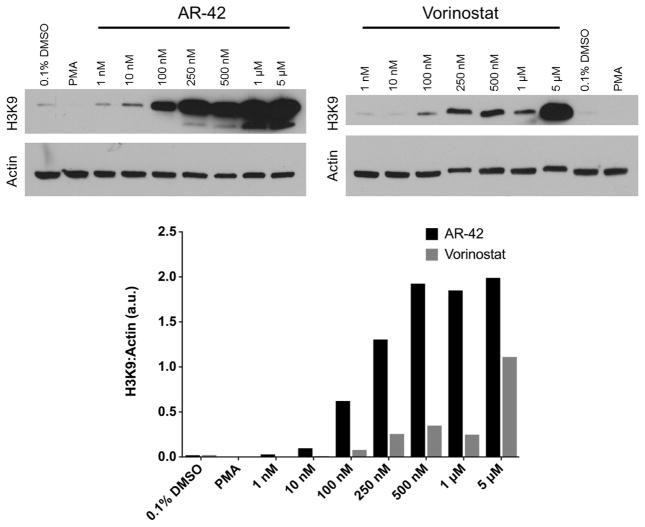
Published data indicated that AR-42 induces histone H3 acetylation in mouse and canine mast cells.16 To determine if AR-42 induces acetylation in cells harboring a HIV-1 provirus, we treated chronically and latently infected ACH-2 cells17 (obtained from Dr. Thomas Folks through the NIH AIDS Research and Reference Reagent Program) with a range of AR-42 (1 nM–5 μM). Following the treatment, cell lysates (15 μg) were electrophoresed on a 10% SDS-PAGE gel and transferred to nitrocellulose. Histone H3 acetylation on lysine 9 was assayed by western blot with the AcH3K9 antibody (Santa Cruz Biotechnology, Inc., 1:1500 dilution) and goat–anti-rabbit immunoglobulin/horseradish peroxidase secondary antibody (cell signaling, 1:5000 dilution). Equivalent protein loading was verified by western blot against actin (cell signaling 4967, 1:1500). Histone acetylation was quantified as a ratio to actin loading control by ImageJ densitometry analysis.
At 10 nM, AR-42 treatment increased histone 3 acetylation, while vorinostat induced acetylation at ~100 nM (Fig. 1). Within the concentrations tested, AR-42-induced histone 3 acetylation was more robust than vorinostat-induced acetylation. As expected, phorbol 12-myristate 13-acetate (PMA)-mediated HIV reactivation did not increase histone 3-acetylation.
Figure 1.
Vorinostat and AR-42 increase histone acetylation. Cellular lysates (15 μg) from ACH-2 cells were loaded per lane and probed with antibodies against acetylated histone H3 and actin. PMA treatments (0.1% DMSO and 100 ng/mL) were negative controls. AR-42 and vorinostat concentrations range from 1 nM to 5000 nM. Densitometry quantification of the actin-loading control and histone 3 acetylation (ImageJ) is displayed as the ratio of histone acetylation intensity to actin-loading control intensity.
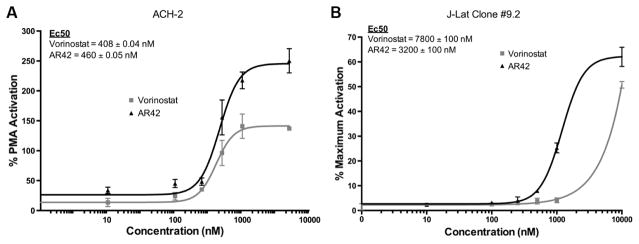
An outcome of histone acetylation in latently and chronically infected CD4+ T-cells is the reactivation of HIV-1. Expanding on AR-42’s ability to acetylate histone 3 (Fig. 1), we determined AR-42-induced HIV-1 reactivation within two well-established CD4+ T-cell models of HIV-1 latency.17,18 ACH-2 cells were maintained in Roswell Park Memorial Institute medium with 10% fetal bovine serum and penicillin–streptomycin at 37°C under 5% CO2. ACH-2 cells were treated with the indicated concentrations of vorinostat or AR-42 for 48 hours, in triplicate, at a final dimethyl sulfoxide (DMSO) concentration of 0.1%. A total of 100 ng/mL PMA (Sigma-Aldrich), also in 0.1% DMSO, was used as a positive control. After incubation, 10 μL of culture supernatant was removed, frozen at −80°C, thawed at room temperature, and then assayed for reverse transcriptase (RT) activity assays as described in Ball et al.19 HIV-1 reactivation was quantified using density (counts/mm2) counts computed by the Typhoon Scanner (GE Healthcare Life Sciences) and the Quantity One software (Bio-Rad Life Science Research). In the ACH-2 cell model, AR-42 reactivated HIV-1 in a dose-dependent manner, while vorinostat achieved a plateau at 500 nM (Fig. 2A). Although both AR-42 and vorinostat have similar potency (460 ± 0.05 nM and 408 ± 0.04 nM, respectively), at higher concentrations, AR-42 is twofold more efficacious than vorinostat in ACH-2 cells.
Figure 2.
AR-42 more effectively induces HIV-1 reactivation and expression from latently infected CD4+ T-cells than vorinostat. (A) RT activity of treatment over% PMA activation after 48 hours (average ± SD, n = 3). Calculated EC50 values for both AR-42 and vorinostat are depicted. (B) HIV-1 latently infected J-Lat cells (clone 9.2) were treated with AR-42 or vorinostat at the indicated concentrations for 24 hours, and GFP-positive cells were scored by flow cytometry. The maximum% of GFP-positive cells was determined with the positive control TNF-α (10 ng/ml), which was set to 100%, and the percentage of activation induced by each drug relative to TNF-α is presented.
The second T-cell model, Jurkat CD4+ T-cell-derived J-Lat cells (full length clone 9.2),18 was obtained from Dr. Eric Verdin through the NIH AIDS Research and Reference Reagent Program. J-Lat cells (clone 9.2) were cultured for 24 hours in the presence of 0.1% DMSO with or without AR-42 or vorinostat. Treatment with tumor necrosis factor alpha (TNF-α) (10 ng/mL) served as a positive control.18 Following the treatment, the cells were washed, fixed in 4% paraformaldehyde, and quantified by flow cytometry using Guava EasyCyte Mini (EMD Millipore). HIV-1 reactivation [green fluorescent protein (GFP) expression] was determined using the FlowJo software (Tree Star) with the gate equivalent to 0.1% DMSO-treated control cells. Additionally, the PRISM software was used to determine the half maximal effective concentration (EC50) for AR-42 and vorinostat. Flow cytometry analysis determined that in the J-Lat (clone 9.2) cell model, AR-42 is 2.4-fold more potent at HIV-1 reaction than vorinostat (EC50 values of 3200 ± 100 nM and 7800 ± 100 nM, respectively; Fig. 2B). Together, the ACH-2 and J-Lat (clone 9.2) data demonstrate that AR-42 can be more potent and efficacious than vorinostat in these HIV-1 reactivation cell line models.
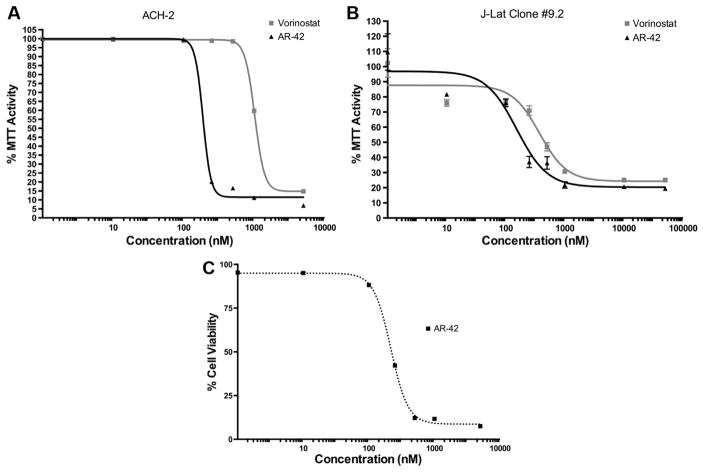
To determine the effect of treatments on cell viability, AR-42-treated cells were assayed using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)/3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay. The effects of AR-42 and vorinostat were tested for 48 hours and 24 hours, respectively, in ACH-2 and J-Lat (clone 9.2) cells. In ACH-2 cells, both vorinostat and AR-42 caused approximately similar reduction in MTT/MTS activity at 5 μM; although at lower treatment concentrations, vorinostat did not lower MTT/MTS activity >0.1% DMSO after 48 hours (Fig. 3A). In the J-Lat cells (clone 9.2), after 24 hours of treatment, the half cytotoxicity concentration (CC50) of AR-42 was 300 ± 100 nM, while that of vorinostat was 1300 ± 100 nM (Fig. 3B).
Figure 3.
AR-42 reduces the viability of latently infected CD4+ T-cells. (A) ACH-2 latently infected cells (48 hours). (B) J-Lat (clone 9.2) latently infected cells (24 hours). MTT or MTS cell viability assays were tested using vorinostat (SAHA) as a positive control. DMSO (0.1%) was used as a vehicle control. (C) Early apoptosis and necrosis (annexin V and propidium iodide staining) were tested in ACH-2 latently infected cells (black dotted) treated with 0.1% DMSO ± AR-42 for 48 hours.
In addition to MTT/MTS cell viability analysis, early apoptosis and necrosis studies were performed on AR-42-treated ACH-2 cells using annexin V and propidium iodide staining. Flow cytometry parameters for annexin V and propidium iodide were set based on heat-killed cells (incubated at 50°C for one hour) and performed using Beckman Coulter Cytomics FC500. Similar to the MTT/MTS results, AR-42 reduced the cell viability of ACH-2 cells at the CC50 of 217 ± 1 nM (Fig. 3C). These data suggest that AR-42 is more toxic than vorinostat in these two HIV-infected cell lines.
This study was designed to assess the ability of a novel HDAC inhibitor (AR-42) to reactivate HIV-1. We observed the following: AR-42 more potently induces histone 3 acetylation than vorinostat, AR-42 is more efficacious and equipotent than vorinostat in its ability to induce HIV-1 gene expression, and AR-42 is more toxic than vorinostat in two CD4+ T-cell line models of HIV-1 latency.
In the cellular models of schwannoma and meningioma, AR-42 inhibited cellular growth (IC50 values between 250 nM and 1 μM, depending on the cell line).20 In several models of non-Hodgkin’s lymphoma, AR-42 significantly enhanced the anti-tumor activity of HB22.7, an anti-CD22 monoclonal biologic.21 AR-42 is currently in two clinical trials: one for the treatment of non-Hodgkin’s lymphoma (NCT01798901) and the other for multiple myeloma (NCT01129193, www.clinicaltrials.gov). In the multiple myeloma phase I trial, a 40-mg dose of AR-42 achieved a maximum concentration (Cmax) of 1 μM, a concentration that is sufficient to reactivate HIV in the ACH-2 model.22,23 In the MT-2 and C8166 cellular models of cancers associated with the deltaretrovirus human T-lymphotropic virus type 1 (HTLV-1), AR-42 induces both histone acetylation and apoptosis; this study did not assess the ability of AR-42 to reactivate HTLV-1 gene expression.11 Furthermore, in a mouse model of HTVL-1-associated adult T-cell leukemia/lymphoma, AR-42 significantly increased animal survival compared to vehicle-treated control animals.11 Thus, AR-42 has promising activity against the cancers of various etiologies.
AR-42 treatment decreased MTT activity and cell viability at the treatment concentrations of 250 nM–1000 nM, although the cellular damage would not be attributed solely to drug treatment, because AR-42-induced HIV-1 release can also result in cell death. Previous studies have indicated that activated latently infected cells are presumed to die due to viral pathogenic effects, apoptosis, or pyroptosis.4,24 A strength of this study is that rather than assessing the supernatant-associated HIV RNA concentration following the reactivation, we assessed either intracellular GFP production (J-Lat cells clone 9.2) or RT activity deposited into the supernatant (ACH-2); both of these assays would not be confounded by HIV RNA or DNA, which could be liberated by cell death.
HIV-1 latently infected cell line models, as used in this study, have proven to be useful in investigating the induced reactivation of HIV from latently infected cells.25 Recognizing that individual HIV-1 latently infected cell models have limitations, we tested the ability of AR-42 to reactivate the HIV-1 gene expression in both the J-Lat cells (clone 9.2) and the ACH-2 models. Although there are slight differences between the results from the two cell lines, compared to vorinostat, AR-42 had at least one favorable pharmacological attribute in each model [ie, better efficacy in ACH-2 and better potency in J-Lat cells (clone 9.2)].
In summary, AR-42 potently induces histone acetylation in the ACH-2 cells and HIV-1 gene expression in the two models of latently infected CD4+ T-cells. These results (ie, favorable efficacy and toxicity profiles), combined with the ongoing AR-42 clinical studies, suggest that AR-42 should be tested in the primary cell models of HIV-1 latency.26
Acknowledgments
FUNDING: This work was supported in part by a research agreement from Arno Therapeutics, Inc. to the Center for Microbial Interface Biology, the infectious diseases AR-12 drug discovery program at the The Ohio State University (OSU). Arno Therapeutics, Inc. played no role in the study design, the interpretation of the results, or the decision to publish the results. LW is supported in part by the Public Health Preparedness for Infectious Diseases Program of the OSU and the NIH grants AI104483 and AI102822. JJK is supported in part by an NIH grant AI090644. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
Footnotes
COMPETING INTERESTS: Authors should state competing interests here in compliance with ICMJE conflict of interest rules (http://www.icmje.org/ethical_4conflicts.html). All authors should review the disclosure form (http://www.icmje.org/coi_instructions.html), returning it if there are disclosures to be made. If none exist, this text will be shown: “Author(s) disclose no potential conflicts of interest.”
PEER REVIEW: Four peer reviewers contributed to the peer review report. Reviewers’ reports totaled 575 words, excluding any confidential comments to the academic editor.
Author Contributions
Conceived and designed the experiments: JMM, SdeS, LW, JJK. Analyzed the data: JMM, SdeS, LW, JJK. Wrote the first draft of the manuscript: JMM, SdeS, LW, JJK. Contributed to the writing of the manuscript: ML, KVD, RAB. Agree with manuscript results and conclusions: JMM, SdeS, ML, KVD, RAB, LW, JJK. Jointly developed the structure and arguments for the paper: LW, JJK. Made critical revisions and approved final version: JMM, SdeS, ML, KVD, RAB, LW, JJK. All authors reviewed and approved of the final manuscript.
References
- 1.Chun TW, Engel D, Berrey MM, Shea T, Corey L, Fauci AS. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc Nat Acad Sci U S A. 1998;95:8869–8873. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choudhary SK, Margolis DM. Curing HIV: pharmacologic approaches to target HIV-1 latency. Annu Rev Pharmacol Toxicol. 2011;51:397–418. doi: 10.1146/annurev-pharmtox-010510-100237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. The challenge of finding a cure for HIV infection. Science. 2009;323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 4.Siliciano RF, Greene WC. HIV latency. Cold Spring Harbor Perspect Med. 2011;1:a007096. doi: 10.1101/cshperspect.a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasmussen TA, Schmeltz Søgaard O, Brinkmann C, et al. Comparison of HDAC inhibitors in clinical development: effect on HIV production in latently infected cells and T-cell activation. Hum Vaccin Immunother. 2013;9:993–1001. doi: 10.4161/hv.23800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Routy JP, Tremblay CL, Angel JB, et al. Valproic acid in association with highly active antiretroviral therapy for reducing systemic HIV-1 reservoirs: results from a multicentre randomized clinical study. HIV Med. 2012;13:291–296. doi: 10.1111/j.1468-1293.2011.00975.x. [DOI] [PubMed] [Google Scholar]
- 7.Archin NM, Liberty AL, Kashuba AD, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blazkova J, Chun TW, Belay BW, et al. Effect of histone deacetylase inhibitors on HIV production in latently infected, resting CD4(+) T cells from infected individuals receiving effective antiretroviral therapy. J Infect Dis. 2012;206:765–769. doi: 10.1093/infdis/jis412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discovery. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 10.Marks PA, Dokmanovic M, et al. Histone deacetylase inhibitors: discovery and development as anticancer agents. Expert Opin Investig Drugs. 2005;14:1497–1511. doi: 10.1517/13543784.14.12.1497. [DOI] [PubMed] [Google Scholar]
- 11.Zimmerman B, Sargeant A, Landes K, Fernandez SA, Chen CS, Lairmore MD. Efficacy of novel histone deacetylase inhibitor, AR42, in a mouse model of, human T-lymphotropic virus type 1 adult T cell lymphoma. Leuk Res. 2011;35:1491–1497. doi: 10.1016/j.leukres.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drummond DC, Noble CO, Kirpotin DB, Guo Z, Scott GK, Benz CC. Clinical development of histone deacetylase inhibitors as anticancer agents. Annu Rev Pharmacol Toxicol. 2005;45:495–528. doi: 10.1146/annurev.pharmtox.45.120403.095825. [DOI] [PubMed] [Google Scholar]
- 14.Yang XJ, Seto E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene. 2007;26:5310–5318. doi: 10.1038/sj.onc.1210599. [DOI] [PubMed] [Google Scholar]
- 15.Lu Q, Wang DS, Chen CS, Hu YD, Chen CS. Structure-based optimization of phenylbutyrate-derived histone deacetylase inhibitors. J Med Chem. 2005;48:5530–5535. doi: 10.1021/jm0503749. [DOI] [PubMed] [Google Scholar]
- 16.Lin TY, Fenger J, Murahari S, et al. AR-42, a novel HDAC inhibitor, exhibits biologic activity against malignant mast cell lines via down-regulation of constitutively activated Kit. Blood. 2010;115:4217–4225. doi: 10.1182/blood-2009-07-231985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Folks TM, Clouse KA, Justement J, et al. Tumor necrosis factor alpha induces expression of human immunodeficiency virus in a chronically infected T-cell clone. Proc Natl Acad Sci U S A. 1989;86:2365–2368. doi: 10.1073/pnas.86.7.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jordan A, Bisgrove D, Verdin E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO Journal. 2003;22:1868–1877. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ball SC, Abraha A, Collins KR, et al. Comparing the ex vivo fitness of CCR5-tropic human immunodeficiency virus type 1 isolates of subtypes B and C. J Virol. 2003;77:1021–1038. doi: 10.1128/JVI.77.2.1021-1038.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bush ML, Oblinger J, Brendel V, et al. AR42, a novel histone deacetylase inhibitor, as a potential therapy for vestibular schwannomas and meningiomas. Neuro Oncol. 2011;13:983–999. doi: 10.1093/neuonc/nor072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong Y, Barisone GA, Abuhay M, et al. Histone deacetylase inhibition enhances the lymphomacidal activity of the anti-CD22 monoclonal antibody HB22.7. Leuk Res. 2014:1320–1326. doi: 10.1016/j.leukres.2014.08.014. [DOI] [PubMed] [Google Scholar]
- 22.Cordero-Nieves H, Sborov DW, Canella A, et al. HDAC inhibitor AR-42 decreases CD44 expression and sensitizes myeloma cells to lenalidomide. Blood. 2014;124:3377–3377. doi: 10.18632/oncotarget.5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofmeister CC, Liu Z, Bowers MA, et al. Phase I study of AR-42 in relapsed multiple myeloma and lymphoma. Blood. 2012;120(21):2955. [Google Scholar]
- 24.Doitsh G, Galloway NL, Geng X, et al. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature. 2014;505:509–514. doi: 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spina CA, Anderson J, Archin NM, et al. An in-depth comparison of latent HIV-1 reactivation in multiple cell model systems and resting CD4+ T cells from aviremic patients. PLoS Pathog. 2013;9:e1003834. doi: 10.1371/journal.ppat.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tyagi M, Romerio F. Models of HIV-1 persistence in the CD4+ T cell compartment: past, present and future. Curr HIV Res. 2011;9:579–587. doi: 10.2174/157016211798998754. [DOI] [PubMed] [Google Scholar]