Abstract
CHARMM-GUI, http://www.charmm-gui.org, is a web-based graphical user interface to prepare molecular simulation systems and input files to facilitate the usage of common and advanced simulation techniques. Since its original development in 2006, CHARMM-GUI has been widely adopted for various purposes and now contains a number of different modules designed to setup a broad range of simulations including free energy calculation and large-scale coarse-grained representation. Here, we describe functionalities that have recently been integrated into CHARMM-GUI PDB Manipulator, such as ligand force field generation, incorporation of methanethiosulfonate (MTS) spin labels and chemical modifiers, and substitution of amino acids with unnatural amino acids. These new features are expected to be useful in advanced biomolecular modeling and simulation of proteins.
1. INTRODUCTION
Computer modeling and simulation of biologically important molecules have come a long way since its inception. For the last two decades, researchers have made considerable efforts to developing modeling and simulation methodologies to elucidate the mysteries in biological phenomena (Benoît Roux & MacKinnon, 1999; Bernèche & Roux, 2001; MacKerell Jr, 2004; Rhee, Sorin, Jayachandran, Lindahl, & Pande, 2004; Dror, Dirks, Grossman, Xu, & Shaw, 2012). The advances in biomolecular modeling and simulation allow researchers not only to explain why certain phenomena occur, but also to predict what would happen in different situations and design new experiments to test the developed hypothesis (Kuhlman et al., 2003; Ostmeyer, Chakrapani, Pan, Perozo, & Roux, 2013; Shukla, Meng, Roux, & Pande, 2014). Combined with tremendous increase in computing power, biomolecular modeling and simulation are becoming routine tools used in many research laboratories.
The rapid development of computational methodologies also made a large number of utilities available for both experts and beginners. As an illustration, more than 700 utilities are currently listed in a directory of computer-aided drug design tools maintained by Swiss Institute of Bioinformatics (http://www.click2drug.org). Among these tools, the emergence of web-based modeling tools is notable. Compared to downloadable software, web-based tools do not require program installation or upgrades, and can be operated on a web browser, making the tool platform-independent and easy to use. Among various web-based modeling tools, the WHAT IF server particularly stands out. It was designed in 1997 and has continued to operate until now. The WHAT IF server provides various services to users ranging from structural analyses, such as solvent accessibility and coordinate manipulation, to homology modeling and ligand docking (Hekkelman et al., 2010).
After the success of the WHAT IF server, various web-based modeling and simulation services have been developed and made publically available. CHARMM-GUI is a unique web-based set of tools developed by Im and co-workers (Sunhwan Jo, Kim, Iyer, & Im, 2008). It was originally developed in 2006 and its primary goal was to prepare molecules for molecular modeling and provide a set of CHARMM inputs for molecular dynamics (MD) simulations, so that users can download the inputs and perform simulations on their local machines or remote computational resources. CHARMM-GUI is powered by CHARMM (Brooks et al., 2009), a highly flexible academic research program used world-wide for simulations of many-particle systems, particularly macromolecules of biological interest. What makes CHARMM-GUI unique is that it sets out to simplify and generalize the protocol for building simulation systems. Molecular simulation systems are often comprised of several components, and in-depth knowledge on modeling software is necessary to understand and build a sophisticated simulation system. In addition, generalizing the system building protocols is challenging because customization is often necessary. For example, a protein-membrane complex system contains water, ions, protein, and lipid components, and it would require considerable efforts even for an expert to build a realistic and physiologically relevant simulation system. CHARMM-GUI Membrane Builder successfully encapsulates such a sophisticated process into a well-defined protocol that can reproducibly generate a realistic protein-membrane complex or a membrane-only system within minutes (Sunhwan Jo, Kim, & Im, 2007; S. Jo, Lim, Klauda, & Im, 2009). Currently, there are several web-based tools that provide similar functionality (Miller et al., 2008; Staritzbichler, Anselmi, Forrest, & Faraldo-Gómez, 2011; Schmidt & Kandt, 2012; Skjevik, Madej, Walker, & Teigen, 2012; Ghahremanpour, Arab, Aghazadeh, Zhang, & van der Spoel, 2014), but CHARMM-GUI Membrane Builder remains the interface containing the most comprehensive list of lipid molecules available for generating realistic biological membrane simulation system.
Since its original development, CHARMM-GUI has been widely used in the biomolecular modeling and simulation community, and it has grown into a platform of web-based tools for simulations: Ligand Binder for free energy perturbation molecular dynamics (FEP/MD) simulations for protein-ligand binding affinity calculations (Sunhwan Jo, Jiang, Lee, Roux, & Im, 2013), Micelle Builder for protein-micelle complex simulation system generation (Cheng, Jo, Lee, Klauda, & Im, 2013), GCMC/BD ion simulator for Brownian dynamics of ions across ion channels (K. I. Lee et al., 2012), Glycan Reader for preparation of simulation systems containing carbohydrates or proteoglycans (Sunhwan Jo, Song, Desaire, MacKerell Jr, & Im, 2011), and recently, PACE CG Builder for coarse-grained simulation system preparation (Qi et al., 2014).
Here, we describe the newest functionalities that have been integrated into CHARMM-GUI PDB Manipulator. First, ligand force field (FF) generation is discussed in detail. Generating a consistent and accurate FF for a new compound is often one of the major hurdles in computational modeling of protein-ligand interactions. Thus, the ability to read or automatically generate ligand FF parameters would be undoubtedly useful to study protein-ligand interactions. Second, incorporation of methanethiosulfonate (MTS) spin-labels and chemical modifiers is discussed. Distance measurements using spin labels and introducing MTS reagents to modify chemical environments around specific residues are widely used in protein structure and function studies, so that making spin labeled and/or chemically modified models would be useful for related computational studies. Third, substituting regular amino acids with unnatural amino acids is discussed. As researchers are more interested in more complex biological problems, these functions would also be useful. Each of these new functionalities is elaborated in a step-wise manner and further discussed with common issues (or mistakes) in using these functionalities.
2. LIGAND FORCE FIELD GENERATION
A molecular mechanics FF is one of the most important components of modern biomolecular modeling and simulation (MacKerell Jr, 2004). These FFs are typically derived and validated using physicochemical properties of (small) target molecules, which are obtained either experimentally or computationally. Parameterizing molecular mechanics FFs is often challenging because it requires careful curation of experimental and computational data as well as optimization of a large number of parameters.
There are several popular contemporary FFs that are readily available, such as AMBER (Weiner et al., 1984; Cornell et al., 1995), CHARMM (MacKerell et al., 2002; Best et al., 2012), GROMOS (Christen et al., 2005), and OPLS (Jorgensen, Maxwell, & Tirado-Rives, 1996). They often cover key biological macromolecules, such as proteins, nucleic acids, carbohydrates, and phospholipids. However, these FFs often do not treat important small molecules. Accurate representation of small molecules in modeling and simulation of protein-ligand interactions is crucial for the studies of rational computer-aided drug discovery as well as proteins with coenzymes and so on. General FFs, such as MM2 (N L Allinger, 1977), MM3 (N L Allinger, Yuh, & Lii, 1989; Lii & Allinger, 1989b, 1989a), MM4 (Norman L Allinger, Chen, Lii, & Durkin, 2003), MMFF94 (Halgren, 1996), and CFF93 (Rappé, Casewit, Colwell, Goddard III, & Skiff, 1992), were developed specifically for small organic compounds, but performed poorly on biomolecules and intermolecular interactions in bulk phase, making them ill-suited for studying protein-ligand interactions. Combining one of the former biomolecular FFs with one of the latter general FFs is not an appropriate solution due to imbalances in the nonbonded interactions, as previously discussed (MacKerell Jr, 2004).
The number of possible drug-like molecules is estimated to be roughly 1060 (Bohacek, McMartin, & Guida, 1996). Hence, it is not currently feasible to develop a comprehensive molecular mechanics FF set that accurately covers such a large chemical space. However, to account for important small molecules in a biomolecular context, typically those of medicinal chemistry interest, generalized FFs for small molecules have been developed such as the CHARMM general force field (CGenFF) (Vanommeslaeghe et al., 2010) and the general AMBER force field (GAFF) (Wang, Wolf, Caldwell, Kollman, & Case, 2004). In addition, algorithms that can suggest topologies and FF parameters have also been developed based on such a generalized FF. PRODRG (Schüttelkopf & van Aalten, 2004) is a web-based interface that provides topology and parameters compatible with the GROMOS FF. The CGenFF program (Vanommeslaeghe & MacKerell Jr, 2012; Vanommeslaeghe, Raman, & MacKerell Jr, 2012) (available to non-profit users through the cgenff.paramchem.org interface) and SwissParam (Zoete, Cuendet, Grosdidier, & Michielin, 2011) provide the same function for the CHARMM FF. Antechamber (Wang, Wang, Kollman, & Case, 2006) and MATCH (Yesselman, Price, Knight, & Brooks III, 2012) provide a command-line utility for automated topology and parameter generation for the AMBER and CHARMM FFs, respectively. There are also new tools such as the VMD plugin ffTK (Mayne, Saam, Schulten, Tajkhorshid, & Gumbart, 2013) and the general automated atomic model parameterization (GAAMP) web server (Huang & Roux, 2013). The GAAMP method is described in Sections 2.3 and 4.1.
CHARMM-GUI adopts the standard CHARMM36 FF. Although the CHARMM36 FF currently covers a large array of biomolecules (Guvench et al., 2008; Klauda et al., 2010; Denning, Priyakumar, Nilsson, & MacKerell Jr, 2011; Best et al., 2012) and various small organic molecules (Vanommeslaeghe et al., 2010), it is still far short to cover a huge number of small molecules of interest. To remedy this situation, CHARMM-GUI provides several options to generate or read a small molecule FF. In this section, we describe how to use CHARMM-GUI to prepare simulations when FF parameters are missing for small molecules of interest.
2.1 Reading Small Molecules that Do Not Exist in the CHARMM Force Field
As shown in Figure 1, CHARMM-GUI provides three options for small molecule FF parameters: (1) CGenFF, (2) Antechamber, and (3) custom CHARMM FF. When a custom CHARMM FF is available to users, it can be uploaded into CHARMM-GUI. Note that the uploaded CHARMM FF must have separate topology and parameter files. In addition, if new atoms are defined, the atom numbers in the MASS section should not overlap with those of the standard CHARMM36 FF.
Figure 1.
(A) A flowchart for reading small molecules that do not exist in the CHARMM FF. (B) A screenshot of options provided by CHARMM-GUI for reading small molecules that do not exist in the CHARMM FF.
If custom CHARMM FF parameters are not available for a small molecule of interest, either the CGenFF or Antechamber option can be used to automatically generate customized FF parameters for the molecule. To use the automatic FF generation option, the molecular representation of the small molecule in Mol2 file format is required. Users can upload a Mol2 format file, or, if a Mol2 format file is not available, a SDF format file can be used instead (which is then converted to a Mol2 format file using OpenBabel software (O'Boyle et al., 2011)). If users do not have those files, a SDF file could be obtained from the RCSB ligand database. However, careful attention must be paid because the ligand residue name in the original PDB file is used to match the small molecule in the RCSB database, and they are not necessarily the same molecule (see Section 2.2).
The CGenFF program is based on the CGenFF, which is part of the standard CHARMM36 FF parameter set, and Antechamber provides GAFF-based FF parameters for small molecules that can be used in CHARMM. Although Antechamber-generated GAFF FF can reproduce hydration energies reasonably well, it is generally desirable to remain internally consistent and avoid mixing the parameters of different FF. Thus, using the CGenFF option is recommended if the small molecule is studied together with other biomolecules using the CHARMM36 FF.
2.2 Issues in Using Automated Ligand Force Field Generation Procedures
When the automatic FF generation option is chosen to combine the small molecule FF with the existing FF parameters, there are three main issues that users need to be cautious about. First, the order of atoms listed in the PDB and the Mol2 (or SDF) files must match. CHARMM-GUI reorders the atoms in the PDB file based on the list of atoms in the Mol2 (or SDF) file, after the small molecule FF is generated by either the CGenFF program or Antechamber. If the atom names in the topology and the PDB files do not match, atomic positions can be mixed when CHARMM reads the coordinate. Note that, if no minimizations are performed, the atoms might look normal even with such a problem, but the problem becomes obvious after a minimization, often leading to failure of the minimization.
Second, if a user chooses to use a SDF format file, CHARMM-GUI downloads a SDF file from the RCSB ligand database (Rose et al., 2011). In this case, the residue name in the originally uploaded PDB and the RCSB database’s entry ID must match. For example, there is an entry for R-wafarin in RCSB (http://www.rcsb.org/pdb/ligand/ligandsummary.do?hetId=RWF), which has an entry ID of “RWF”. If a PDB file containing residue “RWF” and the option for downloading a SDF file from RCSB is selected, CHARMM-GUI downloads the R-wafarin structure from RCSB. Currently, however, there is no way for users to specify which SDF file to download from RCSB, and the residue name used in the original PDB file is used to fetch a SDF file from RCSB. If the residue name and the RCSB ligand entry ID do not match, users need to either change the residue name in the PDB file or manually download a SDF file from other sources (RCSB, PubChem, etc), modify it, and upload the corrected SDF file.
Third, the CHARMM-GUI automatic parameter generation option takes a Mol2 (or SDF) file for molecular representation without any further modification. Therefore, it is very important for users to explicitly add hydrogen atoms according to the desired protonation state. In addition, the bond orders provided by users are also important because they are used to determine proper atom types by the CGenFF program and Antechamber.
2.3 Further Optimization Strategy
The automatic parameter generation is not perfect. For example, the approach in the CGenFF program is based on the similarity between the atom types that define each required parameter and those in existing parameters. The dissimilarity is quantified in terms of a penalty score that is returned to the user in the output toppar stream file. Thus, a lower penalty score indicates better quality parameters, though users should be aware that the transferability of empirical FF parameters is limited and care should be taken to test the assigned parameters. Antechamber uses the AM1-BCC protocol to assign atomic charges, but there is still room for improvement in atomic charges and internal energy (bond/angle/torsion) parameters. A recent benchmark by Brooks III and co-workers (Knight, Yesselman, & Brooks III, 2013) shows that the performance of parameters generated by Antechamber and the CGenFF program is similar on average in terms of hydration free energies estimated using implicit solvent models, but also demonstrates areas where improvements are possible.
The initial force field parameters obtained from the automatic algorithms can be improved in a step-wise manner as detailed in Vanommeslaeghe et al. (2010). Briefly, the optimal geometry of the target molecule is first generated in a quantum mechanics (QM) calculation. Second, given the geometry, water molecules are placed around hydrogen bond donors and acceptors, and the interaction energy is calculated on a QM level. Third, the optimal molecular mechanics charges are determined to reproduce the QM interaction energy following the appropriate scaling. Lastly, bonded parameters such as bonds, angles and dihedrals are optimized using a QM geometry and vibrational analysis and/or QM-level potential energy scans. Note that the optimization is typically performed to only those parameters that are guessed (i.e., those with penalty scores) as the original CGenFF parameters are already optimized.
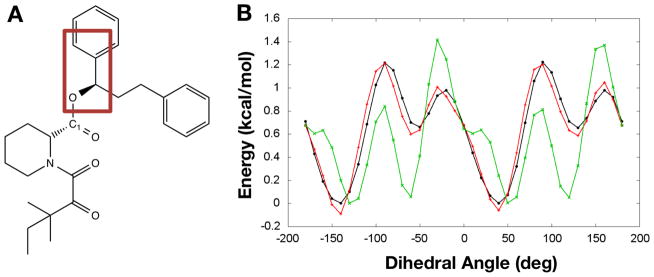
Optimization of FF parameters requires considerable efforts even for a small molecule. There are several tools available that help users to perform the optimization. Two promising tools are ffTK (Mayne et al., 2013) and GAAMP (Huang & Roux, 2013). ffTK is a VMD plugin that provides a GUI for each and every aspect of the aforementioned optimization process, so users do not have to write necessary inputs manually. The GAAMP gateway, http://gaamp.lcrc.anl.gov, is a web-based interface that starts with initial FF parameters generated by the CGenFF program or Antechamber, and automatically performs the FF optimization without any user intervention. Figure 2 shows an example of a dihedral angle optimization of a small molecule (SB3; CHEMBL ID 126568) performed by the GAAMP server. The optimization for this size of molecule took more than a day, which is relatively long considering the fact that the CGenFF program returns the topology and parameters for the small molecule almost instantly. However, GAAMP can improve the atomic charges and torsion angle parameters considerably without any user intervention (Figure 2B).
Figure 2.
(A) Structure of the FKBP ligand SB3. (B) Example optimization result from the GAAMP server for the dihedral angle highlighted in (A): QM (black), optimized (red), and initial (green).
2.4 Practical Applications to Binding Free Energy Calculations
The efficacy of the automatic FF generation option integrated into CHARMM-GUI is illustrated by a recent study of large scale computational docking and simulation. In this study, Im and co-workers (Lee, Jo, Lim, & Im, 2012) have developed an integrated methodology for accurate prediction of the binding mode and affinity of drug-like molecules (Figure 3B). They have applied a computational docking approach to generate a large number of decoy structures and selected a few docking pose candidates using clustering approaches. Because these molecules are not available in the standard CHARMM36 FF, the authors first used the CHARMM-GUI automatic parameterization option to obtain ligand FFs. The selected docking poses were then subjected to short MD simulations to filter out irrelevant poses, followed by the FEP/MD simulations prepared by CHARMM-GUI Ligand Binder (Sunhwan Jo et al., 2013) for further ranking of the poses (Figure 3).
Figure 3.
(A) Ligand structures. (B) Schematic of the docking and FEP/MD protocol used by Im and co-workers (H. S. Lee et al., 2012). (C) The correlation between binding affinity of near-native poses and the non-native poses. The FEP/MD method can discriminate near-native and non-native poses better than a docking score. The figures are reproduced with permission from the Journal of Chemical Information and Modeling.
The target small molecules are antagonists of MDM2 and MDMX. Figure 3A shows the chemical structures of the small molecules used in their study, and the FF parameters were generated using the CGenFF option without any further modification. The calculated binding free energies for MDM2 complexes were overestimated compared to experimental measurements (Figure 3C) mainly due to the difficulties in sampling highly flexible apo-MDM2 conformations within the simulation timescale. Nonetheless, the FEP/MD binding free energy calculations are more promising in discriminating binders from nonbinders than commonly used docking scores (Figures 3B–C). In addition, the FEP/MD calculations provide detailed information on the different energetic contributions to ligand binding, leading to a better understanding of the sensitivity and specificity of protein-ligand interactions. Therefore, CHARMM-GUI PDB Manipulator is expected to be useful as a platform that can rapidly prepare necessary FF parameters of small molecules of interest with help of other tools. Setting up such sophisticated simulations can allow researchers to tackle more complex biological problems of protein-ligand interactions.
3. MTS REAGENTS
MTS reagents are often used for protein structure and function studies. Their use includes labeling and blocking groups, cross-linking groups, affinity-labeling groups, and reporter groups for chemical modification of peptides and proteins. MTS reagents are introduced to a specific site in a protein through site-directed mutagenesis (Hubbell, Mchaourab, Altenbach, & Lietzow, 1996). These reagents react very rapidly and specifically with cysteine residues, converting cysteine sulfhydryls to cysteine disulfide bonds. MTS reagents of cysteine residues may produce a measurable change in different protein functional states, which can be measured by various biophysical techniques. For example, MTSSL (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl methanethiosulfonate; CYR1 in Figure 4) is an MTS reagent that is widely used as a spin-label probe in ESR (electron spin resonance) spectroscopy. MTSSL has an unpaired electron, which offers a very strong signal in the ESR spectrum that provides valuable information about the structure, dynamics, and function of a protein system. In particular, site-specific mutagenesis with MTS reagents has proved to be a very useful technique in characterizing the structure-function relationship of membrane proteins, such as ion channels and transporter proteins, as well as enzymes and receptors (D. D. Roberts, Lewis, Ballou, Olson, & Shafer, 1986; Chen, LiuChen, & Rudnick, 1997; Perozo, Cortes, & Cuello, 1998; Choi et al., 2000; Tombola, Pathak, & Isacoff, 2006; Hvorup et al., 2007; Forrest et al., 2008; J. A. Roberts et al., 2008; Jeschke, 2012; Kazmier et al., 2014; Raghuraman, Islam, Mukherjee, Roux, & Perozo, 2014). Since many biophysical experiments are routinely performed with these MTS reagents, it is often necessary to introduce them into proteins for the purpose of MD simulation. Keeping this in mind, the FF parameters for a number of MTS reagents have been incorporated into CHARMM-GUI, which is expected to help users to readily prepare initial systems and simulation input files for MD simulation with selected MTS reagents.
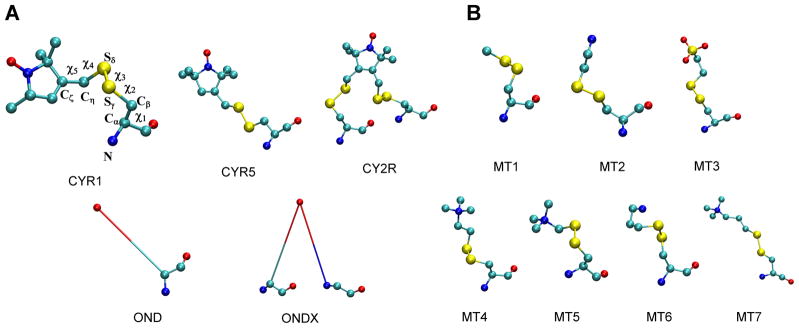
Figure 4.
MTS side chains available in CHARMM-GUI. For clarity, hydrogen atoms are not shown and the oxygen, nitrogen, carbon and sulfur atoms are represented in red, blue, cyan and yellow colors, respectively. (A) Monofunctional spin-label side chains, CYR1 and CYR5, resulting from linking MTSSL (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl methanethiosulfonate) and PROXYL-MTS (1-oxyl-2,2,5,5-tetramethylpyrrolidin-3-yl) methyl methanethiosulfonate) to cysteine through a disulfide bond, a bifunctional spin-label side chain, CY2R, resulting from selectively introducing MTSSL (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl methanethiosulfonate) to two nearby cysteines through disulfide bonds, and the dummy spin labels, OND and ONDX, that mimic the dynamics of the CYR1 and CY2R spin-label side chains, respectively. In the case of ONDX, X = 3 or 4 depending on the position of the 2nd residue to which it is attached. The five dihedral angles connecting Cα atom of the protein backbone to nitroxide ring are also shown in CYR1. (B) The chemically modified MTS side chains, MT1, MT2, MT3, MT4, MT5, MT6 and MT7, resulting from linking methyl methanethiolsulfonate (MMTS), 2-aminoethyl methanethiosulfonate (MTSEA), sodium-(2-sulfonatoethyl) methanethiosulfonate (MTSES), 2-trimethylammonium-ethyl methanethiosulfonate (MTSET), (trimethylammonium)methyl methanethiosulfonate (MTSMT), 3-aminopropyl methanethiosulfonate (MTSPA) and 3-(trimethylammonium)propyl methanethiosulfonate (MTSPT) reagents, respectively, to cysteine through a disulfide bond.
3.1 MTS Reagents Available in CHARMM-GUI
There are two sets of MTS reagents available in CHARMM-GUI: the nitroxide spin-labels (Figure 4A) and the traditional chemical modifiers (Figure 4B). There are two natural monofunctional spin-label side chains (CYR1 (Berliner, Grunwald, Hankovszky, & Hideg, 1982) and CYR5 (McHaourab, Kalai, Hideg, & Hubbell, 1999)), a natural bifunctional (two-residue) spin-label side chain (CY2R (Fleissner et al., 2011)), and a monofunctional and two bifunctional dummy spin-label side chains (OND (Islam, Stein, McHaourab, & Roux, 2013) and ONDX (Islam & Roux, 2014) with X = 3 or 4). All these spin-label side chains were parameterized by Roux and coworkers (Sezer, Freed, & Roux, 2008; Islam et al., 2013; Islam & Roux, 2014). In addition, a total of seven MTS chemical modifiers (MT1, MT2, MT3, MT4, MT5, MT6, and MT7) parameterized by the GAAMP method (Huang & Roux, 2013) are currently available.
The step-by-step procedure to introduce MTS reagents into a protein in solution follows:
-
1
Go to CHARMM-GUI, http://www.charmm-gui.org
-
2
Select “Input Generator”
-
3
Select “Quick MD Simulator”
-
4
Download or upload your PDB file, and then “Next Step”
-
5
Select “Next Step” if you do not need to change anything
-
6
In PDB Manipulator, select “Add MTS reagents: nitroxide spin labels” and/or “Add MTS reagents: chemical modifier” (Figure 5A).
-
6.1
With “Add MTS reagents: nitroxide spin labels”, choose the spin-labels (CYR1, CYR5, CY2R, OND, OND3 and OND4) and “SEGID”, “RESID”, and “Amino acid” from the drop-down menu. This will modify the selected residue(s) with the natural MTS spin-label side chain(s) or add the dummy spin-label(s) to the selected residue(s). For CY2R, select i+4, i+3, i+2, or Custom to attach this spin-label to a second residue. Two more options are also available:
Option 1: Selecting “Mutate the selected residue(s) to Ala for dummy spin label(s)” will mutate all the selected wild type residues to alanine before attachment of the dummy spin-label(s).
Option 2: Selecting “Add dummy spin label(s) (OND) to all the protein residue(s)” will add dummy spin labels to all the residues. This option is particularly useful to conduct a MD simulation of the dummy spin-labels (MDDS) (Islam et al., 2013; Islam & Roux, 2014) to calculate spin-pair distance distributions between two dummy spin-labels.
-
6.2
With “Add MTS reagents: chemical modifier”, select the MTS reagents (MT1, MT2, MT3, MT4, MT5, MT6, and MT7) and “SEGID”, “RESID”, and “Amino acid” from the drop-down menu. This will modify the selected residue(s) with the MTS reagent(s).
-
7
Follow the instructions (essentially the same procedure of “Quick MD Simulator”) to build a system in solution.
Figure 5.
(A) A snapshot to introduce MTS spin-labels and chemical modifiers into some specific residues in CHARMM-GUI. (B) Cartoon representations of T4 lysozyme with nine spin-label side chains at position 65, 72, 75, 76, 82, 115, 119, 131, and 151. (C) Cartoon representations of the diagonal subunits of KcsA K+channel with two spin-label side chains at position 51 and 82 on each subunit. Only diagonal subunits of tetrameric KcsA K+channel are shown for clarity.
Note that the help windows are available in both step 6.1 and step 6.2 of the above illustration. The MTS reagents are also available in Membrane Builder (Sunhwan Jo et al., 2007; S. Jo et al., 2009) and other CHARMM-GUI modules. In particular, the dummy spin labels are available in PACE CG Builder (Qi et al., 2014).
3.2 Initial Conformation Generation of the MTS Reagents in CHARMM-GUI
MTS reagents are highly flexible because they have multiple dihedral angles that can accommodate a number of conformational states. For example, as shown in Figure 4, CYR1 spin-label side chain has five dihedral angles denoted by χ1, χ2, χ3, χ4, and χ5 along the flexible bonds, Cα-Cβ-Sγ-Sδ-Cη-Cζ. From a careful study of the spin-labels inserted in 37 positions in T4 lysozyme, Roux and co-workers recently proposed the most important rotamers for this spin-label side chain (Islam et al., 2013). The dihedral angles χ1 and χ2 can adopt three-fold conformations, +60° (or gauche+), 180° (or trans), and −60° (or gauche), which are denoted by p, t, and m, respectively, and the dihedral angle χ3 can adopt only two stable conformations, p (+90°) and m (−90°). Therefore, eight stable rotamers of the spin-label side chain have been identified along the dihedral angles χ1, χ2, and χ3: tpp, mtp, mmp, mmm, mtm, ttm, tpm, and ptm. These observations are consistent with the available information from X-ray crystallography. χ4 and χ5 have been found extremely flexible, which is also consistent with the fact that no reliable X-ray crystallographic information is available for these dihedral angles.
Finding suitable MTS spin-label side chain conformations in a specific protein site may require a substantially long MD simulation to sample each conformational degree of freedom along its dihedral angles. Therefore, we have implemented an efficient approach in CHARMM-GUI to build a reasonable initial conformation for the MTS reagents (except CY2R, OND, and ONDX) in a protein structure. This approach is discussed below:
The conformations of the MTS spin-label side chains and the MTS chemical modifiers are characterized by three dihedral angles, χ1, χ2, and χ3, along the flexible side-chain Cα-Cβ-Sγ-Sδ, as shown in Figure 4. χ4 and χ5 dihedral angles are ignored due to the fact that they are extremely flexible and they will most likely converge to the right rotameric state with a relatively short MD simulation.
Each of the eight rotameric states (tpp, mtp, mmp, mmm, mtm, ttm, tpm, and ptm) is sequentially added to a selected site in a protein, followed by 200-step minimization with steepest descent (SD) and adopted basis Newton-Raphson (ABNR) algorithms, respectively. During the minimization, all the protein atoms are kept fixed and only the selected MTS reagent atoms are allowed to move with flat bottom harmonic restraints on χ1, χ2, and χ3 of the MTS side chain with a force constant of 500 kcal/(mol·rad2) and a width of 2.5°. The minimum energy conformation of the MTS side chain (among the eight rotameric states) is then taken for further processing.
Step 2 is repeated for all the selected spin label sites. Finally, all the MTS side chains with their minimum energy conformations (determined in step 2) and the surrounding atoms (up to 5 Å from the MTS reagent) are minimized for 200 steps with the SD and ABNR energy minimization, respectively.
This approach has shown to provide very reliable starting conformations of the MTS side chains in proteins. The performance of the above approach is examined by using two protein systems, T4 lysozyme (Figure 5B) and KcsA K+ channel (Figure 5C).
3.3 Comparison with Available Rotameric Data from X-ray Crystallography
Comparison with the available information from X-ray crystallographic structures offers a good route to examine the validity of the initial rotamers generated by CHARMM-GUI. Several crystallographic structures of MTSSL spin-labels inserted into position 65, 72, 75, 76, 82, 115, 119, 131, and 151 in T4 lysozyme (Figure 5B) are available from the literature (Toledo Warshaviak; Langen, Oh, Cascio, & Hubbell, 2000; Fleissner, 2007; Guo, Cascio, Hideg, Kalai, & Hubbell, 2007; Guo, Cascio, Hideg, & Hubbell, 2008; Fleissner, Cascio, & Hubbell, 2009; Sezer, Freed, & Roux, 2009; Fleissner et al., 2011). However, the X-ray data are mostly limited to dihedrals χ1 and χ2, since the electron density along the flexible side-chain Cα-Cβ-Sγ-Sδ-Cη-Cζ is missing beyond the Sδ atom. Using CHARMM-GUI, CYR1 was introduced individually at the same positions in T4 lysozyme. The comparison of spin-label structures from the PDB and CHARMM-GUI is given in Table 1.
Table 1.
Comparison of rotamers of the CYR1 spin-label side chain along χ1 and χ2 obtained from X-ray structures and CHARMM-GUI.
| Residue | PDB ID | Expl. Rotamer | CHARMM-GUI |
|---|---|---|---|
| 65 | 3K2Ra,b,c | tp | mt |
| 72 | MDd | mm, tp | tp |
| 75 | Fleissnera | mt | mt |
| 76 | 3K2Rc | tm | tt |
| 82 | 1ZYTa,e | mm | mm |
| 115 | 2IGC, 2OU8 a,f | mm, tp | tp |
| 119 | 3L2Xf | mm | tp |
| 131 | 2CUU, 3G3Ve,g | mm, tp | mm |
| MDd | mm, tp, mt, tt | ||
| 151 | 3G3Xe | mm | mt |
Toledo Warshaviak et al. ,
The χ1 and χ2 rotamers from CHARMM-GUI are broadly consistent with the information from the X-ray crystal structures and previous MD simulations for five positions: a tp rotamer is observed at 72 and 115, a mt rotamer is observed at 75, and a mm rotamer is observed at 82 and 131. The clearest disagreement is shown in spin-labels at positions 65, 76, 119, and 151 with tp, tm, mm, and mm rotamers, respectively, in the X-ray structure, while CHARMM-GUI provides mt, tt, tp, and mt rotamers, respectively. However, X-ray crystallographic structure of the spin-label at position 65 may not be right because the spin-label is involved in crystal contacts (Langen et al., 2000). Also, CHARMM-GUI provides the rotameric states of t and m along the χ1 dihedral angle of the spin labels at positions 76 and 151, respectively, which agree with their X-ray crystallographic structure. Overall, CHARMM-GUI can provide reliable initial conformations of the MTS side chains, which can be used in further MD simulations.
KcsA K+ channel has four equivalent subunits forming a tetramer. We found that simple minimization with no rotameric search approach yielded multiple (different) conformations of the spin-label side chains at positions 51 and 82 in four subunits of KcsA. However, the rotameric search approach implemented in CHARMM-GUI was able to provide very similar initial conformations of the spin labels at all four subunits (Figure 5B). All four spin-label side chains in position 51 were in the mtp rotamer along the dihedrals χ1, χ2, and χ3. Spin-labels introduced in position 82 were also in the same rotamer conformation (mtm) in all four subunits. This result shows that CHARMM-GUI is able to provide very good starting conformation of the MTS side chains even for a complex system such as KcsA K+ channel.
3.4 Practical Applications to Protein Structure Refinement by Using ESR Data
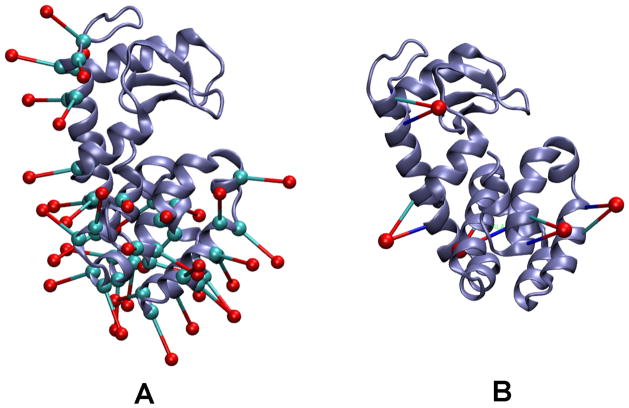
Recently, two state-of-the-art MD simulation methods, the conventional MD simulation of the dummy spin-labels (MDDS) (Islam et al., 2013; Islam & Roux, 2014) and the restrained-ensemble (RE) simulation method (Islam et al., 2013; B. Roux & Islam, 2013), have been developed to analyze ESR/DEER (double electron electron resonance) data. The MDDS simulation provides distance distributions between spin-labels that can be directly compared to the distance distributions obtained from ESR/DEER spectroscopy and be used to assess the reliability or correctness of an already existing model structure of the system. On the other hand, the RE method drives a model structure towards the desired, refined structure by using a global ensemble-based energy restraint that forces the spin-spin distance distribution histograms calculated from a multiple-copy MD simulation to match those obtained from ESR/DEER. The simplified nitroxide dummy spin-labels, shown in Figure 4A, are used mostly for both MDDS and RE simulations. Since the dummy spin-labels do not interact with each other, multiple dummy atoms can be introduced to a single protein structure. CHARMM-GUI can be used to prepare the required files for both MDDS and RE simulations. For example, 51 distance distributions are available from 37 spin labeled positions in T4 lysozyme. CHARMM-GUI was used to attach the dummy spin-labels at these sites in T4 lysozyme (Figure 6A). The generated files are then used for MDDS simulation from which the spin-label distance distributions can be calculated to compare with those obtained from the ESR/DEER spectroscopy. Bifunctional dummy spin-labels (OND4) were also inserted into five positions, 60–64, 72–76, 108–112, 131–135, and 151–155, in T4 lysozyme (Figure 6B). The generated structures can be used for the purpose of MDDS or RE simulations. Thus, CHARMM-GUI PDB Manipulator is expected to be useful in preparing necessary input files that can be used to understand protein structure and function in conjunction with ESR/DEER spectroscopy.
Figure 6.
Cartoon representation of T4 lysozyme with dummy spin-labels. (A) T4 lysozyme with 37 OND dummy spin-labels at positions 59, 60, 61, 62, 64, 65, 72, 75, 76, 79, 82, 83, 85, 86, 89, 90, 93, 94, 108, 109, 112, 115, 116, 119, 122, 123, 127, 128, 131, 132, 134, 135, 140, 151, 154, 155, and 159. (B) T4 lysozyme with 5 OND4 dummy spin-labels at positions 60–64, 72–76, 108–112, 131–135, and 151–155.
4. UNNATURAL AMINO ACIDS
Introducing unnatural amino acids (UAAs) into a protein is a powerful method to investigate the role of specific residues in protein structure and function, as well as to design proteins with novel functions or enhanced catalytic activity (Noren, Anthonycahill, Griffith, & Schultz, 1989; Chin et al., 2003; Hackenberger & Schwarzer, 2008; Lu, Yeung, Sieracki, & Marshall, 2009; Liu & Schultz, 2010). As demonstrated by Ahern and co-workers (Pless, Galpin, Niciforovic, & Ahern, 2011), UAA mutagenesis combined with electrophysiology and voltage-clamp fluorometry could be employed to study the effects of negative electrostatic potentials from Glu293 and Asp316 on the channel gating and voltage sensing of Shaker K+ channels. More applications can be found in a review by Pless and Ahern (Pless & Ahern, 2013). In principle, simulation studies of any of these chemically modified systems could be carried out to complement the experimental information. However, accurate molecular mechanics models are needed for an ever-growing number of possible UAAs.
As mentioned in Section 2, several FFs, including AMBER (Weiner et al., 1984; Cornell et al., 1995), CHARMM (Best et al., 2012), GROMOS (Christen et al., 2005), and OPLS (Jorgensen et al., 1996), are widely used in protein simulations, but there are generally no parameter sets specifically optimized for UAAs. This is becoming a major obstacle to model and study proteins containing UAAs in MD simulations. Although one can use Antechamber or the CGenFF program to assign parameters for an arbitrary compound, as discussed in Section 2.3, the obtained FF parameters often need further optimization as partial atomic charges and torsion parameters are not readily transferable across various molecules. The assigned parameters from analogs may be reasonably good in some cases, but they may not be so reliable in others. Another issue is that the topology and parameter files have to be manually edited to attach the unnatural side chain and protein backbone together. To overcome these difficulties, the GAAMP method was used to develop the FF parameters for UAAs (with negligible human efforts), and CHARMM-GUI PDB Manipulator now provides a facile way to model proteins containing UAAs.
4.1 Unnatural Amino Acids Available in CHARMM-GUI
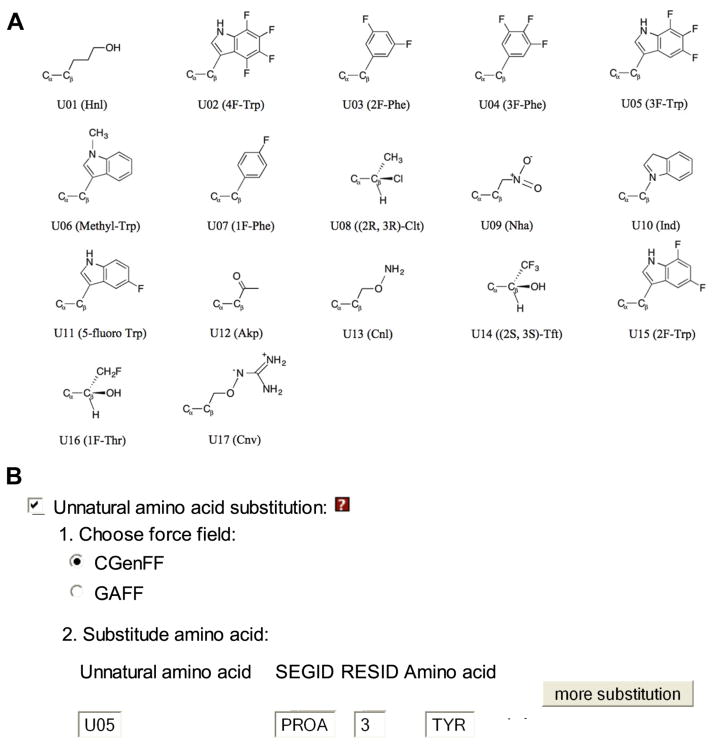
The details about the GAAMP method can be found in Huang and Roux’s work (Huang & Roux, 2013). In the GAAMP method, GAFF or CGenFF is taken as the initial FF. Partial atomic charges and dihedral parameters are optimized to reproduce QM target data. With minor adjustments, such an optimization algorithm has been extended to parameterize arbitrary amino acids, including UAAs, in a manner that is consistent with the backbone from the parent FF. Briefly, the partial atomic charges of the sidechain compound (the side chain capped with one hydrogen atom) are first determined using the GAAMP charge fitting procedure under the constraint that the charge carried by the hydrogen atom is fixed at zero. As a second step, CHARMM format topology, parameter, and coordinate files for the full molecule, comprising the sidechain molecule and the backbone of an alanine dipeptide, are generated automatically. As a third step, the soft dihedrals around rotatable bonds within the side chain are identified and the parameters are optimized. During the sidechain dihedral fitting, the backbone atoms are fixed with the and φ backbone dihedrals in an α-helical conformation (−60° and −45° for ϕ and φ). The parameters of the resulting model have the CHARMM36 FF for the backbone and the GAAMP-optimized side chain. There are two UAA parameter sets available: one uses GAFF and the other uses CGenFF as initial parameters, respectively. A total of 17 UAAs commonly used in experiments (communicated with Dr. Christopher Ahern) have been parameterized and they are shown in
The step-by-step procedure of incorporating UAAs into a protein in solution follows:
Go to CHARMM-GUI, http://www.charmm-gui.org
Select “Input Generator”
Select “Quick MD Simulator”
Download or upload your PDB file, and then “Next Step”
Select “Next Step” if you do not need to change anything
As illustrated Figure 7B, select “Unnatural amino acid substitution”, and then new menu for UAA modeling shows up. Choose the parameters based on “CGenFF” or “GAFF”, and then choose the UAA type and the index of amino acid to be substituted by the selected UAA.
Follow the instructions (essentially the same procedure of “Quick MD Simulator”) to build a system in solution.
Figure 7.
(A) Chemical structures of 17 UAAs available in CHARMM-GUI. (B) A snapshot of CHARMM-GUI to introduce a UAA into a specific residue in CHARMM-GUI.
Note that in step 6, one can view the UAA structures in the help window. In addition, the UAA substitution is also available in Membrane Builder (Sunhwan Jo et al., 2007; S. Jo et al., 2009) and other CHARMM-GUI modules.
4.2 Performance Evaluation
4.2.1. Testing the parameterization strategy with natural amino acids
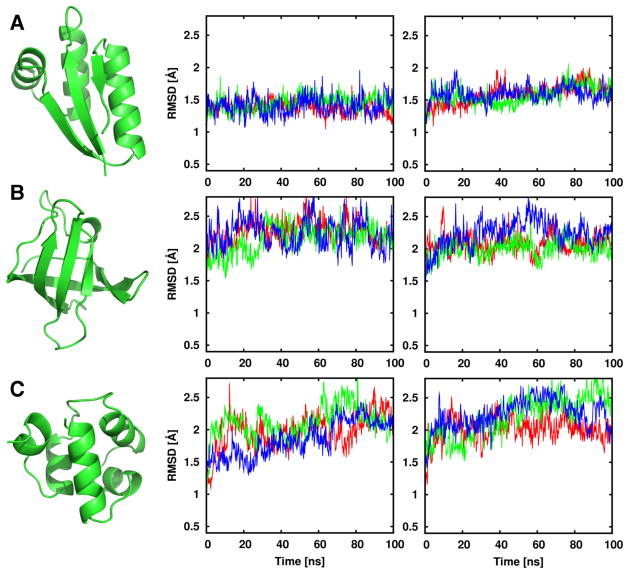
To test the performances of the amino acid parameters obtained by GAAMP, we re-parameterize all amino acids except glycine and proline to be consistent with the CHARMM36 FF. Three proteins with diverse topologies, shown in Figure 8, are used to compare the resulting FF (denoted as GAAMP) with the CHARMM36 FF: PDB:1CTF (mixed α-helices and β-sheets), PDB:1MJC (all β-sheets), and PDB:1R69 (all α-helices). Three independent 100-ns MD simulations in aqueous solution were conducted starting from the crystal structure of each protein. These three proteins are stable in 100-ns simulations with both the CHARMM36 and GAAMP FFs with similar conformational fluctuations. The simulation results suggest that the parameters of amino acids generated by GAAMP are compatible with the CHARMM36 FF.
Figure 8.
Three proteins with diverse structures (PDBs (A) 1CTF, (B) 1MJC, and (C) 1R69) are shown as ribbon representation generated by PyMol (Schrödinger, 2010). The traces of Cα-RMSD of 100-ns MD simulations for three replicas (with different colors) are compared between using the CHARMM36 FF (middle) and using the GAAMP FF (right) for each protein.
4.2.2. Test cases of UAA-containing systems
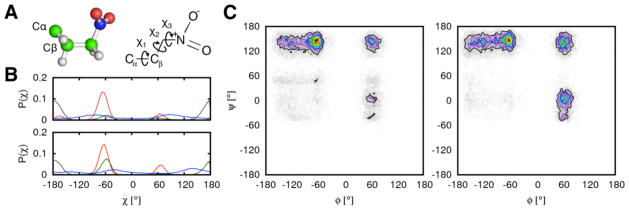
Two separate approaches are taken to illustrate the efficacy of UAA functionality in CHARMM-GUI PDB Manipulator. The first approach involves simulating UAA dipeptides in solution with the two FF parameter options (i.e., GAFF and CGenFF) currently available in CHARMM-GUI. For each of 17 UAAs, a dipeptide is formed with the same UAA species. This could be performed with CHARMM-GUI by substituting both alanine residues in an alanine dipeptide. Afterwards, the dipeptide is put into a water box and simulated for 35 ns using the simulation options provided in CHARMM-GUI Quick MD Simulator. Peptide properties such as torsion angle distributions and hydrogen bonding patterns are calculated and compared between the two FF parameters. A sample result of the torsion angle distributions calculated from the U09 dipeptide simulation is shown in Figure 9. The two parameter sets produce similar structural behaviors of the dipeptides and the differences are subtle in the case of U09. In spite of the similarity in the simulation outcome, the CGenFF is recommended as the protein-UAA system is treated with the CHARMM36 FF.
Figure 9.
(A) Structure (left) and chemical formula (right) of the unnatural amino acid nitrohomoalanine (residue ID U09 in CHARMM-GUI). The three sidechain torsion angles, χ1, χ2, and χ3, are shown on top of the chemical formula. (B) The torsion angle distributions for χ1 (red), χ2 (green), and χ3 (blue) calculated using parameters from GAFF (top) and CGenFF (bottom). (C) The backbone ϕ-ψ angle distributions calculated using parameters from GAFF (left) and CGenFF (right).
In the second approach, protein systems with UAA substitutions are generated and short MD simulations are performed to examine that it is indeed possible to generate a system with UAA substitutions in an automatic fashion using CHARMM-GUI and such systems are relatively stable on a nanosecond timescale. The tested protein-UAA systems include UAA substitutions at different ion coordination sites in the Na+,K+-ATPase. More specifically, the Glu residues at the binding site are replaced by nitrohomoalanine (U09, Figure 9A) one at a time. The protein is then embedded in a POPC bilayer with KCl and NaCl, 0.15 M each, in the aqueous solution. These systems are prepared using Membrane Builder (Sunhwan Jo et al., 2007; S. Jo et al., 2009). After the protein-membrane complex systems are generated, each of them is subjected to a 675-ps equilibration and 5-ns production with options provided by Membrane Builder. The trajectory analyses show that the systems are stable during the course of the simulation and the binding site heavy atom RMSD is below 1.2 Å most of the time. This indicates that the current CHARMM-GUI UAA functionality could be used in conjunction with other modules in CHARMM-GUI, such as Membrane Builder, and it is expected to be a useful tool in the study of complex biological systems with UAA substitutions.
5. PERSPECTIVES
CHARMM-GUI has been developed to provide a web-based user interface to build various molecular systems and generate input files for CHARMM and NAMD simulations. CHARMM-GUI has recently added a Drude Prepper facility that allows the conversion of CHARMM36 PSF and coordinate files into the electronically polarizable Drude FF (Lopes et al., 2013; Savelyev & MacKerell Jr, 2014), which includes parameters for lipids, proteins, DNA, and carbohydrates. PDB Manipulator in CHARMM-GUI aims to help users handle the PDB files easily such as mutating/protonating/phosphorylating specific residues, reading ligands or small molecules, adding MTS reagents and/or unnatural amino acids. In this work, we have described some of these functionalities in detail with a hope that users can use them widely for various biomolecular modeling and simulation. As PDB Manipulator is the first step of most input generation modules in CHARMM-GUI, these functionalities can be used for system building in solution and membrane (bilayer or micelle) environments, as well as in coarse-grained PACE CG Builder. With most sidechain modifications covered (together with future addition of more spectroscopic probes such as FRET and LRET) as well as the ability to handle small molecules, it is our hope that CHARMM-GUI PDB Manipulator becomes a useful tool for structure refinement of biomolecules with spectroscopic probes, structure and dynamics studies with sidechain modifications, and biomolecules’ interactions with small molecules in realistic biological solution and membrane environments.
Acknowledgments
We wish to acknowledge the help from Christopher Ahern with the unnatural amino acids, and the help from Eduardo Perozo and Hassane Mchaourab with the spin-labels. This work was carried out in the context of the Membrane Protein Structural Dynamics Consortium funded by grant U54-GM087519 (BR and WI) from the National Institute of Health (NIH), NIH grants GM072558 (BR and ADM), GM070855 (ADM), GM051501 (ADM), and XSEDE grant MCB070009 (WI).
References
- Allinger NL. Conformational analysis. 130. MM2. A hydrocarbon force field utilizing V1 and V2 torsional terms. Journal of the American Chemical Society. 1977;99(25):8127–8134. doi: 10.1021/ja00467a001. [DOI] [Google Scholar]
- Allinger NL, Chen KH, Lii JH, Durkin KA. Alcohols, ethers, carbohydrates, and related compounds. I. The MM4 force field for simple compounds. Journal of Computational Chemistry. 2003;24(12):1447–1472. doi: 10.1002/jcc.10268. [DOI] [PubMed] [Google Scholar]
- Allinger NL, Yuh YH, Lii JH. Molecular mechanics. The MM3 force field for hydrocarbons. 1. Journal of the American Chemical Society. 1989;111(23):8551–8566. doi: 10.1021/ja3112719. [DOI] [Google Scholar]
- Berliner LJ, Grunwald J, Hankovszky HO, Hideg K. A novel reversible thiol-specific spin label: papain active site labeling and inhibition. Anal Biochem. 1982;119(2):450–455. doi: 10.1016/0003-2697(82)90612-1. [DOI] [PubMed] [Google Scholar]
- Bernèche S, Roux B. Energetics of ion conduction through the K+ channel. Nature. 2001;414(6859):73–77. doi: 10.1038/35102067. [DOI] [PubMed] [Google Scholar]
- Best RB, Zhu X, Shim J, Lopes PEM, Mittal J, Feig M, MacKerell AD., Jr Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, φ and side-chain x (1) and x (2) dihedral angles. Journal of Chemical Theory and Computation. 2012;8(9):3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohacek RS, McMartin C, Guida WC. The art and practice of structure-based drug design: a molecular modeling perspective. Medicinal research reviews. 1996;16(1):3–50. doi: 10.1002/(SICI)1098-1128(199601)16:1<3::AID-MED1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Brooks BR, Brooks CL, III, MacKerell AD, Jr, Nilsson L, Petrella RJ, Roux B, … Karplus M. CHARMM: the biomolecular simulation program. Journal of Computational Chemistry. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, LiuChen S, Rudnick G. External cysteine residues in the serotonin transporter. Biochemistry. 1997;36(6):1479–1486. doi: 10.1021/Bi962256g. [DOI] [PubMed] [Google Scholar]
- Cheng X, Jo S, Lee HS, Klauda JB, Im W. CHARMM-GUI Micelle Builder for Pure/Mixed Micelle and Protein/Micelle Complex Systems. Journal of Chemical Information and Modeling. 2013;53(8):2171–2180. doi: 10.1021/ci4002684. [DOI] [PubMed] [Google Scholar]
- Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang ZW, Schultz PG. An expanded eukaryotic genetic code. Science. 2003;301(5635):964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- Choi YB, Tenneti L, Le DA, Ortiz J, Bai G, Chen HSV, Lipton SA. Molecular basis of NMDA receptor-coupled ion channel modulation by S-nitrosylation. Nature Neuroscience. 2000;3(1):15–21. doi: 10.1038/71090. [DOI] [PubMed] [Google Scholar]
- Christen M, Huenenberger PH, Bakowies D, Baron R, Bürgi R, Geerke DP, … van Gunsteren WF. The GROMOS software for biomolecular simulation: GROMOS05. Journal of Computational Chemistry. 2005;26(16):1719–1751. doi: 10.1002/jcc.20303. [DOI] [PubMed] [Google Scholar]
- Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Jr, Ferguson DM, … Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. Journal of the American Chemical Society. 1995;117(19):5179–5197. [Google Scholar]
- Denning EJ, Priyakumar UD, Nilsson L, MacKerell AD., Jr Impact of 2'-hydroxyl sampling on the conformational properties of RNA: update of the CHARMM all-atom additive force field for RNA. Journal of Computational Chemistry. 2011;32(9):1929–1943. doi: 10.1002/jcc.21777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror RO, Dirks RM, Grossman JP, Xu H, Shaw DE. Biomolecular simulation: a computational microscope for molecular biology. Annual review of biophysics. 2012;41:429–452. doi: 10.1146/annurev-biophys-042910-155245. [DOI] [PubMed] [Google Scholar]
- Fleissner MR. PhD Thesis. University of California; Los Angeles, Los Angeles: 2007. [Google Scholar]
- Fleissner MR, Bridges MD, Brooks EK, Cascio D, Kalai T, Hideg K, Hubbell WL. Structure and dynamics of a conformationally constrained nitroxide side chain and applications in EPR spectroscopy. Proc Natl Acad Sci U S A. 2011;108(39):16241–16246. doi: 10.1073/pnas.1111420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleissner MR, Cascio D, Hubbell WL. Structural origin of weakly ordered nitroxide motion in spin-labeled proteins. Protein Science. 2009;18(5):893–908. doi: 10.1002/Pro.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest LR, Zhang YW, Jacobs MT, Gesmonde J, Xie L, Honig BH, Rudnick G. Mechanism for alternating access in neurotransmitter transporters. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(30):10338–10343. doi: 10.1073/Pnas.0804659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghahremanpour MM, Arab SS, Aghazadeh SB, Zhang J, van der Spoel D. MemBuilder: a web-based graphical interface to build heterogeneously mixed membrane bilayers for the GROMACS biomolecular simulation program. Bioinformatics (Oxford, England) 2014;30(3):439–441. doi: 10.1093/bioinformatics/btt680. [DOI] [PubMed] [Google Scholar]
- Guo ZF, Cascio D, Hideg K, Hubbell WL. Structural determinants of nitroxide motion in spin-labeled proteins: Solvent-exposed sites in helix B of T4 lysozyme. Protein Science. 2008;17(2):228–239. doi: 10.1110/Ps.073174008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo ZF, Cascio D, Hideg K, Kalai T, Hubbell WL. Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme. Protein Science. 2007;16(6):1069–1086. doi: 10.1110/Ps.062739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guvench O, Greene SN, Kamath G, Brady JW, Venable RM, Pastor RW, MacKerell AD., Jr Additive empirical force field for hexopyranose monosaccharides. Journal of Computational Chemistry. 2008;29(15):2543–2564. doi: 10.1002/jcc.21004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberger CPR, Schwarzer D. Chemoselective Ligation and Modification Strategies for Peptides and Proteins. Angewandte Chemie-International Edition. 2008;47(52):10030–10074. doi: 10.1002/anie.200801313. [DOI] [PubMed] [Google Scholar]
- Halgren TA. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. Journal of Computational Chemistry. 1996;17(5–6):490–519. doi: 10.1002/(SICI)1096-987X(199604)17:5/6<490::AID-JCC1>3.0.CO;2-P. [DOI] [Google Scholar]
- Hekkelman ML, te Beek TAH, Pettifer SR, Thorne D, Attwood TK, Vriend G. WIWS: A protein structure bioinformatics web service collection. Nucleic Acid Research. 2010;38(SUPPL 2):W719–W723. doi: 10.1093/nar/gkq453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Roux B. Automated Force Field Parameterization for Nonpolarizable and Polarizable Atomic Models Based on Ab Initio Target Data. Journal of Chemical Theory and Computation. 2013;9(8):3543–3556. doi: 10.1021/Ct4003477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbell WL, Mchaourab HS, Altenbach C, Lietzow MA. Watching proteins move using site-directed spin labeling. Structure. 1996;4(7):779–783. doi: 10.1016/S0969-2126(96)00085-8. [DOI] [PubMed] [Google Scholar]
- Hvorup RN, Goetz BA, Niederer M, Hollenstein K, Perozo E, Locher KP. Asymmetry in the structure of the ABC transporter-binding protein complex BtuCD-BtuF. Science. 2007;317(5843):1387–1390. doi: 10.1126/science.1145950. [DOI] [PubMed] [Google Scholar]
- Islam SM, Roux B. Molecular dynamics of dummy spin-labels (MDDS) for structure and function study of proteins 2014 [Google Scholar]
- Islam SM, Stein RA, McHaourab HS, Roux B. Structural refinement from restrained-ensemble simulations based on EPR/DEER data: application to T4 lysozyme. J Phys Chem B. 2013;117(17):4740–4754. doi: 10.1021/jp311723a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeschke G. DEER Distance Measurements on Proteins. Annual Review of Physical Chemistry. 2012;63:419–446. doi: 10.1146/Annurev-Physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- Jo S, Jiang W, Lee HS, Roux B, Im W. CHARMM-GUI Ligand Binder for Absolute Binding Free Energy Calculations and Its Application. Journal of Chemical Information and Modeling. 2013;53(1):267–277. doi: 10.1021/ci300505n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Kim T, Im W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PloS ONE. 2007;2(9):e880. doi: 10.1371/journal.pone.0000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. Journal of Computational Chemistry. 2008;29(11):1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- Jo S, Lim JB, Klauda JB, Im W. CHARMM-GUI Membrane Builder for Mixed Bilayers and Its Application to Yeast Membranes. Biophysical Journal. 2009;97(1):50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Song KC, Desaire H, MacKerell AD, Jr, Im W. Glycan Reader: automated sugar identification and simulation preparation for carbohydrates and glycoproteins. Journal of Computational Chemistry. 2011;32(14):3135–3141. doi: 10.1002/jcc.21886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. Journal of the American Chemical Society. 1996;118(45):11225–11236. doi: 10.1021/ja9621760. [DOI] [Google Scholar]
- Kazmier K, Sharma S, Quick M, Islam SM, Roux B, Weinstein H, … McHaourab HS. Conformational dynamics of ligand-dependent alternating access in LeuT. Nat Struct Mol Biol. 2014;21(5):472–479. doi: 10.1038/nsmb.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauda JB, Venable RM, Freites JA, O'Connor JW, Tobias DJ, Mondragon-Ramirez C, … Pastor RW. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. Journal of Physical Chemistry B. 2010;114(23):7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JL, Yesselman JD, Brooks CL., III Assessing the quality of absolute hydration free energies among CHARMM-compatible ligand parameterization schemes. Journal of Computational Chemistry. 2013 doi: 10.1002/jcc.23199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a Novel Globular Protein Fold with Atomic-Level Accuracy. Science (New York, NY) 2003;302(5649):1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- Langen R, Oh KJ, Cascio D, Hubbell WL. Crystal structures of spin labeled T4 lysozyme mutants: Implications for the interpretation of EPR spectra in terms of structure. Biochemistry. 2000;39(29):8396–8405. doi: 10.1021/Bi000604f. [DOI] [PubMed] [Google Scholar]
- Lee HS, Jo S, Lim H-S, Im W. Application of Binding Free Energy Calculations to Prediction of Binding Modes and Affinities of MDM2 and MDMX Inhibitors. Journal of Chemical Information and Modeling. 2012 doi: 10.1021/ci3000997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KI, Jo S, Rui H, Egwolf B, Roux B, Pastor RW, Im W. Web interface for brownian dynamics simulation of ion transport and its applications to beta-barrel pores. Journal of Computational Chemistry. 2012;33(3):331–339. doi: 10.1002/jcc.21952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lii JH, Allinger NL. Molecular mechanics. The MM3 force field for hydrocarbons. 2. Vibrational frequencies and thermodynamics. Journal of the American Chemical Society. 1989a;111(23):8566–8575. doi: 10.1021/ja00205a002. [DOI] [Google Scholar]
- Lii JH, Allinger NL. Molecular Mechanics. The MM3 force field for hydrocarbons. 3. The van der Waals' potentials and crystal data for aliphatic and aromatic hydrocarbons. Journal of the American Chemical Society. 1989b;111(23):8576–8582. doi: 10.1021/ja00205a003. [DOI] [Google Scholar]
- Liu CC, Schultz PG. Adding New Chemistries to the Genetic Code. Annual Review of Biochemistry. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- Lopes PEM, Huang J, Shim J, Luo Y, Li H, Roux B, MacKerell AD., Jr Force Field for Peptides and Proteins based on the Classical Drude Oscillator. Journal of Chemical Theory and Computation. 2013;9(12):5430–5449. doi: 10.1021/ct400781b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Yeung N, Sieracki N, Marshall NM. Design of functional metalloproteins. Nature. 2009;460(7257):855–862. doi: 10.1038/nature08304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell AD, Brooks B, Brooks CL, Nilsson L, Roux B, Won Y, Karplus M. Encyclopedia of Computational Chemistry. John Wiley & Sons, Ltd; 2002. CHARMM: The Energy Function and Its Parameterization. [Google Scholar]
- MacKerell AD., Jr Empirical force fields for biological macromolecules: overview and issues. Journal of Computational Chemistry. 2004;25(13):1584–1604. doi: 10.1002/jcc.20082. [DOI] [PubMed] [Google Scholar]
- Mayne CG, Saam J, Schulten K, Tajkhorshid E, Gumbart JC. Rapid parameterization of small molecules using the force field toolkit. Journal of Computational Chemistry. 2013;34(32):2757–2770. doi: 10.1002/jcc.23422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHaourab HS, Kalai T, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme: effect of side chain structure. Biochemistry. 1999;38(10):2947–2955. doi: 10.1021/bi9826310. [DOI] [PubMed] [Google Scholar]
- Miller BT, Singh RP, Klauda JB, Hodoscek M, Brooks BR, Woodcock HL. CHARMMing: a new, flexible web portal for CHARMM. Journal of Chemical Information and Modeling. 2008;48(9):1920–1929. doi: 10.1021/ci800133b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren CJ, Anthonycahill SJ, Griffith MC, Schultz PG. A General-Method for Site-Specific Incorporation of Unnatural Amino-Acids into Proteins. Science. 1989;244(4901):182–188. doi: 10.1126/science.2649980. [DOI] [PubMed] [Google Scholar]
- O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: An open chemical toolbox. Journal of cheminformatics. 2011;3(10):33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostmeyer J, Chakrapani S, Pan AC, Perozo E, Roux B. Recovery from slow inactivation in K+ channels is controlled by water molecules. Nature. 2013;501(7465):121–124. doi: 10.1038/nature12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perozo E, Cortes DM, Cuello LG. Three-dimensional architecture and gating mechanism of a K+ channel studied by EPR spectroscopy. Nature Structural Biology. 1998;5(6):459–469. doi: 10.1038/Nsb0698-459. [DOI] [PubMed] [Google Scholar]
- Pless SA, Ahern CA. Unnatural Amino Acids as Probes of Ligand-Receptor Interactions and Their Conformational Consequences. Annual Review of Pharmacology and Toxicology. 2013;53:211–229. doi: 10.1146/annurev-pharmtox-011112-140343. [DOI] [PubMed] [Google Scholar]
- Pless SA, Galpin JD, Niciforovic AP, Ahern CA. Contributions of counter-charge in a potassium channel voltage-sensor domain. Nature Chemical Biology. 2011;7(9):617–623. doi: 10.1038/nchembio.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Cheng X, Han W, Jo S, Schulten K, Im W. CHARMM-GUI PACE CG Builder for Solution, Micelle, and Bilayer Coarse-Grained Simulations. Journal of Chemical Information and Modeling. 2014;54(3):1003–1009. doi: 10.1021/ci500007n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman H, Islam SM, Mukherjee S, Roux B, Perozo E. Dynamics transitions at the outer vestibule of the KcsA potassium channel during gating. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(5):1831–1836. doi: 10.1073/Pnas.1314875111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappé AK, Casewit CJ, Colwell KS, Goddard WA, III, Skiff WM. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. Journal of the American Chemical Society. 1992;114(25):10024–10035. doi: 10.1021/ja00051a040. [DOI] [Google Scholar]
- Rhee YM, Sorin EJ, Jayachandran G, Lindahl E, Pande VS. Simulations of the role of water in the protein-folding mechanism. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(17):6456–6461. doi: 10.1073/pnas.0307898101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DD, Lewis SD, Ballou DP, Olson ST, Shafer JA. Reactivity of small thiolate anions and cysteine-25 in papain toward methyl methanethiosulfonate. Biochemistry. 1986;25(19):5595–5601. doi: 10.1021/bi00367a038. [DOI] [PubMed] [Google Scholar]
- Roberts JA, Digby HR, Kara M, El Ajouz S, Sutcliffe MJ, Evans RJ. Cysteine substitution mutagenesis and the effects of methanethiosulfonate reagents at P2X2 and P2X4 receptors support a core common mode of ATP action at P2X receptors. J Biol Chem. 2008;283(29):20126–20136. doi: 10.1074/jbc.M800294200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, … Bourne PE. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acid Research. 2011;39(Database issue):D392–401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B, Islam SM. Restrained-Ensemble Molecular Dynamics Simulations Based on Distance Histograms from Double Electron-Electron Resonance Spectroscopy. Journal of Physical Chemistry B. 2013;117(17):4733–4739. doi: 10.1021/Jp3110369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B, MacKinnon R. The cavity and pore helices in the KcsA K+ channel: Electrostatic stabilization of monovalent cations. Science (New York, NY) 1999;285(5424):100–102. doi: 10.1126/science.285.5424.100. [DOI] [PubMed] [Google Scholar]
- Savelyev A, MacKerell AD., Jr All-atom polarizable force field for DNA based on the classical drude oscillator model. Journal of Computational Chemistry. 2014;35(16):1219–1239. doi: 10.1002/jcc.23611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt TH, Kandt C. LAMBADA and InflateGRO2: Efficient membrane alignment and insertion of membrane proteins for molecular dynamics simulations. Journal of Chemical Information and Modeling. 2012;52(10):2657–2669. doi: 10.1021/ci3000453. [DOI] [PubMed] [Google Scholar]
- Schrödinger, LLC. The PyMOL Molecular Graphics System Version 1.3r1. 2010. [Google Scholar]
- Schüttelkopf AW, van Aalten DMF. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 8):1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- Sezer D, Freed JH, Roux B. Parametrization, molecular dynamics simulation, and calculation of electron spin resonance spectra of a nitroxide spin label on a polyalanine alpha-helix. J Phys Chem B. 2008;112(18):5755–5767. doi: 10.1021/jp711375x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezer D, Freed JH, Roux B. Multifrequency Electron Spin Resonance Spectra of a Spin-Labeled Protein Calculated from Molecular Dynamics Simulations. Journal of the American Chemical Society. 2009;131(7):2597–2605. doi: 10.1021/Ja8073819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla D, Meng Y, Roux B, Pande VS. Activation pathway of Src kinase reveals intermediate states as targets for drug design. Nature Communications. 2014;5:3397. doi: 10.1038/ncomms4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skjevik AA, Madej BD, Walker RC, Teigen K. LIPID11: A modular framework for lipid simulations using amber. Journal of Physical Chemistry B. 2012;116(36):11124–11136. doi: 10.1021/jp3059992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staritzbichler R, Anselmi C, Forrest LR, Faraldo-Gómez JD. GRIFFIN: A versatile methodology for optimization of protein-lipid interfaces for membrane protein simulations. Journal of Chemical Theory and Computation. 2011;7(4):1167–1176. doi: 10.1021/ct100576m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo Warshaviak D, Cascio D, Khramtsov VV, Hubbell WL. Crystal Structure of Spin Labeled T4 Lysozyme Mutant K65V1/R76V1. PDB Data Bank; [Google Scholar]
- Tombola F, Pathak MM, Isacoff EY. How does voltage open an ion channel? Annual Review of Cell and Developmental Biology. 2006;22:23–52. doi: 10.1146/Annurev.Cellbio.21.020404.145837. [DOI] [PubMed] [Google Scholar]
- Vanommeslaeghe K, Hatcher ER, Acharya C, Kundu S, Zhong S, Shim J, … MacKerell AD., Jr CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. Journal of Computational Chemistry. 2010;31(4):671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe K, MacKerell AD., Jr Automation of the CHARMM General Force Field (CGenFF) I: bond perception and atom typing. Journal of Chemical Information and Modeling. 2012;52(12):3144–3154. doi: 10.1021/ci300363c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe K, Raman EP, MacKerell AD., Jr Automation of the CHARMM General Force Field (CGenFF) II: assignment of bonded parameters and partial atomic charges. Journal of Chemical Information and Modeling. 2012;52(12):3155–3168. doi: 10.1021/ci3003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang W, Kollman PA, Case DA. Automatic atom type and bond type perception in molecular mechanical calculations. Journal of Molecular Graphics & Modelling. 2006;25(2):247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. Journal of Computational Chemistry. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- Weiner SJ, Kollman PA, Case DA, Singh UC, Ghio C, Alagona G, … Weiner P. A new force field for molecular mechanical simulation of nucleic acids and proteins. Journal of the American Chemical Society. 1984;106(3):765–784. doi: 10.1021/ja00315a051. [DOI] [Google Scholar]
- Yesselman JD, Price DJ, Knight JL, Brooks CL., III MATCH: an atom-typing toolset for molecular mechanics force fields. Journal of Computational Chemistry. 2012;33(2):189–202. doi: 10.1002/jcc.21963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoete V, Cuendet MA, Grosdidier A, Michielin O. SwissParam: a fast force field generation tool for small organic molecules. Journal of Computational Chemistry. 2011;32(11):2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]