Summary
While previous results of genetic association studies for common, complex diseases (eg., coronary artery disease, CAD) have been disappointing, examination of multiple related genes within a physiologic pathway may provide improved resolution. This paper describes a method of calculating a genetic risk score (GRS) for a clinical endpoint by integrating data from many candidate genes and multiple intermediate phenotypes (IPs). First, the association of all single nucleotide polymorphisms (SNPs) to an IP is determined and regression β-coefficients are used to calculate an IP-specific GRS for each individual, repeating this analysis for every IP. Next, the IPs are assessed by a second regression as predictors of the clinical endpoint. Each IP’s individual GRS is then weighted by the regression β-coefficients from the second step, creating a single, composite GRS. As an example, 3,172 patients undergoing coronary angiography were evaluated for 3 SNPs from the cholesterol metabolism pathway. Although these data provide only a preliminary example, the GRS method detected significant differences in CAD by GRS group, whereas separate genotypes did not. These results illustrate the potential of the GRS methodology for multigenic risk evaluation and suggest that such approaches deserve further examination in common, complex diseases such as CAD.
Keywords: Genetic Burden, Polygenic Traits
Introduction
Molecular genetic data provide the potential for probabilistic disease diagnosis and guidance of clinical prevention and treatment for prevalent, complex diseases such as coronary artery disease (CAD). As yet, though, genetic association studies have only provided glimpses of potential risk relationships within susceptibility genes in populations of unrelated individuals (Lander, 1996; Risch & Merikangas, 1996; Collins et al. 1997). Various difficulties have caused the failure of validation studies to replicate an initially positive candidate gene study, including poor study design, difficulty in determining/failure to evaluate clinical endpoints (eg., CAD, myocardial infarction [MI], mortality), differences between study populations, and low statistical power (Anonymous, 1999; Risch, 2000). Other potential problems with association studies may include evaluation of a sole candidate gene, difficulties in modelling many genes with the exponentially-increasing number of possible gene-gene interactions, and failure to consider the effect of multiple genes that are pathophysiologically related through common risk-altering intermediate phenotypes (IPs).
While candidate SNPs are frequently shown to predict an IP - often the quantitative trait used to discover the gene - it is less common to find a study with a significant finding for a clinical endpoint. The most probable effect of any gene on a clinical endpoint is through regulation of an IP, or several IPs. For a prevalent, complex disease (eg., CAD) the effect size for any one gene on a clinical endpoint may be small due to potentially many other genes that may regulate the same IP, and the even larger number of genes influencing multiple IPs acting on a clinical endpoint.
For example, higher HDL and lower LDL cholesterol have well-established associations with lower CAD risk (Gotto, 2002a; 2002b), but studies of SNPs that predict these IPs have failed to demonstrate an association between genotype and CAD risk (Couture et al. 2000; Ordovas et al. 2000). This may be due to low statistical power, mischaracterization of intragenic variance, and exclusion of other physiologically-related genes (i.e., failure to account for underlying pathophysiology). While determination of tagging SNPs may increase power and account for intragenic variance in a single gene (Johnson et al. 2001), a polygenic model of complex disease may also be important to consider, such as that used by Pharoah et al. (2002) for known breast cancer genes with dominant expression. Examination of multiple related genes using a modular risk score based on risk-related associations (i.e., to the IPs) may increase power and provide a more robust estimation of risk across populations.
In this paper we present an approach to polygenic modelling using a genetic risk score (GRS) to characterize the genetics of a physiological pathway based on the genes’ shared IP(s). The goal of this approach is to increase the consistency of findings by creating a robust metric from the combination of statistical, biological, genetic, and clinical methodologies. We demonstrate our approach with a preliminary example using SNPs in three genes from the cholesterol metabolism pathway to examine risk of CAD diagnosis among patients undergoing coronary angiography.
Materials and Methods
Modelling a Physiologic Pathway
Genes selected for a GRS analysis should include those related to each other through a shared IP or set of IPs. Ideally, these genes should include both those that up-regulate the pathway or increase the IP, and those that down-regulate the pathway, decrease the IP, or compete with the other genes’ products.
If multiple markers (i.e., SNPs) are available in a single gene, or in multiple genes that are physically close, the first step in constructing a GRS is to determine whether the SNPs can be considered independent, or if they account for the same variation and only a few tagging SNPs are required. Such a procedure lends itself well to use of principal component analysis, as previously described (Horne & Camp, 2004).
With the independent SNPs selected for a GRS analysis, two analyses are performed. The first determines the relative weightings of each independent SNP to each IP considered (SNP-analysis). The second determines the relative weight of each IP to the clinical endpoint (IP-analysis).
The SNP-analyses involve linear regression with each continuous-valued IP, in turn, as the dependent variable and SNP genotypes being the independent variables. In each regression all of the SNPs are evaluated simultaneously, together with any environmental factors that influence the IP. SNP variables found to significantly predict (p < 0.05) change in IP are selected for inclusion in that IP-specific GRS (GRSi for the ith IP) with weight βij, where βij is the standardized linear regression β-coefficient for the ith IP and the jth SNP variable (βij = 0 for non-significant SNP variables). That is:
where Sj is the jth SNP variable. As in all regression modelling, due to the potential for type II error, investigators can consider non-significant SNPs for potential inclusion in an IP model if substantial a priori knowledge of the role is acknowledged and the statistical significance in the current data is reasonably supportive.
The IP-analysis involves the computation of the relative effects of all of the IPs on the clinical endpoint. This analysis is performed in logistic regression (or other appropriate method: eg., Cox regression for survival) with the clinical endpoint as the dependent variable and the IPs as the simultaneously-evaluated independent variables. The regression coefficients (which we will call Bi here for clarity) for the independent IPs are then used to weight each GRSi in the overall pathway’s GRS (GRStot), with each Bi scaled by dividing it by the modulus of the largest Bi (|Bmax|). In this way, the scale of the GRS remains reasonable and the sign of the correlations between each IP and the endpoint is preserved. That is:
where Bi is the regression coefficient for the ith IP (Bi = 0 for non-significant IPs). The derived GRStot can then be used as an independent variable to investigate a polygenic effect in predicting the clinical endpoint. With inclusion of a larger number of SNPs, the GRStot will approach a continuous distribution and can be entered into an analysis as a continuous variable, or categorized into quartiles. However, with fewer SNPs evaluated it is likely that the score may cluster on a limited scale, and thus the distribution should be inspected for reasonable groupings for classification and further study.
Example Data
The example genetic data evaluated herein are limited by the small number of genes that are included, use of only one SNP per gene, and for other reasons (see Limitations section), but do provide an initial demonstration of the method’s application.
Patients
Patients considered for inclusion in this study were those undergoing coronary angiography from 1994 to 2001 who provided written informed consent for participation in the cardiac catheterization registry of the Intermountain Heart Collaborative Study. Patient consent was obtained prior to catheterization in accordance with the guidelines of the local institutional review board.
Patient data were recorded following the Coronary Artery Surgery Study protocol (Anonymous, 1984). Coronary stenoses and degree of stenosis were reported by the patient’s cardiologist from angiographic evidence. Potential study patients (N = 3,731) either had significant CAD (≥1 coronary lesion of ≥70% stenosis; i.e., a clinically flow-limiting stenosis) or were disease-free (no coronary lesion of ≥10% stenosis). Intermediate disease (10%–69% stenosis) was excluded as indeterminate to provide a clear phenotypic distinction.
IP Data
Low-density lipoprotein (LDL) cholesterol, high-density lipoprotein (HDL) cholesterol, and triglyceride (TG) levels were assayed from fasting blood samples using a clinical assay (Vitros System, Johnson & Johnson Clinical Diagnostics) and were available for 72% of patients (missing data were missing at random).
Genetic Data
Patients were primarily of British/Northern European descent and drawn from a population that is genetically similar to the U.S. Caucasian population (McLellan et al. 1984). Genotypes were available for a single SNP in each of three genes, all primary components of the reverse cholesterol transport pathway (Attie et al. 2001): the cholesteryl ester transfer protein (CETP) gene, the ATP-binding cassette A1 (ABCA1) gene, and hepatic lipase (HL) gene. Variation at the different gene loci, particularly associated with the SNPs used here, has been shown previously to be associated with HDL (Couture et al. 2000; Ordovas et al. 2000; Kuivenhoven et al. 1998; Zambon et al. 2001; Talmud et al. 2002; Kakko et al. 2003). Of the potential study patients, genotyping data were available for 3,172 patients (85%), and for the others the missing data were consistent with the assumption of missing at random.
Covariables
Patient demographic (age, sex) and cardiac risk variables were recorded electronically at the time of cardiac catheterization. Physician-reported cardiac risk factors included: diabetes (untreated fasting glucose ≥126 mg/dl or use of hypoglycemic medication), hypertension (untreated systolic blood pressure ≥140 mmHg or diastolic ≥90 mmHg, or anti-hypertensive use), and hyperlipidemia (untreated total cholesterol≥200 mg/dl or LDL≥130 mg/dl, or cholesterol-lowering medication use). Self-reported measures were: family history of early CAD (first-degree relative suffering cardiovascular death, MI, or revascularization prior to 65 years of age) and tobacco use (current smoker or history of ≥10 pack-years). C-reactive protein (CRP) was measured using a medium-sensitivity assay shown to stratify risk in this population similarly well compared to high-sensitivity CRP assays (Clarke et al. 2005). Body mass index was measured (patient height divided by the square of their mass) but was not included, except as a descriptive, since it was not associated with the genotypes, GRStot, or CAD.
Further Statistical Considerations
Comparisons of patient characteristics to GRS values were evaluated using the chi-square test, t-test, or analysis of variance, as appropriate. Natural log transformation was used (median reported) when normality assumptions were violated. Linear regression was used to evaluate the association of SNPs to IPs. Logistic regression was used to evaluate whether GRStot significantly predicted the clinical CAD endpoint. Note that IPs must be on the same scale in all analyses. Due to the small number of genes included in the example data, and the evaluation of only one SNP per gene, an a priori decision was made to utilize the largest GRS group as the reference group for comparisons, so that the resulting risk estimates would have the least random fluctuation due to inadequate sample sizes. In each IP- and SNP-analysis, multivariable adjustments were performed for age, gender, hypertension, hyperlipidemia, diabetes, smoking, family history, and CRP concentration. In non-GRS analyses, statistical SNP interactions were modelled as X1*X2 for the three combinations of homozygous variant genotypes. Statistical significances are presented as two-tailed p-values, with nominal significance set at p<0.05 for all analyses.
Results
SNPs
The genotype data considered here are from one SNP in each of three genes (CETP, ABCA1, HL) that are located on different chromosomes. Hence, we have considered all SNP variables to be independent.
Coronary Disease
Analyses of lipid measurements demonstrated that TC, LDL, HDL, and TG were each associated with CAD risk, both as continuously valued variables (p = 0.039, 0.015, 0.007, 0.001, respectively) and for the first vs. fourth quartiles (p = 0.022, 0.020, 0.007, 0.032, respectively).
Minimal differences in CAD were found for each SNP, with 74%, 76%, and 71% of patients having CAD in CETP B1B1, B1B2, and B2B2; 74%, 75%, and 74% in ABCA1 GG, AG, and AA; and 74%, 75%, and 72% in HL CC, CT, TT, respectively. These differences were not significant, and a model simultaneously entering all SNPs showed no improvement, even after adjustment (Table 1). Interactions in this model were also not significant for B2B2*AA (p = 0.11), B2B2*TT (p = 0.61), or AA*TT (p = 0.12).
Table 1.
The association of each genotype with CAD, with all three SNPs entered into the same regression model
| Genotype (freq.) | Model: SNPs only
|
Model: SNPs & covariables†
|
||
|---|---|---|---|---|
| Odds Ratio (CI*) | p-value | Odds Ratio (CI*) | p-value | |
| CETP | ||||
| B1B1 (0.33) | 1.0 | 1.0 | ||
| B1B2 (0.50) | 1.1 (0.96, 1.4) | 0.13 | 1.05 (0.86, 1.3) | 0.66 |
| B2B2 (0.18) | 0.86 (0.68, 1.1) | 0.19 | 0.86 (0.67, 1.1) | 0.26 |
| ABCA1 | ||||
| GG (0.53) | 1.0 | 1.0 | ||
| AG (0.39) | 1.05 (0.89, 1.2) | 0.58 | 1.03 (0.85, 1.2) | 0.79 |
| AA (0.08) | 1.0 (0.74, 1.2) | 0.99 | 0.98 (0.70, 1.4) | 0.92 |
| HL | ||||
| CC (0.58) | 1.0 | 1.0 | ||
| CT (0.36) | 1.1 (0.89, 1.3) | 0.54 | 1.1 (0.93, 1.4) | 0.21 |
| TT (0.06) | 0.90 (0.65, 1.3) | 0.54 | 1.01 (0.70, 1.5) | 0.96 |
CI = 95% confidence interval,
Adjusted model entering the three SNPs, age, gender, five cardiac risk factors, and C-reactive protein.
GRS-analyses
Linear regression results for LDL, HDL, and TG are shown in Table 2. HDL was predicted strongly by CETP B2B2 (p < 0.001), and by CETP B1B2 (p = 0.006), ABCA1 AA (p = 0.014), and HL TT (p = 0.033) after adjustment for covariables (i.e., age, gender, 6 risk factors). No genotype was associated with differences in LDL or TG level, and hence all βij and Bi for these IPs are zero in Table 2. Thus for these example data, the GRSHDL coefficient (B1) is the only component in GRStot (i.e., no GRSLDL or GRSTG component) which when divided by its modulus naturally yielded −1. Hence,
Table 2.
Results from linear regression modelling (adjusted for age, gender, risk factors and CRP) evaluating the association of each SNP to HDL cholesterol, LDL cholesterol, and TG. (βij-coefficients indicates the weighting of the jth marker for the ith IP)
| IP | I | Bi | βij | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CETP | genotypes
|
ABCA1 | genotypes
|
HL | genotypes
|
||||||
| B1B1* | B1B2 | B2B2 | GG* | GA | AA | CC* | CT | TT | |||
| HDL | 1 | −1† | 0 | 0.067‡ | 0.104|| | 0 | 0 | 0.049¶ | 0 | 0 | 0.055# |
| LDL | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TG | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
βij = 0 for all referent categories, and βij = 0 for all non-significant variables (p > 0.05),
HDL was the only IP found to be associated with genotypes, and therefore is the only IP with a non-zero Bi in GRStot,
p = 0.006,
p < 0.001,
p = 0.014,
p = 0.033.
where the indicator variable (I) is 1 if a patient has the subscripted genotype, and 0 if not.
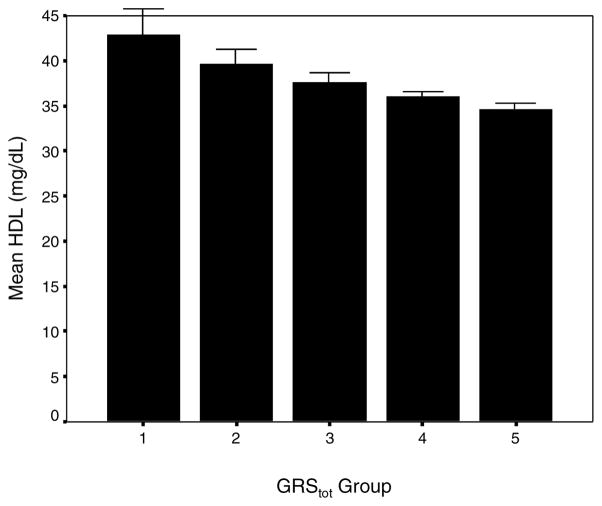
The risk scores calculated for each patient from the above equation resulted in a discontinuous distribution of clustered GRStot values with range −2.09 to 0.00. Given the non-continuous nature, we assigned patients to 5 groups, with groupings determined from clustering of values (within of the average value). Group 1 had the lowest genetic loading and lowest GRStot values and group 5 the highest. We inspected the mean HDL in these groups and, as expected, observed a monotonically lower mean HDL level with group (Figure 1). The linear trend across HDL was highly significant (p < 0.001).
Figure 1.
Mean HDL cholesterol level by GRStot (bars: standard error), demonstrating the trend in decreasing values for a higher genetic loading.
Baseline characteristics
Table 3 presents the baseline characteristics stratified by GRStot group. Characteristics were generally similar across all groups, and similar to overall. Some exceptions were noted in groups 1 and 2, although these groups also had the smallest sample size and should be interpreted with caution.
Table 3.
Baseline characteristics of the study population overall and by GRStot
| Characteristic | GRStot group
|
|||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | Overall | |
| GRStot (mean) | −1.58 | −1.27 | −1.00 | −0.70 | 0.00 | −0.61 |
| Sample size, N = 3,172 | n = 90 | 197 | 484 | 1,495 | 906 | 3,172 |
| Percent of sample | 3% | 6% | 15% | 47% | 29% | 100% |
| Age (years), mean (SD) | 64(12) | 62(13) | 63(12) | 64(12) | 63(12) | 63.5(12) |
| Sex (male) | 55%* | 71% | 70% | 71% | 69% | 70% |
| Hypertension | 61% | 64% | 54% | 56% | 55% | 56% |
| Hyperlipidemia | 47% | 58% | 49% | 51% | 49% | 50% |
| Diabetes | 17% | 25%† | 16% | 19% | 16% | 18% |
| Smoking | 17% | 24% | 22% | 23% | 24% | 23% |
| Family History | 33% | 36% | 34% | 36% | 35% | 35% |
| Body Mass Index (kg/m2) | 28.7 | 29.0 | 28.9 | 28.5 | 28.7 | 28.7 |
| CRP§ (mg/L), median | 13.6‡ | 13.0 | 12.8 | 12.8 | 12.8 | 12.8 |
p = 0.001,
p = 0.046,
p = 0.044,
CRP: C-reactive protein. For variables for which the overall test statistic was significant, pair-wise tests compared individual GRStot categories to group 4.
Association analyses
Initial analysis of GRStot by chi-square (4 d.f.) showed an association (p = 0.019) to CAD, suggesting potential differences between GRS groups. In subsequent univariate logistic regression (Table 4), a significantly lower risk (OR = 0.70, CI = 0.55–0.88, p = 0.003) was seen for GRS group 3 compared to the largest group (group 4, n = 1495). After adjustment (i.e., for age, gender, six risk factors), the results were largely unchanged compared to group 4, with a significantly lower risk still existing for group 3 (OR = 0.73, CI = 0.56–0.96, p = 0.02), and borderline for group 2 (OR = 0.69, CI = 0.47–1.02, p = 0.06).
Table 4.
GRStot results for CAD prediction (unadjusted)
| GRStot: | Group | OR (CI) | p-value |
|---|---|---|---|
| Groupings | 1 | 0.81 (0.50–1.3) | 0.41 |
| 2 | 0.75 (0.53–1.05) | 0.09 | |
| 3 | 0.70 (0.55–0.88) | 0.003 | |
| 4 | 1.0 (referent) | — | |
| 5 | 0.85 (0.70–1.03) | 0.10 |
Secondary association analysis
While analysis of many groups may provide a biologically meaningful picture of clinical endpoint across many genetic loadings, from the perspective of clinical utility the data may be better viewed by categorization into a dichotomous variable to provide a threshold that is useful for clinical decision-making. Such dichotomization was performed by examination of group similarities in covariable levels, differences in population sizes, and clinical results (IP results may also be used, although in this study no threshold was evident). Three groups composed a low genetic loading set (GRS groups 1, 2, and 3) and the other two a high genetic loading set (GRS groups 4 and 5). In univariate (Table 4), a significant difference in CAD was found with lower risk for patients with lower genetic loading (OR = 0.77, CI = 0.64–0.93, p = 0.006). After multiple regression adjustment, the effect size remained (OR = 0.78, CI = 0.63–0.96, p = 0.02).
Exploratory subanalysis
A post hoc analysis was performed to evaluate the GRS score among patients with no history of hyperlipidemia (n = 1,577), who likely never took a lipid-lowering medication and thus wherein the unobserved past medication history would be less of an unaccountable confounder. This analysis showed (Table 5) that the effect of the GRS was more linear than in the overall group (with the exception of group 1). Multivariate ORs for groups 1–5 were 1.3, 0.59, 0.81, 1.0, and 1.05, respectively.
Table 5.
Post-hoc GRStot subanalysis (unadjusted) for CAD among patients with no history of hyperlipidemia (and likely no past use of lipid-lowering medications)
| GRStot: | Group | OR (CI) | p-value |
|---|---|---|---|
| Groupings | 1 | 1.06 (0.56–2.0) | 0.87 |
| 2 | 0.61 (0.38–0.97) | 0.035 | |
| 3 | 0.73 (0.54–0.98) | 0.039 | |
| 4 | 1.0 (referent) | — | |
| 5 | 0.93 (0.72–1.2) | 0.56 |
Discussion
The Polygenic Risk Scoring Model
For prevalent diseases, such as CAD, genetic risk stratification could have an important clinical impact for the general population. GRS methods that can be generalized may provide improved primary and secondary risk assessment, the ability to personalize preventive care and medical treatments, and the opportunity to exclude low-risk patients from screening tests or interventions given an appropriate risk:benefit ratio.
Unfortunately, study of genetic risk among the general population for common, complex diseases is not as straight-forward as past evaluations of rare diseases with Mendelian characteristics (Risch, 2000). Gene discovery/linkage studies usually focus on high-risk pedigrees, not the general population, and usually discover rare mutations present in a very limited set of pedigrees, although the mutations that are discovered often account for a relatively large proportion of disease in the affected pedigrees (Winkelmann et al. 2000). However, linkage studies of loci for quantitative phenotypes (eg., LDL, HDL) often report disparate findings despite the best efforts to control extraneous factors (Klos et al. 2001; Coon et al. 2001), suggesting that diseases such as CAD may be polygenic. Further, association studies for clinical endpoints often fail to replicate initially positive findings (Ludwig et al. 1995; Anderson et al. 1998; Keavney et al. 2000), suggesting that genotypes individually have small effects, as expected for a polygenic disease. The prediction of IPs but not clinical endpoints by individual SNPs in this study provides further evidence of this, suggesting that the use of IPs as surrogates for clinical endpoints may not be appropriate in genetic association studies of complex diseases.
Since common, chronic diseases are likely polygenic, evaluation of genetic risk will require both the evaluation of many candidate genes, and the complete characterization of each gene’s intragenic variation, to isolate the best SNP or combination of SNPs to tag the disease-causing variant(s) in the gene. While the latter is a major focus of genetics today, little effort is focused on genotype combinations from multiple genes. This may be because examination of many variants from multiple genes using traditional statistical methodologies is well nigh impossible, since the number of possible genetic interactions increases exponentially with the number of SNPs considered. However, because of the potential increase in consistency and power that can result from combining many small-effect SNPs from related genes, analyses using polygenic models represent a potentially important advance in genetic risk determination for complex diseases.
One method to reduce the exponential number of genotype combinations is to examine only the important, risk-influencing genotype subcategories. For example, the Multifactor Dimensionality Reduction method (Ritchie et al. 2001) examines differences in risk for multiple SNPs based on relative numbers of cases and controls, in a contingency table of all possible genotype combinations. Other largely untried methods in genetics such as classification trees, logic regression, or neural networking analysis may also provide improved categorization. However, these methods each require the use of the clinical disease endpoint to determine which genotype categories are important in a subset of the data, and then reverse the process by using those categories to analyze risk associations with the disease endpoint in the remaining data. Training and test set analyses run the risk of reduced power because the sample must be split for the analyses. Further, with large numbers of SNPs there may be insufficient data to populate the high-dimensional contingency tables in the training set, in which case the analysis cannot be performed (Multi-Dimensionality Reduction) or over-fitting may occur (other methods).
Stratifying the Genetic Burden of Disease
The current study introduces a polygenic model, defined as a GRS metric, to stratify CAD by integrating genetic and biological information into the statistical model. The GRS method may better categorize individual clinical risk by examining many SNPs from multiple genes in a common pathway and reducing them into a simple, biologically-relevant model. The GRS method defines genotype groupings by risk-related IP, and not by clinical endpoint, under the concept that gene variants from many genes likely influence an IP, while it is the IP that usually influences clinical risk. The GRS method addresses the concept that a more integrated approach may be needed to model the effects of SNPs on clinical endpoints than is required to observe their effects on IPs.
The example GRS data supported this concept. In those data, the GRS score was highly associated with HDL (p < 0.001), but the GRS metric was also able to find an association with the clinical endpoint (p = 0.006, p = 0.02 with and without covariate correction, respectively). The association with clinical endpoint was not as strong, as would be expected if multiple SNPs influence HDL and HDL influences risk, but the GRS method uncovered larger relative CAD differences (Table 4) than the individual SNPs (Table 1).
Another strength of the GRS method is that all risk-related IPs within a pathway are modeled together, potentially increasing the utility of the GRS over simple measurement of a plasma risk marker (eg., LDL, HDL). Further, the GRS model employs a generalized genetic risk model with each genotype’s effect weighted by its individual influence on IPs.
Herein we have introduced a basic concept that provides a framework for the GRS, but that can be expanded to more complex data issues. In particular, GRS scoring schemes including more than one tagging SNP per gene, or haplotypes of these SNPs, to describe intragenic risk will need to be considered as such data become available, since in many cases utilization of one SNP per gene is inadequate for full characterization of the intragenic variation. In that case, intragenic weighting will also likely be required. An additional step could also be used to create a separate GRS for each physiological pathway that is modelled (eg., blood pressure control, glucose metabolism). It may also be that methods such as neural networks could be used sequentially for IPs and clinical phenotypes to integrate biological knowledge into a GRS model in a similar manner as regression was used in this study.
Findings for Reverse Cholesterol Transport
This study showed significant risk stratification for three SNPs that, on their own, were not significantly predictive of CAD in this population, and that other studies have failed to find as significantly predictive of clinical phenotype (Agerholm-Larsen et al. 2000a; 2000b; Ji et al. 2002; Andersen et al. 2003). While the three SNPs did not predict LDL or TG levels, and thus the general GRS method was not demonstrated for the reverse cholesterol transport pathway, GRStot did provide significant risk stratification of the clinical CAD endpoint based on the HDL IP.
Interestingly, the GRS risk model was not linear, but appeared J-shaped, with the highest GRS group potentially having lower risk. This was not expected, and is likely due to several factors: noise in the system due to random fluctuation; the inaccurate modelling of a SNP’s mode of inheritance in the IP-specific GRS (recessive/dominant); the absence of an important SNP(s) in the analysis; and over-simplification of the interaction between genes (epistasis). Theoretically, if all necessary variables (all important SNPs from all related and relevant genes) are incorporated into the model and modelled appropriately, then a monotonic relationship should occur, with minor variation from a linear pattern occurring only due to random fluctuation (note that the anomalies in our example were either non-significant or borderline significant). A subanalysis of only patients with no history of hyperlipidemia, among whom past lipid-lowering medication use is unlikely, suggested a more linear risk curve, suggesting that noise due to unaccounted factors may exist in the example data. All of these findings provide motivation for additional evaluations of the combined genetics of this pathway, including studies of many more genes that predict HDL and those that predict LDL and TG.
For the lowest genetic loaders, an unexpected finding related to inflammation was described, wherein patients with the highest HDL had a significantly elevated CRP level; this finding is supported by experimental evidence (Navab et al. 2001), but requires further study.
Limitations
This study’s use of one SNP for each candidate gene assumes that each SNP is a reasonable marker of the intragenic risk locus. The SNPs used in the example data herein probably are not the most efficient for tagging each gene’s disease-causing locus (or loci), but previous studies’ results for these SNPs and IP suggest that they are reasonable to use (Couture et al. 2000; Ordovas et al. 2000; Kuivenhoven et al. 1998; Zambon et al. 2001; Talmud et al. 2002; Kakko et al. 2003). Interactions of SNPs also were not modelled, while if a priori specific interactions between SNPs are known or suspected in a system they could be incorporated into the GRS.
This study was also limited by its observational nature. Various unobserved or uncontrolled biases may be present, including that the population represents higher-risk patients who presented for coronary angiography. However, genetic factors were not considered in patient admission decisions, the observed genotypes were randomly assorted among the population, and many known risk factors for CAD were controlled for in the study analysis. Also, while the comparison group was not a random, population-based selection of apparently healthy individuals, non-diseased patients were defined based on the outcome of their disease evaluation, and were known to be absolutely free of CAD by the gold-standard test for CAD diagnosis. In contrast, population controls are only apparently healthy but likely include many individuals with asymptomatic CAD.
In the absence of IP data, a modified version of the GRS procedure can be used, in which results from published literature can be used to weight the SNP variables. Due to problems with comparability across studies in the literature, a simpler counting procedure could be used to weight the SNP variables in each GRSi for the ith IP. However, likely the best approach is for researchers to invest in adequate phenotyping of IPs within their study resource to compose a meaningful GRS.
Conclusions
The GRS approach described here may provide a biologically informed, statistically robust, and clinically relevant method to describe a genetic risk score for meaningful application to clinical endpoints (eg., CAD). In the example data, our GRS method was able to detect significant differences in CAD, whereas no significant results were found when SNPs were considered either separately or simultaneously as independent variables. The results here suggest that our multigenic GRS approach holds promise for improved risk stratification and deserves further evaluation in other populations, for other pathophysiological pathways, and with greater numbers of related candidate genes and more SNPs within those genes.
Acknowledgments
This study was supported in part by grants from the Deseret Foundation (Salt Lake City, UT) and by NIH grants HL073117 (to J.F.C. and N.J.C.), CA099844 & CA098364 (to N.J.C.), and HL071878 (to J.L.A. and J.F.C.).
References
- Agerholm-Larsen B, Nordestgaard BG, Steffensen R, Jensen G, Tybjærg-Hansen A. Elevated HDL cholesterol is a risk factor for ischemic heart disease in white women when caused by a common mutation in the cholesteryl ester transfer protein gene. Circulation. 2000a;101:1907–1912. doi: 10.1161/01.cir.101.16.1907. [DOI] [PubMed] [Google Scholar]
- Agerholm-Larsen B, Tybjærg-Hansen A, Schnohr P, Steffensen R, Nordestgaard BG. Common cholesteryl ester transfer protein mutations, decreased HDL cholesterol, and possible decreased risk of ischemic heart disease. The Copenhagen City Heart Study. Circulation. 2000b;102:2197–2203. doi: 10.1161/01.cir.102.18.2197. [DOI] [PubMed] [Google Scholar]
- Andersen RV, Wittrup HH, Tybjærg-Hansen A, Steffensen R, Schnohr P, Nordestgaard BG. Hepatic lipase mutations, elevated high-density lipoprotein cholesterol, and increased risk of ischemic heart disease. J Am Coll Cardiol. 2003;41:1972–1982. doi: 10.1016/s0735-1097(03)00407-8. [DOI] [PubMed] [Google Scholar]
- Anderson JL, Carlquist JF, King GJ, Morrison L, Thomson MJ, Ludwig EH, Muhlestein JB, Bair TL, Ward RH. Angiotensin-converting enzyme genotypes and risk for myocardial infarction in women. J Am Coll Cardiol. 1998;31:790–6. doi: 10.1016/s0735-1097(98)00007-2. [DOI] [PubMed] [Google Scholar]
- Anonymous. Myocardial infarction and mortality in the coronary artery surgery study (CASS) randomized trial. N Engl J Med. 1984;310(12):750–8. doi: 10.1056/NEJM198403223101204. [DOI] [PubMed] [Google Scholar]
- Anonymous. Freely associating [editorial] Nat Genet. 1999;22:1–2. doi: 10.1038/8702. [DOI] [PubMed] [Google Scholar]
- Attie AD, Kastelein JP, Hayden MR. Pivotal role of ABCA1 in reverse cholesterol transport influencing HDL levels and susceptibility to atherosclerosis. J Lipid Res. 2001;42:1717–1726. [PubMed] [Google Scholar]
- Clarke JL, Anderson JL, Carlquist JF, Roberts RF, Horne BD, Bair TL, Kolek MJ, Mower CP, Crane AM, Roberts WL, Muhlestein JB. Comparison of differing C-reactive protein assay methods and their impact on cardiovascular risk assessment. Am J Cardiol. 2005;95:155–158. doi: 10.1016/j.amjcard.2004.08.087. [DOI] [PubMed] [Google Scholar]
- Collins FS, Guyer MS, Charkravarti A. Variations on a theme: cataloging human DNA sequence variation. Science. 1997;278:1580–1581. doi: 10.1126/science.278.5343.1580. [DOI] [PubMed] [Google Scholar]
- Coon H, Leppert MF, Eckfeldt JH, Oberman A, Myers RH, Peacock JM, Province MA, Hopkins PN, Heiss G. Genome-wide linkage analysis of lipids in the hypertension genetic Epidemiology Network (HyperGEN) blood pressure study. Arterioscler Thromb Vasc Biol. 2001;21:1969–1976. doi: 10.1161/hq1201.100228. [DOI] [PubMed] [Google Scholar]
- Couture P, Otvos JD, Cupples LA, Lahoz C, Wilson PWF, Schaefer EJ, Ordovas JM. Association of the C-514T polymorphism in the hepatic lipase gene with variations in lipoprotein subclass profiles. Arterioscler Thromb Vasc Biol. 2000;20:815–822. doi: 10.1161/01.atv.20.3.815. [DOI] [PubMed] [Google Scholar]
- Gotto AM., Jr Management of dyslipidemia. Am J Med. 2002a;112:10S–18S. doi: 10.1016/s0002-9343(02)01085-9. [DOI] [PubMed] [Google Scholar]
- Gotto AM., Jr High-density lipoprotein cholesterol and triglycerides as therapeutic targets for preventing and treating coronary artery disease. Am Heart J. 2002b;144:S33–S42. doi: 10.1067/mhj.2002.130301. [DOI] [PubMed] [Google Scholar]
- Horne BD, Camp NJ. Principal component analysis for selection of optimal SNP-sets that capture intragenic genetic variation. Genet Epidemiol. 2004;26:11–21. doi: 10.1002/gepi.10292. [DOI] [PubMed] [Google Scholar]
- Ji J, Herbison CE, Mamotte CD, Burke V, Taylor RR, van Bockxmeer FM. Hepatic lipase gene–514 C/T polymorphism and premature coronary heart disease. J Cardiovasc Risk. 2002;9:105–113. doi: 10.1177/174182670200900206. [DOI] [PubMed] [Google Scholar]
- Johnson GCL, Esposito L, Barratt BJ, Smith AN, Heward J, Di Genova G, Ueda H, Cordell HJ, Eaves IA, Dudbridge F, Twells RCJ, Payne F, Hughes W, Nutland S, Stevens H, Carr P, Tuomilehto-Wolf E, Tuomilehto J, Gough SCL, Clayton DG, Todd JA. Haplotype tagging for the identification of common disease genes. Nat Genet. 2001;29:233–237. doi: 10.1038/ng1001-233. [DOI] [PubMed] [Google Scholar]
- Kakko S, Kelloniemi J, von Rohr P, Hoeschele I, Tamminen M, Brousseau ME, Kesaniemi YA, Savolainen MJ. ATP-binding cassette transporter A1 locus is not a major determinant of HDL-C levels in a population at high risk for coronary heart disease. Atherosclerosis. 2003;166:285–290. doi: 10.1016/s0021-9150(02)00232-0. [DOI] [PubMed] [Google Scholar]
- Keavney B, McKenzie C, Parish S, Palmer A, Clark S, Youngman L, Delepine M, Lathrop M, Peto R, Collins R. Large-scale test of hypothesised associations between the angiotensin-converting-enzyme insertion/deletion polymorphism and myocardial infarction in about 5000 cases and 6000 controls. Lancet. 2000;355:434–42. doi: 10.1016/s0140-6736(00)82009-7. [DOI] [PubMed] [Google Scholar]
- Klos KL, Kardia SL, Ferrell RE, Turner ST, Boer-winkle E, Sing CF. Genome-wide linkage analysis reveals evidence of multiple regions that influence variation in plasma lipid and apolipoprotein levels associated with risk of coronary heart disease. Arterioscler Thromb Vasc Biol. 2001;21:971–978. doi: 10.1161/01.atv.21.6.971. [DOI] [PubMed] [Google Scholar]
- Kuivenhoven JA, Jukema JW, Zwinderman AH, de Knijff P, McPherson R, Bruschke AVG, Lie KI, Kastelein JP. The role of a common variant of the cholesteryl ester transfer protein gene in the progression of coronary atherosclerosis. N Engl J Med. 1998;338:86–93. doi: 10.1056/NEJM199801083380203. [DOI] [PubMed] [Google Scholar]
- Lander ES. The new genomics: global views of biology. Science. 1996;274:536–539. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- Ludwig E, Corneli PS, Anderson JL, Marshall HW, Lalouel JM, Ward RH. Angiotensin-converting enzyme gene polymorphism is associated with myocardial infarction but not with development of coronary stenosis. Circulation. 1995;91:2120–4. doi: 10.1161/01.cir.91.8.2120. [DOI] [PubMed] [Google Scholar]
- McLellan T, Jorde LB, Skolnick MH. Genetic distances between the Utah Mormons and related populations. Am J Hum Genet. 1984;36:836–857. [PMC free article] [PubMed] [Google Scholar]
- Navab M, Berliner JA, Subbanagounder G, Hama S, Lusis AJ, Castellani LW, Reddy S, Shih D, Shi W, Watson AD, Van Lenten BJ, Vora D, Fogelman AM. HDL and the inflammatory response induced by LDL-derived oxidized phospholipids. Arterioscler Thromb Vasc Biol. 2001;21:481–488. doi: 10.1161/01.atv.21.4.481. [DOI] [PubMed] [Google Scholar]
- Ordovas JM, Cupples LA, Corella D, Otvos JD, Osgood D, Martinez A, Lahoz C, Coltell O, Wilson PWF, Schaefer EJ. Association of cholesteryl ester transfer protein-Taq1B polymorphism with variations in lipoprotein subclasses and coronary heart disease risk: The Framingham Study. Arterioscler Thromb Vasc Biol. 2000;20:1323–1329. doi: 10.1161/01.atv.20.5.1323. [DOI] [PubMed] [Google Scholar]
- Pharoah PDP, Antoniou A, Bobrow M, Zimmern RL, Easton DF, Ponder BAJ. Polygenic susceptibility to breast cancer and implications for prevention. Nat Genet. 2002;31:33–36. doi: 10.1038/ng853. [DOI] [PubMed] [Google Scholar]
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Risch NJ. Searching for genetic determinants in the new millennium. Nature. 2000;2000:405, 847–56. doi: 10.1038/35015718. [DOI] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69:138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmud PJ, Hawe E, Robertson K, Miller GJ, Miller NE, Humphries SE. Genetic and environmental determinants of plasma high density lipoprotein cholesterol and apolipoprotein AI concentrations in healthy middle-aged men. Ann Hum Genet. 2002;66:111–124. doi: 10.1017/S0003480002001057. [DOI] [PubMed] [Google Scholar]
- Winkelmann BR, Hager J, Kraus WE, Merlini P, Keavney B, Grant PJ, Muhlestein JB, Granger CB. Genetics of coronary heart disease: current knowledge and research principles. Am Heart J. 2000;140:S11–S26. doi: 10.1067/mhj.2000.109636. [DOI] [PubMed] [Google Scholar]
- Zambon A, Deeb SS, Brown BG, Hokanson JE, Brunzell JD. Common hepatic lipase gene promoter variant determines clinical response to intensive lipid-lowering treatment. Circulation. 2001;103:792–798. doi: 10.1161/01.cir.103.6.792. [DOI] [PubMed] [Google Scholar]