Abstract
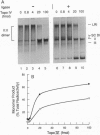
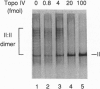
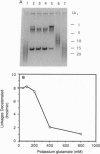
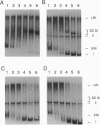
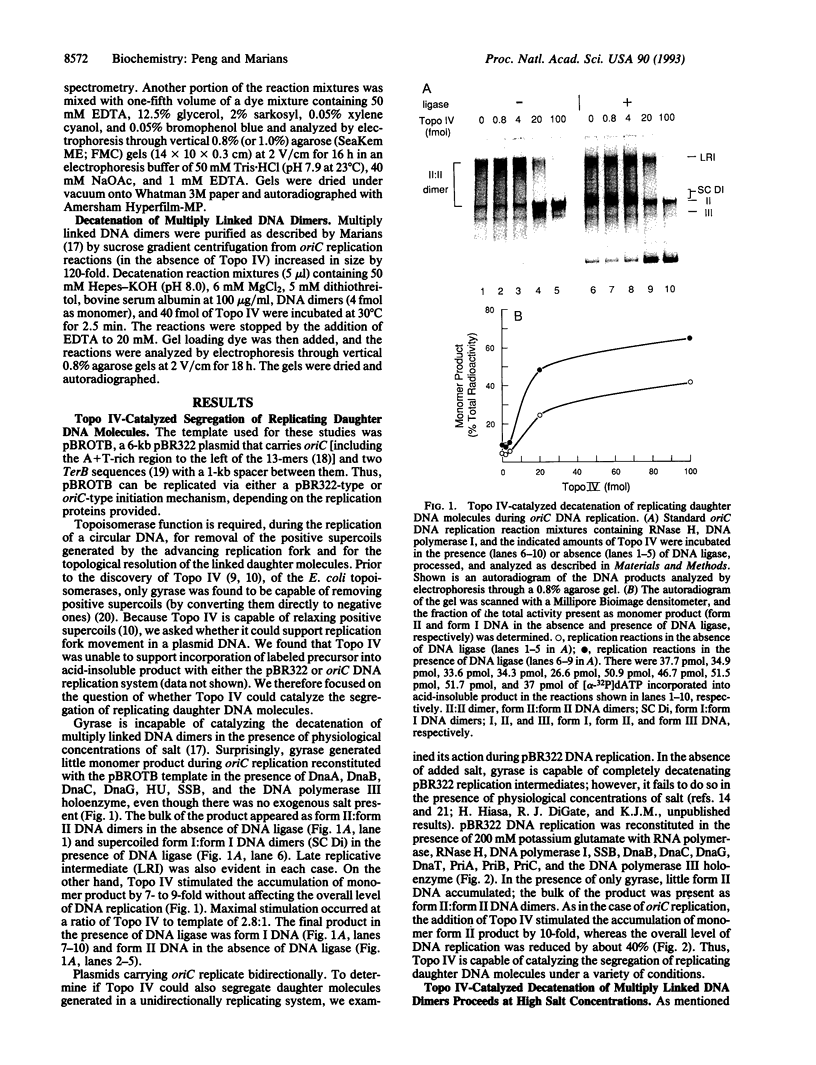
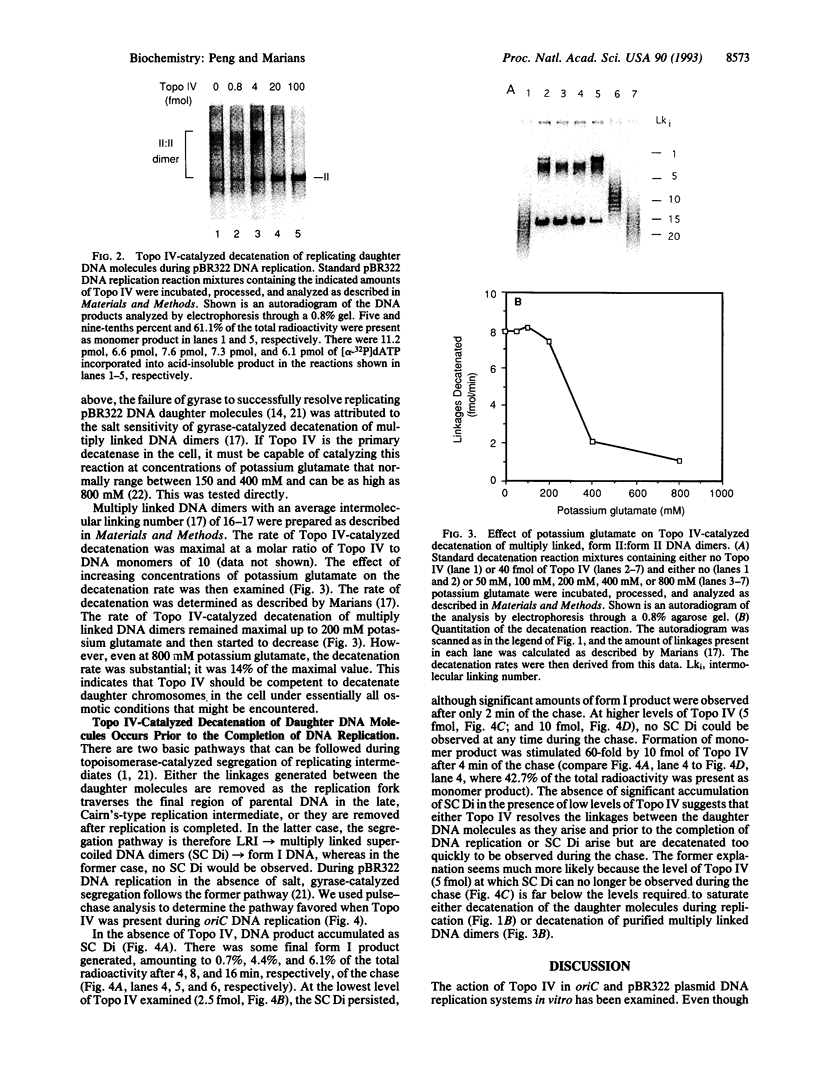
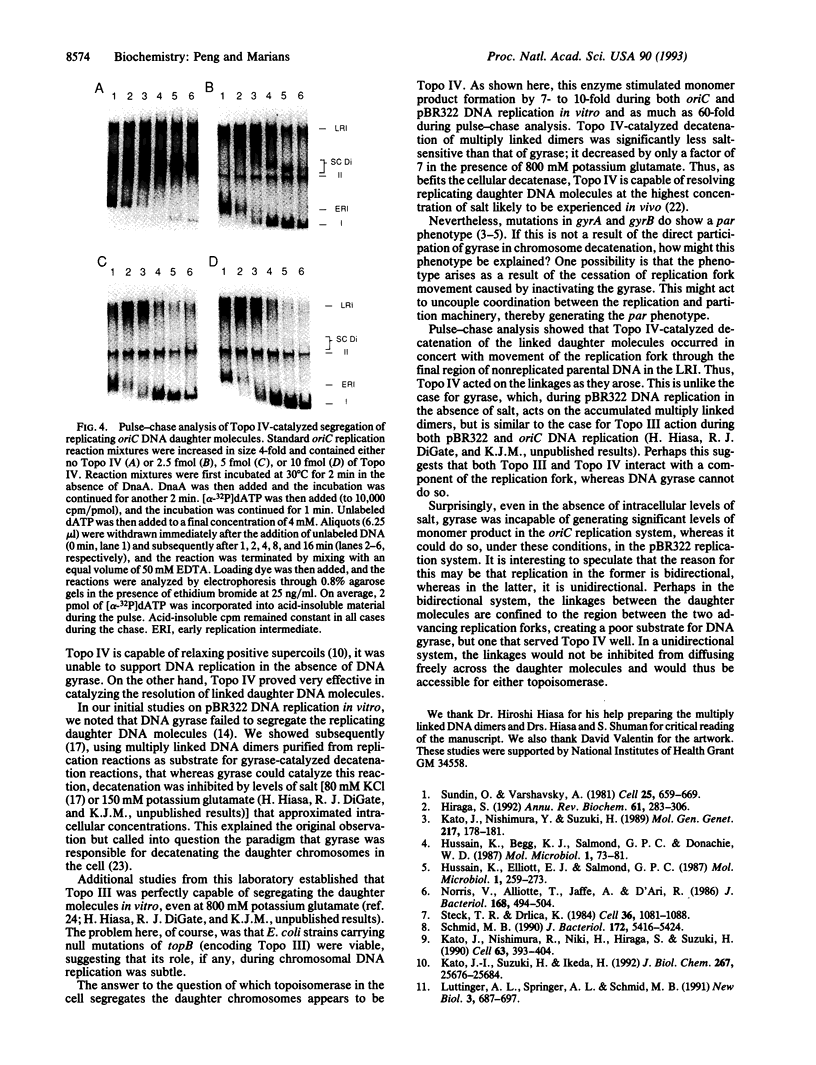
Topoisomerase IV (Topo IV), encoded by parC and parE, is required for partition of the daughter chromosomes in Escherichia coli. This enzyme is likely responsible for decatenating the linked daughter chromosomes after replication. In this report, we have examined the action of Topo IV in both pBR322 and oriC DNA replication reconstituted in vitro with purified proteins. Gyrase fails to decatenate the linked daughter molecules under any condition in the oriC system and at physiological salt concentrations in the pBR322 system, whereas Topo IV stimulates generation of monomer product DNA by 7- to 10-fold. Topo IV-catalyzed decatenation of isolated multiply linked DNA dimers was relatively insensitive to salt; it proceeded at 14% of the maximal rate even in the presence of 800 mM potassium glutamate. In contrast, decatenation in vitro by gyrase was inhibited completely under these conditions. Pulse-chase analysis indicated that Topo IV-catalyzed resolution of linked daughter DNA molecules occurred prior to completion of DNA replication, such that multiply linked daughter molecules did not arise. These results suggest that during DNA replication, gyrase acts primarily to relieve accumulated positive supercoiling and Topo IV acts to segregate the daughter chromosomes.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams D. E., Shekhtman E. M., Zechiedrich E. L., Schmid M. B., Cozzarelli N. R. The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell. 1992 Oct 16;71(2):277–288. doi: 10.1016/0092-8674(92)90356-h. [DOI] [PubMed] [Google Scholar]
- Asai T., Takanami M., Imai M. The AT richness and gid transcription determine the left border of the replication origin of the E. coli chromosome. EMBO J. 1990 Dec;9(12):4065–4072. doi: 10.1002/j.1460-2075.1990.tb07628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliska J. B., Cozzarelli N. R. Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J Mol Biol. 1987 Mar 20;194(2):205–218. doi: 10.1016/0022-2836(87)90369-x. [DOI] [PubMed] [Google Scholar]
- DiGate R. J., Marians K. J. Identification of a potent decatenating enzyme from Escherichia coli. J Biol Chem. 1988 Sep 15;263(26):13366–13373. [PubMed] [Google Scholar]
- Hill T. M., Pelletier A. J., Tecklenburg M. L., Kuempel P. L. Identification of the DNA sequence from the E. coli terminus region that halts replication forks. Cell. 1988 Nov 4;55(3):459–466. doi: 10.1016/0092-8674(88)90032-3. [DOI] [PubMed] [Google Scholar]
- Hussain K., Begg K. J., Salmond G. P., Donachie W. D. ParD: a new gene coding for a protein required for chromosome partitioning and septum localization in Escherichia coli. Mol Microbiol. 1987 Jul;1(1):73–81. doi: 10.1111/j.1365-2958.1987.tb00529.x. [DOI] [PubMed] [Google Scholar]
- Hussain K., Elliott E. J., Salmond G. P. The parD- mutant of Escherichia coli also carries a gyrAam mutation. The complete sequence of gyrA. Mol Microbiol. 1987 Nov;1(3):259–273. doi: 10.1111/j.1365-2958.1987.tb01932.x. [DOI] [PubMed] [Google Scholar]
- Kato J., Nishimura Y., Imamura R., Niki H., Hiraga S., Suzuki H. New topoisomerase essential for chromosome segregation in E. coli. Cell. 1990 Oct 19;63(2):393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- Kato J., Nishimura Y., Suzuki H. Escherichia coli parA is an allele of the gyrB gene. Mol Gen Genet. 1989 May;217(1):178–181. doi: 10.1007/BF00330959. [DOI] [PubMed] [Google Scholar]
- Kato J., Suzuki H., Ikeda H. Purification and characterization of DNA topoisomerase IV in Escherichia coli. J Biol Chem. 1992 Dec 25;267(36):25676–25684. [PubMed] [Google Scholar]
- Luttinger A. L., Springer A. L., Schmid M. B. A cluster of genes that affects nucleoid segregation in Salmonella typhimurium. New Biol. 1991 Jul;3(7):687–697. [PubMed] [Google Scholar]
- Marians K. J. DNA gyrase-catalyzed decatenation of multiply linked DNA dimers. J Biol Chem. 1987 Jul 25;262(21):10362–10368. [PubMed] [Google Scholar]
- Marians K. J., Soeller W., Zipursky S. L. Maximal limits of the Escherichia coli replication factor Y effector site sequences in pBR322 DNA. J Biol Chem. 1982 May 25;257(10):5656–5662. [PubMed] [Google Scholar]
- Minden J. S., Marians K. J. Escherichia coli topoisomerase I can segregate replicating pBR322 daughter DNA molecules in vitro. J Biol Chem. 1986 Sep 5;261(25):11906–11917. [PubMed] [Google Scholar]
- Minden J. S., Marians K. J. Replication of pBR322 DNA in vitro with purified proteins. Requirement for topoisomerase I in the maintenance of template specificity. J Biol Chem. 1985 Aug 5;260(16):9316–9325. [PubMed] [Google Scholar]
- Norris V., Alliotte T., Jaffé A., D'Ari R. DNA replication termination in Escherichia coli parB (a dnaG allele), parA, and gyrB mutants affected in DNA distribution. J Bacteriol. 1986 Nov;168(2):494–504. doi: 10.1128/jb.168.2.494-504.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada C. A., Marians K. J. Mechanism of DNA A protein-dependent pBR322 DNA replication. DNA A protein-mediated trans-strand loading of the DNA B protein at the origin of pBR322 DNA. J Biol Chem. 1991 Oct 5;266(28):18895–18906. [PubMed] [Google Scholar]
- Richey B., Cayley D. S., Mossing M. C., Kolka C., Anderson C. F., Farrar T. C., Record M. T., Jr Variability of the intracellular ionic environment of Escherichia coli. Differences between in vitro and in vivo effects of ion concentrations on protein-DNA interactions and gene expression. J Biol Chem. 1987 May 25;262(15):7157–7164. [PubMed] [Google Scholar]
- Steck T. R., Drlica K. Bacterial chromosome segregation: evidence for DNA gyrase involvement in decatenation. Cell. 1984 Apr;36(4):1081–1088. doi: 10.1016/0092-8674(84)90058-8. [DOI] [PubMed] [Google Scholar]
- Sundin O., Varshavsky A. Arrest of segregation leads to accumulation of highly intertwined catenated dimers: dissection of the final stages of SV40 DNA replication. Cell. 1981 Sep;25(3):659–669. doi: 10.1016/0092-8674(81)90173-2. [DOI] [PubMed] [Google Scholar]
- Wu C. A., Zechner E. L., Marians K. J. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. I. Multiple effectors act to modulate Okazaki fragment size. J Biol Chem. 1992 Feb 25;267(6):4030–4044. [PubMed] [Google Scholar]