Abstract
Aberrant Wnt signaling frequently occurs in pancreatic cancer (PC) and contributes to disease progression/metastases. Likewise, the transmembrane‐mucin MUC4 is expressed de novo in early pancreatic intraepithelial neoplasia (PanINs) and incrementally increases with PC progression, contributing to metastasis. To determine the mechanism of MUC4 upregulation in PC, we examined factors deregulated in early PC progression, such as Wnt/β‐catenin signaling. MUC4 promoter analysis revealed the presence of three putative TCF/LEF‐binding sites, leading us to hypothesize that MUC4 can be regulated by β‐catenin. Immunohistochemical (IHC) analysis of rapid autopsy PC tissues showed a correlation between MUC4 and cytosolic/nuclear β‐catenin expression. Knock down (KD) of β‐catenin in CD18/HPAF and T3M4 cell lines resulted in decreased MUC4 transcript and protein. Three MUC4 promoter luciferase constructs, p3778, p3000, and p2700, were generated. The construct p3778, encompassing the entire MUC4 promoter, elicited increased luciferase activity in the presence of stabilized β‐catenin. Mutation of the TCF/LEF site closest to the transcription start site (i.e., −2629/−2612) and furthest from the start site (i.e., −3425/−3408) reduced MUC4 promoter luciferase activity. Transfection with dominant negative TCF4 decreased MUC4 transcript and protein levels. Chromatin immunoprecipitation confirmed enrichment of β‐catenin on −2629/−2612 and −3425/−3408 of the MUC4 promoter in CD18/HPAF. Functionally, CD18/HPAF and T3M4 β‐catenin KD cells showed decreased migration and decreased Vimentin, N‐cadherin, and pERK1/2 expression. Tumorigenicity studies in athymic nude mice showed CD18/HPAF β‐catenin KD cells significantly reduced primary tumor sizes and metastases compared to scrambled control cells. We show for the first time that β‐catenin directly governs MUC4 in PC.
Keywords: MUC4, Wnt, Pancreatic cancer, β‐Catenin
Highlights
Aberrant β‐catenin and MUC4 are co‐expressed in pancreatic cancer autopsy tissues.
β‐Catenin/TCF4 binds MUC4 promoter and increases transcript and protein expression.
TCF/LEF sites closest to and furthest from the ATG site are critical.
MUC4 contributes to β‐catenin potentiated metastasis in vivo.
Rapid autopsy metastatic tissues co‐express MUC4 and β‐catenin.
Abbreviations
- KD
knock down
- PanIN
Pancreatic intraepithelial neoplasia
- PC
pancreatic cancer
- PDAC
pancreatic ductal adenocarcinoma
- TMA
tissue microarray
- CM
Conditioned medium
1. Introduction
Pancreatic cancer (PC) is a highly lethal disease with a rather dismal prognosis, showing a five‐year survival rate of only 7.2% (SEER Stat Fact Sheet, 2005–2011). While numerous studies have focused on the genetic abnormalities that underpin this disease, much remains unknown regarding this complex, intractable malignancy that unfortunately often only manifests symptoms in patients at advanced, metastatic stages (Rhim et al., 2012; Morris et al., 2010). Most prominent among the mutations driving PC is the Kras oncogene, which is mutated into a constitutively active form (KrasG12D) in around 90% of PC patients (Collins and Pasca di, 2014). In addition to mutations in Kras, the Wnt signaling pathway has been described as one of the 12 pathways most commonly deregulated in pancreatic ductal adenocarcinoma (PDAC), which is the most prevalent type of pancreatic neoplasms (Jones et al., 2008).
A central mediator of the canonical Wnt pathway is β‐catenin, a molecule that plays an important role in both cell adhesion and signaling. There are two distinct pools of β‐catenin – one cytosolic and the other membranous (Fodde et al., 2001). The membranous fraction participates in cell adhesion through interactions with E‐cadherin, while the cytosolic fraction is ordinarily degraded by a destruction complex, which is comprised of Adenomatous polyposis coli (APC), Glycogen Synthase Kinase β, Axin1, and Casein Kinase1 (Fodde et al., 2001). In the presence of a Wnt ligand, which binds the Frizzled/LRP receptor, this complex is abolished and β‐catenin is released, whereupon β‐catenin enters the nucleus and upregulates a host of tissue‐specific target genes, typically partnering with the TCF/LEF family of transcription factors (Fodde et al., 2001). Wnt ligands can also activate the non‐canonical pathway, which is independent of β‐catenin (Fodde et al., 2001). While mutations in this pathway are rare in PDAC, a spate of recent studies demonstrate the importance of both the canonical and non–canonical pathways in PDAC (Zeng et al., 2006; Al‐Aynati et al., 2004; Pasca di et al., 2007; Arensman et al., 2014; Jiang et al., 2014). Specifically, the gene expression signature of the Wnt/β‐catenin pathway as well as aberrant β‐catenin localization are implicated in conferring a poorer prognosis in patients (Qiao et al., 2001), as well as promoting PC metastases (Arensman et al., 2014).
Aberrant cytosolic and nuclear localization of β‐catenin occurs early on in PDAC and steadily increases with disease progression, starting from the earliest stage of pancreatic intraepithelial neoplasia 1 (PanIN‐1) (Al‐Aynati et al., 2004). Sustained low‐grade activation of the canonical Wnt pathway is essential for PDAC progression, subsequent to the Kras mutation, in a mouse model of PDAC (Zhang et al., 2013). Further, the Wnt/β‐catenin pathway is active in most PDAC cell lines and confers increased proliferative and anti‐apoptotic properties to PDAC cells (Pasca di et al., 2007).
MUC4 is a transmembrane mucin that is absent in the normal pancreas but incrementally increases as PDAC advances, with expression commencing at the PanIN‐1 stage (Andrianifahanana et al., 2001; Ansari et al., 2013; Swartz et al., 2002). Importantly, our lab has shown the importance of MUC4 in the invasion and metastases of PDAC (Seshacharyulu et al., 2015; Chaturvedi et al., 2007; Senapati et al., 2012; Singh et al., 2004). A 2008 study by Chaturvedi et al. proposed that the epidermal growth factor (EGF) domains of MUC4 act as ligands for HER2, thereby triggering an intracellular cascade of signaling events involving the MAPK and AKT pathways (Chaturvedi et al., 2008). Other studies have shown that knock down (KD) of MUC4 is sufficient to induce a decrease in mesenchymal markers, such as Vimentin, and increase in epithelial markers, such as E‐cadherin, in PDAC cell lines (Zhi et al., 2014; Rachagani et al., 2012). These alterations in epithelial and mesenchymal markers suggest that MUC4 expression alone induces epithelial–mesenchymal transition (EMT) in PDAC. Notably, MUC4 expression is also a marker for poor prognosis in PDAC (Saitou et al., 2005).
The de novo expression of MUC4 in PDAC has been attributed to factors such as nicotine, retinoic acid, interferon‐γ, CFTR, TGF‐β, and several miRNAs including miR‐200c, miR‐219‐1‐3p, miR‐150 (Andrianifahanana et al., 2005, 2007, 2004, 2012, 2015, 2013, 2007, 2011). The de novo expression of MUC4 has been further attributed to the transcription factor NCOA3 (Kumar et al., 2014) as well as epigenetic mechanisms like hypomethylation and histone acetylation (Vincent et al., 2008).
The MUC4 promoter is well characterized; it is around 3.7 kb long (Perrais et al., 2001), and its TATA box is located at −2672/−2668 (Perrais et al., 2001). The MUC4 proximal promoter contains two highly transcriptionally active regions; −219/−1 and −2781/−2572 (Perrais et al., 2001). Interestingly, MUC4 has been hypothesized to aid in the nuclear localization of β‐catenin in PDAC by inducing dissociation of β‐catenin from E‐cadherin via HER2/Src signaling (Zhi et al., 2014).
The objective of this study was to examine the nature and functional implications of the β‐catenin‐MUC4 relationship and the effect of this relationship on PDAC metastasis. For this purpose, the study herein involved MUC4 promoter analysis, which showed the presence of three putative TCF/LEF sites. This finding implies that MUC4 is a putative transcriptional target of the Wnt/β‐catenin pathway in PDAC. Another piece of evidence suggesting a β‐catenin‐MUC4 relationship was the observation that when β‐catenin was depleted using a pancreas‐specific Cre in the KPC (PDX‐1‐Cre, LSL‐KrasG12D, LSL‐Trp53R172H/‐) mouse model, β‐catenin‐negative cells showed significantly reduced mucin expression, as measured by alcian blue staining (Zhang et al., 2013). Another recent study by Olsen et al. showed genetically ablated β‐catenin by zinc finger nucleases in the PDAC cell line BXPC3 (Olsen et al., 2014). A subsequent microarray showed that MUC4 was one of the most significantly downregulated genes in PDAC (Olsen et al., 2014), giving further impetus to our hypothesis that Wnt/β‐catenin pathway colludes with transcription factors including retinoic acid, interferon‐γ, and NCOA3 to precipitate MUC4 expression in PDAC.
2. Materials and methods
2.1. Cell culture
CD18/HPAF was a metastatic clone derived from the HPAF PDAC cell line (Mullins et al., 1991). T3M4 was derived from lymph node metastasis of a tumor in the exocrine pancreas (Okabe et al., 1983). Other PDAC cell lines, Capan1, BXPC3, Panc1, AsPc1, and MiaPaCa, were cultured in DMEM containing 10% fetal bovine serum supplemented with 100 μg/ml penicillin and streptomycin. Cells transfected with lentiviral constructs were maintained in 5 μg/ml Puromycin as a selection agent. The L Wnt3A cells were a gift from Dr. Jing (aka. Jenny) Wang at the University of Nebraska Medical Center (UNMC). Wnt‐3A conditioned medium was collected as stipulated by American Type Culture Collection (ATCC). All cells were maintained at 37 °C with 5% CO2 in a humidified atmosphere.
2.2. Lentiviral transfection
The lentiviral plasmids used included the following: pLKO.1.sh.beta‐catenin.2279 (Addgene plasmid #19762), pLKO.1.sh.beta‐catenin.1248 (Addgene plasmid #19761), pLKO.1 shSCR (Addgene plasmid #17920), and packaging plasmid pCMV‐dR8.2 dvpr and envelope plasmid pCMV‐VSVG, which were a kind gift from Dr. Yuzuru Shiio (University of Tennessee Health Science Center). The Lenti‐X‐293T cell line #632180 (Clontech; Mountain View, CA, USA) was used for transfection, and the Lenti‐X™ Concentrators #631231 and #631232 from Clontech were used to concentrate the lentiviral supernatant. Briefly, 2 × 106 Lenti‐X‐293T cells were plated in 10 cm plates, and transfection using a calcium phosphate precipitate method with 20 μg transfer vector, 15 μg packaging plasmid, and 6 μg envelope plasmid was performed the next day. Concentration of the supernatant collected after 48 h was done per manufacturer's instructions (Clontech). Following concentration, lentiviral supernatant was used to transfect 2 × 105 cells plated per well of a 12‐well plate. Puromycin (5 μg/ml) was used to select for positive clones.
2.3. Tissues specimens and immunohistochemistry
The UNMC Rapid Autopsy Program was used to obtain tissue microarrays (TMA) containing primary PDAC tissue specimens (IRB‐091‐01) and normal pancreas, kidney, and colon tissue spots as controls (n = 30, 25 PC spots). TMAs containing metastatic spots (n = 26; 25 liver metastatic spots, 1 lung metastatic spot) with normal pancreas, colon, and kidney tissues as controls were also used (IRB‐091‐01). Prior consent was obtained from all participants in the Rapid Autopsy Program, and the UNMC Institutional Review Board (IRB) approved this study. The following antibodies were used: anti‐β‐catenin #610154, (BD Biosciences; San Jose, CA, USA) anti‐MUC4 monoclonal antibody clone 8G7 (developed in our lab). The protocols used for immunohistochemical (IHC) detection of β‐catenin and MUC4 have been described previously (Barker and van den Born, 2008; Kaur et al., 2014). The tissue microarrays (TMAs) were evaluated by a UNMC pathologist and were given a composite score ranging from 0 to 12, which was a product of the intensity of staining (range 1–3) and number of cells stained (range 1–4; 0–25% area stained = score of 1, 26–50% = score 2, 51–75% = score 3, and 76–100% = score 4).
2.4. Transient transfection and luciferase assays
All transient transfections were performed with Lipofectamine 2000 (Life Technologies; Carlsbad, CA, USA). Lysates/RNA/luciferase readings were taken 48 h post‐transfection. The pGL4.17 vector was a gift from Dr. Robert Bennett at UNMC. The dominant‐negative TCF4 plasmid pPGS dnTcf‐4(deltaN41) was a gift from Eric Fearon (Addgene plasmid #19284) (Kolligs et al., 1999). M50 Super 8x TOPFlash and M51 Super 8x FOPFlash (TOPFlash mutant) were gifts from Randall Moon (Addgene plasmids #12456 and #12457) (Veeman et al., 2003). The TOPflash vector contains seven TCF/LEF‐binding sites upstream of a firefly luciferase gene, while the FOPflash vector contains seven mutant TCF/LEF sites and was used as a negative control. The pRenilla‐CMV luciferase vector #E2261 (Promega; Madison, WI, USA) was used as an internal transfection control in all luciferase assays, which were performed in triplicate and repeated twice. Briefly, 2 × 105 cells were seeded per well in 12 well plates and were transfected the following day. Luciferase readings were taken 48 h later, using the Dual‐Glo luciferase assay kit (Promega) as per the manufacturer's instructions.
2.5. RNA isolation and real‐time PCR analysis
RNA was isolated and purified using the QIAGEN RNeasy mini kit (Qiagen; Valenica, CA, USA); the RNA concentration was measured using a NanoDrop ND 1000 Spectrophotometer. The Oligo(dT)12–18 Primer #18418‐012 (Life Technologies) and Super Script II RNase reverse transcriptase (Invitrogen, Life Technologies) were used to obtain cDNA from 1.5 μg of RNA per cell line. The PCR primers used are enumerated in Supplementary Table B1. Real‐time PCR analysis was performed using the 480 Real‐Time PCR System (Roche; Indianapolis, IN, USA). A master mix comprised of 2X Sybr green mix of primers (Life Technologies) and nuclease‐free water was used to constitute a 10 μl reaction mixture consisting of 1 μl cDNA and 9 μl of the master mix.
2.6. Immunofluorescence
For IHC with CD18/HPAF, 2 × 105, cells were seeded on coverslips in a 12‐well plate and processed 48 h later using a previously described protocol described (Seshacharyulu et al., 2015). The antibodies and dilutions used are listed in Supplementary Table A. Images were taken using an LSM 710 Zeiss Confocal Microscope located at UNMC Advanced Microscopy Core Facility.
2.7. Migration assay
For migration assays, 1.5 × 106 cells were plated in 1.5 ml serum‐free DMEM on an 8 μm pore polyethylene cell culture insert in a 6‐well plate (Falcon/VWR #353093; Radnor, PA, USA). The lower chamber contained 2 ml DMEM supplemented with 10% FBS. The inserts were removed 36 h after seeding. Next, the cells at the top of the chamber were scraped off, and the cells that had traversed the membrane were stained with a Diff‐Quick cell staining kit (Dade Behring Inc.; Westwood, MA, USA). Images of inserts were taken using QCapture (Surrey, BC, Canada) software version 2.0.12 at a 10× magnification.
2.8. Cell proliferation and colony formation assays
The cell proliferation reagent WST‐1 (Roche Life Science, #05015944001; Penzberg, Upper Bavaria, Germany) was used to measure the proliferative rate of 1000 cells/well in a 96‐well plate over three days in DMEM that contained 1% fetal bovine serum. The readings were taken as per the manufacturer's instructions. For the colony formation assay, 1000 cells were seeded per well in a 6‐well plate (CD18/HPAF Scr and sh‐β‐catenin) in triplicate. The cells were maintained in 1% DMEM and allowed to form colonies for 21 days. Cells were then fixed with 100% methanol, stained with 0.4% crystal violet in methanol, and colonies were manually counted.
2.9. Western blot analysis
Western blot analysis was performed as previously described (Seshacharyulu et al., 2015). Briefly, 1.5–2 × 106 cells were seeded in a 10 cm plate, and lysates/RNAs were extracted 48 h later, such that cells were 70–80% confluent. After a freeze‐thaw cycle, lysates were thawed and syringe‐passed through a 215/8 gauge needle. Next, cells were quantified using Bio‐Rad protein assay kit (Hercules, CA, USA). A 10% SDS‐PAGE gel was used to resolve 20–40 μg of whole cell lysates for all proteins described, except MUC4, which was resolved on a 2%‐agarose gel owing to its high molecular weight. The proteins were transferred onto a polyvinylidene difluoride membrane (Millipore; Billerica, MA, USA) and probed with primary antibodies overnight at 4 °C. The antibodies used and their respective dilutions are described in Supplementary Table A.
2.10. Quantitative ChIP assay
A total of 2 × 107 cells per cell line were used for the chromatin immunoprecipitation (IP), which was performed as described previously (Kumar et al., 2014). The 1% formaldehyde cross‐linked chromatin was isolated and sheared into 500–1000 bp fragments by sonication (Bioruptor UCD‐200, Diagenode; New York, NY, USA). Prior to IP, 1% of the sonicated DNA was taken as input. The concentrations of antibodies used for overnight incubation at 4 °C were as follows: 2.5 μg of anti‐β‐catenin #610154 (BD Biosciences), −2 μg of IgG (negative control). Primers for TCF/LEF included Site #1, #2, and #3 on the MUC4 promoter; the TCF/LEF site on the c‐myc promoter (positive control) and primers for the promoter of an unrelated gene (negative control) were also used (Supplementary Table B2). Immunoprecipitated qPCR Ct (cycle threshold) values were normalized to input Ct values, and all data are represented as a percentage of input.
2.11. Generation of constructs
For the 4ACAT construct, the β‐catenin transcript was amplified using the appropriate primers (Supplementary Table B3) from the cDNA of a PDAC cell line, T3M4 that expressed the wild‐type transcript. The amplicon was cloned into a p3XFLAG‐CMV10 vector (Sigma–Aldrich; St. Louis, MO) digested with the Not1 enzyme (New England Biolabs; Ipswich, MA). Point mutations were introduced at Ser33, Ser37, Thr41, Ser45, which were mutated to alanine using appropriate primers ( Supplementary Table B3). The MUC4‐promoter fragment was generated from genomic DNA (CD18/HPAF cell line) using primers that incorporated the HindIII and KpnI restriction sites (Supplementary Table B3). The amplicon was cloned into a pGL4.17 vector digested with HindIII and KpnI (New England Biolabs). The p3778 construct encompasses the entire MUC4 promoter (proximal and distal promoter); the p3000 construct encompasses two TCF/LEF sites (proximal promoter and part of distal promoter), and the p2700 construct encompasses one TCF/LEF site (primarily proximal promoter). Primers used to generate these constructs are enumerated in Supplementary Table B3. Mutations at the TCF/LEF sites were introduced using primers enumerated in Supplementary Table B3. The instructions from the QuikChange® Site‐Directed Mutagenesis Kit (Agilent Technologies; Santa Clara, CA) were used for primer design. Platinum® Taq DNA Polymerase High Fidelity Assay (Life Technologies) was used for all site‐directed mutagenesis PCR reactions. Luciferase experiments were performed in triplicate and repeated a minimum of three times. Figure 4B, C represent the average of a minimum of three attempts.
Figure 4.
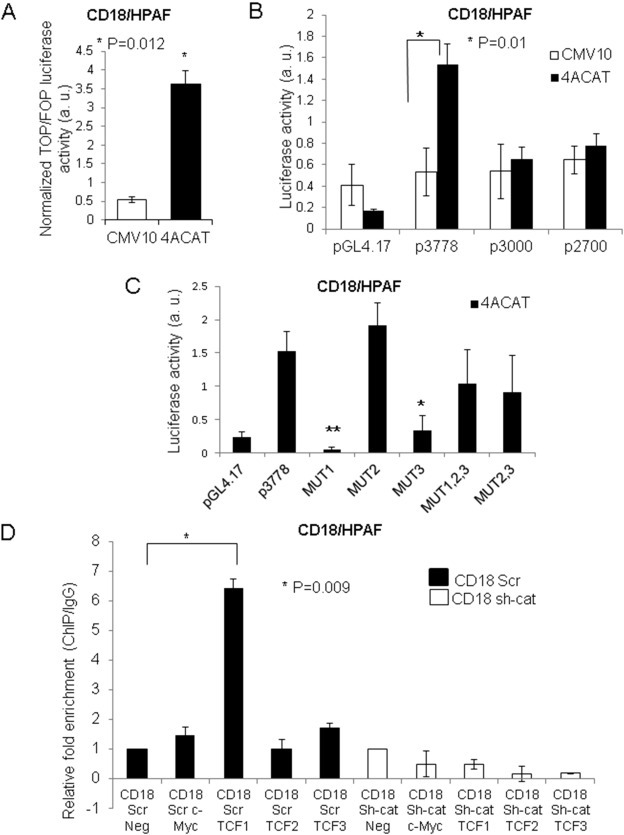
β‐catenin directly regulates MUC4 transcription. (A) A stabilized β‐catenin construct (4ACAT; S33A, S37A, T41A, T45A) elicited increased TOP/FOP luciferase activity in comparison to the empty vector control in CD18/HPAF cells. (B) Luciferase activity for MUC4 promoter fragments p3778, p3000, and p2700 in the presence of 4ACAT compared to the empty vector control. Luciferase readings are the average of three or more separate experiments performed in triplicate; the standard deviation shown is for three or more separate experiments. (C) The p3778 promoter construct with each of the three putative TCF/LEF sites mutated (i.e., −2612:MUT1, −3226:MUT2, −3408: MUT3) was transfected into CD18/HPAF cells in the presence of 4ACAT. The pCMV9‐Renilla vector was used as an internal transfection control; all luciferase experiments were performed in triplicate and repeated a minimum of three times. Images represent the average of at least three experiments, each performed in triplicate. *p = 0.02, **p = 0.002 (D) Quantitative ChIP assay using β‐catenin antibody to pull down sheared chromatin isolated from CD18/HPAF Scr and sh‐cat. Primer pairs specific to the TCF/LEF Sites #1, #2, and #3 on the MUC4 promoter were used. Primers amplifying the TCF/LEF site on the c‐Myc promoter were used as a positive control; an unrelated primer pair was used as a negative control. All real‐time values were normalized to the 1% input control.
2.12. Promoter analysis
Promoter analysis was performed using the MatInspector program (Genomatix GmbH; Munich, Bavaria, Germany). A matrix similarity score of >0.85 was used to screen transcription factor binding sites.
2.13. Tumorigenicity assay
For the tumorigenicity assay, sub‐confluent cultures of CD18/HPAF Scr/sh‐β‐catenin cells were trypsinized and then counted using the Countess™ Automated Cell Counter (Life Technologies) after their viability had been ascertained (>95%). Next, 0.25 × 106 cells/50 μl PBS were orthotopically implanted in the head of the pancreas of 14 female athymic nude mice obtained from Harlan Sprague Dawley in Indianapolis, IN (7 per group). Mice were observed for five weeks, and were subsequently sacrificed and weighed. The animals were treated in accordance with guidelines from the UNMC Institutional Animal Care and Use Committee (IACUC). The primary tumors were excised and weighed. Tumor metastases sites were counted and dissected after a thorough physical examination. Both primary tumors and metastases were kept in 10% formalin for 48 h, after which they were embedded in paraffin blocks that were sectioned into 0.5 micron‐thick sections.
2.14. Statistical analysis
All data were analyzed using two‐tailed T test with unequal variance.
3. Results
3.1. Nuclear/cytosolic β‐catenin was associated with MUC4 expression in PDAC
Western blot analysis was used to screen panels of PDAC cell lines for expressions of MUC4 and β‐catenin (Figure 1A). MUC4 was shown to be expressed in all β‐catenin‐expressing cell lines (i.e., BXPC3, Capan1, CD18/HPAF, T3M4) but was absent in β‐catenin‐non‐expressing cell lines (i.e., MiaPaCa, Panc1), with the exception of AsPc1. Next, IHC was performed on a PDAC TMA, and it was found that both MUC4 and cytosolic/nuclear β‐catenin, which is a hallmark of active Wnt/β‐catenin signaling (Barker and van den Born, 2008), were present in 80% of the PDAC tissue spots examined (n = 25, mean composite score for MUC4 = 3.52 [21/25] and for β‐catenin = 7.92 [24/25]) (Figure 1B). Further, tissue immunofluorescence of human PDAC showed that MUC4 was expressed in cells that expressed nuclear/cytosolic β‐catenin (Figure 1C, Supplementary Figure 1A).
Figure 1.
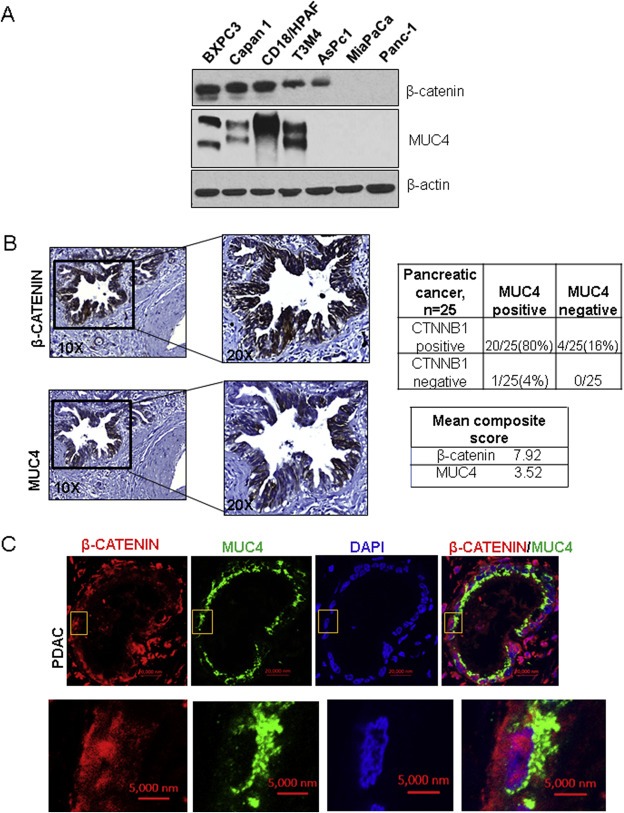
The expression pattern of MUC4 and β‐catenin in PC tissue and cell lines. (A) Western blot analysis of protein lysates from a panel of PDAC cell lines showed that β‐catenin (upper panel) was expressed in cell lines that expressed MUC4 (except the AsPc1 cell line). β‐actin was used as a loading control. (B) Immunohistochemistry (IHC) of a tissue microarray (TMA) obtained from the UNMC Rapid Autopsy Program (n = 25) showed that both β‐catenin (average composite score 7.92) and MUC4 (average composite score 3.52) were co‐expressed in 80% of the PDAC spots examined. (C) Tissue immunofluorescence in pancreatic ductal adenocarcinoma (PDAC) tissue showed that MUC4 (green) was expressed in cells that also expressed nuclear/cytosolic β‐catenin (red).
3.2. Wnt/β‐catenin regulated MUC4 expression in PDAC
Two lentiviral shRNAs that targeted β‐catenin were used to stably KD expression of β‐catenin in PDAC cell lines T3M4 and CD18/HPAF. It was found that upon the KD of β‐catenin, expressions of MUC4 transcripts and proteins were reduced in comparison to the scrambled control‐transfected cells in both cell lines (Figure 2A, Supplementary Figure 1B). Confocal microscopy was used to confirm the decrease in MUC4 expression in CD18/HPAF and T3M4 cell lines (Figure 2B, Supplementary Figure 1C). For further confirmation, Lithium Chloride (LiCl) was used; LiCl inhibits the GSK3‐β enzyme (Cai et al., 2007) and potentiates increased nuclear β‐catenin due to the prevention of its N‐terminal phosphorylation‐mediated degradation (Cai et al., 2007). Next, 20 mM and 50 mM LiCl treatment was performed for 48 h in CD18/HPAF, resulting in increased MUC4 expression in a dose‐dependent manner (Figure 2C). These findings are concurrent with the increase in the inhibitory expression of pSer9GSK3‐β. In order to employ a more specific inducer of Wnt/β‐catenin signaling, CD18/HPAF cells were treated with Wnt‐3A conditioned medium. Compared to untreated control cells, increasing doses of Wnt‐3A conditioned medium resulted in increased MUC4 protein expression (Figure 2D). The level of c‐Myc, which is a Wnt/β‐catenin target gene in PDAC (Li et al., 2005), was used as a positive control.
Figure 2.
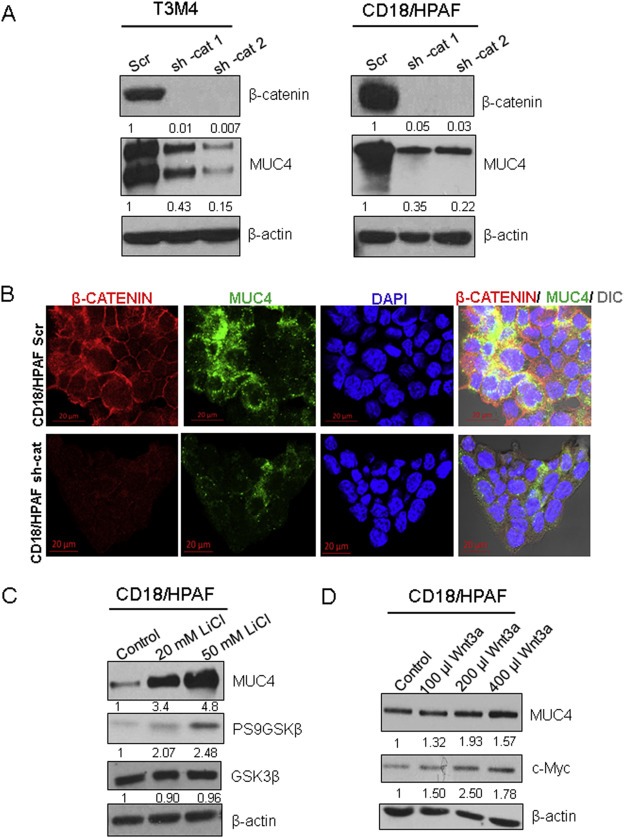
MUC4 protein and RNA expression are governed by β‐catenin. (A) CD18/HPAF and T3M4 pancreatic ductal adenocarcinoma (PDAC) cell lines were transfected with two lentiviral shRNAs that targeted β‐catenin (shRNA‐cat1, shRNA‐cat2) or a scrambled sequence in the PLKO.1 vector. Further, knock down of β‐catenin resulted in reduced MUC4 protein expression. (B) Confocal microscopy analysis was used to analyze MUC4 (green) and β‐catenin (red) levels in CD18/HPAF Scr and CD18/HPAF shRNA‐β‐catenin (shRNA‐cat) cells. (C) Treatment with lithium chloride (LiCl, a GSK3‐β inhibitor) at 20 mM and 50 mM concentrations was used to induce nuclear β‐catenin. Western blot analysis showed a dose‐dependent effect on MUC4 levels and an increase in phosphorylation of the inhibitory Ser9 residue of GSK3‐β, while total GSK3‐β levels were unaffected. (D) CD18/HPAF cells were treated with increasing amounts of Wnt‐3A‐conditioned medium; levels of c‐Myc were used as a positive control.
3.3. β‐catenin directly regulated MUC4 transcript expression
In light of observations for a correlation between aberrant β‐catenin and MUC4 expression in PDAC, and the decrease in MUC4 protein and RNA levels upon β‐catenin KD, it was next determined whether β‐catenin can directly regulate MUC4 transcription. MUC4‐promoter analysis by the MatInspector program (genomatix.de) revealed the presence of three putative TCF/LEF sites at the following positions: −3408 (Site #3), −3226 (Site #2), and −2612 (Site #1), with matrix similarity scores of 0.91, 0.84, and 0.907, respectively (Figure 3, Table 1). Next, the following three MUC4 promoter constructs were generated: (i) p3778, which encompasses the entire MUC4 promoter and has three TCF/LEF sites, (ii) p3000, which encompasses the proximal promoter as well as part of the distal promoter and has two TCF/LEF sites, and (iii) p2700, which encompasses the proximal promoter as well as TATA box and has one TCF/LEF site (Figure 3). Following the generation of promoter constructs, a stabilized β‐catenin construct, 4ACAT, was generated with four mutations (S33A, S37A, T41A, and S45A) that prevent degradation; the TOPflash/FOPflash assay was used to test the efficacy of this construct. Notably, CD18/HPAF that was transiently transfected with either 4ACAT or the empty vector showed a significantly elevated TOPflash/FOPflash luciferase activity in the 4ACAT cells (Figure 4A), indicating that 4ACAT elicits an increased β‐catenin/TCF‐mediated transcription activity. Next, the luciferase activities were compared for all three promoter constructs in the presence of 4ACAT and the empty vector. Compared to the other two constructs p3000 and p2700, the promoter construct p3778 elicited significantly increased luciferase activity compared to the empty vector (Figure 4B). On the other hand, the p2700 construct displayed a higher basal level of luciferase activity, but no significant increase in luciferase activity was seen in the presence of 4ACAT.
Figure 3.
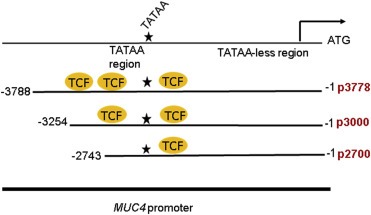
Schematic representation of the MUC4 promoter constructs generated. Three MUC4 promoter constructs were generated and cloned into pGL4.7; p3778 encompasses the full promoter and incorporates all three putative TCF/LEF sites, p3000 encompasses the proximal promoter and part of the distal promoter and two putative TCF/LEF sites, while p2700 incorporates mainly the proximal promoter and one putative TCF/LEF site.
Table Table 1.
The putative TCF/LEF sites unearthed by MUC4 promoter analysis. Given below is the sequence of the TCF/LEF sites, with the core sequence in bold letters, and the mutations introduced into each site, as well as the position with respect to the transcription start site.
| Putative TCF/LEF site | From | To | Sequence | Mutated sequence |
|---|---|---|---|---|
| Site 1 | −2612 | −2629 | TAAGACTCAAAGGGGAC | TAAGACGTGCGAGGGAC (MUT1) |
| Site 2 | −3226 | −3243 | GACAAGATTGAACTTTA | GACAAGCCAGCGCTTTA (MUT2) |
| Site 3 | −3408 | −3425 | CAGCGCTTTGTACTTCA | CAGCGACCCAGGCTTCA (MUT3) |
Following these findings from the luciferase assays for 4ACAT cells, luciferase assays were then performed for T3M4 Scr and KD cells. Subsequently, p3778‐driven luciferase activity was shown to be reduced in the KD cells but not in the T3M4 Scr cells (Supplementary Figure 2A). Each TCF/LEF site on the p3778 construct was mutated individually as well as in combination; the following five different constructs were then generated: MUT1, MUT2, MUT3, MUT2,3 and MUT1,2,3, which are enumerated in Table 1. Interestingly, after CD18/HPAF cells were transfected with p3778, MUT1, MUT2, and MUT3 in the presence of 4ACAT, it was seen that MUT1 elicited significantly reduced luciferase activity in comparison to the un‐mutated promoter (Figure 4C). Surprisingly, in comparison to p3778, MUT2 elicited an increased luciferase activity, while MUT3 elicited a decreased luciferase activity. Additionally, when all three sites were mutated, luciferase activity was diminished compared to p3778, but was increased compared to MUT1 and MUT3 (Figure 4C). A similar pattern was observed for T3M4 (Supplementary Figure 2B).
Because Site #1 appeared to be critical for MUC4 transcription, another construct was generated, MUT2,3 for which Sites #2 and #3 were mutated, but Site #1 was retained. The levels of luciferase activity for MUT2,3 were decreased compared to that of p3778, suggesting that Sites #2 and/or #3 also play a role in 4ACAT‐mediated transcription of MUC4. To confirm if a β‐catenin/TCF complex physically occupies the MUC4 promoter at one or more TCF/LEF sites, quantitative Chromatin immunoprecipitation (ChIP) assays were performed in CD18/HPAF Scr and CD18/HPAF β‐catenin KD cells (Figure 4D). A pull down was performed with a β‐catenin antibody and IgG as a negative control. Real‐time PCR was performed using primers that amplified regions containing the TCF/LEF Sites #1, #2, and #3 on the MUC4 promoter. As a positive control, the TCF/LEF site on the c‐Myc promoter was used, which is a β‐catenin target gene in PDAC; a non‐specific primer pair was used as a negative control. In CD18/HPAF Scr, but not CD18/HPAF sh‐β‐catenin, significant enrichment of β‐catenin was seen for TCF Site #1, but not for TCF Site #2; further, roughly two‐fold enrichment was seen for Site #3 compared to the negative control.
3.4. β‐catenin partnered with TCF4 to regulate MUC4 expression
Cell lines used in this study (i.e., CD18/HPAF and T3M4) were profiled for the expression of TCF/LEF factors. Notably, TCF1, 3, and 4 were found to be abundantly expressed in both cell lines (Supplementary Figure 3A). Because TCF4 is the most significantly over‐expressed TCF/LEF factor in PDAC (Uhlen et al., 2015), CD18/HPAF cells were transfected with dominant‐negative TCF4 (i.e., dnTCF4), which lacks an N‐terminal β‐catenin‐binding domain. Decreases in MUC4 protein levels and RNA expression were observed upon transfection with dnTCF4 (Supplementary Figure 3B), suggesting that TCF4 partners with β‐catenin on the MUC4 promoter. Further, confocal microscopy for HPAF/CD18 cells showed that TCF4 primarily co‐localized with β‐catenin in the nucleus (Supplementary Figure 3C).
3.5. β‐catenin contributed to migratory and mesenchymal properties of PDAC cell lines
In order to ascertain the functional implications of the β‐catenin‐MUC4 axis, migration, proliferation, and colony formation assays were performed with CD18/HPAF Scr and KD cells. Because MUC4 can mediate migration, metastasis, and EMT via HER2/RAF/MEK/ERK signaling (Zhi et al., 2014; Ponnusamy et al., 2010; Rachagani et al., 2012), it was hypothesized for the present study that β‐catenin causes EMT partly via MUC4 upregulation. It was seen that, while migration was significantly reduced upon β‐catenin KD in both CD18/HPAF and T3M4 cells (Figure 5A, Supplementary Figure 4C), there was a non‐significant decline in proliferation and colony formation (Supplementary Figure 4A, B). It was further observed that the CD18/HPAF KD cells also assumed a more cobblestone‐like morphology compared to the dispersed, spindle‐shaped Scr cells (Figure 5B).
Figure 5.
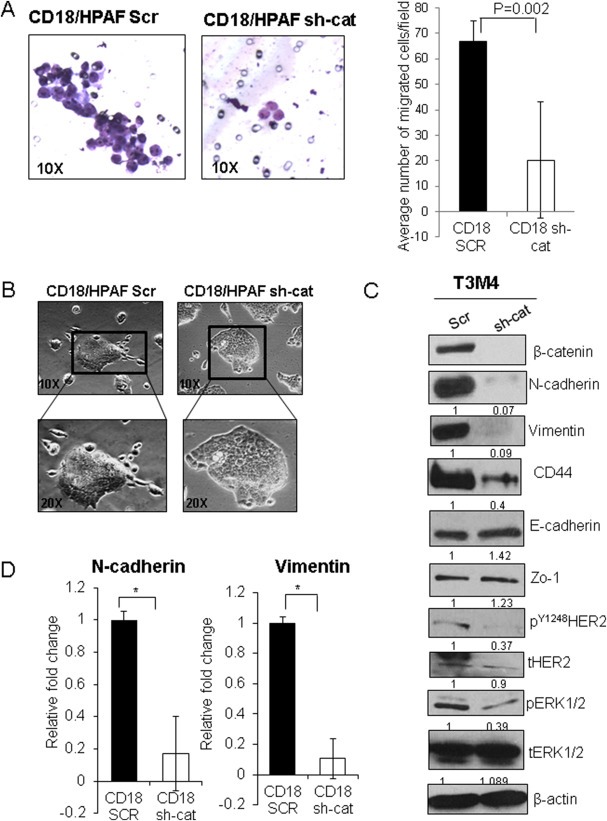
Effect of β‐catenin on migratory properties/EMT. (A) Knock down (KD) of β‐catenin in the CD18/HPAF cell line significantly reduced the migration of cells, as measured by a trans‐well migration assay using 1.5 × 106 cells seeded in uncoated Boyden's chambers (8 μm pore size) in triplicate. Cells were seeded in serum‐free medium in the chamber, and 10% fetal bovine serum containing DMEM was used as a chemoattractant below the chamber in a 6‐well plate. Cells were allowed to traverse the membrane for 36 h, following which the chamber was removed, non‐migrant cells were scraped off, and cells that traversed the membrane were stained. The image on the left is representative of 10 random fields that were analyzed as depicted in the image on the right, which quantifies the number of cells per field in CD18/HPAF Scr and CD18/HPAF sh‐cat. (B) Morphological changes observed in CD18/HPAF sh‐cat cells in comparison to the Scr vector control transfected cells. (C) Western blot analysis showing that KD of β‐catenin in T3M4 cells resulted in reduced N‐cadherin, CD44, Vimentin, pERK1/2, and pHER2 (Y1248), while E‐cadherin levels marginally increased; Zo‐1 levels also increased. (D) Real‐time PCR in CD18/HPAF Scr and sh‐cat cells showed decreased N‐cadherin and Vimentin (*p < 0.05).
Given the change in morphology and the decreased migration upon β‐catenin KD, the markers of EMT were then analyzed. Notably, in T3M4 KD cells, marked decreases at the protein levels were observed for the mesenchymal markers N‐cadherin, CD44, and Vimentin, and increases were observed at the protein level for epithelial markers E‐cadherin and Zo‐1. On the other hand, CD18/HPAF KD cells showed a decrease in the EMT‐regulator Snail and an increase in E‐cadherin, as well as an increase in the molecular weight of Zo‐1 (Figure 5C, Supplementary Figure 5A). Further, a reduction in phospho‐ERK1/2 occurred in the KD cells for both cell lines (Figure 5C, Supplementary Figure 5A ); phospho‐ERK1/2 is a downstream effector of MUC4‐HER2 signaling (Chaturvedi et al., 2008). In contrast, the total ERK levels did not decrease in the KD cells for both cell lines. Next, considering that CD18/HPAF cells express very low protein levels of N‐cadherin and Vimentin, the RNA levels of these mesenchymal makers were examined. Interestingly, RNA levels were decreased in the CD18/HPAF KD cells (Figure 5D). In addition, the T3M4 KD cells showed a decrease in phospho‐HER2 (Figure 5C).
Given that the relationship between β‐catenin and other transmembrane mucins such as MUC1 and MUC16 is well studied, we sought to determine whether β‐catenin KD affects other mucins in addition to MUC4. In the CD18/HPAF KD cells, a large decrease was seen in MUC16 levels (Supplementary Figure 5B), while no significant difference was seen in T3M4 KD cells (data not shown). Interestingly, an increase in MUC1 levels occurred in both KD cell lines (Supplementary Figure 5B). Overall, these observations suggest that β‐catenin contributes to an increase in the migratory and mesenchymal properties of PDAC cells as well as alterations in mucin levels.
3.6. Orthotopic implantation of β‐catenin KD cells affected metastases
In order to examine tumorigenesis and metastases of PDAC, equal amounts of CD18/HPAF Scr and β‐catenin KD cells (0.25 × 106 cells/50 μl) were orthotopically implanted at the head of the pancreases of seven athymic nude mice, who were monitored for the next five weeks. While the Scr group showed high cachexia, mortality, elevated tumor burden, and metastases, the β‐catenin KD group showed significantly lower tumor burden and reduced metastases (Figure 6A and C, Supplementary Figure 6A). Sites of metastases were most significantly reduced for the diaphragm and peritoneal cavity (Figure 6C). IHC and Western blot analysis of tumor lysates from primary tumors confirmed that β‐catenin KD was maintained in vivo. Further, MUC4, pHER2, and tHER2 levels were also reduced in the KD tumor lysates compared to the control cells (Figure 6B, Supplementary Figure 6B).
Figure 6.
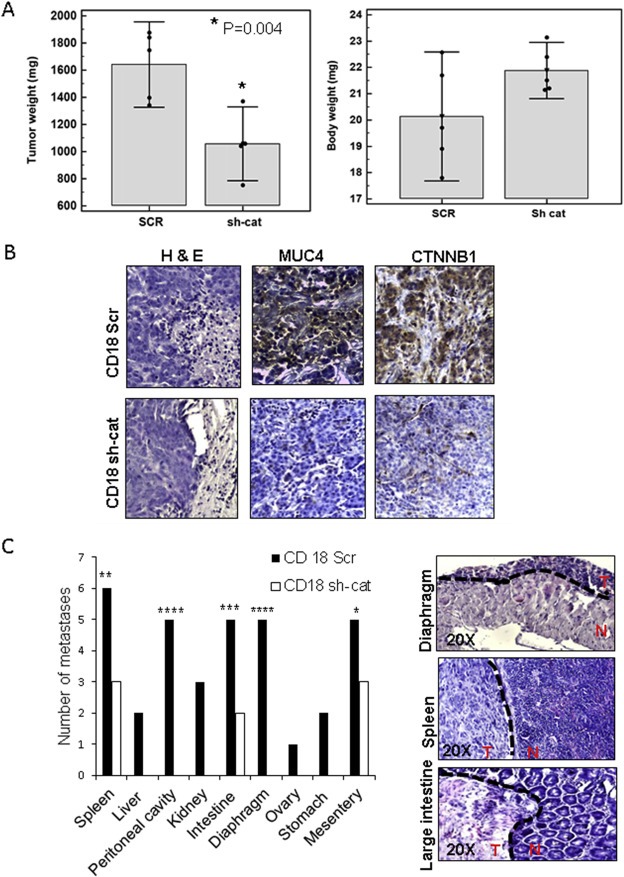
β‐catenin KD reduces tumorigenicity/metastasis. (A) Significantly reduced tumor weight and increased body weight were observed for β‐catenin KD mice. (B) IHC for β‐catenin, MUC4, and Hematoxylin and Eosin staining for the CD18/HPAF Scr and sh‐cat tumors. Images were taken at a 20× magnification. (C) Analysis of metastases to various organs (*p = 0.03, **p = 0.02, ***p = 0.008, ****p = 0.0009). No metastases were detected in organs where the white bars are absent in the CD18 sh‐cat xenografted mice. The panel on the right shows Hematoxylin and Eosin staining for representative metastatic tumors taken from the Scr cohort. Dotted line indicates border between tumor and normal tissue. T = tumor, N = normal. Images were taken at a 20× magnification.
In order to ascertain the effect of the β‐catenin‐MUC4 axis on metastasis in human tissue, two metastatic TMAs were analyzed for β‐catenin and MUC4 expression (25 spots, liver metastasis, UNMC Rapid Autopsy Program). Both aberrant β‐catenin and MUC4 were elevated in spots corresponding to the same patient (Figure 7). Of the liver metastatic spots examined, 40% (10/25) expressed both MUC4 and β‐catenin, while MUC4 was expressed in 40% (10/25) of the spots (mcs = 5.7). Notably, aberrant β‐catenin was universally expressed in all metastatic spots.
Figure 7.
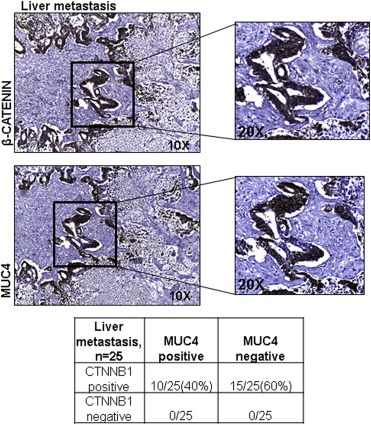
MUC4 and β‐catenin are co‐expressed in a subset of human metastatic lesions. Immunohistochemical (IHC) staining for β‐catenin (upper panel) and MUC4 (lower panel) in serial sections of a liver metastasis taken from the UNMC Rapid Autopsy Program's tissue microarray (TMA) containing metastatic lesions.
4. Discussion
Both aberrant β‐catenin signaling and MUC4 overexpression have been shown to contribute to PDAC progression (Pasca di et al., 2007; Zeng et al., 2006; Zhang et al., 2013; Chaturvedi et al., 2007; Ansari et al., 2013). Mutations in the key components of Wnt signaling, β‐catenin, and APC are uncommon in PDAC (Zeng et al., 2006). Despite this, pronounced aberrant β‐catenin signaling has been observed in over 65% of patients with PDAC (Zeng et al., 2006). Importantly, aberrant β‐catenin signaling has been attributed to factors such as overexpression of ataxia‐telangiectasia group D complementing gene (ATDC) (Wang et al., 2009, 2015), upregulation of Wnt‐7b (Arensman et al., 2014), the c‐met receptor tyrosine kinase (Danilkovitch‐Miagkova et al., 2001), and the chemokine receptor CXCR4 (Wang et al., 2008). While β‐catenin signaling does not appear to drive the formation of PDAC, it was shown to be a strong driver of metastases and tumor cell invasion in mice with a Kras‐mutant background (Zhang et al., 2013). Further, oncogenic Kras has been shown to induce the expression of the ATDC gene (Wang et al., 2015), which activates β‐catenin signaling via stabilization of Disheveled‐2, abolishing the destruction complex. Likewise, the ATDC gene was found to induce EMT and metastases via β‐catenin in a mouse model that expressed both transgenic ATDC and mutant Kras, which was driven by a pancreas‐specific promoter p48‐Cre (Wang et al., 2015).
While MUC4 expression incrementally increases from PanIN‐1A to PDAC (Andrianifahanana et al., 2001), aberrant (cytosolic/nuclear) β‐catenin first occurs in the PanIN‐1 stage, with increasing aberrant localization occurring in advanced PanIN lesions and PDAC (Al‐Aynati et al., 2004). Our expression analysis of primary PDAC tissues suggests that aberrant β‐catenin localization and MUC4 occur in most (80%) cases of PDAC. In a 2014 study, Olsen et al. performed a microarray analysis of the human PDAC cell line BXPC3, which was completely depleted of β‐catenin using zinc‐finger nucleases. Their analysis showed that β‐catenin potentially regulates a wide array of cell adhesion molecules, including integrins, laminins, tight junction proteins, and other members of the adherens junction (Olsen et al., 2014). Interestingly, MUC4, which is a molecule shown to contribute to EMT by its ability to act as a ligand for HER2 was included in their list of the most significantly downregulated transcripts upon depletion of β‐catenin. These observations, as well as the fact that PDAC cell lines that express the β‐catenin protein also express MUC4, spurred further analyses of the MUC4 promoter, which was found to contain three putative TCF/LEF sites.
The MUC4 promoter has been shown to contain binding sites for numerous transcription factors, such as STATs, GATA, Sp1, and GR (Perrais et al., 2001). Further, the first TATA box is located at −2672/−2668 (Perrais et al., 2001). Our analysis showed that the first putative TCF/LEF site was located just proximal to the TATA box, at −2629/−2612, while the other two putative sites were located distal to the TATA box, at −3226 and −3408. Notably, the TCF/LEF binding site is a conserved sequence in the minor groove of DNA, with the consensus sequence being (A/T) (A/T)CAA(A/T)G (Gustavson et al., 2004). Having confirmed that β‐catenin can regulate MUC4 protein and RNA levels, we generated a construct that encompasses the entire MUC4 promoter (p3778), which contains all three TCF/LEF sites. We further generated two other constructs (p3000 and p2700) that contain two and one TCF/LEF sites, respectively. Promoter luciferase studies with 4ACAT showed that, despite the presence of TCF/LEF sites in p2700 and p3000, there was significantly increased MUC4 promoter luciferase activity only in the presence of all three sites (i.e., p3778; −3408 [Site #3], −3226 [Site #2], and −2612 [Site #1]). Our promoter luciferase studies indicated that (i) the MUC4 transcript is directly regulated by β‐catenin when all three TCF/LEF sites are present, and (ii) the TCF/LEF site proximal to the TATA box is critical for β‐catenin mediated MUC4 regulation. The latter result was surprising given that the p2700 construct, which containins the first TCF/LEF site, was unable to elicit significantly increased MUC4 promoter luciferase activity in the presence of 4ACAT. However, we also observed that MUT3 reduces MUC4 promoter luciferase activity, and that some β‐catenin/TCF binding also takes place at Site #3, suggesting that Site #3 also regulates MUC4 transcription. Likewise, MUT2 increased MUC4 promoter luciferase activity, suggesting that Site #2 ordinarily represses MUC4 transcription. However, we did not observe any discernible β‐catenin/TCF‐binding at Site #2 via ChIP analysis. Thus, it appears that while TCF/LEF Site #1 is the most critical for MUC4 transcription, Site #3 is also required for β‐catenin‐mediated MUC4 regulation. Further, there appears to be a combinatorial enhancement of transcription via Sites #1 and #3. Another factor to be considered is that TCF/LEF Site #1 is in extreme proximity to, but does not overlap with, the TATA box. As such, mutating this site may also affect numerous other factors that bind in this very active promoter region (Perrais et al., 2001). However, our ChIP results suggest that β‐catenin does indeed bind this site. Given that MUC4 transcription has been shown to be governed by several other factors, such as STAT1 (Seshacharyulu et al., 2015), NCOA3 (Kumar et al., 2014), IFNγ, and retinoic acid (Kunigal et al., 2012), it is likely that multiple disease‐stage specific factors govern MUC4 expression, and may collude to increase MUC4 expression.
Functionally, several studies have shown that aberrant β‐catenin signaling contributes to the migratory and metastatic properties of PDAC cells (Pasca di et al., 2007; Wang et al., 2015). Further, it has been suggested that β‐catenin signaling is epistatic to the MAPK/ERK pathway in PDAC (Zhang et al., 2013), although the precise mechanism has not been delineated. Notably, the ERK pathway has been implicated in PDAC metastases and invasion (He et al., 2012). Incidentally, KD of MUC4 in PDAC has been shown to decrease pERK1/2 levels (Rachagani et al., 2012). The mesenchymal marker Vimentin has been shown to be a direct target of β‐catenin/TCF‐LEF in breast cancer (Gilles et al., 2003). The reduction in Vimentin protein and RNA in our KD cells suggests that Vimentin may also be a target in PDAC. Furthermore, Zo‐1 is considered an epithelial marker, but some studies indicate that Zo‐1 is overexpressed in PDAC and contributes to metastasis (Kleeff et al., 2001). While we saw an increase in Zo‐1 levels in T3M4 KD cells, we observed a shift to a slightly higher molecular weight Zo‐1 in CD18/HPAF KD cells. The occurrence of two distinct isoforms of Zo‐1 is documented for different cell types (Balda and Anderson, 1993). Although we did not determine whether there was indeed a shift to a different isoform in the KD cells, in light of the changes in morphology observed in the KD cells, we surmise that this could be a possibility. What we did observe in CD18/HPAF KD cells was a reduction in the EMT regulator Snail, which been shown to repress E‐cadherin and MUC1 expression, as well as promote transcription of Vimentin (Nieto, 2002). This could be an explanation for the increased levels of MUC1 observed in both our KD cell lines. Alternatively, the increase in MUC1 could represent a compensatory mechanism for the loss of other transmembrane mucins such as MUC16 (in CD18/HPAF) and MUC4. Given that the MUC1‐β‐catenin interaction is well studied; these interesting observations with regard to MUC1 and β‐catenin warrant further study. Notably, MUC16 has also been shown to interact with β‐catenin (Giannakouros et al., 2014). However, while we observed a significant reduction in MUC16 protein expression in the CD18/HPAF KD cells, no significant change was seen in T3M4 KD cells. These observations also warrant further studies.
KD of β‐catenin in T3M4 cells resulted in decreased levels of MUC4, accompanied by reduced pHER2, suggesting that MUC4/HER2‐driven oncogenic signaling was reduced in this cell line. While we did not observe a significant difference in pHER2 in the CD18/HPAF KD cells in vitro, we observed reduced pHER2 in the tumor lysates from the KD cells that were orthotopically implanted. Our earlier studies (Lakshmanan et al., 2015) showed that MUC4 is promiscuous and can also partner with other Erbb family members such as HER3 and HER4, whose expression increases as a compensatory mechanism when HER2 is knocked down. However, given that the MUC4 levels were significantly reduced in both our KD cell lines, the likelihood of any MUC4‐HER3/HER4 coupling is low.
All the aforementioned studies indicate that β‐catenin acts an EMT/metastasis driver in the context of a Kras mutation (Wang et al., 2015; Zhang et al., 2013). In addition to up‐regulating conventional EMT‐related genes, such as CD44 (Wang et al., 2015), it is likely that Vimentin, which has been shown to be a direct target of β‐catenin signaling in breast cancer (Gilles et al., 2003), and MUC4 represent other genes that are up‐regulated by β‐catenin as part of a larger EMT program. Our tumorigenicity studies showed that both β‐catenin and MUC4 expression contribute to enhanced tumorigenicity. Specifically, IHC analysis of metastatic human tissues showed that β‐catenin and MUC4 were expressed in 40% of the liver metastatic tissues examined.
MUC4 is theorized to play an important role in EMT in PC (Rachagani et al., 2012) as well as in ovarian (Ponnusamy et al., 2010) and breast cancers (Mukhopadhyay et al., 2013). For example, MUC4 stabilizes fibroblast growth factor receptor 1 (FGFR), thereby stabilizing N‐cadherin (Rachagani et al., 2012). The N‐cadherin and FGFR complex then potentiates activation of the AKT and ERK pathways and stabilizes NF‐ΚB and AP‐1 transcription factors (Rachagani et al., 2012). Interestingly, it has also been posited that MUC4 induces nuclear localization of β‐catenin in PDAC (Zhi et al., 2014), indicating the existence of a possible feed‐forward loop. Given the importance of both MUC4 and β‐catenin in the progression and metastasis PDAC, as well as their interdependence, a two‐pronged, targeted therapy approach for both molecules merits examination.
In conclusion, our study demonstrates, for the first time, that β‐catenin directly regulates MUC4 transcription in PC and that MUC4 may exacerbate the β‐catenin‐induced invasive and metastatic phenotype of PDAC cells by contributing to the upregulation of several EMT markers.
Conflicts of interest
The authors declare no conflicts of interest.
Supporting information
The following is the supplementary data related to this article:
Supplementary data
Supplementary data
Acknowledgments
This work was supported, in part, by the grants from National Institutes of Health (UO1 CA111294, P50 CA127297, U54 CA163120 and RO1 CA183459). The authors would like to thank Ms. Kavita Mallya, Dr. Parthasarathy Seshacharyulu, Dr. Sushil Kumar, Dr. Arokiapriyanka Vaz and Suprit Gupta for technical support; Ms. Michelle Varney for the lentiviral transfection protocol; Drs. Robert Bennett (UNMC) and Yuzuru Shiio (UTHSC) for plasmids donated; Dr. Jing (Jenny) Wang (UNMC) for the L Wnt3A cells; Drs. Angie Rizzino and Shilpa Buch for letting us use their facilities; UNMC DNA Sequencing Core Facility, and Janice A. Taylor and James R. Talaska of the UNMC confocal laser scanning facility for assistance with taking confocal images. Finally, we kindly thank Melody A. Montgomery, BS, MFA, for the professional editing of this manuscript.
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2015.10.005.
Pai Priya, Rachagani Satyanarayana, Lakshmanan Imayavaramban, Macha Muzafar A., Sheinin Yuri, Smith Lynette M., Ponnusamy Moorthy P., Batra Surinder K., (2016), The canonical Wnt pathway regulates the metastasis‐promoting mucin MUC4 in pancreatic ductal adenocarcinoma, Molecular Oncology, 10, doi: 10.1016/j.molonc.2015.10.005.
References
- Al-Aynati, M.M. , Radulovich, N. , Riddell, R.H. , Tsao, M.S. , 2004. Epithelial-cadherin and beta-catenin expression changes in pancreatic intraepithelial neoplasia. Clin. Cancer Res. (4) 1235–1240. [DOI] [PubMed] [Google Scholar]
- Andrianifahanana, M. , Agrawal, A. , Singh, A.P. , Moniaux, N. , Van Seuningen, I. , Aubert, J.P. , Meza, J. , Batra, S.K. , 2005. Synergistic induction of the MUC4 mucin gene by interferon-gamma and retinoic acid in human pancreatic tumour cells involves a reprogramming of signalling pathways. Oncogene. (40) 6143–6154. [DOI] [PubMed] [Google Scholar]
- Andrianifahanana, M. , Moniaux, N. , Schmied, B.M. , Ringel, J. , Friess, H. , Hollingsworth, M.A. , Buchler, M.W. , Aubert, J.P. , Batra, S.K. , 2001. Mucin (MUC) gene expression in human pancreatic adenocarcinoma and chronic pancreatitis: a potential role of MUC4 as a tumor marker of diagnostic significance. Clin. Cancer Res. (12) 4033–4040. [PubMed] [Google Scholar]
- Andrianifahanana, M. , Singh, A.P. , Nemos, C. , Ponnusamy, M.P. , Moniaux, N. , Mehta, P.P. , Varshney, G.C. , Batra, S.K. , 2007. IFN-gamma-induced expression of MUC4 in pancreatic cancer cells is mediated by STAT-1 upregulation: a novel mechanism for IFN-gamma response. Oncogene. (51) 7251–7261. [DOI] [PubMed] [Google Scholar]
- Ansari, D. , Urey, C. , Gundewar, C. , Bauden, M.P. , Andersson, R. , 2013. Comparison of MUC4 expression in primary pancreatic cancer and paired lymph node metastases. Scand. J. Gastroenterol. (10) 1183–1187. [DOI] [PubMed] [Google Scholar]
- Arensman, M.D. , Kovochich, A.N. , Kulikauskas, R.M. , Lay, A.R. , Yang, P.T. , Li, X. , Donahue, T. , Major, M.B. , Moon, R.T. , Chien, A.J. , Dawson, D.W. , 2014. WNT7B mediates autocrine Wnt/beta-catenin signaling and anchorage-independent growth in pancreatic adenocarcinoma. Oncogene. (7) 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balda, M.S. , Anderson, J.M. , 1993. Two classes of tight junctions are revealed by ZO-1 isoforms. Am. J. Physiol. (4 Pt 1) C918–C924. [DOI] [PubMed] [Google Scholar]
- Barker, N. , van den Born, M. , 2008. Detection of beta-catenin localization by immunohistochemistry. Methods Mol. Biol. 91–98. [DOI] [PubMed] [Google Scholar]
- Cai, G. , Wang, J. , Xin, X. , Ke, Z. , Luo, J. , 2007. Phosphorylation of glycogen synthase kinase-3 beta at serine 9 confers cisplatin resistance in ovarian cancer cells. Int. J. Oncol. (3) 657–662. [PubMed] [Google Scholar]
- Chaturvedi, P. , Singh, A.P. , Chakraborty, S. , Chauhan, S.C. , Bafna, S. , Meza, J.L. , Singh, P.K. , Hollingsworth, M.A. , Mehta, P.P. , Batra, S.K. , 2008. MUC4 mucin interacts with and stabilizes the HER2 oncoprotein in human pancreatic cancer cells. Cancer Res. (7) 2065–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi, P. , Singh, A.P. , Moniaux, N. , Senapati, S. , Chakraborty, S. , Meza, J.L. , Batra, S.K. , 2007. MUC4 mucin potentiates pancreatic tumor cell proliferation, survival, and invasive properties and interferes with its interaction to extracellular matrix proteins. Mol. Cancer Res. (4) 309–320. [DOI] [PubMed] [Google Scholar]
- Collins, M.A. , Pasca di, M.M. , 2014. Kras as a key oncogene and therapeutic target in pancreatic cancer. Front Physiol. (407) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilkovitch-Miagkova, A. , Miagkov, A. , Skeel, A. , Nakaigawa, N. , Zbar, B. , Leonard, E.J. , 2001. Oncogenic mutants of RON and MET receptor tyrosine kinases cause activation of the beta-catenin pathway. Mol. Cell Biol. (17) 5857–5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodde, R. , Smits, R. , Clevers, H. , 2001. APC, signal transduction and genetic instability in colorectal cancer. Nat. Rev. Cancer. (1) 55–67. [DOI] [PubMed] [Google Scholar]
- Giannakouros, P. , Comamala, M. , Matte, I. , Rancourt, C. , Piche, A. , 2014. MUC16 mucin (CA125) regulates the formation of multicellular aggregates by altering beta-catenin signaling. Am. J. Cancer Res. (1) 219–230. [PMC free article] [PubMed] [Google Scholar]
- Gilles, C. , Polette, M. , Mestdagt, M. , Nawrocki-Raby, B. , Ruggeri, P. , Birembaut, P. , Foidart, J.M. , 2003. Transactivation of vimentin by beta-catenin in human breast cancer cells. Cancer Res. (10) 2658–2664. [PubMed] [Google Scholar]
- Gustavson, M.D. , Crawford, H.C. , Fingleton, B. , Matrisian, L.M. , 2004. Tcf binding sequence and position determines beta-catenin and Lef-1 responsiveness of MMP-7 promoters. Mol. Carcinog. (3) 125–139. [DOI] [PubMed] [Google Scholar]
- He, X. , Zheng, Z. , Li, J. , Ben, Q. , Liu, J. , Zhang, J. , Ji, J. , Yu, B. , Chen, X. , Su, L. , Zhou, L. , Liu, B. , Yuan, Y. , 2012. DJ-1 promotes invasion and metastasis of pancreatic cancer cells by activating SRC/ERK/uPA. Carcinogenesis. (3) 555–562. [DOI] [PubMed] [Google Scholar]
- Jiang, H. , Li, Q. , He, C. , Li, F. , Sheng, H. , Shen, X. , Zhang, X. , Zhu, S. , Chen, H. , Chen, X. , Yang, C. , Gao, H. , 2014. Activation of the Wnt pathway through Wnt2 promotes metastasis in pancreatic cancer. Am. J. Cancer Res. (5) 537–544. [PMC free article] [PubMed] [Google Scholar]
- Jonckheere, N. , Perrais, M. , Mariette, C. , Batra, S.K. , Aubert, J.P. , Pigny, P. , Van Seuningen, I. , 2004. A role for human MUC4 mucin gene, the ErbB2 ligand, as a target of TGF-beta in pancreatic carcinogenesis. Oncogene. (34) 5729–5738. [DOI] [PubMed] [Google Scholar]
- Jones, S. , Zhang, X. , Parsons, D.W. , Lin, J.C. , Leary, R.J. , Angenendt, P. , Mankoo, P. , Carter, H. , Kamiyama, H. , Jimeno, A. , Hong, S.M. , Fu, B. , Lin, M.T. , Calhoun, E.S. , Kamiyama, M. , Walter, K. , Nikolskaya, T. , Nikolsky, Y. , Hartigan, J. , Smith, D.R. , Hidalgo, M. , Leach, S.D. , Klein, A.P. , Jaffee, E.M. , Goggins, M. , Maitra, A. , Iacobuzio-Donahue, C. , Eshleman, J.R. , Kern, S.E. , Hruban, R.H. , Karchin, R. , Papadopoulos, N. , Parmigiani, G. , Vogelstein, B. , Velculescu, V.E. , Kinzler, K.W. , 2008. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. (5897) 1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, S. , Momi, N. , Chakraborty, S. , Wagner, D.G. , Horn, A.J. , Lele, S.M. , Theodorescu, D. , Batra, S.K. , 2014. Altered expression of transmembrane mucins, MUC1 and MUC4, in bladder cancer: pathological implications in diagnosis. PLoS One. (3) e92742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleeff, J. , Shi, X. , Bode, H.P. , Hoover, K. , Shrikhande, S. , Bryant, P.J. , Korc, M. , Buchler, M.W. , Friess, H. , 2001. Altered expression and localization of the tight junction protein ZO-1 in primary and metastatic pancreatic cancer. Pancreas. (3) 259–265. [DOI] [PubMed] [Google Scholar]
- Kolligs, F.T. , Hu, G. , Dang, C.V. , Fearon, E.R. , 1999. Neoplastic transformation of RK3E by mutant beta-catenin requires deregulation of Tcf/Lef transcription but not activation of c-myc expression. Mol. Cell Biol. (8) 5696–5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S. , Das, S. , Rachagani, S. , Kaur, S. , Joshi, S. , Johansson, S.L. , Ponnusamy, M.P. , Jain, M. , Batra, S.K. , 2014. NCOA3-mediated upregulation of mucin expression via transcriptional and post-translational changes during the development of pancreatic cancer. Oncogene. (34) 4879–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunigal, S. , Ponnusamy, M.P. , Momi, N. , Batra, S.K. , Chellappan, S.P. , 2012. Nicotine, IFN-gamma and retinoic acid mediated induction of MUC4 in pancreatic cancer requires E2F1 and STAT-1 transcription factors and utilize different signaling cascades. Mol. Cancer. (26) 11:24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahdaoui, F. , Delpu, Y. , Vincent, A. , Renaud, F. , Messager, M. , Duchene, B. , Leteurtre, E. , Mariette, C. , Torrisani, J. , Jonckheere, N. , Van Seuningen, I. , 2015. miR-219-1-3p is a negative regulator of the mucin MUC4 expression and is a tumor suppressor in pancreatic cancer. Oncogene. (6) 780–788. [DOI] [PubMed] [Google Scholar]
- Lakshmanan, I. , Seshacharyulu, P. , Haridas, D. , Rachagani, S. , Gupta, S. , Joshi, S. , Guda, C. , Yan, Y. , Jain, M. , Ganti, A.K. , Ponnusamy, M.P. , Batra, S.K. , 2015. Novel HER3/MUC4 oncogenic signaling aggravates the tumorigenic phenotypes of pancreatic cancer cells. Oncotarget. (25) 21085–21099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.J. , Wei, Z.M. , Meng, Y.X. , Ji, X.R. , 2005. Beta-catenin up-regulates the expression of cyclinD1, c-myc and MMP-7 in human pancreatic cancer: relationships with carcinogenesis and metastasis. World J. Gastroenterol. (14) 2117–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, J.P. , Wang, S.C. , Hebrok, M. , 2010. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat. Rev. Cancer. (10) 683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay, P. , Lakshmanan, I. , Ponnusamy, M.P. , Chakraborty, S. , Jain, M. , Pai, P. , Smith, L.M. , Lele, S.M. , Batra, S.K. , 2013. MUC4 overexpression augments cell migration and metastasis through EGFR family proteins in triple negative breast cancer cells. PLoS One. (2) e54455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins, T.D. , Kern, H.F. , Metzgar, R.S. , 1991. Ultrastructural differentiation of sodium butyrate-treated human pancreatic adenocarcinoma cell lines. Pancreas. 5, 578–587. [DOI] [PubMed] [Google Scholar]
- Nieto, M.A. , 2002. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell Biol. (3) 155–166. [DOI] [PubMed] [Google Scholar]
- Okabe, T. , Yamaguchi, N. , Ohsawa, N. , 1983. Establishment and characterization of a carcinoembryonic antigen (CEA)-producing cell line from a human carcinoma of the exocrine pancreas. Cancer. (4) 662–668. [DOI] [PubMed] [Google Scholar]
- Olsen, P.A. , Solberg, N.T. , Lund, K. , Vehus, T. , Gelazauskaite, M. , Wilson, S.R. , Krauss, S. , 2014. Implications of targeted genomic disruption of beta-catenin in BxPC-3 pancreatic adenocarcinoma cells. PLoS One. (12) e115496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca di, M.M. , Biankin, A.V. , Heiser, P.W. , Cano, D.A. , Gutierrez, P.J. , Deramaudt, T. , Segara, D. , Dawson, A.C. , Kench, J.G. , Henshall, S.M. , Sutherland, R.L. , Dlugosz, A. , Rustgi, A.K. , Hebrok, M. , 2007. Common activation of canonical Wnt signaling in pancreatic adenocarcinoma. PLoS One. (11) e1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrais, M. , Pigny, P. , Ducourouble, M.P. , Petitprez, D. , Porchet, N. , Aubert, J.P. , Van Seuningen, I. , 2001. Characterization of human mucin gene MUC4 promoter: importance of growth factors and proinflammatory cytokines for its regulation in pancreatic cancer cells. J. Biol. Chem. (33) 30923–30933. [DOI] [PubMed] [Google Scholar]
- Ponnusamy, M.P. , Lakshmanan, I. , Jain, M. , Das, S. , Chakraborty, S. , Dey, P. , Batra, S.K. , 2010. MUC4 mucin-induced epithelial to mesenchymal transition: a novel mechanism for metastasis of human ovarian cancer cells. Oncogene. (42) 5741–5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, Q. , Ramadani, M. , Gansauge, S. , Gansauge, F. , Leder, G. , Beger, H.G. , 2001. Reduced membranous and ectopic cytoplasmic expression of beta -catenin correlate with cyclin D1 overexpression and poor prognosis in pancreatic cancer. Int. J. Cancer. (3) 194–197. [DOI] [PubMed] [Google Scholar]
- Rachagani, S. , Macha, M.A. , Ponnusamy, M.P. , Haridas, D. , Kaur, S. , Jain, M. , Batra, S.K. , 2012. MUC4 potentiates invasion and metastasis of pancreatic cancer cells through stabilization of fibroblast growth factor receptor 1. Carcinogenesis. (10) 1953–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan, P. , Mohr, A.M. , Grandgenett, P.M. , Steele, M.M. , Batra, S.K. , Hollingsworth, M.A. , 2013. MicroRNA-200c modulates the expression of MUC4 and MUC16 by directly targeting their coding sequences in human pancreatic cancer. PLoS One. (10) e73356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhim, A.D. , Mirek, E.T. , Aiello, N.M. , Maitra, A. , Bailey, J.M. , McAllister, F. , Reichert, M. , Beatty, G.L. , Rustgi, A.K. , Vonderheide, R.H. , Leach, S.D. , Stanger, B.Z. , 2012. EMT and dissemination precede pancreatic tumor formation. Cell. (1–2) 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou, M. , Goto, M. , Horinouchi, M. , Tamada, S. , Nagata, K. , Hamada, T. , Osako, M. , Takao, S. , Batra, S.K. , Aikou, T. , Imai, K. , Yonezawa, S. , 2005. MUC4 expression is a novel prognostic factor in patients with invasive ductal carcinoma of the pancreas. J. Clin. Pathol. (8) 845–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapati, S. , Gnanapragassam, V.S. , Moniaux, N. , Momi, N. , Batra, S.K. , 2012. Role of MUC4-NIDO domain in the MUC4-mediated metastasis of pancreatic cancer cells. Oncogene. (28) 3346–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshacharyulu, P. , Ponnusamy, M.P. , Rachagani, S. , Lakshmanan, I. , Haridas, D. , Yan, Y. , Ganti, A.K. , Batra, S.K. , 2015. Targeting EGF-receptor(s) – STAT1 axis attenuates tumor growth and metastasis through downregulation of MUC4 mucin in human pancreatic cancer. Oncotarget. (7) 5164–5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, A.P. , Chauhan, S.C. , Andrianifahanana, M. , Moniaux, N. , Meza, J.L. , Copin, M.C. , Van Seuningen, I. , Hollingsworth, M.A. , Aubert, J.P. , Batra, S.K. , 2007. MUC4 expression is regulated by cystic fibrosis transmembrane conductance regulator in pancreatic adenocarcinoma cells via transcriptional and post-translational mechanisms. Oncogene. (1) 30–41. [DOI] [PubMed] [Google Scholar]
- Singh, A.P. , Moniaux, N. , Chauhan, S.C. , Meza, J.L. , Batra, S.K. , 2004. Inhibition of MUC4 expression suppresses pancreatic tumor cell growth and metastasis. Cancer Res. (2) 622–630. [DOI] [PubMed] [Google Scholar]
- Srivastava, S.K. , Bhardwaj, A. , Singh, S. , Arora, S. , Wang, B. , Grizzle, W.E. , Singh, A.P. , 2011. MicroRNA-150 directly targets MUC4 and suppresses growth and malignant behavior of pancreatic cancer cells. Carcinogenesis. (12) 1832–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz, M.J. , Batra, S.K. , Varshney, G.C. , Hollingsworth, M.A. , Yeo, C.J. , Cameron, J.L. , Wilentz, R.E. , Hruban, R.H. , Argani, P. , 2002. MUC4 expression increases progressively in pancreatic intraepithelial neoplasia. Am. J. Clin. Pathol. (5) 791–796. [DOI] [PubMed] [Google Scholar]
- Uhlen, M. , Fagerberg, L. , Hallstrom, B.M. , Lindskog, C. , Oksvold, P. , Mardinoglu, A. , Sivertsson, A. , Kampf, C. , Sjostedt, E. , Asplund, A. , Olsson, I. , Edlund, K. , Lundberg, E. , Navani, S. , Szigyarto, C.A. , Odeberg, J. , Djureinovic, D. , Takanen, J.O. , Hober, S. , Alm, T. , Edqvist, P.H. , Berling, H. , Tegel, H. , Mulder, J. , Rockberg, J. , Nilsson, P. , Schwenk, J.M. , Hamsten, M. , von, F.K. , Forsberg, M. , Persson, L. , Johansson, F. , Zwahlen, M. , von, H.G. , Nielsen, J. , Ponten, F. , 2015. Proteomics. Tissue-based map of the human proteome. Science. (6220) 1260419 [DOI] [PubMed] [Google Scholar]
- Veeman, M.T. , Slusarski, D.C. , Kaykas, A. , Louie, S.H. , Moon, R.T. , 2003. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr. Biol. (8) 680–685. [DOI] [PubMed] [Google Scholar]
- Vincent, A. , Ducourouble, M.P. , Van Seuningen, I. , 2008. Epigenetic regulation of the human mucin gene MUC4 in epithelial cancer cell lines involves both DNA methylation and histone modifications mediated by DNA methyltransferases and histone deacetylases. FASEB J. (8) 3035–3045. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Heidt, D.G. , Lee, C.J. , Yang, H. , Logsdon, C.D. , Zhang, L. , Fearon, E.R. , Ljungman, M. , Simeone, D.M. , 2009. Oncogenic function of ATDC in pancreatic cancer through Wnt pathway activation and beta-catenin stabilization. Cancer Cell. (3) 207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Yang, H. , Abel, E.V. , Ney, G.M. , Palmbos, P.L. , Bednar, F. , Zhang, Y. , Leflein, J. , Waghray, M. , Owens, S. , Wilkinson, J.E. , Prasad, J. , Ljungman, M. , Rhim, A.D. , Pasca di, M.M. , Simeone, D.M. , 2015. ATDC induces an invasive switch in KRAS-induced pancreatic tumorigenesis. Genes Dev. (2) 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Ma, Q. , Liu, Q. , Yu, H. , Zhao, L. , Shen, S. , Yao, J. , 2008. Blockade of SDF-1/CXCR4 signalling inhibits pancreatic cancer progression in vitro via inactivation of canonical Wnt pathway. Br. J. Cancer. (10) 1695–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, G. , Germinaro, M. , Micsenyi, A. , Monga, N.K. , Bell, A. , Sood, A. , Malhotra, V. , Sood, N. , Midda, V. , Monga, D.K. , Kokkinakis, D.M. , Monga, S.P. , 2006. Aberrant Wnt/beta-catenin signaling in pancreatic adenocarcinoma. Neoplasia. (4) 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Morris, J.P. , Yan, W. , Schofield, H.K. , Gurney, A. , Simeone, D.M. , Millar, S.E. , Hoey, T. , Hebrok, M. , Pasca di, M.M. , 2013. Canonical Wnt signaling is required for pancreatic carcinogenesis. Cancer Res. (15) 4909–4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi, X. , Tao, J. , Xie, K. , Zhu, Y. , Li, Z. , Tang, J. , Wang, W. , Xu, H. , Zhang, J. , Xu, Z. , 2014. MUC4-induced nuclear translocation of beta-catenin: a novel mechanism for growth, metastasis and angiogenesis in pancreatic cancer. Cancer Lett. (1) 104–113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following is the supplementary data related to this article:
Supplementary data
Supplementary data


