Abstract
High-grade epithelial ovarian cancer (OvCa) kills more women than any other gynecologic cancer and is rarely diagnosed at an early stage. We sought to identify tumor-associated antigens (TAA) as candidate diagnostic and/or immunotherapeutic targets by taking advantage of tumor autoantibody responses in individuals with OvCa. Plasma-derived IgG from a pool of five patients with advanced OvCa was subjected to iterative biopanning using a library of bacteriophage MS2 virus-like particles (MS2-VLPs) displaying diverse short random peptides. After two rounds of biopanning, we analyzed the selectant population of MS2-VLPs by Ion Torrent deep-sequencing. One of the top 25 most abundant peptides identified (DISGTNTSRA) had sequence similarity to cancer antigen 125 (CA125/MUC16), a well-known OvCa-associated antigen. Mice immunized with MS2-DISGTNTSRA generated antibodies that cross-reacted with purified soluble CA125 from OvCa cells but not membrane-bound CA125, indicating that the DISGTNTSRA peptide was a CA125/MUC16 peptide mimic of soluble CA125. Pre-operative OvCa patient plasma (n = 100) was assessed for anti-DISGTNTSRA, anti-CA125, and CA125. Patients with normal CA125 (< 35 IU/mL) at time of diagnosis had significantly more antibodies to DISGTNTSRA and to CA125 than those patients who had high CA125 (> 35 IU/mL). A statistically significant survival advantage was observed for patients who had either normal CA125 and/or higher concentrations of antibodies to CA125 at time of diagnosis. These data show the feasibility of using deep sequence-coupled biopanning to identify TAA autoantibody responses from cancer patient plasma and suggest a possible antibody-mediated mechanism for low CA125 plasma concentrations in some OvCa patients.
INTRODUCTION
There are more deaths from epithelial ovarian cancer (OvCa) than any other gynecological cancer and it is one of the top five causes of cancer death in women in the United States (1). OvCa is usually diagnosed after the disease has disseminated. Despite aggressive surgical and chemotherapeutic interventions, dissemination is associated with poor outcomes (2). Because of this, developing diagnostic tests for early stage disease and more effective and better-tolerated treatments for OvCa are high research priorities (3).
Many cancers are associated with autoantibody responses to tumor associated antigens (anti-TAAs). Anti-TAAs are attractive candidates for the detection of preclinical disease because they often occur early in disease and are less prone to variation from confounding factors than other circulating protein biomarkers (4–9). Furthermore, the ability to induce anti-TAAs suggests that the tumor antigen is immunogenic in at least some patients and is a potential target for immunotherapy.
In this study, we took an unbiased approach to identifying the targets of anti-TAAs in OvCa patients. Our lab has developed a novel affinity selection technology based on virus-like particles (VLPs) of the RNA bacteriophage MS2 (10). Because VLPs are highly immunogenic, we have used this technology to identify vaccines that elicit high-titer antibody responses mimicking the activity of the selecting monoclonal antibody (mAb) (11–13). Here, we report a novel application of the MS2-VLP affinity selection technology to identify anti-TAAs in OvCa patients. By coupling the affinity selection capabilities of the VLP platform with highly sensitive Ion Torrent deep-sequencing, we identified immunoepitopes recognized by OvCa patient antibodies, including the well-known OvCa antigen CA125. Patients with antibodies to this peptide had less serum CA125 and better outcomes.
MATERIALS AND METHODS
Patient plasma samples and IgG isolation
Patients (n = 100) with OvCa stages I, II, and III were recruited at the Johns Hopkins Hospital. Patient blood was collected into heparin-treated tubes prior to surgery. Plasma was obtained and stored at −80°C. Written informed consent was provided by each participant and this study was approved by the Johns Hopkins Institutional Review Board. The p53 autoantibodies in the plasma were measured using the commercial p53 ELISAPLUS (autoantibody) kit from Calbiochem (QIA53) following the manufacturer’s instructions. The MILLIPLEX™MAP Human Cancer Biomarker Panel kit (Millipore) was used to measure the CA125 in human plasma according to the manufacturer’s protocol. The plates were washed with a Bio-Plex Pro II Wash Station (Bio-Rad, Hercules, CA). The samples were read with Bio-Plex Array Reader (Bio-Rad, Hercules, CA) and the data were analyzed with Bio-Plex Manager Software 5.0. Immunoglobulin G (IgG) was isolated from a pool of 5 patient plasma samples (5 μL/patient) using Dynabeads protein G (Invitrogen), following the manufacturer’s protocol.
Affinity selection with MS2-VLPs
Affinity selections were done overnight at 4°C using a pool of patient IgG (500ng) and mixtures of 6-, 7-, 8-, and 10-mer random peptide MS2-VLP libraries (10ug each), generated as previously described (13), in 100uL total volume with PBS.
Antibody/VLP complexes were mixed with 10μL Dynabeads Protein G, incubated for 4h at 4°C on a rotator, washed 6 times in PBS, eluted with 50μL 0.1M glycine pH 2.7 for 10min at RT, and immediately neutralized with 5μL 1M Tris pH 9.0. Selections were done in triplicate, and eluates pooled for RT-PCR amplification of corresponding VLP RNA.
RT reactions were prepared using M-MLV RT (Invitrogen) and a primer specific for the MS2-coat protein transcript. The RT reaction mixture contained the following in a 20μL total reaction: 1μL 10mM dNTP mix (Invitrogen), 4μL 5X First Strand Buffer (Invitrogen), 2μL 0.1M DTT, 200U M-MLV RT (1μL), 8μL eluted VLPs, and 4μL 10uM E2 primer (5′-TCAGCGGTGGCAGCAGCCAA-3′). Primer, dNTPs, and eluted VLPs were heated to 65°C for 5min and quick-chilled on ice. DTT and 5X First Strand Buffer were added and incubated at 37°C for 2min, and then M-MLV RT was added. This reaction was incubated at 37°C for 50min, and then 70°C for 15min. Resultant cDNA was used in subsequent PCR reactions to amplify the recovered sequences. High Fidelity Platinum Taq polymerase (Invitrogen) was used in 50μL total reactions according to manufacturer’s recommended conditions. Primers for PCR amplification were E3.2 (5′-CGGGCTTTGTTAGCAGCCGG-3′) and 62up (5′-CTATGCAGGGGTTGTTGAAG-3′), with 35 cycles of PCR at 60°C annealing temperature. RT reaction from above (2μL) was used as template for PCR in 8 replicates. PCR reactions were pooled and purified by Qiaquick PCR purification columns (Qiagen) following manufacturer’s protocol. Purified PCR products were digested with BamHI and SalI (New England Biolabs) for 1h at 37°C, and purified using Qiaquick Gel Extraction Kit (Qiagen). Digested PCR products were cloned into BamHI/SalI digested pDSP62(am) vector plasmid (13) with T4 DNA ligase. Ethanol precipitated ligation reactions were transformed into electrocompetent 10G E. coli cells and grown overnight in LB. Plasmid libraries were isolated with Qiafilter Plasmid Purification Maxiprep Kit (Qiagen), digested with KpnI to eliminate wild-type plasmid background, ethanol precipitated and transformed into C41(pSupA) cells for generation of corresponding selectant MS2-VLP libraries (13). VLPs were isolated as indicated above and used in another round of affinity selection with patient plasma IgG as described above.
Ion Torrent sequencing of selectant populations
Affinity selected plasmid libraries were used in PCR reactions with primers containing Ion Torrent adapter sequences and 4 nucleotide barcodes. PCR reaction mixes were carried out in 50μL total volume, with High Fidelity Platinum Taq polymerase (Invitrogen) using the manufacturer’s protocol. These primers are as follows: ITBC-rev (5′-CCTCTCTATGGGCAGTCGGTGATGTGAACGCGAGTTAGAGC-3′), and ITBC-X (5′CCATCTCATCCCTGCGTGTCTCCGACTCAGXXXXCTCTACGGCAACTTTACTCAG-3′) where XXXX corresponds to a 4 nucleotide barcode sequence of ITBC-1 (AGTC), ITBC-2 (CGTA), ITBC-3 (CTAG), and ITBC-4 (TAGC). Plasmid template (50ng) was added to the PCR reaction with 20 cycles of amplification at 55°C annealing temperature. PCR products were separated on a 1.5% agarose TBE gel, purified by Qiaquick Gel Extraction Kit following the manufacturer’s recommended protocol, and sequenced with an Ion Torrent deep-sequencer.
Data analysis and selection of candidate peptides
Raw sequencing data was quality controlled and processed using custom MATLAB scripts (details in Supplemental Material). Final data sets were analyzed using Microsoft Excel and NCBI BLAST to identify peptides with sequence similarity to human protein sequences. The top 25 peptides identified for the second round of affinity selections with OvCa patient IgG were analyzed with NCBI BLAST, limiting the query to the non-redundant human protein sequences. The corresponding reports were analyzed for potential hits of interest.
Generation of MS2-DISGTNTSRA
Primers were generated for site-directed mutagenesis of the pDSP62 plasmid to construct MS2-DISGTNTSRA VLPs. Plasmid identity was confirmed by sequencing and used to generate corresponding MS2-VLPs displaying the DISGTNTSRA peptide in the AB loop of the MS2-coat protein dimer in C41 E. coli cells. Cultures of C41 cells harboring the expression plasmids were grown to OD600nm of ~1.0 and induced with 0.4mM IPTG for 3h. Cells were pelleted, frozen at −80°C, and VLPs were isolated as previously described (13).
Immunization of mice with MS2 and MS2-DISGTNTSRA
Animal work was carried out with the approval of the UNM IACUC. BALB/c mice (3–8 mice/group) were immunized with 5μg VLP intramuscularly 3X at 2-week intervals in PBS. Two weeks after the final immunization, mice were sacrificed and blood collected by cardiac puncture under appropriate anesthetic and analgesic.
ELISA
ELISA plates were coated with purified CA125 protein (US Biological, 100U/well) or streptavidin (10ug/mL) in 100μL volumes by incubating at 37°C for 2h or overnight at 4°C. Blank wells (not coated with CA125) were used as a negative control. After washing with PBS, streptavidin-coated plates were subsequently treated with succinimidyl 6((betamaleimidopropionamido)hexanoate) (SMPH) and synthetic peptide DISGTNTSRAGGGC (Genscript). Plates were then blocked with 0.5% dry milk/PBS (300μL/well) for 2h at RT. Serum samples were added in triplicate at 1:100 dilutions (CA125 assay) or 1:200 dilutions (peptide assay) in 0.5% dry milk/PBS and incubated for 2h at room temp. Plates were washed in PBS and HRP-conjugated goat anti-mouse IgG (1:5000 dilution) or HRP-conjugated donkey anti-human IgG (1:5000 dilution) was added in 0.5% milk/PBS in a total volume of 100μL (CA125 assay) or 200uL (peptide assay). Plates were incubated for 1h at RT, washed with PBS, and then TMB substrate was added. Plates were incubated at RT with shaking until sufficient color was developed. For CA125 assay, 100 μL HCl stop solution was added and mixed prior to reading at 450nm. For peptide assay, plates were read at 630nm.
Statistical analysis
Demographic and clinical characteristics of the study patients were summarized with descriptive statistics. Spearman correlation coefficient was used to assess the association between CA125, anti-CA125 and anti-DISGTNTSRA plasma levels. Patients were divided into two groups by CA125 (normal: < 35 IU/ml vs. elevated: > 35 IU/mL), anti-DISGTNTSRA (cutoff at median, positive if ABS 630nm > 1.0380 vs. negative if ABS 630nm < 1.0380) and anti-CA125 (positive if ABS 630nm reading > 0.295 vs. negative if ABS 630nm reading <0.295). For those dichotomized groups, the exact Pearson’s chi-square test with Monte Carlo estimates was used to assess the associations of anti-CA125 or anti-DISGTNTSRA with CA125. McNemar’s chi-square test was used to test whether the proportion of pairs for elevated CA125 is the same as the proportion of pairs for anti-CA125 positive (also for that of anti-DISGTNTSRA positive). Survival data were available for 60 patients (Stage I/II, n = 9; Stage III, n =51). Kaplan-Meier curve along with log-rank test was used to compare the median survival time between sub-groups of CA125, anti-CA125 or anti-DISGTNTSRA. The hazard ratios (HRs) were estimated by Cox proportional hazard models. The proportional hazard assumption was checked by testing time-dependent covariates. All models were met for the assumption. All tests were two-sided and considered statistically significant at the alpha level of 0.05. Statistical analysis was performed using R Statistical Software (version 3.1.3).
RESULTS
Deep sequence-coupled biopanning for autoantibodies
We have developed an affinity selection platform in which random peptides are displayed on bacteriophage MS2-VLPs (13). This platform can identify the epitopes of mAbs and identify vaccines that elicit mAb-like responses (11–13). In this study we asked whether this approach could also be used to identify the epitopes targeted by a complex, polyclonal mixture of serum antibodies from OvCa patients. We obtained serum samples from five OvCa patients diagnosed with advanced high-grade ovarian serous carcinoma (Table 1). Serum samples were obtained prior to treatment. All of the serum samples selected had detectable antibodies to p53, indicating that the patients were able to mount antibody responses to at least one tumor antigen.
Table 1.
Patient samples used for deep sequence-coupled biopanning.
| Patient ID | Disease | Serum CA125 (IU/mL) | p53 Antibody (U) |
|---|---|---|---|
| DM672 | III | 749.06 | 128.8 |
| DM684 | III | 542.45 | 104.7 |
| DM689 | III | 159.02 | 276.4 |
| DM778 | III | 19.85 | 155.2 |
| DM538 | III | 289.89 | 195.8 |
We performed two rounds of biopanning with pooled patient IgG to a VLP-library displaying random 6-, 7-, 8-, and 10-amino acid sequences (and containing > 1011 individual members) following a selection protocol (Fig. 1 schematic). After each round of selection, Ion Torrent deep-sequencing was used to identify selected peptide sequences. For each unique peptide, the absolute number of sequences and its percentage of the total population in the sample was determined. For each peptide identified in the second round of selection, the corresponding rank and percent abundance in the first round of selection were determined (Supplemental Table 1).
Figure 1. Schematic of deep sequence-coupled biopanning using MS2-VLPs and human serum.
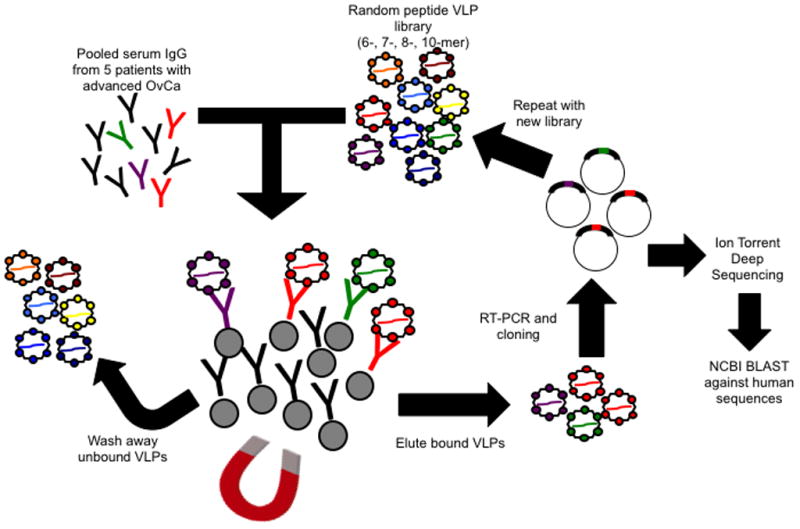
Immunoglobulin isolated from pooled OvCa patient plasma or normal plasma was incubated with a mixed MS2-VLP library displaying 6–10 amino acid random peptides. Antibody/VLP complexes are pulled down with magnetic Protein G Dynabeads, unbound VLPs are washed away, and bound VLPs are eluted. RT-PCR recovers the coding sequences encapsidated by the VLPs, and cloning and expression of the VLPs results in an enriched library. Biopanning is repeated and the resulting cloned coding sequences are used as template for Ion Torrent deep-sequencing.
In order to identify candidate proteins corresponding to the most commonly selected peptides, we performed BLAST queries with the top-ranked peptide sequences. Because peptides were short and polyclonal serum from OvCa patients almost certainly includes antibodies that are specific for non-TAAs, we limited queries to human proteins (non-redundant database). We were particularly interested in peptide DISGTNTSRA, the 16th ranked hit in our screen, which was identified in our BLAST analysis as having sequence similarity to CA125/MUC16, a well-established OvCa marker. Using NCBI BLAST, we identified a number of positions at which the DISGTNTSRA peptide aligns to CA125/MUC16 (Supplemental Table 2). These alignments map primarily to the highly glycosylated N-terminal domain (Fig. 2).
Figure 2. BLAST alignment hits of DISGTNTSRA against CA125/MUC16.
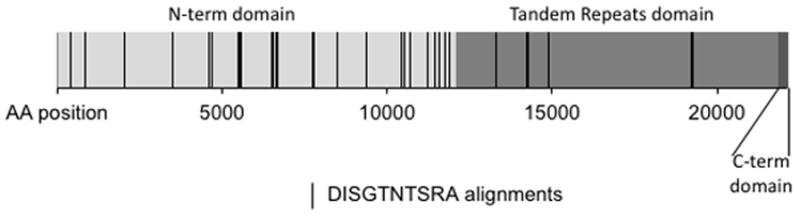
DISGTNTSRA was used as query against the MUC16 protein (reference #: Q8WXI7). Alignments shown contained at least four amino acid identity matches and had no gaps. N-terminal domain, tandem-repeat domain, and C-terminal domain (including membrane proximal region, trans-membrane domain, and cytoplasmic domain) are indicated.
MS2-DISGTNTSRA is a peptide mimic of CA125
To investigate whether DISGTNTSRA was an immunologic mimic of CA125/MUC16, we generated VLPs displaying the DISGTNTSRA peptide (MS2-DISGTNTSRA), immunized BALB/c mice, and assessed their serum for reactivity to purified human CA125 by ELISA. Sera of mice immunized with MS2-DISGTNTSRA showed significantly higher reactivity to purified soluble human CA125 than mice immunized with control MS2-VLPs (Fig. 3). Thus, antibodies raised to the DISGTNTSRA peptide can cross-react with soluble CA125. However, flow cytometric analysis of antibodies from mice immunized with MS2-DISGTNTSRA showed no detectable binding to membrane-bound CA125 on the surface of OvCa cell line OVCAR-3 (data not shown).
Figure 3. MS2-DISGTNTSRA is a CA125 epitope mimic.
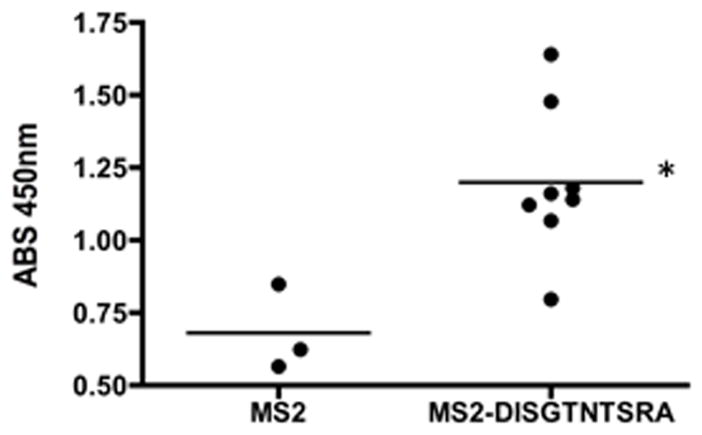
Mice were immunized three times at 2 week intervals with MS2-VLP (displaying no foreign peptide) (n = 3) or MS2-DISGTNTSRA (n = 8) and assessed for reactivity to purified CA125. *P-value = 0.0105 by Student’s t-test.
Human plasma reactivity to DISGTNTSRA
Having found that deep sequence-coupled biopanning with OvCa patient plasma identified a CA125 mimic, we were interested in investigating the extent to which OvCa patients have antibodies to DISGTNTSRA and CA125. First, we determined whether plasma reactivity to DISGTNTSRA differed between OvCa patients and normal individuals. We selected 40 normal and 47 pre-operative stage III OvCa patient samples. The OvCa samples included the five patients used for the deep sequence-coupled biopanning. Normal plasma showed significantly higher reactivity to the DISGTNTRSA peptide than OvCa patient plasma (Supplemental Fig. S1). However, a subset of OvCa patients had sera with strong reactivity to DISGTNTSRA. Indeed, the patient DM778, whose sera was used in the biopanning experiment that identified this peptide, showed the highest reactivity to DISGTNTSRA (this patient is denoted by the open circle in Supplemental Fig. S1). Interestingly, this patient also had normal plasma CA125 levels (Table 1). This led us to hypothesize that patients with anti-DISGTNTSRA or anti-CA125 antibodies would have lower CA125 in their plasma.
In order to test this hypothesis, we identified 99 OvCa patients that had either normal (< 35 IU/mL; n = 33) or elevated (> 35 IU/mL; n = 66) serum CA125 and assessed these samples for anti-DISGTNTSRA antibodies. These samples included patients with stage I, II, or III OvCa (Supplemental Table 3) and were collected at time of diagnosis, prior to treatment. Patient serum CA125 was negatively associated with anti-DISGTNTSRA (Fig. 4A, Spearman’s rank correlation coefficient: −0.286, P-value: 0.004) and patients with elevated serum CA125 had significantly lower anti-DISGTNTSRA than patients with normal serum CA125 (Fig. 4B, Mann-Whitney, P-value: 0.0033). When we separated patients into positive and negative groups based on the median of the ELISA readings, we found that the proportions of patients who were positive for anti-DISGTNTSRA with normal serum CA125 (n = 19) were significantly different from those patients who were negative for anti-DISGTNTSRA with elevated levels of CA125 (n = 43) (Supplemental Table 4, McNemar test, P-value: 0.041).
Figure 4. Correlation of serum CA125 with anti-DISGTNTSRA or anti-CA125.
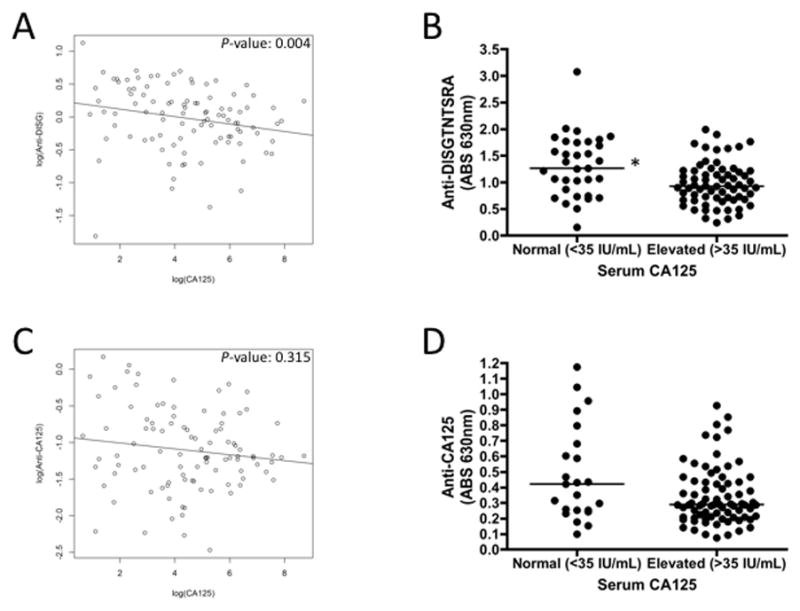
OvCa patient serum from time of diagnosis was assessed for serum CA125 and antibodies to DISGTNTSRA and purified CA125. Scatter plots of log(CA125) vs log(anti-DISGTNTSRA) (A) or log(anti-CA125) (C) are shown, with P-values determined for Spearman’s rank correlation. Patients with grouped by normal or elevated serum CA125 and anti-DISGTNTSRA (B) or anti-CA125 (D) was determined. The line is presented at the median for each group. * P-value = 0.0033 (Mann-Whitney).
Given the negative association between serum CA125 and anti-DISGTNTSRA in OvCa patients, we used an ELISA to test whether this was indicative of overall CA125 autoantibody responses in patients. We did not detect a statistically significant correlation between overall CA125 autoantibody responses and serum CA125 (Fig. 4C, Spearman’s rank correlation coefficient: −0.102, P-value: 0.315) and patients with elevated serum CA125 did not have significantly different CA125 autoantibody responses than patients with normal serum CA125 (Fig. 4D). However, using a cutoff value based on the median of the ELISA readings, we found that the proportion of patients who were positive for CA125 autoantibodies with normal serum CA125 (n = 20) were significantly different from those patients who were negative for CA125 autoantibodies with elevated serum CA125 (n = 38 ) (Supplemental Table 5, McNemar test, P-value: 0.026). These observations suggest that either the presence of free CA125 in sera is interfering with the detection of CA125-specific antibodies, or potentially that CA125-specific antibodies are protective against CA125+ OvCa.
Associations of antibodies to DISGTNTSRA and CA125 with survival outcomes
Low serum CA125 at time of diagnosis of OvCa is associated with a favorable outcome (9). Knowing that CA125 plasma and antibodies to CA125 or the DISGTNTSRA peptide mimic were negatively correlated, we next wanted to investigate whether there was a correlation between OvCa patient reactivity to CA125 or DISGTNTSRA and patient survival. We generated Kaplan-Meier survival curves for our patient samples by stratifying based on normal (< 35 IU/mL) or elevated (> 35 IU/mL) CA125 in plasma. We did not have survival data on all 99 patients, so our sample set was limited to only 60 patients. Similar to data previously shown with these samples (9), pre-operative elevated serum CA125 was associated with decreased survival in OvCa patients (Supplemental Fig. S2, Table 2). We also generated Kaplan-Meier survival curves by stratifying based on positive vs. negative CA125 autoantibodies and DISGTNTSRA antibodies. Although the median survival time of patients with CA125 autoantibodies was considerably longer than those without (positive: 104 months, negative: 56 months, P-value: 0.586), this difference was not statistically significant (Supplemental Fig. 2, Table 2). This was also the case for survival based on anti-DISGTNTSRA status (positive: 104 months, negative: 52 months, P-value: 0.244, Supplemental Fig. S2, Table 2). Since patient outcome is strongly associated with stage of disease at diagnosis, and we had very few Stage I/II samples, we also assessed survival for OvCa Stage III patients only (n = 51). OvCa Stage III patients also did not show a statistically significant difference in survival for either DISGTNTSRA or CA125 antibodies (Supplemental Fig. S2, Table 2, P-value: 0.385, P-value: 0.454 respectively). Given our sample sizes and the number of events, our statistical power is not sufficient to rule out an effect of anti-DISGTNTSRA or CA125 autoantibodies on patient survival.
Table 2.
Cox univariate analysis of OvCa patient survival
| Patients | Variable | Hazard Ratio | 95% CI | P-value (Wald) |
|---|---|---|---|---|
| Stages I/II and III | CA125 | 4.651 | (1.053, 20.539) | 0.043 |
| Anti-CA125 | 0.758 | (0.279, 2.055) | 0.586 | |
| Anti-DISGTNTSRA | 0.551 | (0.199, 1.521) | 0.250 | |
| Stage III | CA125 | 2.851 | (0.642, 12.657) | 0.168 |
| Anti-CA125 | 0.678 | (0.241, 1.910) | 0.462 | |
| Anti-DISGTNTSRA | 0.620 | (0.210, 1.831) | 0.387 | |
| Stages I/II and III | CA125 normal/anti-CA125 positive | 0.357 | (0.132, 0.965) | 0.042 |
| Stage III | CA125 normal/anti-CA125 positive | 0.461 | (0.163, 1.302) | 0.144 |
Normal serum CA125 (< 35 IU/mL) at time of diagnosis is a predictor of improved patient outcome (9). We asked whether adding anti-CA125 status would further improve the prediction of patient survival. To this purpose, we divided patients into two groups: (a) patients with normal CA125 and/or positive for CA125 antibodies (n = 38), and (b) patients with elevated CA125 but not positive for anti-CA125 (n = 21) (Fig. 5, Table 2). In analyses including all patients (Stage I/II, and III), but not Stage III patients alone, we saw a statistically significant difference in the survival curves of the two groups (Fig. 5, Table 2, P-value: 0.034). These data suggest that assessing patient antibodies to CA125 at the time of diagnosis may provide additional predictive value for survival.
Figure 5. Normal serum CA125 and/or anti-CA125 positive at time of diagnosis are correlated with increased OvCa patient survival.
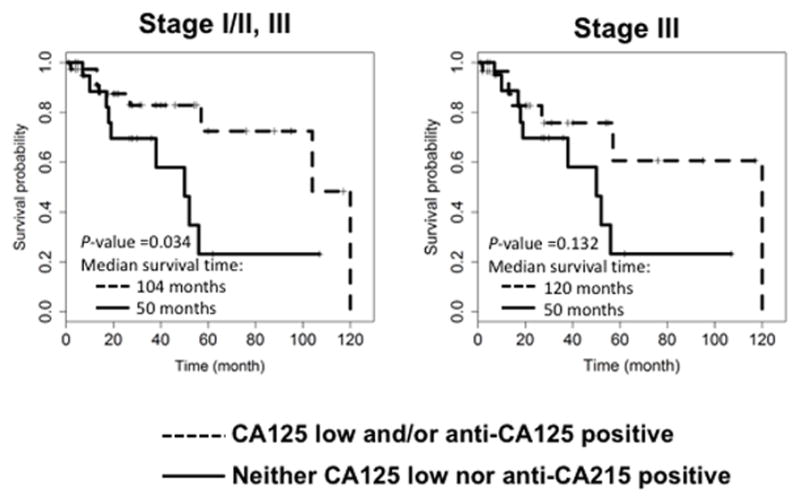
Patients with survival data (n = 9 for Stage I/II, n = 50 for Stage III) were divided into groups based on CA125 (normal <35 IU/mL) and/or anti-CA125 (median cutoff), or compared to remaining patients. Kaplan-Meier survival curves were generated using all patients or Stage III patients only. P-values are reported for Log-rank test and median survival time is indicated for each group.
DISCUSSION
Here, we report a novel application of the MS2-VLP affinity selection technology for the identification of candidate OvCa antigens. Coupling next-generation sequencing with iterative biopanning, we examined epitopes recognized by a complex mixture of antibodies in human plasma. We show the feasibility of using this approach for identifying anti-TAAs in OvCa and identified an interesting target using publicly available bioinformatics tools. We identified a peptide, DISGTNTSRA, which showed sequence similarity at a number of positions to CA125/MUC16, we confirmed that DISGTNTSRA is indeed a peptide mimic of CA125, and showed that reactivity of OvCa patient plasma to this peptide and CA125 was inversely correlated with CA125 plasma levels at time of diagnosis.
Affinity selection has been used to identify both linear and conformational epitopes of mAbs. However, its utility for comprehensively examining epitopes recognized by complex mixtures of antibodies (polyclonal serum) is technically limiting. Next-generation sequencing technologies can expand the capability of the traditional affinity selection approach (14–18). One other recent report has described the use of deep-sequence coupled affinity selection for interrogating the antibody response from polyclonal serum, using polyclonal serum from HIV-infected individuals in affinity selection with filamentous phage and Illumina sequencing (17). A similar approach was recently used by Larman et al., who performed iterative biopanning against filamentous phage libraries with cerebrospinal fluid-derived IgG and used Illumina sequencing to investigate the selectant populations (14). Interrogating polyclonal sera for antibody specificity using phage-display technologies was also recently reported by Xu et al., who used phage-displayed human virus-associated peptides to characterize the virus exposure history of humans with a very small volume of blood (19). Our approach is similar in that we can use phage VLPs to identify anti-TAAs from small volumes of sera. Our approach expands the use of deep sequence-coupled biopanning of phage-displayed peptides for the identification of TAAs in cancer.
There has been interest in therapeutically targeting CA125 in OvCa because (a) it is expressed in > 95% of all non-mucinous advanced stage epithelial OvCa, (b) it may contribute to evasion of anti-tumor immunity, and (c) it may be involved in seeding of the peritoneum and metastasis. Several mAbs targeting CA125 are being tested for therapeutic efficacy in patients, but, as observed with Oregovomab (a mAb that targets CA125), passive immunization may not be sufficient (20, 21). An active immunization approach against CA125 could be more potent. Our data suggest that patients who naturally develop an antibody response against CA125 have lower plasma CA125. This provides support for the immunogenicity of CA125 itself and the activity of these antibodies for decreasing serum CA125 levels.
A recent study shows significantly lower preoperative serum CA125 in OvCa patients with a history of puerperal mastitis, and significantly higher anti-CA125 antibodies in healthy controls (22). Limitations on the samples available in that study did not allow the authors to directly investigate the preoperative levels of anti-CA125 for OvCa patients who had a history of puerperal mastitis. The data we present here supports the hypothesis of the authors of that study, who suggest that key reproductive events (such as puerperal mastitis) may lower OvCa risk by inducing immune reactions to mucins (such as CA125/MUC16) and could be detected by assessing for elevated concentrations of antibodies to CA125. In our study, we show a statistically significant association between OvCa patients with normal or elevated serum CA125 and their autoantibodies to CA125, and an interesting trend toward better survival in patients with either low serum CA125 or anti-CA125 responses. This may indicate that anti-CA125 antibodies are directly protective against OvCa, or it may be a surrogate marker of an overall increased anti-tumor immune response. However, it would be important to rule out simple competition in the ELISA format by free CA125 in serum.
Several prior studies have detected CA125 autoantibodies in OvCa patients. Taylor and colleagues detected CA125 autoantibodies in advanced OvCa patients, but did not examine whether these antibodies were correlated with survival (23). Budiu et al. measured CA125 antibodies in a cohort of 28 OvCa patients that were treated intraperitoneally with IL-2 after completing chemotherapy, but neither CA125 nor CA125 autoantibodies were significantly associated with a clinical response to a platinum/taxane regimen or overall survival (24). Although we did not detect a statistically significant survival difference between patients based on their anti-DISGTNTSRA or CA125 autoantibodies status, the association between anti-DISGTNTSRA antibodies and serum CA125 suggest a possible mechanism for variation in CA125 levels among OvCa patients, whereby patients with CA125 autoantibodies have less serum CA125 due to antibody-mediated immune clearance of CA125, by direct killing of OvCa cells via antibody-dependent cytotoxicity and reduced tumor burden (or both). The status of anti-CA125 response in the patient may be of prognostic value when combined with CA125 status, as shown in Fig. 5. In our analysis we used the median of the ELISA values for anti-DISGTNTSRA and anti-CA125 in order to dichotomize into positive and negative patients for these antibodies. However, given the low statistical power of our analysis, future studies should investigate a larger set of samples in order to assess what if any values of anti-DISGTNTSRA and anti-CA125 have relevance for prognosis of OvCa patients.
Our data suggest that prophylactic anti-CA125 responses may be protective against OvCa. If patients with CA125 autoantibodies have a better prognosis, then it follows that eliciting similar antibodies through an active immunization strategy may provide an advantage for women at high risk of OvCa. Indeed, the reported lowered risk for OvCa in women with CA125 antibodies supports this hypothesis (22). MS2-DISGTNTSRA is just one candidate antigen that may elicit such prophylactic CA125 antibodies in vivo. Further, VLPs typically induce a more balanced response than the Th2-dominated response to alum-adjuvanted antigens, including Abagovomab, and this may be important for effective ADCC against OvCa.
In this report, we identified a unique epitope of CA125. The DISGTNTSRA peptide had multiple positions for possible alignment to the CA125/MUC16 protein (Supplemental Table 2 & Fig. 2). The DISGTNTSRA peptide does not appear to correspond with the binding site of any currently known anti-CA125 mAb (25). This suggests that the epitopes targeted by CA125 autoantibodies in OvCa patients could be very different from those elicited artificially in mice. Although we showed that MS2-DISGTNTSRA was able to elicit antibodies in mice that bind to purified CA125 by ELISA (Fig. 3), we were unable to show binding of these mouse sera to CA125 positive OvCa cancer cell line OVCAR3 by flow cytometry (data not shown). This result may have been due to low titers or avidity of the antibody in the mouse sera or it could be that the antibodies elicited by MS2-DISGTNTSRA recognize a buried epitope of CA125 presented on cells, e.g. by glycosylation. The structure CA125/MUC16 and the importance of glycosylation are poorly understood, including if there are distinct structural features of the membrane-bound vs. soluble CA125. It may be that the DISGTNTSRA epitope of CA125/MUC16 is only exposed on soluble CA125.
It is interesting to note that our identification of DISGTNTSRA seemed to be driven largely by a single OvCa patient (DM778) from the pool of 5 that were chosen. This sample showed high reactivity to DISGTNTSRA and CA125 and was the only sample included in our pool of 5 that had normal serum CA125, although not by design (Table 1). This indicates that, although we sought to limit the importance of intrasample variation by pooling samples for the deep sequence-coupled iterative biopanning protocol, individual samples can drive the outcome of this protocol and should be taken into account in future use of this technology.
In summary, we present here a novel deep sequence-coupled VLP-based affinity selection technology for identifying anti-TAA responses in a polyclonal serum sample. By coupling next-generation sequencing technology with the MS2-VLP affinity selection platform, we expand the opportunities for understanding the antibody response in cancer as well as infectious disease and autoimmunity. Additionally, we present data suggesting a role for CA125 antibody responses in OvCa patient outcome. This supports exploration of active immunization approaches targeting CA125 in OvCa patients, either in combination with current treatment strategies, prophylactically for women at high risk for OvCa, or to prevent or delay recurrence in the setting of minimal residual disease.
Supplementary Material
Acknowledgments
Financial Support: KMF was supported by a University of New Mexico Cancer Center Postdoctoral Matching Grant. This study was funded, in part, by NIH grant R01 AI083305 to BC.
We thank Jeremy Edwards, Norah Torrez-Martinez, and John O’Rourke for Ion Torrent Deep Sequencing technical advice and assistance.
Footnotes
Conflicts of Interest: BC and DSP have an equity interest in Agilvax, which has licensed the bacteriophage affinity selection platform used in this project. KF has served as a consultant for Aglivax.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59(4):225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Rosenthal AN, Jacobs IJ. The role of CA 125 in screening for ovarian cancer. Int J Biol Markers. 1998;13(4):216–20. doi: 10.1177/172460089801300408. [DOI] [PubMed] [Google Scholar]
- 3.Vaughan S, Coward JI, Bast RC, Jr, Berchuck A, Berek JS, Brenton JD, et al. Rethinking ovarian cancer: recommendations for improving outcomes. Nat Rev Cancer. 2011;11(10):719–25. doi: 10.1038/nrc3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatterjee M, Tainsky MA. Autoantibodies as biomarkers for ovarian cancer. Cancer Biomark. 2010;8(4–5):187–201. doi: 10.3233/CBM-2011-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodell V, Salazar LG, Urban N, Drescher CW, Gray H, Swensen RE, et al. Antibody immunity to the p53 oncogenic protein is a prognostic indicator in ovarian cancer. Journal of Clinical Oncology. 2006;24(5):762–8. doi: 10.1200/JCO.2005.03.2813. [DOI] [PubMed] [Google Scholar]
- 6.Gunawardana CG, Memari N, Diamandis EP. Identifying novel autoantibody signatures in ovarian cancer using high-density protein microarrays. Clin Biochem. 2009;42(4–5):426–9. doi: 10.1016/j.clinbiochem.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 7.Hudson ME, Pozdnyakova I, Haines K, Mor G, Snyder M. Identification of differentially expressed proteins in ovarian cancer using high-density protein microarrays. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(44):17494–9. doi: 10.1073/pnas.0708572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soussi T. p53 Antibodies in the sera of patients with various types of cancer: a review. Cancer research. 2000;60(7):1777–88. [PubMed] [Google Scholar]
- 9.Tsai-Turton M, Santillan A, Lu D, Bristow RE, Chan KC, Shih Ie M, et al. p53 autoantibodies, cytokine levels and ovarian carcinogenesis. Gynecologic oncology. 2009;114(1):12–7. doi: 10.1016/j.ygyno.2009.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peabody DS, Manifold-Wheeler B, Medford A, Jordan SK, do Carmo Caldeira J, Chackerian B. Immunogenic display of diverse peptides on virus-like particles of RNA phage MS2. J Mol Biol. 2008;380(1):252–63. doi: 10.1016/j.jmb.2008.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ord RL, Caldeira JC, Rodriguez M, Noe A, Chackerian B, Peabody DS, et al. A malaria vaccine candidate based on an epitope of the Plasmodium falciparum RH5 protein. Malaria journal. 2014;13:326. doi: 10.1186/1475-2875-13-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Rourke JP, Daly SM, Triplett KD, Peabody D, Chackerian B, Hall PR. Development of a mimotope vaccine targeting the Staphylococcus aureus quorum sensing pathway. PloS one. 2014;9(11):e111198. doi: 10.1371/journal.pone.0111198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chackerian B, do Caldeira JC, Peabody J, Peabody DS. Peptide epitope identification by affinity selection on bacteriophage MS2 virus-like particles. J Mol Biol. 2011;409(2):225–37. doi: 10.1016/j.jmb.2011.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larman HB, Zhao Z, Laserson U, Li MZ, Ciccia A, Gakidis MA, et al. Autoantigen discovery with a synthetic human peptidome. Nat Biotechnol. 2011;29(6):535–41. doi: 10.1038/nbt.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matochko WL, Chu K, Jin B, Lee SW, Whitesides GM, Derda R. Deep sequencing analysis of phage libraries using Illumina platform. Methods. 2012;58(1):47–55. doi: 10.1016/j.ymeth.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 16.Ravn U, Didelot G, Venet S, Ng KT, Gueneau F, Rousseau F, et al. Deep sequencing of phage display libraries to support antibody discovery. Methods. 2013;60(1):99–110. doi: 10.1016/j.ymeth.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Ryvkin A, Ashkenazy H, Smelyanski L, Kaplan G, Penn O, Weiss-Ottolenghi Y, et al. Deep Panning: steps towards probing the IgOme. PloS one. 2012;7(8):e41469. doi: 10.1371/journal.pone.0041469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.t Hoen PA, Jirka SM, Ten Broeke BR, Schultes EA, Aguilera B, Pang KH, et al. Phage display screening without repetitious selection rounds. Anal Biochem. 2011;421(2):622–31. doi: 10.1016/j.ab.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 19.Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, Petalcorin MI, et al. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature. 2015;518(7538):258–62. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berek JS, Taylor PT, Gordon A, Cunningham MJ, Finkler N, Orr J, Jr, et al. Randomized, placebo-controlled study of oregovomab for consolidation of clinical remission in patients with advanced ovarian cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2004;22(17):3507–16. doi: 10.1200/JCO.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 21.Berek J, Taylor P, McGuire W, Smith LM, Schultes B, Nicodemus CF. Oregovomab maintenance monoimmunotherapy does not improve outcomes in advanced ovarian cancer. Journal of Clinical Oncology. 2009;27(3):418–25. doi: 10.1200/JCO.2008.17.8400. [DOI] [PubMed] [Google Scholar]
- 22.Cramer DW, Williams K, Vitonis AF, Yamamoto HS, Stuebe A, Welch WR, et al. Puerperal mastitis: a reproductive event of importance affecting anti-mucin antibody levels and ovarian cancer risk. Cancer causes & control : CCC. 2013;24(11):1911–23. doi: 10.1007/s10552-013-0266-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor DD, Gercel-Taylor C, Parker LP. Patient-derived tumor-reactive antibodies as diagnostic markers for ovarian cancer. Gynecologic oncology. 2009;115(1):112–20. doi: 10.1016/j.ygyno.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 24.Budiu RA, Mantia-Smaldone G, Elishaev E, Chu T, Thaller J, McCabe K, et al. Soluble MUC1 and serum MUC1-specific antibodies are potential prognostic biomarkers for platinum-resistant ovarian cancer. Cancer Immunology, Immunotherapy : CII. 2011;60(7):975–84. doi: 10.1007/s00262-011-1010-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcos-Silva L, Narimatsu Y, Halim A, Campos D, Yang Z, Tarp MA, et al. Characterization of binding epitopes of CA125 monoclonal antibodies. Journal of Proteome Research. 2014;13(7):3349–59. doi: 10.1021/pr500215g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


