Abstract
Molecular machines have previously been designed that are propelled by DNAzymes1–3, protein enzymes4–6 and strand-displacement7–9. These engineered machines typically move along precisely defined one- and two-dimensional tracks. Here, we report a DNA walker that uses hybridisation to drive walking on DNA-coated microparticle surfaces. Through purely DNA:DNA hybridisation reactions, the nanoscale movements of the walker can lead to the generation of a single-stranded product and the subsequent immobilisation of fluorescent labels on the microparticle surface. This suggests that the system could be of use in analytical and diagnostic applications, similar to how strand exchange reactions in solution have been used for transducing and quantifying signals from isothermal molecular amplification assays10,11. The walking behaviour is robust and the walker can take more than 30 continuous steps. The traversal of an unprogrammed, inhomogeneous surface is also due entirely to autonomous decisions made by the walker, behaviour analogous to amorphous chemical reaction network computations12,13 that have been shown to lead to pattern formation14–17.
We have adapted a simple nucleic acid circuit, catalytic hairpin assembly (CHA)7,18 to a microparticle surface as a basis for developing a novel DNA walker that does not covalently alter its substrates (unlike a DNAzyme-based DNA walker that cleaves substrates as it walks) (Fig. 1a). This CHA circuit was originally developed by Pierce and Yin and has proven to be extraordinarily versatile7,18. In CHA, two hairpins are kinetically trapped, but in the presence of a single-stranded catalyst can undergo strand exchange reactions that lead to the formation of a double-stranded nucleic acid and the recycling of the catalyst. In greater detail, a linear single-stranded DNA catalyst can interact with a toehold on surface-bound H1 and open the hairpin via toehold-mediated strand exchange. A newly exposed ssDNA region within H1 can then hybridise to a toehold on H2 and trigger branch migration, ultimately forming a tripartite complex between H1, H2, and the catalyst. As strand displacement proceeds, this complex will resolve into the most thermodynamically favourable configuration, the H1:H2 duplex, with displacement of the free catalyst, which can then participate in subsequent reaction cycles. The formation of the duplex reaction product potentially avoids the necessity of using DNAzymes1–3, protein enzymes4–6, or a chemical fuel such as Hg2+/cysteine19 to drive the movement of the walker. The DNA walker traversed the irregular surface of a microparticle (see SEM image, Supplementary Fig. 1). By using a colloidal substrate rather than a more defined or confined 1D or 2D track, such as DNA origami2,5,6, we open the way to a variety of useful applications, including microparticle-based signal amplification. Microscopy experiments also show that our walker can travel between individual particles within 3D clusters when microparticles are in close contact.
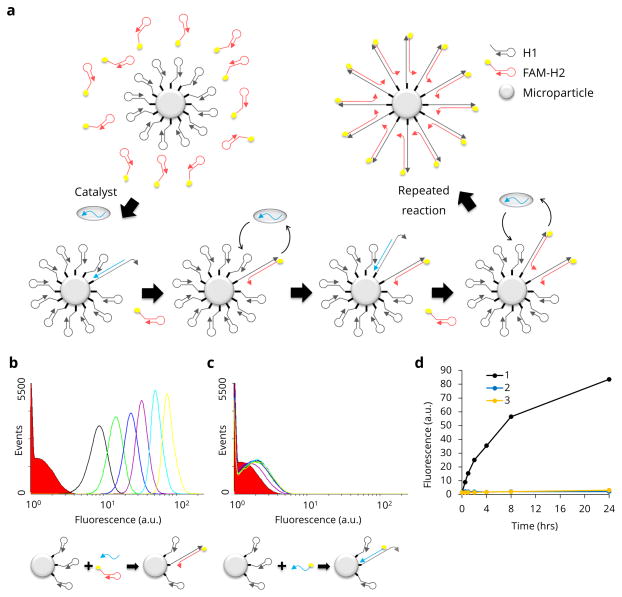
Figure 1. Schematic and proof-of-concept for CHA on microparticles.
a, A schematic of the CHA reaction on the microparticle surface shows how fluorescent FAM-H2 (red hairpins) are catalytically hybridised to the surface-immobilised H1 (gray hairpins). b, Flow cytometry data of the positive control including FAM-H2 and catalyst (catalytic reaction) show the change in microparticles’ fluorescence over time (red solid: 0 hr, black line: 0.5 hr, green line: 1 hr, blue line: 2 hrs, purple line: 4 hrs, cyan line: 8 hrs and yellow line: 24 hrs). c, Flow cytometry data of the negative control without H2 (non-catalytic reaction) show the low level of immobilised fluorescence on H1-particles incubated only with a fluorescent catalyst. d, Median fluorescence values were extracted from the flow cytometry data and plotted over time (black: catalytic, blue: non-catalytic, yellow: no-catalyst control).
We initially performed a feasibility test to determine whether CHA could proceed on microparticle surfaces using a reliable CHA reaction system described in an earlier work18. H1-containing microparticles were mixed with a H2 molecule derivatised with fluorescein (H2-FAM) and the catalyst. A successful reaction should result in the capture of H2-FAM on the microparticle surface, and thus to increasing microparticle fluorescence (Fig. 1b). A control, non-catalytic reaction was prepared with H1-microparticles and FAM-catalyst. In this instance, only simple hybridisation should occur, without turnover (Fig. 1c). A no-catalyst control was also performed and is shown in Fig. 1d. In the CHA reaction the fluorescence intensity increased by a factor of 126 relative to the non-catalytic reaction, where there was little reaction even after 24 hours (Fig. 1b–d and Supplementary Section 1.).
Since the CHA reaction was initially performed in a standard CHA buffer7,18 that contained only 140 mM NaCl it was possible that walking might have been limited by charge repulsion. We attempted to optimise the concentration of NaCl to improve the CHA reaction on the microparticle surface (Supplementary Fig. 2a), and based on the signal-to-background ratio 300 mM NaCl was chosen as the optimal reaction condition for further experiments (Supplementary Fig. 2b). Supplementary Fig. 2 also shows that the CHA reaction on the microparticle surface is incredibly robust across a range of reaction conditions, and can be reliably measured by flow cytometry.
Given that surface-based reactions were robust, we hypothesised that it might be possible to catalyse proximal hairpin assembly reactions by combining two catalytic domains in a single walker. A similar walker system was demonstrated by Pierce and Yin on a 1D DNA track that performed three walking steps7. To the extent that one ‘leg’ of the catalyst is freed from a surface-bound duplex before the other, that ‘leg’ may be able to ‘walk’ and begin the reaction cycle with a new hairpin on the surface. Multiple cycles of catalysis would overall lead to the localised generation of double-stranded DNA, and the concomitant immobilisation of fluorophores. To prove that the double-catalyst was walking, we monitored both the double-catalyst’s locality (to its track over a population of particles) and its processivity (via the number of fluorophores that became bound to these particles).
The double-catalyst was simple but elegant in design, being comprised of a longer single-stranded DNA oligonucleotide that contained two catalyst domains separated by a linear, non-hybridising spacer sequence. We anticipated that the double-catalyst would persist on the surface of a single microparticle for a longer period of time than the single-catalyst. While the single-catalyst should be released from the H1-microparticle after only one turnover of the catalytic cycle, the double-catalyst walker should undergo multiple cycles of surface-bound catalysis.
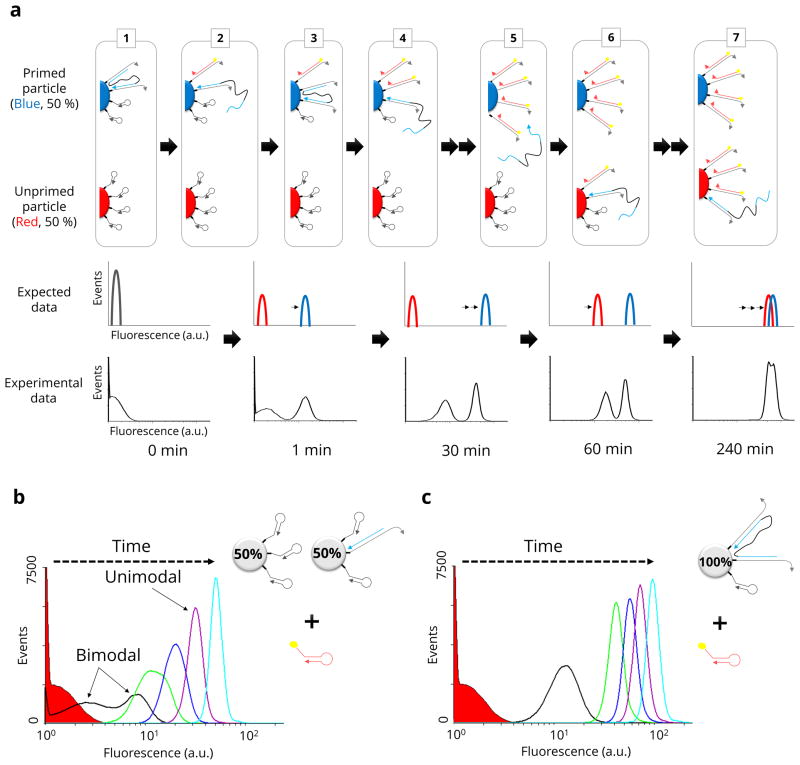
To confirm the general prediction that double-catalysts walk on a surface, two different types of microparticles were prepared: H1-microparticles and H1-microparticles primed or pre-hybridised with catalysts. About 1% of the available H1 sites were initially occupied by catalyst (Supplementary Fig. 3 and 4). Primed and unprimed microparticles were mixed in a 50%:50% ratio, FAM-H2 was added, and flow cytometry was performed at different time points. If the double-catalyst was walking the surface of the microparticle, then the sub-population of primed microparticles should ‘light up’ relative to non-primed microparticles and relative to microparticles mixed with single-catalysts. In fact, a bimodal distribution of microparticle fluorescence was observed that persisted for several hours (compare Fig. 2a and 2b). While single-catalysts initially yielded a bimodal distribution, this rapidly devolved to a unimodal distribution within only 30 minutes. We also performed a control experiment using a homogeneous suspension of microparticles that were all primed with the double-catalyst (rather than 1:1 mixed) (Fig. 2c). As expected, this experiment showed a unimodal distribution over time.
Figure 2. Double-catalyst walking behaviour detected with flow cytometry.
a, A schematic of the double-catalyst reaction shows how the walking behaviour was detected with flow cytometry as a bimodal distribution of fluorescence values. In the initial state (step 1) a population (blue) of H1-particles is primed with double-catalyst; a second population of H1-particles in the same suspension is unprimed (red). In subsequent steps 2–7, the two populations were predicted to evolve in time as indicated (expected data). Experimental flow cytometry data are shown below this qualitative prediction. b, Flow cytometry data generated with a single-domain catalyst (non-walking control) initially shows a bimodal distribution but this degenerates to a unimodal distribution within 30 min (green histogram). c, Flow cytometry data of a single population of microparticles primed with double-catalyst (walking control) shows a unimodal distribution over time (red solid: 0 min (without catalyst), black line: 1 min, green line: 30 min, blue line: 60 min, purple line: 120 min, cyan line: 240 min).
To further prove that walking was the mechanism underlying the bimodal distribution of fluorescence, we created different fractions of primed and unprimed microparticles (100%:0%, 80%:20% and 20%:80%). The greater the fraction of primed microparticles, the greater the fraction of fluorescent microparticles that should be observed, since the walkers would more quickly light up the primed microparticles relative to the unprimed. In fact, the greater fraction of primed microparticles eventually gave greater fractions with brighter signals (Supplementary Fig. 5). Conversely, we demonstrated the difference between walking by the single- and double-catalyst by introducing “trap-microparticles” that would bind any released catalyst. When primed, H1-coated microparticles were mixed with trap-microparticles at a 1:2 ratio and we observed that the activity of the single-catalyst was markedly reduced, but that the double-catalyst remained highly active (Supplementary Fig. 6).
Other parameters can potentially be altered to further change the functionality of the walkers. There was no effect of spacer length (28 bp, 41 bp and 53 bp) on walking, likely because the density of hairpins on the microparticle (estimated to be 3.6 molecules per 10 nm2 based on information provided by the manufacturer) should have been close enough that any length would work well (Supplementary Fig. 7). Previous experiments with ‘spiders’ that cleaved their DNA substrates as they walked had shown the utility of having multiple legs that remained attached to the surface at any one time2. Therefore, we similarly used avidin to simultaneously immobilise four catalyst legs. The tetra-catalyst indeed appears to have higher adherence to microparticles (Supplementary Fig. 8a). Overall, these results also pave the way to analytical applications in which the walker can be used to detect molecular events that initiate on individual microparticles.
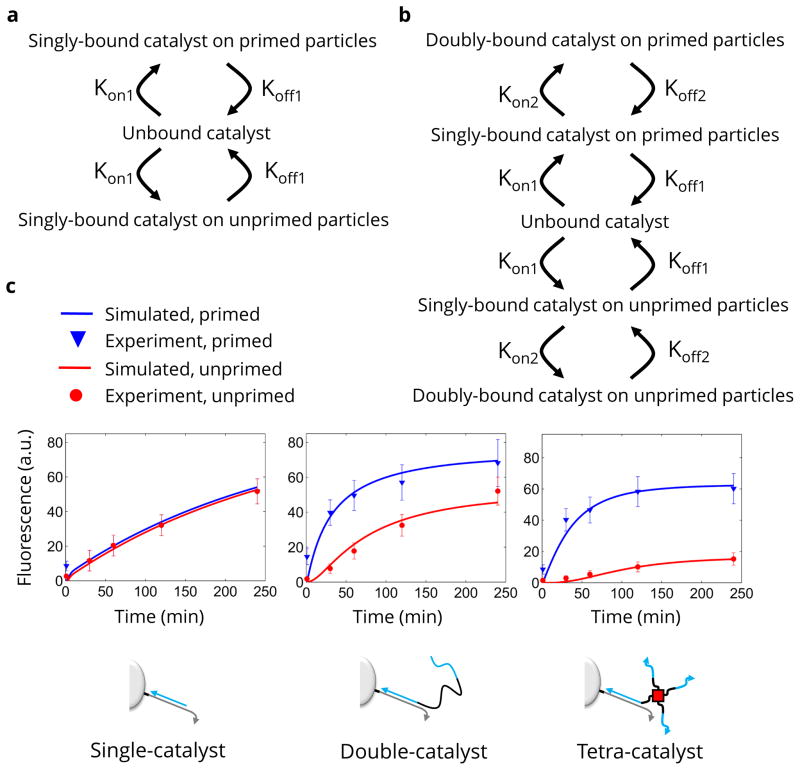
A mathematical model that fit the distribution of catalysts on the microparticles was used to determine the average number of steps ‘walked’ by different catalysts. The basic form of the model used for single-catalysts is shown in Fig. 3a. The single-catalyst can exist in one of three states: 1) bound to the primed microparticles (initially all catalysts exist in this state); 2) unbound from any microparticle (taken to exist in well mixed solution around the microparticles); or 3) bound to the unprimed microparticles. Transition from bound to unbound is governed by the Koff1 rate constant and the concentration of bound catalyst. Transition from unbound to bound is governed by the Kon1 rate constant, unbound catalyst concentration, and uncatalysed H1 concentration (remaining fuel on the microparticle). In conventional flow cytometry assays, cells are often found to rebind antibody labels after dissociation20. Our assumption that the solution was well-mixed (no preference for binding to the particle of origin) is reasonable based on our single-catalyst results. The single-catalyst equilibrated across all particles in the system within minutes in the cytometry experiments. In our microscopy experiments (below) the single-catalyst appeared to diffuse away without significant rebinding. There is also a global loss rate that accounts for catalyst that may become stuck to the microparticle surface or tube wall. The model for the double-catalyst was an expanded version of the single-catalyst model (Fig. 3b) that also included separate rate coefficients for transitions between singly-bound and doubly-bound states (Kon2 and Koff2). In all cases, catalyst release was assumed to take place when H1 and H2 bound and produced a fluorescent signal. The sum of all catalyst release over time was taken to be the fluorescence observed on the microparticles. We optimised the rate constants in the model in order to fit the model’s behaviour to the experimentally observed behaviour (Fig. 3c).
Figure 3. Interpretation of walking behaviour with numerical simulation.
a, A schematic shows how the single-catalyst simulation considers catalyst molecules to be in one of three states. The transitions between these states are indicated by arrows. b, A schematic of the double-catalyst simulation shows how a second, doubly-bound state is added for primed and unprimed particles. A separate rate constant is assumed for the transition between singly-bound and doubly-bound catalyst. c, The median experimental microparticle fluorescence values (discrete points with error bars) are shown as an overlay with the simulation results using the best-fit parameters for the rate constants (solid lines).
The parameters derived from the experimental data and ODE model can be used to generate a single-molecule Monte Carlo model for how many steps the catalyst takes on average. Monte Carlo analysis based on all extracted rate constants (including the rate of irreversible binding) indicated that the double-catalyst took an average of 10 steps before being released or becoming irreversibly bound. The tetra-catalyst took 5 steps. The number of steps taken in the Monte Carlo simulation was primarily limited by the rate of irreversible binding to the particle or tube wall. When the kinetic rate constant for irreversible binding was set to zero (i.e., simulating an ideal case without nonspecific binding), Monte Carlo analysis indicated that the double-catalyst would take 48 steps and the tetra-catalyst 49 steps.
In order to confirm these predictions, we designed an experimental measure of the number of steps. In this experiment catalysts were deactivated after release. One population of particles was coated with H1 and primed with a catalyst. A second population of particles was designed to trap any catalysts that were released (Supplementary Fig. 6). When the trap-particle experiment was compared with appropriate controls an experimental estimate of the number of steps walked could be made (see Supplementary Fig. 9). The final fluorescence values in this experiment indicated that the double-catalyst took 36 steps. The difference of 12 steps between experiment (36 steps) and the ideal theoretical case (48 steps) can be accounted for by a small amount of irreversible binding.
In addition, we performed experiments to determine the effect of temperature on the processivity of our walker (Supplementary Fig. 9). We carried out the trap-particle experiment as described in Supplementary Fig. 8, but measured the fluorescence increase in a temperature controlled real-time PCR instrument, rather than in a plate reader. At 37 °C the performance of the walker (i.e. number of steps taken in the first 10 min and the overall signal-to-background ratio, SBR) was comparable to the experiment performed in the plate reader. Overall, the reaction seems to be quite tolerant of temperatures between 25 °C and 37 °C, although with different processivities. Over the initial 10 minutes of the reaction the walker takes 12 and 26 steps, respectively. In contrast, as the temperature was increased to near the melting point of the hairpin (50 °C) background reactions begin to occur even in the absence of catalyst. At 50 °C the catalyst took 15 steps but the SBR was significantly lower (10) than at 25 °C or 37 °C (16 and 23, respectively).
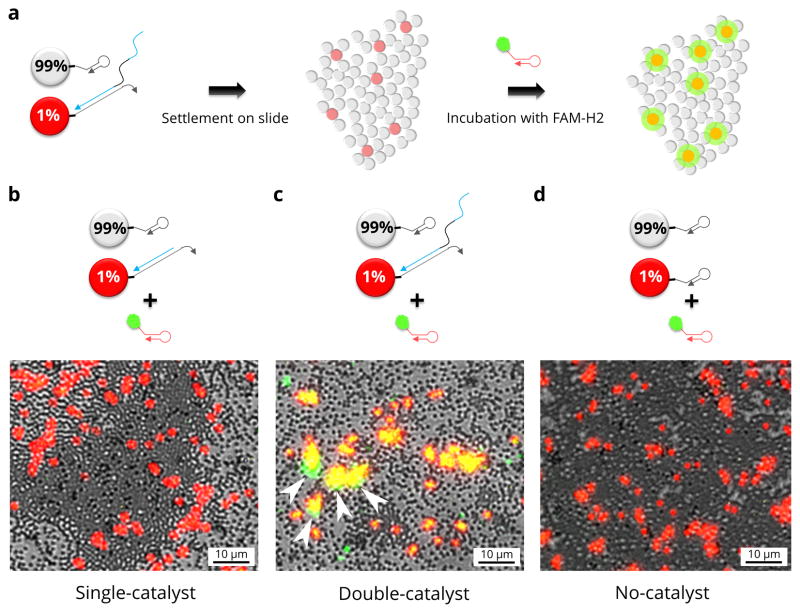
As with other undirected DNA walkers, it is anticipated that the CHA walkers should remain in a local area. To visually demonstrate that the walkers remain bound to their tracks, we performed fluorescence microscopy on a lawn of primed and unprimed microparticles. Microparticles bearing H1 were distributed onto the surface of a well plate (see experimental outline Fig. 4a). The lawn of microparticles included a small number (1%) of red fluorescent, catalyst-primed microparticles that were distributed randomly among the unprimed, clear microparticles. Walking was initiated by adding the green-fluorescent hairpin, FAM-H2, in solution. Background FAM-H2 was washed away and fluorescence visualised by microscopy. As expected, red microparticles initially primed with single- or double-catalyst became fluorescent. What was interesting to observe, though, was what happened to adjacent microparticles. For single-domain catalysts no adjacent microparticles lit up (Fig. 4b). This shows that the no significant amount of catalyst could transfer from the (red) particles primed with single-catalyst to adjacent (clear) unprimed particles. However, for double-catalysts (the walker) there were multiple green microparticles adjacent to the red microparticles, indicating that the walkers had crossed between the microparticles, but only in the local, 3D landscape (Fig. 4c). Arrows show regions where clear particles have taken on green fluorescence. Co-localised green and red fluorescence is expected (yellow in the false-coloured micrographs). Green colour outside of the yellow regions indicates that double-catalysts have transferred away from their red microparticles of origin. This appears in the false-coloured micrographs as green colour outside of the yellow regions (red, primed particles that also develop green fluorescence). There would seem to be two most likely explanations: first, the double-catalysts may walk from primed to unprimed particles via the ‘valleys’ between particles, and second the walkers may release from primed particles and rebind to adjacent particles more efficiently than do single-catalysts. While we cannot eliminate either of these mechanisms, it is nonetheless clear that the walkers can immobilise fluorophores on unprimed particles in close proximity to primed particles: that is, they either walk directly or make very short ‘jumps’ more efficiently than do single-catalysts. In the control case, red microparticles not primed with catalyst remained non-fluorescent in the green channel (Fig. 4d). Magnified and component colour data are included in Supplementary Fig. S11 and S12, respectively. Arrows in Figure 4 and Supplementary Fig. S11 highlight the clearest regions of green fluorescence that extend beyond the red fluorescent primed particles. These green locations are due to fluorescent product that has been immobilised on unprimed particles. Since our quantitative model indicated that only 1% of the double-catalyst was released from the primed particles in the short incubation allowed, and since the comparison to the single-catalyst control suggests that rebinding is inefficient (most single-catalyst was rapidly lost to the bulk solution), we hypothesise that walking between particles in contact is a likely mechanism for green cluster formation.
Figure 4. Fluorescence microscopy of CHA on microparticles.
a, A schematic outline shows how we performed microscopy experiments. b, False colour fluorescence micrograph shows brightfield (gray) of a lawn of microparticles; red fluorescence (red) indicates the microparticles that were primed with single-catalyst; green fluorescence (green) shows the location of fluorescein modified H2 immobilised by CHA. Arrows indicate regions where green fluorescence clearly extends outside of primed particles (red fluorescence). c, False colour fluorescence micrograph shows the results when red microparticles were primed with double-catalyst. d, False colour micrograph of the results when red microparticles were not primed with catalyst.
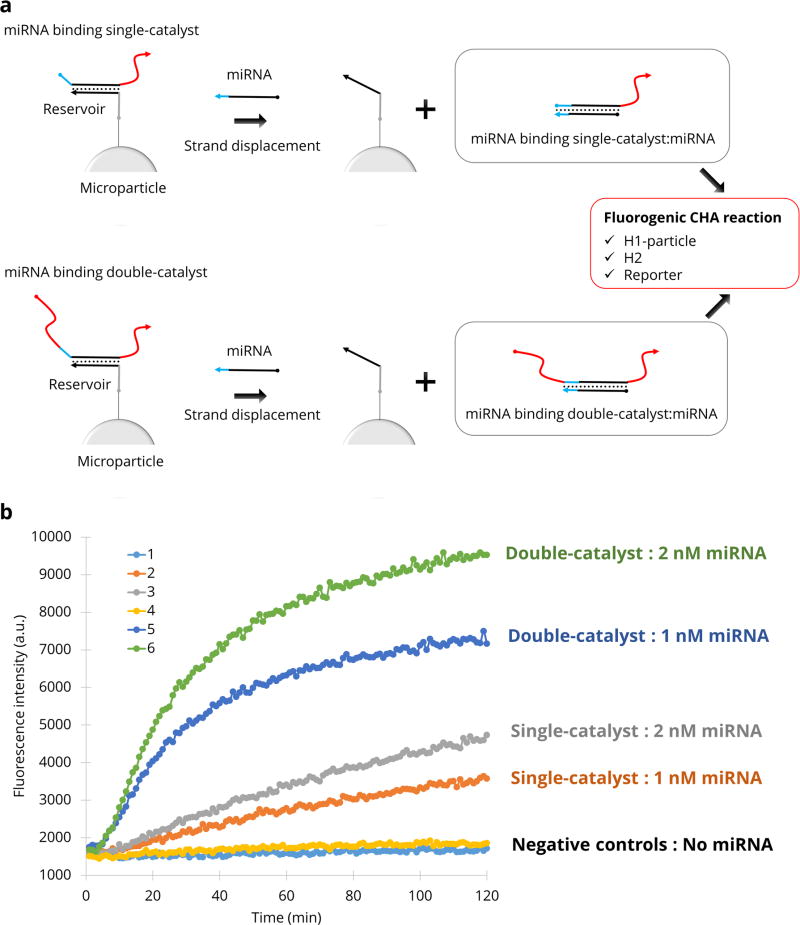
In order to apply this technology to an analytical target, we employed a microRNA (miRNA 122) to displace the double-catalyst. The microRNA releases the catalyst molecule from “reservoir-particles” in the reaction buffer. The free catalyst can then serve as an amplifier of the initial signal. Following addition of the target, reservoir-particles are centrifuged and the supernatant is applied to the reaction mixture including the H1-particles, H2, and reporter complex in free solution (Figure 5a). As before, the presence of walkers immobilises fluorophores on the surface of particles. Figure 5b shows that the performance of this analytical system is improved by using a displaced double-catalyst (walker) rather than a displaced single-catalyst. To demonstrate that such displacement can potentially be used to quantitate analytes, we added 2 nM of miRNA 122 to the reservoir particles, which should displace 2 nM of double-catalyst. The response was in fact comparable to our previous experiment where 2 nM of double-catalyst was added directly (Supplementary Fig. 13).
Figure 5. Amplification of miRNA detection using a walker.
a, A miRNA displaces either a single- or double-catalyst hybridised to reservoir-microparticles. b, Fluorescence accumulation is measured as a function of time for single-catalysts (line 1: no miRNA, line 2: 1 nM miRNA, line 3: 2 nM miRNA) and double-catalysts (line 4: no miRNA, line 5: 1 nM miRNA, line 6: 2 nM miRNA).
We have shown that a catalytic DNA molecule can locally traverse a microparticle surface on which it is immobilised. The modelled behaviour of the double-catalyst walker matched the experimentally observed behaviour, and suggested that the walker could take up to 36 steps before dissociating. This compares well with other published nanorobot demonstrations listed in Supplementary Table 2.
While molecular walkers have been demonstrated these walkers usually navigate very precise tracks, such as long DNA molecules or DNA origami-based tiles. A walker that uses an unprogrammed track is uniquely scalable. Microparticles or other surfaces can be coated randomly and in bulk with DNA by many methods21, and are inherently more scalable than attempting to fabricate all components of a track using molecularly-precise building blocks. The fidelity of the double-catalyst system is such that it is more likely to run out of uncatalysed H1 and be released than it is to be randomly released. Thus, it may be possible to attempt larger and larger surfaces, as we have currently shown by merely aggregating microparticles with substrates to create ‘valleys’ and ‘hills’ for the walkers to traverse.
A walker that can move extensively over microparticle surfaces opens many new application areas for DNA nanotechnology. For instance, digital PCR uses micro-scale colloidal droplets to restrict the volume of a PCR reaction. The product of the catalyst is activated (by revealing a new toehold) rather than cleaved (as in a DNAzyme-based walker). The newly revealed toehold potentially allows for integration with downstream computations and fluorescence-based detection schemes or other DNA circuits. For example, the double-catalyst walker could in principle be used to also count the number of amplicons on the surface, potentially melding digital and analogue technologies. This could prove to be especially useful if combined with devices that can monitor microparticles, such as flow cytometers. For example, Luminex XMAP microparticles are encoded for their specific analyte identity by far-red fluorescence so that ‘addressing’ and analytes are detected simultaneously by green fluorescence22. CHA walkers would be initiated by programmed interactions with nucleic acid analytes and leave behind trails of green fluors. The walkers might also be used as mobile inputs and outputs for more complex strand exchange circuits23,24. Finally, our results have implications for biomaterials development, since circuits such as CHA can be implemented on the surfaces of colloidal microparticles25,26. By activating H1-like substrates to interact with their hairpin partners not in solution but on other particles, nucleic acid walkers could selectively assemble particles into programmed amalgams27.
Methods
Reagents
All chemicals were of analytical grade and were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise indicated. All oligonucleotides were ordered from Integrated DNA Technology (IDT, Coralville, IA) and the sequences are summarised in Supplementary Table 1. Biotin-H1 and FAM-H2 were purified via denaturing PAGE (7 M Urea, 1X TBE). Streptavidin- and carboxylate- coated microparticles (1 μm) were purchased from Bangs Laboratories (Fishers, IN).
Preparation of biotin-H1, FAM-H2, H1-microparticles and catalyst-H1-microparticles
Purified biotin-H1 and FAM-H2 at a concentration of 20 μM in TNaK buffer (20 mM Tris, 140 mM NaCl, 5 mM KCl, pH 7.5) were refolded by heating to 95°C for 5 min followed by slowly decreasing the temperature to 23°C at a rate of 0.1°C/second.
50 μl of streptavidin coated microparticles from stock solution was washed two times with 50 μl of binding buffer (20 mM Tris, 1 M NaCl, 1 mM EDTA, 0.0005% Triton™ X-100, pH 7.5) by centrifuging the microparticles at 10K rpm for 3 minutes followed by decanting the supernatant. After resuspension with 50 μl of binding buffer, 5 μl of H1 of 20 μM in TNaK buffer (20 mM Tris, 140 mM NaCl, 5 mM KCl, pH 7.5) was added into the microparticles. After incubation for 15 minutes at room temperature on a rotator, the sample was washed two times with 50 μl of TNaKT buffer with one of 4 different NaCl concentrations (20 mM Tris, 140/300/500/1000 mM NaCl, 5 mM KCl, 0.01% Triton X-100, pH 7.5) by centrifuging the microparticles at 10K rpm for 3 minutes followed by resuspending in 50 μl of TNaKT buffer.
5 μl of the suspension of prepared H1-microparticles were incubated without any catalyst and with single-catalyst (final concentration: 5 nM) and double-catalyst (final concentration: 2.5 nM) in a total of 50 μl TNaKT buffer with 300 mM NaCl. The samples were incubated at 37°C for 1 hour followed by washing with TNaKT buffer with 300 mM NaCl.
Catalytic hairpin assembly reaction on microparticle surface
For the feasibility test and optimisation of NaCl concentration for CHA on the surface of microparticles, 5 μl of the prepared H1-microparticles was mixed with FAM-H2 (final concentration: 400 nM) in TNaKT buffer with different NaCl concentrations (140, 300, 500 and 1000 mM). Then the catalyst was added into the mixture to make total 50 μl, followed by incubation for the appropriate time at 37°C. Catalyst was omitted in the 0 hr incubation case (for this and in all further experiments).
For monitoring the behaviour of single- and double-catalyst on microparticle surface, unprimed H1-microparticles and catalyst primed (single/double catalyst)-H1-microparticles were mixed in different ratios (0%:100%, 20%:80%, 50%:50%, 80%:20% and 100%:0%). Then, FAM-H2 (final concentration: 400 nM) was added to a total of 50 μl, followed by incubation at 37°C for the appropriate time.
Evaluation of microparticle-immobilised CHA by flow cytometry
Reactions were prepared in advance using a mixture of catalyst-primed and unprimed microparticles. Fluorescein-linked H2 (FAM-H2, 400 nM) was added to each sample of the H1-microparticle suspension. This was incubated for the appropriate amount of time. These microparticles were then washed three times with TNaKT with 300 mM NaCl to stop the reaction. The quantity of immobilised fluorescein-modified DNA (FAM-H2 hybridised to immobilised H1) was measured using flow cytometry. Flow cytometry was performed using a FACSCalibur (BD Biosciences, San Jose, CA) using laser excitation at 488 nm and emission at 530 nm.
Modelling of surface immobilised CHA
Modelling was performed in MATLAB. A multi-state model was constructed using partial differential equations (PDE) which were then solved using a simple Euler method solver. Several parameters were predetermined using experimental data. The maximum fluorescence of the microparticles was estimated by using the fluorescence at 24 hours (the model ran for only 240 minutes). The initial quantity of catalyst linked to the primed microparticles was estimated by reference to experiment (see Supplementary Section 2. and Supplementary Fig. 2). Parameters in the model for association rate and dissociation rate of the catalyst (or double-catalyst) were fit to the experimental data using the MATLAB fminsearch optimisation routine.
Fluorescence microscopy imaging of the CHA reaction on microparticle surface
Two varieties of microparticles were coated with H1 using the carboxylate/EDC/amine-DNA reaction as per manufacturer’s instructions (Bangs Technical Note 205). Both types of microparticles were carboxylate coated, 1 μm in diameter. The first variety was red-fluorescent whereas the second variety was non-dyed. The red-fluorescent microparticles were treated with the appropriate catalyst (1 μM in PBS for 30 min) then washed with PBS by centrifugation and resuspension. Red-fluorescent, primed H1-microparticles were mixed with a 100 fold excess of the undyed, unprimed H1-microparticles. This mixture of microparticles was then added to a coverslip-bottom well slide and allowed to settle overnight at 4°C. Once in contact with the floor of the well, the microparticles were largely immobile. The microparticles were then washed by perfusion four times in order to remove any mobile microparticles. FAM-H2 was added in TNaK and allowed to react for one minute. Microparticles were then washed three times with TNaK to remove any unbound FAM-H2. The resulting fluorescent microparticles were imaged via a standard epi-fluorescence microscope (IX-81, Olympus, Center Valley, PA).
miRNA detection using a fluorogenic CHA reaction
A fluorogenic CHA reaction was performed to detect miRNA (Supplementary Fig. 6a). Extended H1-microparticles and reservoir-particles were prepared by the same process as the H1-particles mentioned above. An amount of 2.5 μM stock of reporter-F:reporter-Q complex was prepared by annealing 2.5 μM reporter-F and 5 μM reporter-Q in 1× TNaK buffer (20 mM Tris, 140 mM NaCl, 5 mM KCl, pH 7.5). An excess of reporter-Q ensures efficient quenching of reporter-F but does not interfere with the readout of an extended H1:H2.
The single- and double-catalysts were released from reservoir-particles by a specific RNA, a synthetic RNA oligonucleotide with the sequence of miRNA 122 acquired from IDT (Coralville, IA). Reservoir particles were incubated with miRNA 122 at 25°C for 1 hour. The samples were centrifuged and the supernatants were transferred to a new reaction tube including extended H1-microparticles, H2 (final concentration: 400 nM), and reporter complex (final concentration: 50 nM). Then a fluorogenic CHA reaction was performed and the fluorescence intensities were monitored at 37°C for 2 hours using a fluorescence plate reader (M200, Tecan, Männedorf, Switzerland).
Supplementary Material
Acknowledgments
This work was funded by the National Institutes of Health (EUREKA, 1-R01-GM094933), The Welch Foundation (F-1654), and a National Security Science and Engineering Faculty Fellowship (FA9550-10-1-0169). We also gratefully acknowledge Darko Stefanovic for his helpful discussion of the modelling system.
Footnotes
Author contributions
C.J. and P.B.A. conceived the walker scheme. C.J. carried out most of experiments. P.B.A. carried out a fluorescence microscopy experiment and performed modelling with analysis. A.D.E. supervised the project. C.J., P.B.A. and A.D.E. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Yin P, Yan H, Daniell XG, Turberfield AJ, Reif JH. A Unidirectional DNA Walker That Moves Autonomously along a Track. Angew Chem Int Ed. 2004;43:4906–4911. doi: 10.1002/anie.200460522. [DOI] [PubMed] [Google Scholar]
- 2.Lund K, et al. Molecular robots guided by prescriptive landscapes. Nature. 2010;465:206–210. doi: 10.1038/nature09012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cha TG, et al. A synthetic DNA motor that transports nanoparticles along carbon nanotubes. Nat Nanotechnol. 2014;9:39–43. doi: 10.1038/nnano.2013.257. [DOI] [PubMed] [Google Scholar]
- 4.Bath J, Green SJ, Turberfield AJ. A Free-Running DNA Motor Powered by a Nicking Enzyme. Angew Chem. 2005;117:4432–4435. doi: 10.1002/anie.200501262. [DOI] [PubMed] [Google Scholar]
- 5.Wickham SFJ, et al. Direct observation of stepwise movement of a synthetic molecular transporter. Nat Nanotechnol. 2011;6:166–169. doi: 10.1038/nnano.2010.284. [DOI] [PubMed] [Google Scholar]
- 6.Wickham SFJ, et al. A DNA-based molecular motor that can navigate a network of tracks. Nat Nanotechnol. 2012;7:169–173. doi: 10.1038/nnano.2011.253. [DOI] [PubMed] [Google Scholar]
- 7.Yin P, Choi HMT, Calvert CR, Pierce NA. Programming biomolecular self-assembly pathways. Nature. 2008;451:318–322. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- 8.Green S, Bath J, Turberfield A. Coordinated Chemomechanical Cycles: A Mechanism for Autonomous Molecular Motion. Phys Rev Lett. 2008;101:238101. doi: 10.1103/PhysRevLett.101.238101. [DOI] [PubMed] [Google Scholar]
- 9.Omabegho T, Sha R, Seeman NC. A Bipedal DNA Brownian Motor with Coordinated Legs. Science. 2009;324:67–71. doi: 10.1126/science.1170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li B, Chen X, Ellington AD. Adapting Enzyme-Free DNA Circuits to the Detection of Loop-Mediated Isothermal Amplification Reactions. Anal Chem. 2012;84:8371–8377. doi: 10.1021/ac301944v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang Y (Sherry), Li B, Milligan JN, Bhadra S, Ellington AD. Real-Time Detection of Isothermal Amplification Reactions with Thermostable Catalytic Hairpin Assembly. J Am Chem Soc. 2013;135:7430–7433. doi: 10.1021/ja4023978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soloveichik D, Seelig G, Winfree E. DNA as a universal substrate for chemical kinetics. Proc Natl Acad Sci. 2010;107:5393–5398. doi: 10.1073/pnas.0909380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen YJ, et al. Programmable chemical controllers made from DNA. Nat Nanotechnol. 2013;8:755–762. doi: 10.1038/nnano.2013.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isalan M, Lemerle C, Serrano L. Engineering Gene Networks to Emulate Drosophila Embryonic Pattern Formation. PLoS Biol. 2005;3:e64. doi: 10.1371/journal.pbio.0030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simpson ZB, Tsai TL, Nguyen N, Chen X, Ellington AD. Modelling amorphous computations with transcription networks. J R Soc Interface. 2009;6:S523–S533. doi: 10.1098/rsif.2009.0014.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Padirac A, Fujii T, Estévez-Torres A, Rondelez Y. Spatial Waves in Synthetic Biochemical Networks. J Am Chem Soc. 2013;135:14586–14592. doi: 10.1021/ja403584p. [DOI] [PubMed] [Google Scholar]
- 17.Chirieleison SM, Allen PB, Simpson ZB, Ellington AD, Chen X. Pattern transformation with DNA circuits. Nat Chem. 2013;5:1000–1005. doi: 10.1038/nchem.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li B, Ellington AD, Chen X. Rational, modular adaptation of enzyme-free DNA circuits to multiple detection methods. Nucleic Acids Res. 2011;gkr504 doi: 10.1093/nar/gkr504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang ZG, Elbaz J, Willner I. DNA Machines: Bipedal Walker and Stepper. Nano Lett. 2011;11:304–309. doi: 10.1021/nl104088s. [DOI] [PubMed] [Google Scholar]
- 20.Goldstein B, et al. Competition between solution and cell surface receptors for ligand. Dissociation of hapten bound to surface antibody in the presence of solution antibody. Biophys J. 1989;56:955–966. doi: 10.1016/S0006-3495(89)82741-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walsh MK, Wang X, Weimer BC. Optimizing the immobilization of single-stranded DNA onto glass beads. J Biochem Biophys Methods. 2001;47:221–231. doi: 10.1016/s0165-022x(00)00146-9. [DOI] [PubMed] [Google Scholar]
- 22.Dunbar SA. Applications of Luminex® xMAP™ technology for rapid, high-throughput multiplexed nucleic acid detection. Clin Chim Acta. 2006;363:71–82. doi: 10.1016/j.cccn.2005.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian L, Winfree E. Scaling Up Digital Circuit Computation with DNA Strand Displacement Cascades. Science. 2011;332:1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- 24.Qian L, Winfree E, Bruck J. Neural network computation with DNA strand displacement cascades. Nature. 2011;475:368–372. doi: 10.1038/nature10262. [DOI] [PubMed] [Google Scholar]
- 25.Wu Y, Zhang DY, Yin P, Vollmer F. Ultraspecific and Highly Sensitive Nucleic Acid Detection by Integrating a DNA Catalytic Network with a Label-Free Microcavity. Small. 2014;10:2067–2076. doi: 10.1002/smll.201303558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang B, et al. Intelligent layered nanoflare: ‘lab-on-a-nanoparticle’ for multiple DNA logic gate operations and efficient intracellular delivery. Nanoscale. 2014;6:8990–8996. doi: 10.1039/c4nr01676a. [DOI] [PubMed] [Google Scholar]
- 27.Fernandez JG, Khademhosseini A. Micro-Masonry: Construction of 3D Structures by Microscale Self-Assembly. Adv Mater. 2010;22:2538–2541. doi: 10.1002/adma.200903893. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.