Abstract
DNA mismatch repair (MMR) repairs mispaired bases in DNA generated by replication errors. MutS or MutS homologs recognize mispairs and coordinate with MutL or MutL homologs to direct excision of the newly synthesized DNA strand. In most organisms, the signal that discriminates between the newly synthesized and template DNA strands has not been definitively identified. In contrast, Escherichia coli and some related gammaproteobacteria use a highly elaborated methyl-directed MMR system that recognizes Dam methyltransferase modification sites that are transiently unmethylated on the newly synthesized strand after DNA replication. Evolution of methyl-directed MMR is characterized by the acquisition of Dam and the MutH nuclease and by the loss of the MutL endonuclease activity. Methyl-directed MMR is present in a subset of Gammaproteobacteria belonging to the orders Enterobacteriales, Pasteurellales, Vibrionales, Aeromonadales, and a subset of the Alteromonadales (the EPVAA group) as well as in gammaproteobacteria that have obtained these genes by horizontal gene transfer, including the medically relevant bacteria Fluoribacter, Legionella, and Tatlockia and the marine bacteria Methylophaga and Nitrosococcus.
Keywords: Mismatch repair, Evolution, Dam methylase, MutH endonuclease, MutL endonuclease
Introduction
A critical role of DNA mismatch repair is to recognize and repair mispaired bases generated by DNA replication errors within a large background of properly base-paired DNA [1–3]. For all organisms, the core MMR steps are recognition of the mispair, excision of the newly synthesized strand at least up to the mispair, and resynthesis of the excised strand (Figure 1). A key aspect of this mechanism is the discrimination of the newly synthesized DNA strand from the template DNA strand; excision and resynthesis of the template strand rather than the newly synthesized strand would incorporate the replication error into the genome rather than excise and repair it.
Fig 1. Diagram of methyl-directed MMR.
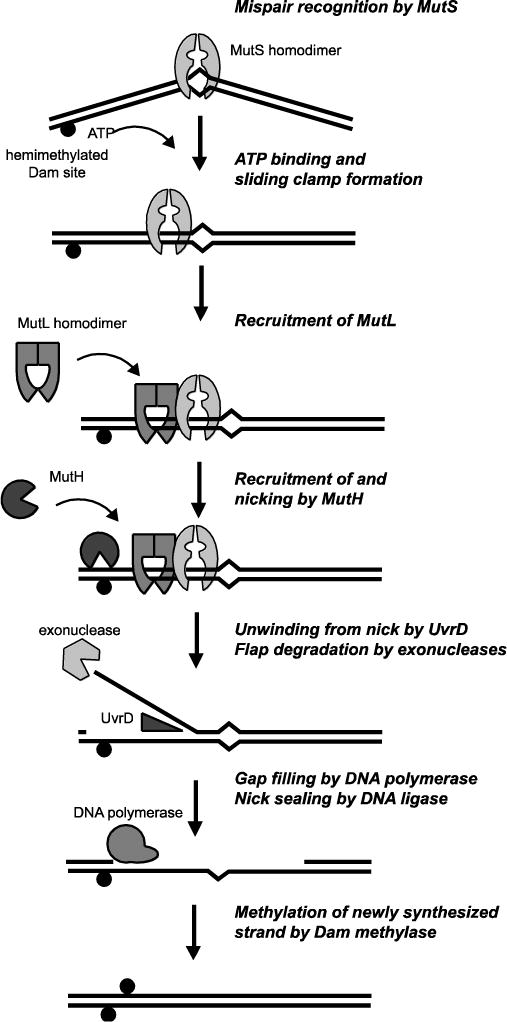
Main steps in the E. coli methyl-directed MMR pathway (see main text). Black circle indicates the presence of a methylated adenosine at a d(GATC) site.
The mechanisms of strand discrimination in MMR are best understood for the bacteria Escherichia coli; however, the phylogenetic distribution of the methyl-directed MMR system found in E. coli is restricted to a set of closely related gammaproteobacteria (Figure 2). Thus, methyl-directed MMR must have evolved from the canonical MMR system present in most other organisms. The novel aspects of methyl-directed MMR involve recognition of and cleavage at the transiently unmethylated strand in hemi-methylated d(GATC) sites that are present after replication but before methylation of the newly synthesized strand (Figure 1) [4]. Remarkably, the elaborations to the canonical MMR system in the methyl-directed MMR system facilitated the identification of the MMR genes required for mispair recognition (mutS), signal propagation (mutL), strand discrimination (mutH), and excision and resynthesis (uvrD/mutU) [5], as mutations in these genes suppress the 2-aminopurine and N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) sensitivity and suppress the lethality of recombination mutations in E. coli mutants with defects in d(GATC) methylation [6–9].
Fig 2. Distribution of methyl-directed MMR in living organisms.
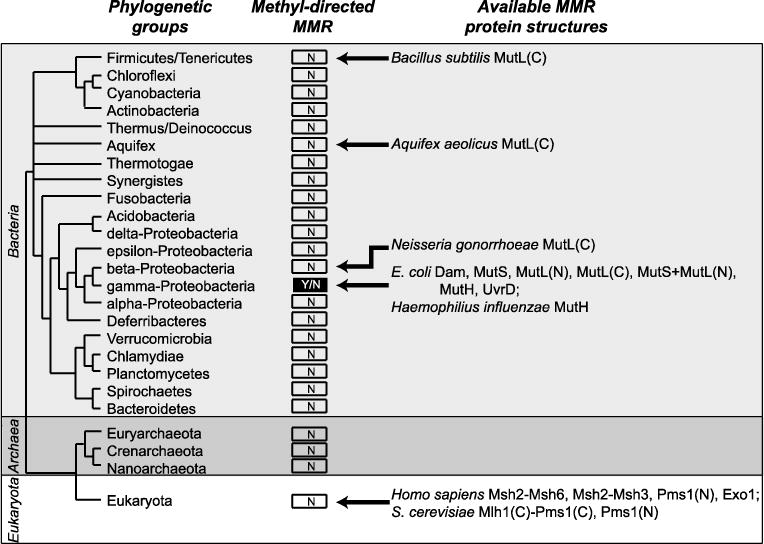
Presence or absence of the methyl-directed MMR is indicated by “Y” for yes and “N” for no. Available structures of MMR proteins for each group are depicted. Relationships between the bacterial groups derived from [92]. For MutL homologs, available structures of N-terminal domains are indicated with (N) and the available structures of C-terminal domains are indicated with (C).
In contrast, the failure of extensive genetic screens in other organisms, such as the budding yeast Saccharomyces cerevisiae, to identify a clear strand discrimination signal suggests that the strand discrimination signal in the canonical MMR system may be fundamental to the DNA replication process, such as the presence of nicks on the lagging strand. Several facts are consistent with this view: (i) the Msh2-Msh6 homologs of MutS are physically associated with the replication fork, though downstream steps are not [10], (ii) loci are only competent for undergoing MMR in a short (10–15 minute) window of time after the locus is replicated [11]; and (iii) biochemical reconstitution of eukaryotic MMR is targeted to substrates having pre-existing nicks [12–14] (see the minireview by Kadyrova and Kadyrov in this issue). The fact that some feature of pre-existing nicks, such as the nick itself or a nick-loaded replicative clamp [15], may function as a signal in the canonical MMR system echoes observations made over 30 years ago that MMR-mediated heteroduplex repair in the bacterium Streptococcus pneumoniae, which has a canonical MMR system, is targeted to the incompletely integrated donor strand [16–18].
Roles of the E. coli DNA adenine methyltransferase Dam
The acquisition of the DNA adenine methyltransferase Dam in gammaproteobacteria related to E. coli was the key evolutionary innovation that created a novel mechanism for identifying newly replicated DNA and discriminating the newly synthesized strand from the template strand. The Dam methyltransferase catalyzes post-replication methylation of the adenosine N6 position at palindromic d(GATC) sites using S-adenosylmethionine as a substrate [19]. DNA replication of a fully methylated template gives rise to hemi-methylated d(GATC) sites, in which the template strand is methylated and the newly synthesized strand is unmodified. This transient hemi-methylated status only lasts on the order of minutes [19, 20]. Despite its relatively recent acquisition, Dam has key roles in bacterial genome maintenance, and E. coli dam mutants have defects in replication initiation, chromosome partitioning, nucleoid structure, and mismatch repair [21].
In addition to roles in strand discrimination during MMR described below, hemi-methylated d(GATC) sites play important roles in identifying newly synthesized DNA. The SeqA protein binds hemi-methylated d(GATC) sites after replication [22, 23], with high-affinity binding of SeqA requiring at least two sites on the same face of DNA [24]. Near the E. coli chromosomal origin, oriC, SeqA binding sequesters oriC into a membrane-protein complex, which ensures that DNA replication initiates once per cell cycle by preventing binding by the DNA replication initiation protein DnaA [19, 25, 26]. Sites near oriC are protected from being fully methylated by Dam for up to a third of the cell cycle [19]. SeqA also reduces expression from genes near oriC; overexpression of Dam or loss of SeqA causes increased expression from these genes, including dnaA [19, 27]. An additional hemi-methylated d(GATC) binding factor, yccV/hspQ, has been isolated that suppresses the temperature sensitivity of dnaA mutants and suppresses dnaA transcription [28], although yccV/hspQ has also been implicated as a heat shock protein that stabilizes mutant DnaA proteins [29]. SeqA binding is also important for forming nucleoid structure in E. coli [30], potentially through its interactions with the chromosome partitioning complex made up of MukF, MukE, and MukB, which replaces the chromosome partitioning complex involving Smc-ScpAB that is present in most other bacteria [31, 32].
MMR in E. coli
MMR is initiated by the recognition of a pro-mutagenic mispair generated by replication or chemical modification of DNA (Figure 1). These mispairs are bound in E. coli by MutS, which is a homodimeric ABC-family ATPase that can bind to single base mismatches and insertion/deletions of up to four nucleotides in the absence of nucleotide or the presence of ADP [33, 34] (see the minireview by Hingorani in this issue). Crystal structures of the MutS homodimer from Thermus aquaticus and E. coli revealed that the mispair-recognition complex binds the DNA at the site of the mispair, bends the DNA by ~60 degrees, and opens the DNA base stack so that one face of a base in the mispair is exposed for recognition by a conserved phenylalanine side chain [35, 36] (see the minireview by Groothuizen and Sixma in this issue). The mispair recognition complex is functionally asymmetric; one subunit interacts with bases at the site of the mispair, whereas the other subunit binds dsDNA. For eukaryotic MutS homologs, this functional asymmetry is reflected by specialization of gene-duplicated homologs [37–39]. A subtle feature of the MutS-mispaired DNA structures is that these complexes stack the phenylalanine side chain onto the same base in the mispair, such as the thymidine base in the T:G mispair, regardless of whether this thymidine base is on the template or the newly synthesized strand. Hence, MutS and its eukaryotic homologs do not perform strand discrimination.
Upon binding to a mispair, the MutS ABC ATPase domains bind ATP or exchange ADP for ATP and undergoes a conformational change that allows the MutS dimer to rapidly slide along the DNA, the so-called “sliding clamp”, and to recruit MutL to DNA (Figure 1) [40, 41] (see minireviews by Hingorani, by Kadyrova and Kadyrov, and by Groothuizen and Sixma in this issue). This conformational change involves loss of the ~60 degree DNA bend present in the mispair recognition complex [42]. This conformation has been visualized in a crystal structure of a MutS-MutL complex trapped by chemical crosslinking and is dominated by a large scale motion of the ABC ATPase domains, which is propagated through the sides of the MutS ring, exposing the connector domain for interaction with MutL [44]. These conformational changes are consistent with altered deuteration kinetics of backbone amides [45], the exposure of a surface on the MutS connector domain that interacts with MutL [46], and the conformation of related ATP-bound ABC ATPase domains [43]. ATP binding is sufficient for MutL recruitment by MutS and activation of downstream MMR steps; however, ATP hydrolysis by E. coli MutS is also required in vivo and may regulate the activation of MutL [47] or may be necessary to allow MutS to promote multiple rounds of MutL loading.
MutL, like MutS, is a homodimeric ATPase; however, the N-terminal ATPase domain of MutL belongs to the GHKL family [48] and is separated from the C-terminal domain by an unstructured linker. The C-terminal domains of MutL are constitutively dimerized, whereas the N-terminal domains dimerize only upon ATP binding to form a ring. In organisms with a canonical MMR system, the MutL C-terminal domains possess endonuclease motifs that bind two Zn2+ ions [49–52] (see minireviews by Kadyrova and Kadyrov and by Groothuizen and Sixma in this issue), and generate single-stranded breaks in DNA [50, 51, 53–56]. In organisms with a methyl-directed MMR system, the C-terminal domains have similar folds, but the endonuclease motifs and metal binding are absent [57]; however, these domains are involved in binding to and activating downstream components of the methyl-directed MMR pathway [58–60].
The Mg2+-dependent endonuclease MutH is bound and activated by MutL [58, 60] (Figure 1). MutH makes a single-stranded nick 5′ of the G in the unmethylated strand of hemimethylated d(GATC) Dam sites and thereby uses the methyl marker to perform strand-discrimination [64]. The MutH-generated nick serves as the entry point for displacement of the newly synthesized strand by the UvrD helicase and degradation by single-stranded DNA exonucleases [65]. The methyl-directed MMR system is bidirectional; the hemimethylated d(GATC) site can be located either 5′ or 3′ of and up to 1–2 kb away from the mispair on the unmethylated strand [66, 67]. In the E. coli genome, a d(GATC) site is present on average every 242 bp and only around 2% of the sites are separated from other d(GATC) by over 1 kb. However, a genetic assay that used a trinucleotide repeat sequence as a source of 3 bp insertion/deletion mutations revealed that only d(GATC) sites between the mispair and the replication fork are utilized and hence MMR repair is “unidirectional” with regards to the chromosome orientation [67]. Thus, use of sites 5′ or 3′ on the unmethylated strand most likely corresponds to the use of origin-distal and fork-proximal d(GATC) sites for the lagging or leading strands, respectively. The fact that the d(GATC) sites do not have to be immediately adjacent to the mispair also suggests that the MMR machinery must somehow signal over a distance to activate MutH. MMR in vitro requires a continuous and unblocked DNA between the mispair and the hemi-methylated GATC sites for MMR [61]. Models involving sliding of MutS, MutL, or MutS-MutL complexes are attractive given the protein structures and ATP-driven conformational changes (see the minireviews by Hingorani and by Groothuizen and Sixma in this issue). Given the transient nature of the MutS-MutL complex, which required crosslinking for crystallization [44], and that the foci containing MutL homologs in S. cerevisiae, which either had no or substantially substoichiometric levels of MutS homologs [10], it seems likely that MutL is the major mediator for the ability of MMR to act at a distance (see the minireview by Schmidt and Hombauer in this issue).
The 3′–>5′ UvrD DNA helicase is also bound and activated by MutL [59, 68, 69] (Figure 1). Since UvrD has a fixed polarity but mismatch repair is bidirectional, the helicase must be loaded either onto the newly synthesized DNA strand or the template strand depending on the orientation of the hemimethylated site relative to the mispair. The displaced single strand is then a substrate for multiple redundant exonucleases, including RecJ, ExoVII, ExoI, and ExoX [70, 71]. Unwinding and degradation of the displaced strand typically terminate ~100 nucleotides after the mispair [72]. Termination may be a consequence of the need for MutL to mediate UvrD loading combined with the rather short (~50 bp) processivity of UvrD [73, 74]. Reconstitution of the repair reaction in vitro showed that DNA polymerase III could mediate resynthesis of DNA across the gap and that DNA ligase could mediate sealing of the final nick [65].
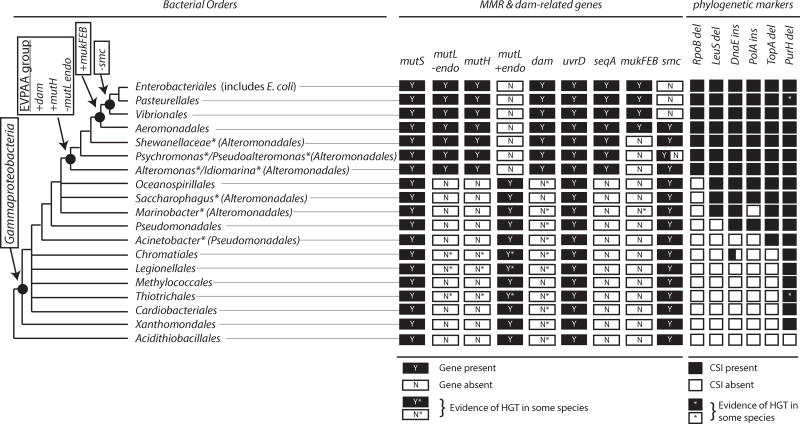
Existence of two MMR systems in the class Gammaproteobacteria
The class Gammaproteobacteria is a very large group containing 14 orders of diverse bacteria [75], but only a subset of the species possess methyl-directed MMR systems. The phylogenetic tree for Gammaproteobacteria (Figure 3) is derived from previous phylogenetic trees generated from alignments of “super-genes” generated by concatenating the sequences of multiple conserved proteins as well as the patterns of conserved signature insertion/deletions (CSIs) in proteins [76–80]. Well-defined protein CSIs are particularly useful for deciphering evolutionary relationships between bacteria as they are less likely to arise from independent mutational events [81], though CSI patterns can be complicated by horizontal gene transfer (HGT) events [82]. For example, a conserved 2 amino acid deletion in the PurH protein is characteristic of Gammaproteobacteria, except for Francisella and Bibersteinia, which likely results from HGT of purH from a bacterium in Alphaproteobacteria and a bacterium in Firmicutes, respectively (Figure 3) [76]. Phylogenetically restricted genes also provide insight into phylogenetic relationships. The dam, seqA, mutH, and mukFEB genes have been recognized as having distributions in Gammaproteobacteria restricted to species related to E. coli [21, 76, 83–85] (Figure 3). Many other genes appear to have similarly restricted distributions, including some genes like tus, priC, and wecF that are essential to E. coli viability [76, 84]; however, most are not known to interact with MMR or Dam.
Fig 3. Distribution of methyl-directed MMR in Gammaproteobacteria.
The distribution of dam-related genes is shown as “Y” or yes and “N” for no for orders within Gammaproteobacteria based on analysis of over 250 bacterial genomes (Supplemental Table 1). Names annotated with asterisks are genera or families, and the orders to which they are assigned are in parentheses. The endonuclease-proficient and endonuclease-deficient types of mutL genes are shown separately, and that only “resident” dam genes are shown with a “Y”. Phylogenetically informative CSIs are also shown. Nodes corresponding to Gammaproteobacteria, the EVPAA group, which includes bacteria containing the methyl-directed MMR system, and the bacteria in which Smc-ScpAB is replaced by the MukFEB chromosome partitioning system are labeled. Relationships between the gammaproteobacterial species are derived from previous phylogenetic analyses [76, 77, 79].
MutS, which is a key player in both the canonical and methyl-directed MMR systems, is widely distributed across Gammaproteobacteria, and is missing only in a subset of species, including endosymbionts like Buchnera (Figure 3; Supplemental Table 1). Gammaproteobacteria lacking MutS also lack MutL (Supplemental Table 1); it is not clear if these bacteria have functional MMR. In addition, some gammaproteobacteria have a number of other protein families that contain some, but not all of the MutS domains; however, these proteins are not known to act in MMR (Supplemental Table 1) [86–89]. For example, the fairly common bacterial MutS2 (not to be confused with the eukaryotic Msh2) is found in the order Acidithiobacillales, which is not a member of Gammaproteobacteria in some phylogenetic reconstructions [79]. MutS2 proteins have a nuclease activity in a C-terminal Smr domain, function in anti-recombination [90, 91], and are often found in bacteria without MutL homologs [86]. Homologs without known function are also observed in some species, including MutS3 (in some bacteria in the order Xanthomondales), MutS5 (in some bacteria in the orders Methylococcales and Chromatiales), and MutS9 (in some bacteria in the orders Acidithiobacillales and Thiotrichales).
MutL also has a wide distribution across Gammaproteobacteria due to a requirement in both the canonical and methyl-directed MMR systems. Unlike MutS, MutL homologs can be divided into two groups: those with endonuclease motifs in the C-terminal domain and those without endonuclease motifs (Figure 3; Supplemental Table 1). A clear observation among the >250 gammaproteobacterial species analyzed here as well as smaller numbers of species analyzed previously [53, 55] is that bacteria with endonuclease-proficient MutL do not have MutH homologs (bacteria possessing the canonical MMR system), and bacteria with endonuclease-deficient MutL have MutH homologs (bacteria possessing the methyl-directed MMR system). Bacteria possessing the methyl-directed MMR system are observed primarily in the orders Enterobacteriales (which includes E. coli), Pasteurellales, Vibrionales, Aeromondales, and some bacteria in the order Alteromonadales (genera Alteromonas, Idiomarina, Pseudoalteromonas, Psychromonas, and Shewanella). For purposes of this review, we term this set of bacterial species the EPVAA group.
Dam is found in many of the orders in Gammaproteobacteria, but the dam gene appears to have been obtained through HGT in many cases. Phylogenetic analysis of the Dam protein matches the phylogenetic branching pattern of the bacterial groups [92] only for the EPVAA group (Figure 3; Supplemental Table 1). The dam genes in these species have been previously been referred to as “resident” dam genes [84]. The distribution of Dam-related genes, including those encoding components of the methyl-directed MMR system, suggests that Dam was incorporated into a gammaproteobacterium that was ancestral to the EPVAA group. This common ancestor also obtained SeqA and MutH and lost the endonuclease activity in MutL (Figure 3). The dramatic shift from one form of MMR to another in EPVAA bacteria is reminiscent of the switch from the Smc-ScpAB to the MukFEB chromosome partitioning systems, which affects a subset of bacteria in the EPVAA group and likely occurred in a common ancestor to the orders Enterobacteriales, Pasteurellales, and Aeromonadales (Figure 3) [83, 93].
HGT of genes involved in the methyl-directed MMR system
Analysis of the pattern of bacterial species with the methyl-directed MMR system identifies several, presumably independent HGT events. For example, Nitrosococcus, a genera of marine aerobic ammonia-oxidizing bacteria that belong to the order Chromatiales (the purple sulfur bacteria) based on CSI patterns (Figure 2) and a 16S rRNA phylogeny [94], have obtained a methyl-directed MMR system by HGT, including the dam, mutL, and mutH genes (Figure 4). Of the mutS and mutL genes in Nitrosococcus, only the mutS genes are most closely related to mutS genes in other bacteria belonging to Chromatiales. HGT involving gain of a mutH homolog and an endonuclease-deficient mutL homolog also appear to have occurred in other orders of Gammaproteobacteria (Figure 4), including the order Legionellales (genera Fluoribacter, Legionella, and Tatlockia) and the order Thiotrichales (genus Methylophaga). In each of these bacteria, only genes involved in methyl-directed MMR were obtained by HGT; other Dam-dependent genes such as seqA, mukF, mukE, and mukF are not present. Dam, MutH, and endonuclease-deficient MutL proteins are also found in the genus Rheinheimera (order Chromatiales) and the genus Kangiella (order Oceanospirillales); however, in these cases CSI patterns and protein homologies suggest that these are not HGT events but rather that these species are misclassified (Rheinheimera and Kangiella have gene conservation and CSI patterns like species with methyl-directed MMR systems in Alteromonadales).
Fig 4. Putative horizontal gene transfer events establish methyl-directed MMR systems.
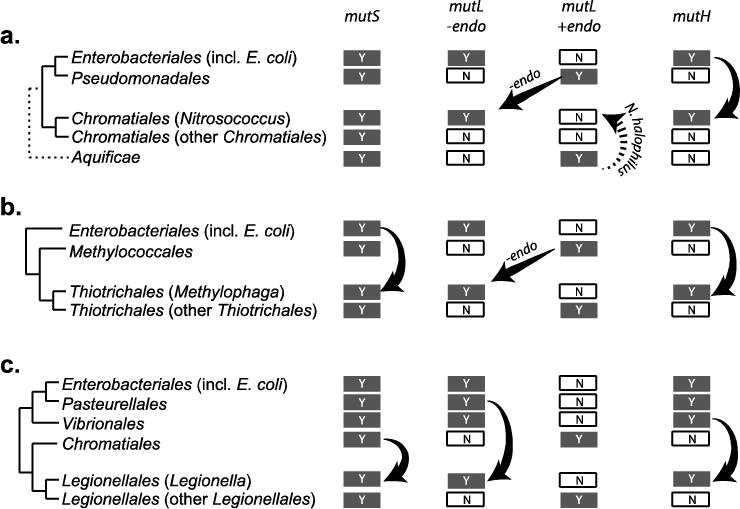
Sources for the mutS, mutL, and mutH genes were derived based on closest homologs from BLAST analyses for methyl-directed MMR genes in Nitrosococcus (a), Methylophaga (b), and Legionella (c). The dashed arrow corresponds to a HGT event specific to N. halophilus Nc 4, which contains two MutL homologs, one is predicted to be endonuclease-proficient and the other is endonuclease-deficient. The endonuclease-proficient MutL is most closely related to MutL proteins from the bacterial phylum Aquificae and not Gammaproteobacteria and likely indicates a separate HGT event, whereas the endonuclease-deficient MutL proteins from multiple Nitrosococcus species are most closely related to MutL proteins in the genus Pseudomonas.
In contrast, there are no clear cases of HGT involving replacement of a methyl-directed MMR system with a canonical MMR system. A few bacteria in the order Alteromonadales (genera Marinobacter, Saccharophagus, and Teredinibacter) lack the methyl-directed MMR system of other species in Alteromonadales (Supplemental Table 1); however, Marinobacter and Saccharophagus were shown previously to group with bacterial species in the orders Pseudomonadales and Oceanospirillales and not with other species in the order Alteromonadales (Teredinibacter was not included in this analysis) [79]. Re-assignment of all three genera to groups other than Alteromonadales would be consistent with (i) the lack of the seqA gene, (ii) the lack of the 4 amino acid deletion in RpoB, and (iii) the lack of the methyl-directed MMR system (Figure 3) and would argue against HGT involving MMR genes in these genera.
Evolution of the methyl-directed MMR system
The evolution of the methyl-directed MMR system from the canonical MMR system can be envisioned to occur in multiple steps. The necessary first step is the acquisition of Dam, which appears to be closely related to methyltransferases from other restriction-modification systems [21]. The subsequent acquisition of mutH and loss of the mutL endonuclease activity would be one pathway to obtain a methyl-directed MMR system. In Pseudomonas aeruginosa (order Pseudomonadales), which has an endonuclease-proficient MutL and lacks the methyl-directed MMR system, mutations in mutS, mutL, and uvrD increase the levels of spontaneous mutations [95], suggesting that the UvrD helicase operates in the canonical MMR system of P. aeruginosa. Thus MMR can be envisioned to be functional throughout the evolutionary transition, with an endonuclease-proficient and dam-independent MutL acting until MutH becomes available. Consistent with this, MutL from P. aeruginosa can also complement a mutL− E. coli strain (although it is unclear if MutH or Dam are required for MMR in this complemented strain) [96]. Other orders of gene addition and modification, however, might be possible depending on the activities of the individual proteins. For example, the endonuclease-deficient MutL from E. coli can surprisingly complement loss of the endonuclease-proficient MutL from P. aeruginosa [96]; under these conditions, it is not clear what provides the strand discrimination function or nicks in the DNA although nicks produced during DNA replication could serve this purpose.
MutH may have evolved from a type II restriction endonuclease with a PD-(D/E)XK domain that was a common ancestor with Sau3AI [97] (Figure 5a). Most restriction endonucleases are homodimers, and each subunit cleaves one DNA strand [98]. In contrast, MutH is monomeric in solution and when bound to DNA (Figure 5b,c), and Sau3AI is monomeric in solution, but dimerizes upon binding DNA [99] (Figure 5d). Both MutH and Sau3AI recognize the Dam-targeted d(GATC) sites and cleave 5′ of the G residue. MutH is sensitive to adenosine methylation and recognizes the unmethylated adenosine at d(GATC) Dam sites using a tyrosine residue (E. coli Y212), which is required for cleavage [97, 100, 101]. In contrast, Sau3AI is sensitive to cytosine methylation. Like MutL-mediated activation of MutH, Sau3AI activity also appears to be inducible, albeit through an allosteric mechanism. Each Sau3AI monomer contains two copies of the MutH fold, although only the N-terminal domain has a functional PD-(D/E)XK motif [99, 102, 103] (Figure 5a,b). Based on the ability of Sau3AI dimers to generate DNA loops observed by electron microscopy and studies of the isolated C-terminal domain, the C-terminal domain also binds d(GATC) sites [99, 103]. Sau3AI is most active on substrates with two sites, although the first double-stranded cleavage is rapid and the second is slow, suggesting that the DNA binding by the C-terminal domain acts as an allosteric activator for the N-terminal domain [99] (Figure 5d). Thus, the common ancestor to both MutH and Sau3AI had numerous properties that were advantageous for MutH evolution: (i) stability as a monomer to allow for single-stranded DNA nicking, (ii) inducible enzymatic activity to allow for activation by MutL, and (iii) recognition of d(GATC) for strand-specific inhibition by Dam methylation.
Fig 5. Relationship of MutH with Sau3AI.
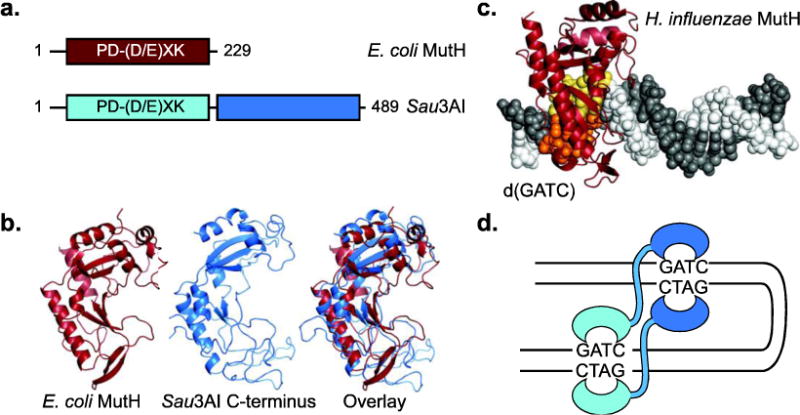
a. Sau3AI contains two copies of the MutH PD-(D/E)XK domain, but only the first has a functional nuclease motif. b. E. coli MutH (red; PDB id 2azo; [97]) and the C-terminal domain of Sau3AI (blue; PDB id 2reu; [103]) have a common fold. C. Structure of Haemophilus influenzae MutH (red) in complex with a d(GATC) site (yellow and orange; PDB id 2aoq; [100]). D. Model of DNA looping by a Sau3AI dimer on a DNA with two d(GATC) sites.
Of the evolutionary steps necessary to generate a methyl-directed MMR system, loss of the endonuclease active sites from MutL is probably the simplest, and in principle could have occurred multiple times. Conserved C-terminal motifs in endonuclease-proficient MutL homologs are constrained by the need to bind metals and catalyze strand cleavage and are lost in endonuclease-deficient MutL (Figure 6a); however, nuclease-proficient and nuclease-deficient MutL homologs have the same folds for both the N- and C-terminal domains (Figure 6b,c; also see the minireview by Groothuizen and Sixma in this issue). Two of the key endonuclease motifs, DxHxxxER and CHG are present on conserved structural elements. The third motif containing the sequence CNHGRPT, however, is present on an extended metal-binding loop where the cysteine and histidine side chains are metal ligands (Figure 6d). Remarkably, MutL proteins from Enterobacteriales retain precisely the same number of residues as the metal-binding loop, and the structure of this region in E. coli MutL terminates helix E′ with P589 and uses this proline to begin a new, short, α-helix (F′) which is terminated by P595 and P596 (Figure 6e). These three proline residues are highly conserved in MutL proteins from Enterobacteriales (Figure 6a). This loop, which contains CNHGRPT in endonuclease-proficient MutL, is highly divergent in other endonuclease-deficient MutL proteins; the features of this loop that are conserved in E. coli and the Enterobacteriales are not conserved in endonuclease-deficient MutL from Pasteurellales, Vibrionales, Aeromondales, Alteromondales, and Legionellales. This dramatic divergence could, in principle, be due to loss of evolutionary constraints on this region or due to multiple independent events in which the endonuclease function was lost.
Fig 6. Loss of endonuclease motifs in E. coli MutL.
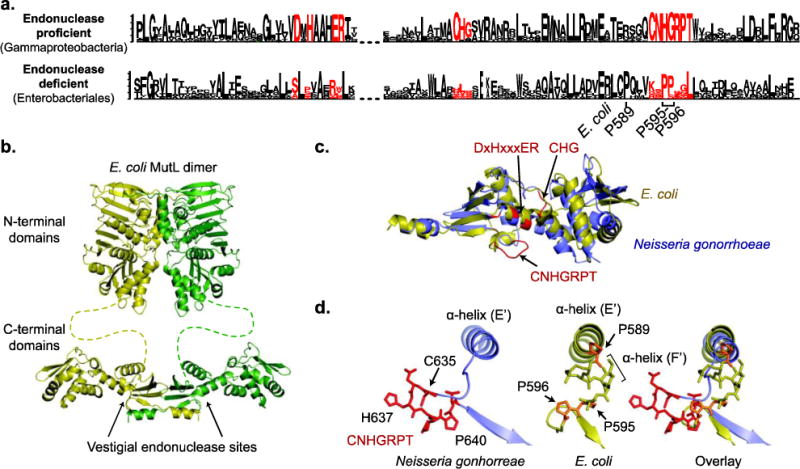
a. Sequence logos, where the height of the letter indicates of its degree of conservation, were generated by Seq2Logo [107] for the endonuclease-proficient MutL in Gammaproteobacteria and the endonuclease-deficient MutL in Enterobacteriales, which includes E. coli. The endonuclease motifs are shown in red. b. Modeled structure of full length MutL based on the N-terminal domain structure (PDB id 1b62; [108]) and the C-terminal domain structure (PDB id 1×9z; [57]). c. Overlay of the E. coli (yellow) and Neisseria gonorrhoeae (green) MutL C-terminal domains reveals that the folds are the same; N. gonorrhoeae is the nuclease-proficient domain structure that is most closely related to E. coli (Fig. 1). Residues in red correspond to the endonuclease motifs. d. Changes in the CNHGRPT motif-containing loop are depicted for the nuclease-proficient N. gonorrhoeae (left) and E. coli (middle) with an overlay of the two structures (right).
Conclusions
In principle, any transient state affecting the newly synthesized strand that exists immediately after replication could be used to mediate strand discrimination during DNA MMR. Although the advantage of switching from the canonical MMR system to methyl-directed MMR system is not entirely clear, Dam methylation has several advantages over other DNA modifications. First, the target of Dam methylase, d(GATC), is short and found frequently in the genome. Other DNA-modifying enzymes target longer sequences that are less frequent in the genome, such as the E. coli Dcm cytosine N5-methyltransferase that modifies the second cytosine at d(CC(A/T)GG) sequences. Second, unlike 6-methyladenine generated by Dam, 5-methylcytosine readily deaminates and generates pro-mutagenic G:T mispairs that are repaired by base-excision repair, very short patch (VSP) repair, or MMR [104]. However, the evolution of the E. coli version of the methyl-directed MMR system may not have been due to the advantages of 6-methyladenine or the d(GATC) sites but rather due to the favorable features of the Dam and MutH ancestors combined with historical contingency. If we could perform Stephen Jay Gould’s Gedanken experiment of “replaying life’s tape” [105], it seems equally possible that the E. coli methyl-directed MMR system might not have arisen at all or may have coopted some other available DNA modification, such as β-glucosyl-5-hydroxymethylcytosine via HGT of genes from bacteriophage T4 [106].
Supplementary Material
HIGHLIGHTS.
E. coli contains an unusual methyl-directed mismatch repair (mdMMR) pathway.
The mdMMR arose in an ancestor to a subset of Gammaproteobacteria (the EPVAA group).
mdMMR features are gain of Dam and MutH and loss of the MutL endonuclease activity.
Gammaproteobacteria either have the canonical MMR or mdMMR pathway but not both.
Gammaproteobacteria like Legionella obtained mdMMR by horizontal gene transfer.
Acknowledgments
I thank Anjana Srivastsan, Eva M. Goellner, William J. Graham V, and Richard D. Kolodner for helpful discussions. This work was supported by NIH Grant GM50006 and the Ludwig Institute for Cancer Research.
Abbreviations
- CSI
conserved signature insertion/deletions
- EPVAA
Enterobacteriales, Pasteurellales, Vibrionales, Aeromonadales, and Altermonoadales
- HGT
horizontal gene transfer
- MMR
mismatch repair
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chemical reviews. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 2.Jiricny J. Postreplicative mismatch repair. Cold Spring Harbor perspectives in biology. 2013;5:a012633. doi: 10.1101/cshperspect.a012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Current opinion in genetics & development. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 4.Langle-Rouault F, Maenhaut-Michel G, Radman M. GATC sequences, DNA nicks and the MutH function in Escherichia coli mismatch repair. The EMBO journal. 1987;6:1121–1127. doi: 10.1002/j.1460-2075.1987.tb04867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horst JP, Wu TH, Marinus MG. Escherichia coli mutator genes. Trends in microbiology. 1999;7:29–36. doi: 10.1016/s0966-842x(98)01424-3. [DOI] [PubMed] [Google Scholar]
- 6.Glickman BW, Radman M. Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proceedings of the National Academy of Sciences of the United States of America. 1980;77:1063–1067. doi: 10.1073/pnas.77.2.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karran P, Marinus MG. Mismatch correction at O6-methylguanine residues in E. coli DNA. Nature. 1982;296:868–869. doi: 10.1038/296868a0. [DOI] [PubMed] [Google Scholar]
- 8.Glickman B, van den Elsen P, Radman M. Induced mutagenesis in dam-mutants of Escherichia coli: a role for 6-methyladenine residues in mutation avoidance. Molecular & general genetics: MGG. 1978;163:307–312. doi: 10.1007/BF00271960. [DOI] [PubMed] [Google Scholar]
- 9.McGraw BR, Marinus MG. Isolation and characterization of Dam+ revertants and suppressor mutations that modify secondary phenotypes of dam-3 strains of Escherichia coli K-12. Molecular & general genetics: MGG. 1980;178:309–315. doi: 10.1007/BF00270477. [DOI] [PubMed] [Google Scholar]
- 10.Hombauer H, Campbell CS, Smith CE, Desai A, Kolodner RD. Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell. 2011;147:1040–1053. doi: 10.1016/j.cell.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hombauer H, Srivatsan A, Putnam CD, Kolodner RD. Mismatch repair, but not heteroduplex rejection, is temporally coupled to DNA replication. Science. 2011;334:1713–1716. doi: 10.1126/science.1210770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Constantin N, Dzantiev L, Kadyrov FA, Modrich P. Human mismatch repair: reconstitution of a nick-directed bidirectional reaction. The Journal of biological chemistry. 2005;280:39752–39761. doi: 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, Gu L, Li GM. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 14.Bowen N, Smith CE, Srivatsan A, Willcox S, Griffith JD, Kolodner RD. Reconstitution of long and short patch mismatch repair reactions using Saccharomyces cerevisiae proteins. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:18472–18477. doi: 10.1073/pnas.1318971110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georgescu R, Langston L, O’Donnell M. A proposal: Evolution of PCNA’s role as a marker of newly replicated DNA. DNA repair. 2015;29:4–15. doi: 10.1016/j.dnarep.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Claverys JP, Roger M, Sicard AM. Excision and repair of mismatched base pairs in transformation of Streptococcus pneumoniae. Molecular & general genetics: MGG. 1980;178:191–201. doi: 10.1007/BF00267229. [DOI] [PubMed] [Google Scholar]
- 17.Guild WR, Shoemaker NB. Mismatch correction in pneumococcal transformation: donor length and hex-dependent marker efficiency. Journal of bacteriology. 1976;125:125–135. doi: 10.1128/jb.125.1.125-135.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacks SA, Dunn JJ, Greenberg B. Identification of base mismatches recognized by the heteroduplex-DNA-repair system of Streptococcus pneumoniae. Cell. 1982;31:327–336. doi: 10.1016/0092-8674(82)90126-x. [DOI] [PubMed] [Google Scholar]
- 19.Campbell JL, Kleckner N. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell. 1990;62:967–979. doi: 10.1016/0092-8674(90)90271-f. [DOI] [PubMed] [Google Scholar]
- 20.Ogden GB, Pratt MJ, Schaechter M. The replicative origin of the E. coli chromosome binds to cell membranes only when hemimethylated. Cell. 1988;54:127–135. doi: 10.1016/0092-8674(88)90186-9. [DOI] [PubMed] [Google Scholar]
- 21.Lobner-Olesen A, Skovgaard O, Marinus MG. Dam methylation: coordinating cellular processes. Current opinion in microbiology. 2005;8:154–160. doi: 10.1016/j.mib.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 22.Yamazoe M, Adachi S, Kanaya S, Ohsumi K, Hiraga S. Sequential binding of SeqA protein to nascent DNA segments at replication forks in synchronized cultures of Escherichia coli. Molecular microbiology. 2005;55:289–298. doi: 10.1111/j.1365-2958.2004.04389.x. [DOI] [PubMed] [Google Scholar]
- 23.Waldminghaus T, Weigel C, Skarstad K. Replication fork movement and methylation govern SeqA binding to the Escherichia coli chromosome. Nucleic acids research. 2012;40:5465–5476. doi: 10.1093/nar/gks187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brendler T, Sawitzke J, Sergueev K, Austin S. A case for sliding SeqA tracts at anchored replication forks during Escherichia coli chromosome replication and segregation. The EMBO journal. 2000;19:6249–6258. doi: 10.1093/emboj/19.22.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slater S, Wold S, Lu M, Boye E, Skarstad K, Kleckner N. E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell. 1995;82:927–936. doi: 10.1016/0092-8674(95)90272-4. [DOI] [PubMed] [Google Scholar]
- 26.Taghbalout A, Landoulsi A, Kern R, Yamazoe M, Hiraga S, Holland B, Kohiyama M, Malki A. Competition between the replication initiator DnaA and the sequestration factor SeqA for binding to the hemimethylated chromosomal origin of E. coli in vitro. Genes to cells: devoted to molecular & cellular mechanisms. 2000;5:873–884. doi: 10.1046/j.1365-2443.2000.00380.x. [DOI] [PubMed] [Google Scholar]
- 27.Lobner-Olesen A, Marinus MG, Hansen FG. Role of SeqA and Dam in Escherichia coli gene expression: a global/microarray analysis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4672–4677. doi: 10.1073/pnas.0538053100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.d’Alencon E, Taghbalout A, Bristow C, Kern R, Aflalo R, Kohiyama M. Isolation of a new hemimethylated DNA binding protein which regulates dnaA gene expression. Journal of bacteriology. 2003;185:2967–2971. doi: 10.1128/JB.185.9.2967-2971.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimuta TR, Nakano K, Yamaguchi Y, Ozaki S, Fujimitsu K, Matsunaga C, Noguchi K, Emoto A, Katayama T. Novel heat shock protein HspQ stimulates the degradation of mutant DnaA protein in Escherichia coli. Genes to cells: devoted to molecular & cellular mechanisms. 2004;9:1151–1166. doi: 10.1111/j.1365-2443.2004.00800.x. [DOI] [PubMed] [Google Scholar]
- 30.Weitao T, Nordstrom K, Dasgupta S. Escherichia coli cell cycle control genes affect chromosome superhelicity. EMBO reports. 2000;1:494–499. doi: 10.1093/embo-reports/kvd106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamazoe M, Onogi T, Sunako Y, Niki H, Yamanaka K, Ichimura T, Hiraga S. Complex formation of MukB, MukE and MukF proteins involved in chromosome partitioning in Escherichia coli. The EMBO journal. 1999;18:5873–5884. doi: 10.1093/emboj/18.21.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohsumi K, Yamazoe M, Hiraga S. Different localization of SeqA-bound nascent DNA clusters and MukF-MukE-MukB complex in Escherichia coli cells. Molecular microbiology. 2001;40:835–845. doi: 10.1046/j.1365-2958.2001.02447.x. [DOI] [PubMed] [Google Scholar]
- 33.Parker BO, Marinus MG. Repair of DNA heteroduplexes containing small heterologous sequences in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:1730–1734. doi: 10.1073/pnas.89.5.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su SS, Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to a G × T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 36.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 37.Marsischky GT, Filosi N, Kane MF, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes & development. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 38.Gupta S, Gellert M, Yang W. Mechanism of mismatch recognition revealed by human MutSbeta bound to unpaired DNA loops. Nature structural & molecular biology. 2012;19:72–78. doi: 10.1038/nsmb.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warren JJ, Pohlhaus TJ, Changela A, Iyer RR, Modrich PL, Beese LS. Structure of the human MutSalpha DNA lesion recognition complex. Molecular cell. 2007;26:579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 40.Gradia S, Subramanian D, Wilson T, Acharya S, Makhov A, Griffith J, Fishel R. hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Molecular cell. 1999;3:255–261. doi: 10.1016/s1097-2765(00)80316-0. [DOI] [PubMed] [Google Scholar]
- 41.Mendillo ML, Mazur DJ, Kolodner RD. Analysis of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 and MLH1-PMS1 complexes with DNA using a reversible DNA end-blocking system. The Journal of biological chemistry. 2005;280:22245–22257. doi: 10.1074/jbc.M407545200. [DOI] [PubMed] [Google Scholar]
- 42.Hura GL, Tsai CL, Claridge SA, Mendillo ML, Smith JM, Williams GJ, Mastroianni AJ, Alivisatos AP, Putnam CD, Kolodner RD, Tainer JA. DNA conformations in mismatch repair probed in solution by X-ray scattering from gold nanocrystals. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:17308–17313. doi: 10.1073/pnas.1308595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 44.Groothuizen FS, Winkler I, Cristovao M, Fish A, Winterwerp HH, Reumer A, Marx AD, Hermans N, Nicholls RA, Murshudov GN, Lebbink JH, Friedhoff P, Sixma TK. MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA. Elife. 2015;4 doi: 10.7554/eLife.06744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mendillo ML, Putnam CD, Mo AO, Jamison JW, Li S, Woods VL, Jr, Kolodner RD. Probing DNA- and ATP-mediated conformational changes in the MutS family of mispair recognition proteins using deuterium exchange mass spectrometry. The Journal of biological chemistry. 2010;285:13170–13182. doi: 10.1074/jbc.M110.108894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mendillo ML, Hargreaves VV, Jamison JW, Mo AO, Li S, Putnam CD, Woods VL, Jr, Kolodner RD. A conserved MutS homolog connector domain interface interacts with MutL homologs. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:22223–22228. doi: 10.1073/pnas.0912250106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robertson AB, Pattishall SR, Gibbons EA, Matson SW. MutL-catalyzed ATP hydrolysis is required at a post-UvrD loading step in methyl-directed mismatch repair. The Journal of biological chemistry. 2006;281:19949–19959. doi: 10.1074/jbc.M601604200. [DOI] [PubMed] [Google Scholar]
- 48.Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends in biochemical sciences. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 49.Gueneau E, Dherin C, Legrand P, Tellier-Lebegue C, Gilquin B, Bonnesoeur P, Londino F, Quemener C, Le Du MH, Marquez JA, Moutiez M, Gondry M, Boiteux S, Charbonnier JB. Structure of the MutLalpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site. Nature structural & molecular biology. 2013;20:461–468. doi: 10.1038/nsmb.2511. [DOI] [PubMed] [Google Scholar]
- 50.Namadurai S, Jain D, Kulkarni DS, Tabib CR, Friedhoff P, Rao DN, Nair DT. The C-terminal domain of the MutL homolog from Neisseria gonorrhoeae forms an inverted homodimer. PloS one. 2010;5:e13726. doi: 10.1371/journal.pone.0013726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pillon MC, Lorenowicz JJ, Uckelmann M, Klocko AD, Mitchell RR, Chung YS, Modrich P, Walker GC, Simmons LA, Friedhoff P, Guarne A. Structure of the endonuclease domain of MutL: unlicensed to cut. Molecular cell. 2010;39:145–151. doi: 10.1016/j.molcel.2010.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mizushima R, Kim JY, Suetake I, Tanaka H, Takai T, Kamiya N, Takano Y, Mishima Y, Tajima S, Goto Y, Fukui K, Lee YH. NMR characterization of the interaction of the endonuclease domain of MutL with divalent metal ions and ATP. PloS one. 2014;9:e98554. doi: 10.1371/journal.pone.0098554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 54.Kadyrov FA, Holmes SF, Arana ME, Lukianova OA, O’Donnell M, Kunkel TA, Modrich P. Saccharomyces cerevisiae MutLalpha is a mismatch repair endonuclease. The Journal of biological chemistry. 2007;282:37181–37190. doi: 10.1074/jbc.M707617200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kosinski J, Plotz G, Guarne A, Bujnicki JM, Friedhoff P. The PMS2 subunit of human MutLalpha contains a metal ion binding domain of the iron-dependent repressor protein family. Journal of molecular biology. 2008;382:610–627. doi: 10.1016/j.jmb.2008.06.056. [DOI] [PubMed] [Google Scholar]
- 56.Duppatla V, Bodda C, Urbanke C, Friedhoff P, Rao DN. The C-terminal domain is sufficient for endonuclease activity of Neisseria gonorrhoeae MutL. The Biochemical journal. 2009;423:265–277. doi: 10.1042/BJ20090626. [DOI] [PubMed] [Google Scholar]
- 57.Guarne A, Ramon-Maiques S, Wolff EM, Ghirlando R, Hu X, Miller JH, Yang W. Structure of the MutL C-terminal domain: a model of intact MutL and its roles in mismatch repair. The EMBO journal. 2004;23:4134–4145. doi: 10.1038/sj.emboj.7600412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ahrends R, Kosinski J, Kirsch D, Manelyte L, Giron-Monzon L, Hummerich L, Schulz O, Spengler B, Friedhoff P. Identifying an interaction site between MutH and the C-terminal domain of MutL by crosslinking, affinity purification, chemical coding and mass spectrometry. Nucleic acids research. 2006;34:3169–3180. doi: 10.1093/nar/gkl407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hall MC, Jordan JR, Matson SW. Evidence for a physical interaction between the Escherichia coli methyl-directed mismatch repair proteins MutL and UvrD. The EMBO journal. 1998;17:1535–1541. doi: 10.1093/emboj/17.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hall MC, Matson SW. The Escherichia coli MutL protein physically interacts with MutH and stimulates the MutH-associated endonuclease activity. The Journal of biological chemistry. 1999;274:1306–1312. doi: 10.1074/jbc.274.3.1306. [DOI] [PubMed] [Google Scholar]
- 61.Pluciennik A, Modrich P. Protein roadblocks and helix discontinuities are barriers to the initiation of mismatch repair. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12709–12713. doi: 10.1073/pnas.0705129104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gorman J, Plys AJ, Visnapuu ML, Alani E, Greene EC. Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nature structural & molecular biology. 2010;17:932–938. doi: 10.1038/nsmb.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Plys AJ, Rogacheva MV, Greene EC, Alani E. The unstructured linker arms of Mlh1-Pms1 are important for interactions with DNA during mismatch repair. Journal of molecular biology. 2012;422:192–203. doi: 10.1016/j.jmb.2012.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Welsh KM, Lu AL, Clark S, Modrich P. Isolation and characterization of the Escherichia coli mutH gene product. The Journal of biological chemistry. 1987;262:15624–15629. [PubMed] [Google Scholar]
- 65.Lahue RS, Au KG, Modrich P. DNA mismatch correction in a defined system. Science. 1989;245:160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- 66.Au KG, Welsh K, Modrich P. Initiation of methyl-directed mismatch repair. The Journal of biological chemistry. 1992;267:12142–12148. [PubMed] [Google Scholar]
- 67.Hasan AM, Leach DR. Chromosomal directionality of DNA mismatch repair in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:9388–9393. doi: 10.1073/pnas.1505370112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dao V, Modrich P. Mismatch-, MutS-, MutL-, and helicase II-dependent unwinding from the single-strand break of an incised heteroduplex. The Journal of biological chemistry. 1998;273:9202–9207. doi: 10.1074/jbc.273.15.9202. [DOI] [PubMed] [Google Scholar]
- 69.Yamaguchi M, Dao V, Modrich P. MutS and MutL activate DNA helicase II in a mismatch-dependent manner. The Journal of biological chemistry. 1998;273:9197–9201. doi: 10.1074/jbc.273.15.9197. [DOI] [PubMed] [Google Scholar]
- 70.Burdett V, Baitinger C, Viswanathan M, Lovett ST, Modrich P. In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:6765–6770. doi: 10.1073/pnas.121183298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Viswanathan M, Burdett V, Baitinger C, Modrich P, Lovett ST. Redundant exonuclease involvement in Escherichia coli methyl-directed mismatch repair. The Journal of biological chemistry. 2001;276:31053–31058. doi: 10.1074/jbc.M105481200. [DOI] [PubMed] [Google Scholar]
- 72.Grilley M, Griffith J, Modrich P. Bidirectional excision in methyl-directed mismatch repair. The Journal of biological chemistry. 1993;268:11830–11837. [PubMed] [Google Scholar]
- 73.Ali JA, Lohman TM. Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science. 1997;275:377–380. doi: 10.1126/science.275.5298.377. [DOI] [PubMed] [Google Scholar]
- 74.Matson SW, Robertson AB. The UvrD helicase and its modulation by the mismatch repair protein MutL. Nucleic acids research. 2006;34:4089–4097. doi: 10.1093/nar/gkl450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brenner DJ, Krieg NR, Staley JT, Garrity GM. Bergey’s Manual of Systematic Bacteriology, Springer, New York. 2005 [Google Scholar]
- 76.Gao B, Mohan R, Gupta RS. Phylogenomics and protein signatures elucidating the evolutionary relationships among the Gammaproteobacteria. International journal of systematic and evolutionary microbiology. 2009;59:234–247. doi: 10.1099/ijs.0.002741-0. [DOI] [PubMed] [Google Scholar]
- 77.Cutino-Jimenez AM, Martins-Pinheiro M, Lima WC, Martin-Tornet A, Morales OG, Menck CF. Evolutionary placement of Xanthomonadales based on conserved protein signature sequences. Molecular phylogenetics and evolution. 2010;54:524–534. doi: 10.1016/j.ympev.2009.09.026. [DOI] [PubMed] [Google Scholar]
- 78.Naushad HS, Gupta RS. Molecular signatures (conserved indels) in protein sequences that are specific for the order Pasteurellales and distinguish two of its main clades. Antonie van Leeuwenhoek. 2012;101:105–124. doi: 10.1007/s10482-011-9628-4. [DOI] [PubMed] [Google Scholar]
- 79.Williams KP, Gillespie JJ, Sobral BW, Nordberg EK, Snyder EE, Shallom JM, Dickerman AW. Phylogeny of gammaproteobacteria. Journal of bacteriology. 2010;192:2305–2314. doi: 10.1128/JB.01480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rokas A, Williams BL, King N, Carroll SB. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003;425:798–804. doi: 10.1038/nature02053. [DOI] [PubMed] [Google Scholar]
- 81.Gupta RS. Protein phylogenies and signature sequences: A reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiology and molecular biology reviews: MMBR. 1998;62:1435–1491. doi: 10.1128/mmbr.62.4.1435-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2129. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 83.Hiraga S, Ichinose C, Onogi T, Niki H, Yamazoe M. Bidirectional migration of SeqA-bound hemimethylated DNA clusters and pairing of oriC copies in Escherichia coli. Genes to cells: devoted to molecular & cellular mechanisms. 2000;5:327–341. doi: 10.1046/j.1365-2443.2000.00334.x. [DOI] [PubMed] [Google Scholar]
- 84.Brezellec P, Hoebeke M, Hiet MS, Pasek S, Ferat JL. DomainSieve: a protein domain-based screen that led to the identification of dam-associated genes with potential link to DNA maintenance. Bioinformatics. 2006;22:1935–1941. doi: 10.1093/bioinformatics/btl336. [DOI] [PubMed] [Google Scholar]
- 85.Eisen JA, Hanawalt PC. A phylogenomic study of DNA repair genes, proteins, and processes. Mutation research. 1999;435:171–213. doi: 10.1016/s0921-8777(99)00050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Eisen JA. A phylogenomic study of the MutS family of proteins. Nucleic acids research. 1998;26:4291–4300. doi: 10.1093/nar/26.18.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lin Z, Nei M, Ma H. The origins and early evolution of DNA mismatch repair genes–multiple horizontal gene transfers and co-evolution. Nucleic acids research. 2007;35:7591–7603. doi: 10.1093/nar/gkm921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ogata H, Ray J, Toyoda K, Sandaa RA, Nagasaki K, Bratbak G, Claverie JM. Two new subfamilies of DNA mismatch repair proteins (MutS) specifically abundant in the marine environment. The ISME journal. 2011;5:1143–1151. doi: 10.1038/ismej.2010.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sachadyn P. Conservation and diversity of MutS proteins. Mutation research. 2010;694:20–30. doi: 10.1016/j.mrfmmm.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 90.Pinto AV, Mathieu A, Marsin S, Veaute X, Ielpi L, Labigne A, Radicella JP. Suppression of homologous and homeologous recombination by the bacterial MutS2 protein. Molecular cell. 2005;17:113–120. doi: 10.1016/j.molcel.2004.11.035. [DOI] [PubMed] [Google Scholar]
- 91.Kang J, Huang S, Blaser MJ. Structural and functional divergence of MutS2 from bacterial MutS1 and eukaryotic MSH4-MSH5 homologs. Journal of bacteriology. 2005;187:3528–3537. doi: 10.1128/JB.187.10.3528-3537.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lang JM, Darling AE, Eisen JA. Phylogeny of bacterial and archaeal genomes using conserved genes: supertrees and supermatrices. PloS one. 2013;8:e62510. doi: 10.1371/journal.pone.0062510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gruber S. MukBEF on the march: taking over chromosome organization in bacteria? Molecular microbiology. 2011;81:855–859. doi: 10.1111/j.1365-2958.2011.07764.x. [DOI] [PubMed] [Google Scholar]
- 94.Campbell MA, Chain PS, Dang H, El Sheikh AF, Norton JM, Ward NL, Ward BB, Klotz MG. Nitrosococcus watsonii sp. nov., a new species of marine obligate ammonia-oxidizing bacteria that is not omnipresent in the world’s oceans: calls to validate the names ‘Nitrosococcus halophilus’ and ‘Nitrosomonas mobilis’. FEMS microbiology ecology. 2011;76:39–48. doi: 10.1111/j.1574-6941.2010.01027.x. [DOI] [PubMed] [Google Scholar]
- 95.Oliver A, Baquero F, Blazquez J. The mismatch repair system (mutS, mutL and uvrD genes) in Pseudomonas aeruginosa: molecular characterization of naturally occurring mutants. Molecular microbiology. 2002;43:1641–1650. doi: 10.1046/j.1365-2958.2002.02855.x. [DOI] [PubMed] [Google Scholar]
- 96.Jacquelin DK, Filiberti A, Argarana CE, Barra JL. Pseudomonas aeruginosa MutL protein functions in Escherichia coli. The Biochemical journal. 2005;388:879–887. doi: 10.1042/BJ20042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ban C, Yang W. Structural basis for MutH activation in E.coli mismatch repair and relationship of MutH to restriction endonucleases. The EMBO journal. 1998;17:1526–1534. doi: 10.1093/emboj/17.5.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chan SH, Stoddard BL, Xu SY. Natural and engineered nicking endonucleases--from cleavage mechanism to engineering of strand-specificity. Nucleic acids research. 2011;39:1–18. doi: 10.1093/nar/gkq742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Friedhoff P, Lurz R, Luder G, Pingoud A. Sau3AI, a monomeric type II restriction endonuclease that dimerizes on the DNA and thereby induces DNA loops. The Journal of biological chemistry. 2001;276:23581–23588. doi: 10.1074/jbc.M101694200. [DOI] [PubMed] [Google Scholar]
- 100.Lee JY, Chang J, Joseph N, Ghirlando R, Rao DN, Yang W. MutH complexed with hemi- and unmethylated DNAs: coupling base recognition and DNA cleavage. Molecular cell. 2005;20:155–166. doi: 10.1016/j.molcel.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 101.Friedhoff P, Thomas E, Pingoud A. Tyr212: a key residue involved in strand discrimination by the DNA mismatch repair endonuclease MutH. Journal of molecular biology. 2003;325:285–297. doi: 10.1016/s0022-2836(02)01224-x. [DOI] [PubMed] [Google Scholar]
- 102.Bujnicki JM. A model of structure and action of Sau3AI restriction endonuclease that comprises two MutH-like endonuclease domains within a single polypeptide. Acta microbiologica Polonica. 2001;50:219–231. [PubMed] [Google Scholar]
- 103.Xu CY, Yu F, Xu SJ, Ding Y, Sun LH, Tang L, Hu XJ, Zhang ZH, He JH. Crystal structure and function of C-terminal Sau3AI domain. Biochimica et biophysica acta. 2009;1794:118–123. doi: 10.1016/j.bbapap.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 104.Lieb M, Bhagwat AS. Very short patch repair: reducing the cost of cytosine methylation. Molecular microbiology. 1996;20:467–473. doi: 10.1046/j.1365-2958.1996.5291066.x. [DOI] [PubMed] [Google Scholar]
- 105.Gould SJ. Wonderful Life: The Burgess Shale and the Nature of History. W. W. Norton and Company; New York: 1990. [Google Scholar]
- 106.Revel HR. DNA modification: Glucosylation. In: Mathews CK, Kutter EM, Mosig G, Berget PB, editors. Bacteriophage T4. ASM; Washington, DC: 1983. pp. 156–165. [Google Scholar]
- 107.Thomsen MC, Nielsen M. Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic acids research. 2012;40:W281–287. doi: 10.1093/nar/gks469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ban C, Junop M, Yang W. Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell. 1999;97:85–97. doi: 10.1016/s0092-8674(00)80717-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.