Abstract
Opioid dependence, a severe addictive disorder and major societal problem, has been demonstrated to be moderately heritable. We conducted a genome-wide association study in Comorbidity and Trauma Study data comparing opioid dependent daily injectors (N=1167) with opioid misusers who never progressed to daily injection (N=161). The strongest associations, observed for CNIH3 SNPs, were confirmed in two independent samples, the Yale-Penn genetic studies of opioid, cocaine, and alcohol dependence and the Study of Addiction: Genetics and Environment, which both contain non-dependent opioid misusers and opioid dependent individuals. Meta-analyses found 5 genome-wide significant CNIH3 SNPs. The A allele of rs10799590, the most highly associated SNP, was robustly protective [p=4.30E-9; OR 0.64 (95%CI 0.55 – 0.74)]. Epigenetic annotation predicts that this SNP is functional in fetal brain. Neuroimaging data from the Duke Neurogenetics Study (N=312) provide evidence of this SNP’s in vivo functionality; rs10799590 A allele carriers displayed significantly greater right amygdala habituation to threat-related facial expressions, a phenotype associated with resilience to psychopathology. Computational genetic analyses of physical dependence on morphine across 23 mouse strains yielded significant correlations for haplotypes in CNIH3 and functionally-related genes. These convergent findings support CNIH3 involvement in the pathophysiology of opioid dependence complementing prior studies implicating the AMPA glutamate system.
INTRODUCTION
Twin and family studies provide evidence of a genetic contribution to liability for opioid dependence with heritability estimates ranging from 40 to 60%.1–4 However, genetic association studies have produced few consistently replicated findings.5 One important factor contributing to this inconsistency is the lack of a definitive control population6,7 for these investigations.
Studies e.g.,8 have used unassessed controls based on the premise that, for low prevalence disorders, this approach only modestly reduces power.9 However, opioids are among the most highly addictive drugs10,11 with high rates of progression from misuse to dependence12 and thus the main constraint on the prevalence of opioid dependence may be the prevalence of opioid misuse. The extent to which genetic factors contribute to liability at various stages of opioid addiction (e.g., initiation, regular use, and dependence), and are shared between stages, is not well characterized.5 Thus, the use of unassessed, predominately unexposed controls might be problematic for identifying genetic effects expressed after the initiation of opioid misuse. Importantly, significant effects of common SNPs manifesting at intermediate and later stages of addiction would be missed in comparisons to unexposed controls. Similarly, comparisons to assessed, unexposed controls are more useful to examine shared liability for initiation and dependence.
Analyses of candidate gene data in the current report’s discovery sample, the Comorbidity and Trauma Study (CATS), showed that association findings vary substantially depending on the comparison group’s substance exposure status.6,7 The current investigation builds on this observation and draws from genetic studies of licit drugs13–19 that have yielded well-replicated findings by comparing non-dependent, drug-exposed to substance dependent individuals. Unfortunately, no large samples of non-dependent opioid misusers have been collected. Our discovery6,7,20 and confirmation samples19,21–24 contain only modest numbers of non-dependent opioid misusers, but are currently the largest such samples with genome-wide association study (GWAS) data. We hypothesize that genetic polymorphisms in opioid misusers influence progression to the population’s opioid dependence endpoint (ODE). Our analyses of GWAS data observed the strongest association for cornichon family AMPA receptor auxiliary protein 3 (CNIH3) polymorphisms, findings which map nicely onto literature25–29 supporting AMPA glutamate system involvement in the pathophysiology of opioid dependence.
MATERIALS AND METHODS
GWAS sample subjects and assessment
Detailed descriptions of CATS data collection have been reported.6,7,20 Opioid dependent individuals, aged 18 or older, were recruited from opioid substitution therapy (OST) clinics in the greater Sydney region. Neighborhood controls, individuals with little or no lifetime opioid misuse, were recruited from socially disadvantaged neighborhoods in geographic proximity to OST clinics. Written informed consent was obtained from all participants as approved by the institutional review boards (IRBs) of all participating institutions and clinics. Semi-structured psychiatric diagnostic interviews, a modified Semi-Structured Assessment for the Genetics of Alcoholism - Australia (SSAGA-OZ),30 were completed in-person.
Since 94.1% of the CATS6,7,20 opioid dependent participants reported a period of daily injection, we operationalized having had such a period as the population’s normative opioid dependence endpoint (ODE). Comparisons of opioid dependent individuals who differed on daily injection status found substantial phenotypic differences (Supplementary Table 1 and Supplementary Methods). Comparisons of dependent individuals who never injected daily to non-dependent opioid misusers revealed fewer significant differences. Our GWAS analyses compared the ODE group (N=1167 opioid dependent daily injectors) to a group characterized as having opioid use with impeded progression (OUIP) that combined non-dependent opioid misusers (N=88; 69.3% reporting heroin use) and opioid dependent individuals without a history of daily injection (N=73).
The Yale-Penn genetic studies of opioid, cocaine, and alcohol dependence19,23,24 were recruited at 5 U.S. sites. All participants gave written informed consent as approved by each site’s IRB. We addressed design and assessment differences (Supplementary Tables 2 and 3, and Supplementary Methods) that prevented defining phenotypes identical to those in CATS by using an extreme discordant approach. We operationalized the ODE group as opioid dependent individuals whose opioid use had been daily or near daily, included heroin, and injection at least 100 times lifetime and the OUIP group as individuals reporting heroin use who met no lifetime DSM-IV opioid dependence criteria. We limited inclusion to European ancestry (EA) participants to examine confirmation in a sample of comparable ethnicity, retaining 643 ODE and 157 OUIP individuals for analysis.
The Study of Addiction: Genetics and Environment (SAGE)21,22 is an alcohol dependence GWAS that selected cases and controls from large investigations targeting non-opioid substance dependence. Each contributing institution’s IRB approved the recruitment protocols. All participants provided written informed consent. Since SAGE did not ascertain participants on the basis of opioid dependence, it included fewer severely dependent individuals and more participants of unclear affection status. We operationalized the ODE group as DSM-IV opioid dependence and the OUIP group as opioid misusers who met at most one dependence and no abuse criterion. Limiting inclusion to EA participants, we retained 190 ODE and 319 OUIP individuals.
Genotyping and data cleaning
CATS samples were genotyped using the Illumina Human660W-Quad BeadChip at the Johns Hopkins Center for Inherited Disease Research (CIDR). For data cleaning details, see Supplementary Methods. The genotyping rate for the 470,296 SNPs that remained after data cleaning was 99.93%.
The Yale-Penn samples were genotyped on the Illumina HumanOmni1-Quad v1.0 microarray at CIDR and the Yale Center for Genome Analysis. SAGE samples were genotyped at CIDR using Illumina Human 1Mv1_C BeadChips. Genotypic data cleaning and quality control details have been reported for the Yale-Penn studies19,23,24 and SAGE.21,22
Data analyses
Admixture
PCA was conducted using the SmartPCA program31 to provide additional admixture correction. Three PCs were generated via PCA and included as covariates in the regression models. Similar methods were used in the Yale-Penn and SAGE data sets to generate PCs for inclusion in analyses (consistent with their prior publications).19,21,23,24
SNP-based association
The genomic inflation factor for CATS data was calculated in PLINK32 based on the median chi-square value. Logistic regression analyses were performed in PLINK32 to examine the association between the log-additive effects of risk allele dosage and group status (ODE versus OUIP) controlling for sex, age category, and three PCs. Manhattan and qq plots were constructed for results.
Association analyses of confirmation sample data were conducted consistent with prior reports. The Yale-Penn data were analyzed using logistic regression models embedded in generalized estimating equations to correct for correlations of data from related individuals with age, sex, and three PCs included as covariates. Analyses of SAGE data were conducted in PLINK32 with contributing component study, age, sex, and two PCs included as covariates.
Meta analyses were performed using the inverse variance weighting approach of the METAL program.33 The phenotypic variance in ODE status explained by rs10799590 was calculated34 for the meta-analytic results using odds ratios, risk allele frequencies (RAFs), and prevalence estimates ranging from 0.005 (population prevalence of heroin dependence)12,35,36 to 0.25 (approximate prevalence reported for heroin dependence among users).12
Epigenetic annotation (see Supplementary Methods)
Duke Neurogenetics Study (DNS) (see Supplementary Methods)
Participants
DNS participants were in good general health and provided informed written consent approved by the Duke University Medical Center IRB. The EA subsample reported here consisted of 312 participants (age = 19.71 ± 1.23 years; 151 males; 65 with DSM-IV diagnoses).
Genotyping
DNS participants’ DNA was isolated from saliva and genotyped with Illumina HumanOmniExpress BeadChips. The genotyping rate of rs10799590 was 1.0 and was within HWE χ2=0.39, p=0.53.
DNS neuroimaging protocol BOLD fMRI paradigm
A widely used and reliable challenge paradigm was employed to elicit amygdala reactivity. The paradigm consists of 4 task blocks requiring face-matching interleaved with 5 control blocks requiring shape-matching (see Figure S1). All 4 facial expressions convey threat, ambiguity, and/or novelty that robustly recruit corticolimbic circuitry that includes the amygdala.37
BOLD fMRI data analysis
Following the preprocessing steps, linear contrasts employing canonical hemodynamic response functions were used to estimate amygdala habituation as the linear decrease over successive face matching blocks (i.e., block 1 > block 2 > block 3 > block 4). Follow-up analyses evaluated amygdala response differences in block 1 across genotype groups. Individual contrast images (i.e., weighted sum of beta images) were used in second-level random effects models accounting for scan-to-scan and participant-to-participant variability to determine mean contrast-specific responses using one-sample t-tests. A voxel-level statistical threshold of P < 0.05, family wise error corrected for multiple comparisons across the bilateral amygdala regions of interest, and a cluster-level extent threshold of 10 contiguous voxels was applied to these analyses. The bilateral amygdala regions of interest (ROI) were defined using the AAL template. BOLD parameter estimates from maximal voxels in the right and left amygdala ROI exhibiting a main effect for the amygdala habituation contrast were extracted using the VOI tool in SPM8 and exported for regression analyses in SPSS (v.22). Extracting parameter estimates from clusters activated by our fMRI paradigm, rather than those specifically correlated with our independent variables of interest, precludes the possibility of any correlation coefficient inflation that may result when an explanatory covariate is used to select a region of interest.
Statistical Analyses
Statistical analyses of the imaging data were conducted using linear regression in SPSS to test the association between rs10799590 A allele carrier status and amygdala habituation (i.e., declining amygdala response to repeated stimuli). To maintain variability but constrain the influence of extreme outliers, all imaging variables were winsorized prior to analyses. Gender and psychiatric diagnosis (0,1) were entered as covariates for analyses. To determine whether rs1079950 was most strongly associated with amygdala habituation within this genomic region, SNPs ± 100 kbp that had an LD r2 ≥ .50 with rs10799590 within the dataset were identified and binned according to LD (r2 ≥ .80). Similar analyses were then conducted to examine the association between carrier status of each SNP and amygdala habituation.
Genetic analysis of murine interstrain differences
Male mice, aged 7–8 weeks, from 23 inbred strains (Supplementary Table 5) were housed in Stanford University’s animal care facilities. Experimental protocols were approved by the Institutional Animal Care and Use Committee and complied with the Guide for the Care and Use of Laboratory Animals. Details are provided in Supplementary Methods. ANOVA was used to calculate a P value to assess the likelihood that the within-block genetic variation underlies the phenotypic distribution (i.e., the mean number of jumps) observed for the inbred strains examined. Haplotype data were examined for seven genes: CNIH3, CNIH2, GRIA1, GRIA2, CACNG8, GRIP1, and DLG4. P values were adjusted to control the false-discovery rate.38
RESULTS
CATS GWAS
The quantile-quantile (q-q) plot of association results (Figure S2, Panel A) and the genomic inflation factor value (λ=1.01) indicate an absence of test statistic inflation. The strongest association involves a cluster of chromosome 1 SNPs (Manhattan plot, Figure S2, Panel B). The six most highly associated, all located in CNIH3 (Table 1), are in moderate to high LD (r2=0.35–0.97); conditional analyses suggest they represent a single association signal (Figure S2, Panel C). The association reached genome-wide significance (GWS) for rs1436175 [p=2.72 E-8; OR 0.50 (0.39–0.64)] with the risk allele halving the likelihood of progression to ODE.
Table 1.
SNPs associated with ODE in CATS participants (p values<1E-5)
| Gene | SNP | Chr | Genomic coordinates | RA | Risk Allele Frequency | p value | Odds Ratio (95% confidence interval) | |
|---|---|---|---|---|---|---|---|---|
| ODE N=1167 |
OUIP N=161 |
|||||||
| CNIH3 | rs10799590 | 1 | 224822482 | A | 0.42 | 0.56 | 1.51E-6 | 0.55 (0.43 – 0.70) |
| CNIH3 | rs12130499 | 1 | 224836514 | T | 0.42 | 0.56 | 1.15E-6 | 0.54 (0.43 – 0.70) |
| CNIH3 | rs298733 | 1 | 224842251 | A | 0.42 | 0.57 | 1.53E-6 | 0.55 (0.43 – 0.70) |
| CNIH3 | rs1436171 | 1 | 224881828 | A | 0.44 | 0.59 | 6.26E-7 | 0.54 (0.42 – 0.68) |
| CNIH3 | rs1369846 | 1 | 224894095 | C | 0.38 | 0.54 | 9.42E-8 | 0.52 (0.41 – 0.66) |
| CNIH3 | rs1436175 | 1 | 224908366 | T | 0.37 | 0.53 | 2.72E-8 | 0.50 (0.39 – 0.64) |
| STAB2 | rs10861067 | 12 | 104038974 | G | 0.25 | 0.38 | 2.00E-6 | 0.55 (0.43 – 0.71) |
| STAB2 | rs10778270 | 12 | 104045678 | A | 0.20 | 0.31 | 5.16E-6 | 0.55 (0.42 – 0.71) |
| intergenic | rs9521590 | 13 | 110709426 | A | 0.05 | 0.11 | 4.31E-6 | 0.36 (0.23 – 0.55) |
| FUT8 | rs6573615 | 14 | 66046534 | G | 0.36 | 0.49 | 9.68E-6 | 0.58 (0.46 – 0.74) |
Confirmation of association findings
These analyses focused on the CNIH3 SNPs because of the substantially stronger association observed for these polymorphisms and the gene’s obvious biological relevance. In the Yale-Penn data (Table 2), trend-level association was observed for three of the six SNPs examined. In the SAGE data, a stronger association was found for five of the six SNPs. In both data sets, all association signals were in the same direction as in the CATS.
Table 2.
Association of CNIH3 SNPs with ODE in CATS, Yale-Penn, and SAGE data, and in meta-analysis
| SNP | CATS (1167 ODE vs 161 OUIP) | Yale-Penn (643 ODE vs 157 OUIP) | SAGE (190 ODE vs 319 OUIP) | Meta-analysis | |||||
|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | p value | OR (95%CI) | p value | OR (95%CI) | p value | OR (95%CI) | p value | Het X2 (2df) p value | |
| rs10799590 | 0.55 (0.43 – 0.70) | 1.51E-6 | 0.78 (0.60 – 1.01) | 5.76E-2 | 0.62 (0.47– 0.82) | 6.73E-4 | 0.64 (0.55 – 0.74) | 4.30E-9 | 0.15 |
| rs12130499 | 0.54 (0.43 – 0.70) | 1.15E-6 | 0.78 (0.60– 1.01) | 6.14E-2 | 0.62 (0.47 – 0.82) | 7.49E-4 | 0.64 (0.55 – 0.74) | 4.31E-9 | 0.13 |
| rs298733 | 0.55 (0.43 – 0.70) | 1.53E-6 | 0.79 (0.62 – 1.03) | 7.61E-2 | 0.64 (0.49 – 0.84) | 1.25E-3 | 0.65 (0.56 – 0.75) | 1.25E-8 | 0.12 |
| rs1436171 | 0.54 (0.42 – 0.68) | 6.26E-7 | 0.82 (0.64 – 1.04) | 1.07E-1 | 0.65 (0.49 – 0.85) | 2.04E-3 | 0.66 (0.57 – 0.76) | 2.17E-8 | 0.06 |
| rs1369846 | 0.52 (0.41 – 0.66) | 9.42E-8 | 0.85 (0.67 – 1.09) | 1.94E-1 | 0.66 (0.50 – 0.87) | 2.96E-3 | 0.66 (0.57 – 0.77) | 2.60E-8 | 0.02 |
| rs1436175 | 0.50 (0.39 – 0.64) | 2.72E-8 | 0.93 (0.73 – 1.18) | 5.50E-1 | 0.77 (0.59 – 1.01) | 6.14E-2 | 0.71 (0.61 – 0.82) | 3.09E-6 | 0.002 |
Meta-analyses performed on these SNPs using data from the three samples found GWS association for five of the six CNIH3 SNPs (Table 2). The strongest meta-analytic association signal (p=4.30 E-9) was observed for rs10799590; the odds ratio [0.64 (95%CI 0.55 – 0.74)] is indicative of the risk allele’s robust protective effects. Rs1436175, which had the lowest p value in the CATS, failed to reach meta-analytic GWS. This SNP has the lowest LD with the other CNIH3 SNPs (r2=0.35–0.54) and the largest heterogeneity chi-square (p=0.002). The meta-analytic GWS SNPs are in high LD (Figure S2, Panels C and D).
Epigenetic annotation
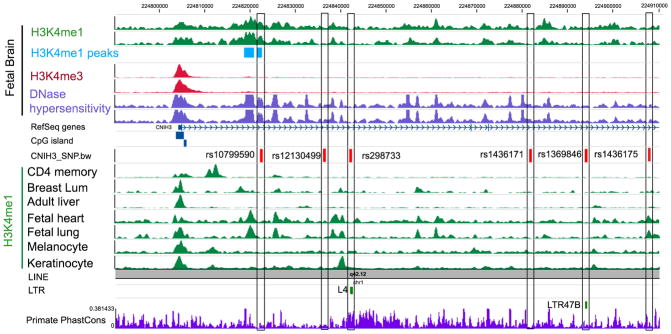
The GWS CNIH3 SNPs are intronic and not highly conserved. Although the observed associations may be due to high LD with a non-genotyped variant, no exonic SNPs in high LD were identified. Epigenetically-mediated changes in gene expression, which have been reported to occur with opioid use,39 are plausible mechanisms for functional associations involving intronic SNPs. Rs10799590 is located within an enhancer that is specific to fetal brain (Figure 1). It is within an H3K4me1peak in fetal brain that DNaseI hypersensitivity data indicate is in an open chromatin state.40–42 It is predicted that rs10799590 is within the binding site of transcription factor (TF) TAL1 (which plays important roles in middle brain GABAergic neuron differentiation68); the G allele has significantly higher binding potential than the A allele (Supplementary Table 6).
Figure 1. Epigenetic Landscape of the Six Intronic CNIH3 SNPs.
Rs1369846 and rs298733 are located within retrotransposons.40,41 Evidence of epigenetic functionality for rs10799590 includes the location of this SNP within a fetal brain specific enhancer.46,47 Fetal brain H3K4me1 data indicate that it is within a H3K4me1 peak; DNaseI hypersensitivity data suggest that it is in an open chromatin state in fetal brain. This enhancer mark on rs10799590 was specific to fetal brain, but was not in CD4+ T cells, breast luminal epithelial cells, adult liver, fetal heart, fetal lung, melanocyte, or keratinocytes. The results of motif analyses (Supplementary Table 6) predict that rs10799590 is within the binding site of transcription factor Tal1 with the G allele having significantly higher binding affinity than the A allele.
Protective allele carrier status predicts greater amygdala habituation
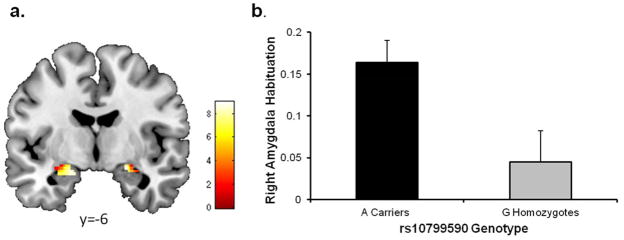
The observation25 that similar changes in AMPAR GluA1 subunits occur in the amygdala with opioid addiction26 and fear conditioning,43–45 coupled with evidence of epigenetically-mediated alterations in gene expression that ensue in both processes after environmental exposures,43,46 provided the rationale for examining amygdala habituation to threat-related facial expressions (a reliable intermediate phenotype linked to psychopathology).46,47 After accounting for sex and the presence of a DSM-IV disorder, rs10799590 A allele carrier status predicted right (stand Beta = 0.147; ΔF1,308=6.93, p < .009, ΔR2=0.022; Figure 2), but not left (stand Beta = 0.031; ΔF1,308=0.302, p > .582, ΔR2<0.001), amygdala habituation. G allele homozygotes (n=102) had blunted right amygdala habituation (0.045±0.374) relative to A allele carriers (n=210; 0.164±0.371). We identified 3 genotyped SNPs (rs1369848, rs12730234, rs1965776) in moderate LD with rs10799590 that tagged SNP blocks; however, none was more strongly associated with amygdala habituation than rs10799590 (Supplementary Table 7). Follow-up analyses revealed that genotype groups did not differ in initial right amygdala responses to stimuli (stand Beta = 0.067; ΔF1,308=1.42, p > .234, ΔR2=0.004).
Figure 2. Amygdala Habituation.
(a) Statistical parametric map illustrating mean bilateral amygdala habituation across all DNS participants (left MNI coordinates: x = −20 y = −8 z = −16, kE = 144, t = 9.03, P < 0.001; right MNI coordinates x = 22 y = −6 z = −14, kE = 83, t = 8.97, P < 0.001) (b) A allele carriers had greater right amygdala habituation (i.e., less persistent activation) relative to G allele homozygotes. The Y axis indicates habituation with greater values indicating a larger decrease in activation over time. See Figure S3 for a depiction of activation across blocks.
Genetic analysis of interstrain differences in physical dependence on morphine
To link our findings further to existing animal literature, we performed computational haplotype-based genetic mapping analyses48–50 of data from 23 inbred mouse strains for a robust measure of opioid physical dependence,48 counts of jumps made by morphine dependent mice after naloxone administration. Correlations were calculated for the distribution of the mean number of jumps per strain with known haplotype blocks across strains for CNIH3 and genes encoding AMPAR subunits and proteins involved in alterations of AMPAR subunit composition in response to opioids. Significant correlation was observed for CNIH3 haplotype, but not for the more widely expressed cornichon family AMPA receptor auxiliary protein 2 (CNIH2) (Table 3). Significant correlations were also noted for haplotypes in glutamate receptor, ionotropic, AMPA 1 (GRIA1), glutamate receptor, ionotropic, AMPA 2 (GRIA2), calcium channel, voltage-dependent, gamma subunit 8 (CACNG8), and glutamate receptor-interacting protein 1 (GRIP1), but not discs, large homolog 4 (DLG4)].
Table 3.
Computational genetic analysis of interstrain differences in physical dependence on morphine
| Gene | P value* |
|---|---|
| CNIH3 | 4.0E-4 |
| CACNG8 | 8.3E-4 |
| GRIA1 | 8.1E-6 |
| GRIP1 | 1.7E-5 |
| GRIA2 | 7.4E-3 |
| CNIH2 | .35 |
| DLG4 | .35 |
Corrected for multiple testing38
DISCUSSION
The current report provides evidence for CNIH3 involvement in the pathophysiology of opioid dependence. CNIH3 encodes a small, highly conserved protein. The AMPA receptor core is formed by tetramers of the GluA1-4 subunits and up to four members of three protein groups: transmembrane AMPAR regulatory proteins (TARPs), cornichon homologs (CNIH3 and CNIH2), and the GSG1l protein.51 The receptor’s periphery contains transmembrane and other proteins [e.g., post-synaptic density protein 95 (PSD-95)] that bind with core proteins and each other in the postsynaptic density (PSD).51–54 CNIH2 and CNIH3 markedly slow AMPAR deactivation and desensitization in heterologous systems.54–56 One study54 suggested that the actions of CNIH2 and CNIH3 are selective for AMPARs containing GluA1 subunits; however, more recent reports51,57 do not support this specificity of binding. An investigation that focused on two hippocampal cell types with markedly different excitatory postsynaptic currents (EPSCs) implicated CNIH2 as largely responsible for the distinction between fast and slow EPSCs. Although this report did not examine whether CNIH3 plays a similar role, prior studies e.g., 55 have found that the two proteins have comparable effects on slowing AMPAR deactivation and desensitization.
Rodent studies26–29 have implicated alterations in the subunit composition of brain AMPARs in diverse aspects of opioid addiction. Increased expression of GluA1-containing/GluA2-lacking AMPARs has been observed in the central nucleus of the amygdala26 and the hippocampal PSD28 in studies of morphine-related context-reward conditioning26 and context-dependent behavioral sensitization.28 The latter 28 found that these changes were mediated via interactions with TARP gamma-8 and GRIP1 proteins. GluA1 knockout mice displayed impaired drug-induced state dependency after operant conditioning with morphine.29 Another study27 implicated down-regulation of GluA2 expression in the prefrontal cortex in reinstatement of heroin self-administration after prolonged abstinence. Interestingly, CNIH3 expression is greatest in the frontal cortex, amygdala, and hippocampus in the adult human brain.58
Our GWAS analyses found that CNIH3 SNPs are associated with protection against progression to ODE in OUIP individuals. Since this effect was observed in analyses limited to opioid misusers, it likely represents liability unrelated to that for initiation of opioid misuse. The discovery and confirmation sample OUIP groups have substantially higher RAFs than an Australian (EA) general population sample59 (Supplementary Table 8) to which the ODE groups’ RAFs are more similar. Post-hoc SNP-based association analyses comparing the CATS OUIP group and this general population sample found substantial differences (p values ≤ 6.4E-5); similar comparisons with the CATS ODE group found more modest differences (p≥2.3E-2) in the opposite direction. The phenotypic variance in ODE explained by rs10799590 in our meta-analysis is estimated at 1.17% to 5.85% (Supplementary Table 9), indicative of a strong effect. Overall, our findings suggest that these CNIH3 SNPs enable greater, but not complete control in the use of these otherwise highly addictive drugs.
An examination of human post-mortem amygdalae reported a strong positive correlation between GluA1 and PSD-95 mRNA expression in heroin dependent cases, but not in controls (CNIH3 expression was not reported).25 Another study55 observed positive correlations for these proteins with CNIH3 (GluA1 0.43; DLG4 0.28) in (unexposed) mammalian brains. Thus, human25 and animal26 studies provide evidence of altered amygdala GluA1 expression in opioid dependence. The post-mortem report noted25 that similar changes in AMPAR GluA1 subunits occur in the amygdala with associative learning of opioid reward26 and fear conditioning.43–45 Importantly, both processes involve epigenetically-mediated changes in gene expression following an environmental exposure.39,46 We thus examined the effects of our most strongly associated SNP using imaging genetics46,60 and observed significantly greater right amygdala habituation to threat-related facial expression in rs10799590 A allele carriers. Consistent with the reduced risk we observed in opioid exposed individuals, the polymorphism’s protective neural effects were apparent with subsequent, but not initial, exposures to environmental stimuli. The association of this SNP with ODE likely represents a similar protective process involving greater habituation to the neural effects of opioids that impacts additional opioid use.
Our focus on opioid misusers is a major strength; the modest size of our OUIP groups is an unavoidable limitation. Our assumption that SNPs can offer protection against transitions at different points of the addictive process is supported by the existing animal literature.26–29 We only examined confirmation of the CNIH3 SNP associations; a broader examination might have confirmed other CATS associations. Since both the epigenetic landscape and motif analysis support its potential functionality, our neuroimaging genetics study focused on rs10799590 with post-hoc analyses confirming it as the CNIH3 SNP most strongly associated with amygdala habituation. Future work should incorporate examination of the other GWS SNPs (e.g., rs298733 also affects TF binding - Supplementary Table 6) and the possibility of more highly associated, non-genotyped polymorphisms in high LD. Although similarly ascertained samples e.g., 61 have comparable rates of daily opioid injection, reports in other populations have noted lower prevalence. e.g.,62 A more detailed characterization of the OUIP groups’ opioid use would have been useful, but was not obtained. We examined somewhat divergent ODE’s in our confirmation samples because of ascertainment and assessment differences (Supplementary Table 3). While these methodological differences are a limitation, the observed confirmation in two multi-site U.S.-based studies supports the generalizability of our findings. The highly comorbid composition of the ODE and OUIP groups may raise concerns that intergroup differences are better attributable to another phenotype. Post-hoc association analyses conducted in CATS to address this possibility (Supplementary Table 10) found few associations for comorbid disorders reaching nominal significance. Comorbidity pattern differences across samples (Supplementary Tables 1 and 2) further argue against this possibility. Finally, our exclusion of participants with non-European ancestry and the lack of genomic inflation (λ=1.01) suggest spurious association due to uncorrected admixture is unlikely.
Our meta-analyses found GWS association with CNIH3 SNPs conferring robust protective effects against ODE, findings that map onto reports of AMPA glutamatergic involvement in opioid dependence.25–29 The finding of significantly greater habituation in the right amygdala rs10799590 A allele carriers supports this SNP’s in vivo functional effects in humans (complementing evidence of epigenetic functionality). The genetic analyses of mouse strain data support the involvement of CNIH3, but not the more highly expressed CNIH2, in murine opioid physical dependence. The significant correlations observed for genes encoding AMPAR subunits and related proteins provide additional evidence for genetic risk mediated via this pathway. These convergent findings implicate CNIH3’s involvement in opioid dependence and could provide a route to target glutamatergic processes for translational research focusing on improving opioid dependence treatments and developing opioid analgesics with lower dependence risk.
Supplementary Material
Acknowledgments
Funding support for the Comorbidity and Trauma Study (CATS) was provided by the National Institute on Drug Abuse (R01 DA17305); GWAS genotyping services at the Center for Inherited Disease Research (CIDR) at The Johns Hopkins University were supported by the National Institutes of Health [contract N01-HG-65403].
Funding for the Yale-Penn genetic studies of opioid, cocaine, or alcohol dependence was provided by the National Institutes of Health [RC2 DA028909, R01 DA12690, R01 DA12849, R01 DA18432, R01 AA11330, R01 AA017535, 5T32GM007205-38, 8UL1TR000142-07]; and the Veterans Affairs Connecticut and Veterans Affairs Philadelphia Mental Illness Research, Education and Clinical Centers. Genotyping services at the Center for Inherited Disease Research (CIDR) at The Johns Hopkins University were supported by the National Institutes of Health [contract N01-HG-65403].
Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment and Health Initiative [GEI] (U01 HG004422). SAGE is one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of datasets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392), and the Family Study of Cocaine Dependence (FSCD; R01 DA013423, R01 DA019963). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse, and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C). The Duke Neurogenetics Study is supported by Duke University and National Institute on Drug Abuse grant DA033369. AA also receives support from R01 DA23668 and K02 DA32573. GWM and NRW are supported by NHMRC Fellowships. TW, BZ, and XZ receive support from DA027995, R01HG007354, R01HG007175, R01ES024992, and ACS grant RSG-14-049-01-DMC. A.R.H. receives support through National Institute on Drug Abuse grants DA033369 and DA031579. RB receives support from the Klingenstein Third Generation Foundation and the National Institutes of Health (NIA R01-AG045231). CHD receives support from T32DA007313. CEC receives support from NSF DGE-1143954.
Footnotes
Supplementary information is available at the Molecular Psychiatry website (http://www.nature.com/mp)
Conflict of Interest
Although unrelated to the current study, Dr. Kranzler has been a consultant or advisory board member for Alkermes, Lilly, Lundbeck, Pfizer, and Roche. He is also a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which is supported by Lilly, Lundbeck, Abbott, and Pfizer. No other authors declare any financial interests.
References
- 1.Merikangas KR, Stolar M, Stevens DE, Goulet J, Preisig MA, Fenton B, et al. Familial transmission of substance use disorders. Arch Gen Psychiatry. 1998;55:973–979. doi: 10.1001/archpsyc.55.11.973. [DOI] [PubMed] [Google Scholar]
- 2.Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J, et al. Co-occurrence of abuse of different drugs in men: the role of drug-specific and shared vulnerabilities. Arch Gen Psychiatry. 1998;55:967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- 3.Kendler KS, Jacobson KC, Prescott CA, Neale MC. Specificity of genetic and environmental risk factors for use and abuse/dependence of cannabis, cocaine, hallucinogens, sedatives, stimulants, and opiates in male twins. Am J Psychiatry. 2003;160:687–695. doi: 10.1176/appi.ajp.160.4.687. [DOI] [PubMed] [Google Scholar]
- 4.Sun J, Bi J, Chan G, Oslin D, Farrer L, Gelernter J, et al. Improved methods to identify stable, highly heritable subtypes of opioid use and related behaviors. Addict Behav. 2012;37:1138–1144. doi: 10.1016/j.addbeh.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agrawal A, Verweij KJ, Gillespie NA, Heath AC, Lessov-Schlaggar CN, Martin NG, et al. The genetics of addiction-a translational perspective. Transl Psychiatry. 2012;2:e140. doi: 10.1038/tp.2012.54. Erratum in: Transl Psychiatry 2012 (2), e193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson EC, Lynskey MT, Heath AC, Wray N, Agrawal A, Shand FL, et al. Association of OPRD1 polymorphisms with heroin dependence in a large case-control series. Addict Biol. 2014;19:111–121. doi: 10.1111/j.1369-1600.2012.00445.x. Epub 2012 Apr 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson EC, Lynskey MT, Heath AC, Wray N, Agrawal A, Shand F, et al. ANKK1, TTC12, and NCAM1 polymorphisms and heroin dependence: importance of considering drug exposure. JAMA Psychiatry. 2013;70:325–333. doi: 10.1001/jamapsychiatry.2013.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng S, Du J, Jiang H, Fu Y, Chen H, Sun H, et al. The dopamine receptor D1 gene is associated with the length of interval between first heroin use and onset of dependence in Chinese Han heroin addicts. J Neural Transm. 2013;120:1591–1598. doi: 10.1007/s00702-013-1029-6. [DOI] [PubMed] [Google Scholar]
- 9.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nutt D, King LA, Saulsbury W, Blakemore C. Development of a rational scale to assess the harm of drugs of potential misuse. Lancet. 2007;369:1047–1053. doi: 10.1016/S0140-6736(07)60464-4. [DOI] [PubMed] [Google Scholar]
- 11.Linares OA. The Linares addictive potential model. Journal of Pharmacy and Nutrition Sciences. 2012;2:132–139. [Google Scholar]
- 12.Anthony JC, Warner LA, Kessler RC. Comparative epidemiology of dependence on tobacco, alcohol, controlled substances, and inhalants basic findings From the National Comorbidity Survey. Exp Clin Psychopharmacol. 1994;2:244–268. [Google Scholar]
- 13.Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PAF, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42:436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bierut LJ, Goate AM, Breslau N, Johnson EO, Bertelsen S, Fox L, et al. ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Mol Psychiatry. 2012;17:445–450. doi: 10.1038/mp.2011.124. Epub 2011 Oct 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, et al. Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry. 2014;19:41–49. doi: 10.1038/mp.2013.145. Epub 2013 Oct 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shand FL, Degenhardt L, Slade T, Nelson EC. Sex differences amongst dependent heroin users: histories, clinical characteristics and predictors of other substance dependence. Addict Behav. 2011;36:27–36. doi: 10.1016/j.addbeh.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, et al. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci U S A. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennett SN, Caporaso N, Fitzpatrick AL, Agrawal A, Barnes K, Boyd HA, et al. Phenotype harmonization and cross-study collaboration in GWAS consortia: the GENEVA experience. Genet Epidemiol. 2011;35:159–173. doi: 10.1002/gepi.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gelernter J, Kranzler HR, Sherva R, Koesterer R, Almasy L, Zhao H, et al. Genome-wide association study of opioid cependence: multiple associations mapped to calcium and potassium pathways. Biol Psychiatry. 2014;76:66–74. doi: 10.1016/j.biopsych.2013.08.034. Epub 2013 Oct 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gelernter J, Sherva R, Koesterer R, Almasy L, Zhao H, Kranzler HR, et al. Genome-wide association study of cocaine dependence and related traits: FAM53B identified as a risk gene. Mol Psychiatry. 2014;19:717–723. doi: 10.1038/mp.2013.99. Epub 2013 Aug 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okvist A, Fagergren P, Whittard J, Garcia-Osta A, Drakenberg K, Horvath MC, et al. Dysregulated postsynaptic density and endocytic zone in the amygdala of human heroin and cocaine abusers. Biol Psychiatry. 2011;69:245–252. doi: 10.1016/j.biopsych.2010.09.037. Epub 2010 Dec 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai YQ, Wang W, Hou YY, Zhang Z, Xie J, Pan ZZ. Central amygdala GluA1 facilitates associative learning of opioid reward. J Neurosci. 2013;33:1577–1588. doi: 10.1523/JNEUROSCI.1749-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van den Oever MC, Goriounova NA, Li KW, Van der Schors RC, Binnekade R, Schoffelmeer AN, et al. Prefrontal cortex AMPA receptor plasticity is crucial for cue-induced relapse to heroin-seeking. Nat Neurosci. 2008;11:1053–1058. doi: 10.1038/nn.2165. [DOI] [PubMed] [Google Scholar]
- 28.Xia Y, Portugal GS, Fakira AK, Melyan Z, Neve R, Lee HT, et al. Hippocampal GluA1-containing AMPA receptors mediate context-dependent sensitization to morphine. J Neurosci. 2011;31:16279–16291. doi: 10.1523/JNEUROSCI.3835-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aitta-aho T, Möykkynen TP, Panhelainen AE, Vekovischeva OY, Bäckström P, Korpi ER. Importance of GluA1 subunit-containing AMPA glutamate receptors for morphine state-dependency. PLoS One. 2012;7:e38325. doi: 10.1371/journal.pone.0038325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr, et al. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- 31.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.So HC, Gui AH, Cherny SS, Sham PC. Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet Epidemiol. 2011;35:310–317. doi: 10.1002/gepi.20579. [DOI] [PubMed] [Google Scholar]
- 35.Hall WD, Ross JE, Lynskey MT, Law MG, Degenhardt LJ. How many dependent heroin users are there in Australia? Med J Aust. 2000;173:528–531. doi: 10.5694/j.1326-5377.2000.tb139321.x. [DOI] [PubMed] [Google Scholar]
- 36.Degenhardt L, Hall W. The extent of illicit drug use and dependence, and their contribution to the global burden of disease. Lancet. 2012;379:55–70. doi: 10.1016/S0140-6736(11)61138-0. [DOI] [PubMed] [Google Scholar]
- 37.Whalen PJ, Phelps EA, editors. The Human Amygdala. Guilford Press; New York: 2009. [Google Scholar]
- 38.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 39.Sun H, Maze I, Dietz DM, Scobie KN, Kennedy PJ, Damez-Werno D, et al. Morphine epigenomically regulates behavior through alterations in histone H3 lysine 9 dimethylation in the nucleus accumbens. J Neurosci. 2012;32:17454–17464. doi: 10.1523/JNEUROSCI.1357-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou X, Maricque B, Xie M, Li D, Sundaram V, Martin EA, et al. The Human Epigenome Browser at Washington University. Nat Methods. 2011;8:989–990. doi: 10.1038/nmeth.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou X, Lowdon RF, Li D, Lawson HA, Madden PAF, Costello JF, et al. Exploring long-range genome interactions using the WashU Epigenome Browser. Nat Methods. 2013;10:375–376. doi: 10.1038/nmeth.2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou X, Li D, Zhang B, Lowdon RF, Rockweiler NB, Sears RL, et al. Epigenomic annotation of genetic variants using the Roadmap Epigenome Browser. Nat Biotechnol. 2015 Feb 18; doi: 10.1038/nbt.3158. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeh SH, Mao SC, Lin HC, Gean PW. Synaptic expression of glutamate receptor after encoding of fear memory in the rat amygdala. Mol Pharmacol. 2006;69:299–308. doi: 10.1124/mol.105.017194. Epub 2005 Oct 11. [DOI] [PubMed] [Google Scholar]
- 44.Dalton GL, Wang YT, Floresco SB, Phillips AG. Disruption of AMPA receptor endocytosis impairs the extinction, but not acquisition of learned fear. Neuropsychopharmacology. 2008;33:2416–2426. doi: 10.1038/sj.npp.1301642. Epub 2007 Nov 28. [DOI] [PubMed] [Google Scholar]
- 45.Nedelescu H, Kelso CM, Lázaro-Muñoz G, Purpura M, Cain CK, Ledoux JE, et al. Endogenous GluR1-containing AMPA receptors translocate to asymmetric synapses in the lateral amygdala during the early phase of fear memory formation: an electron microscopic immunocytochemical study. J Comp Neurol. 2010;518:4723–4739. doi: 10.1002/cne.22472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nikolova YS, Koenen KC, Galea S, Wang CM, Seney ML, Sibille E, et al. Beyond genotype: serotonin transporter epigenetic modification predicts human brain function. Nat Neurosci. 2014;17:1153–1155. doi: 10.1038/nn.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plichta MM, Grimm O, Morgen K, Mier D, Sauer C, Haddad L, et al. Amygdala habituation: A reliable fMRI phenotype. Neuroimage. 2014;103C:383–390. doi: 10.1016/j.neuroimage.2014.09.059. [DOI] [PubMed] [Google Scholar]
- 48.Chu LF, Liang DY, Li X, Sahbaie P, D’arcy N, Liao G, et al. From mouse to man: the 5-HT3 receptor modulates physical dependence on opioid narcotics. Pharmacogenet Genomics. 2009;19:193–205. doi: 10.1097/FPC.0b013e328322e73d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng M, Dill D, Peltz G. A better prognosis for genetic association studies in mice. Trends Genet. 2012;28:62–69. doi: 10.1016/j.tig.2011.10.006. Epub 2011 Nov 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang DY, Zheng M, Sun Y, Sahbaie P, Low SA, Peltz G, et al. The Netrin-1 receptor DCC is a regulator of maladaptive responses to chronic morphine administration. BMC Genomics. 2014;15:345. doi: 10.1186/1471-2164-15-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwenk J, Baehrens D, Haupt A, Bildl W, Boudkkazi S, Roeper J, et al. Regional diversity and developmental dynamics of the AMPA-receptor proteome in the mammalian brain. Neuron. 2014;84:41–54. doi: 10.1016/j.neuron.2014.08.044. [DOI] [PubMed] [Google Scholar]
- 52.Sumioka A, Brown TE, Kato AS, Bredt DS, Kauer JA, Tomita S. PDZ binding of TARPγ-8 controls synaptic transmission but not synaptic plasticity. Nat Neurosci. 2011;14:1410–1412. doi: 10.1038/nn.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harmel N, Cokic B, Zolles G, Berkefeld H, Mauric V, Fakler B, et al. AMPA receptors commandeer an ancient cargo exporter for use as an auxiliary subunit for signaling. PLoS One. 2012;7:e30681. doi: 10.1371/journal.pone.0030681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Herring BE, Shi Y, Suh YH, Zheng CY, Blankenship SM, Roche KW, et al. Cornichon proteins determine the subunit composition of synaptic AMPA receptors. Neuron. 2013;77:1083–1096. doi: 10.1016/j.neuron.2013.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coombs ID, Soto D, Zonouzi M, Renzi M, Shelley C, Farrant M, et al. Cornichons modify channel properties of recombinant and glial AMPA receptors. J Neurosci. 2012;32:9796–9804. doi: 10.1523/JNEUROSCI.0345-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boudkkazi S, Brechet AJS, Fakler B. Cornichon2 dictates the time course of excitatory transmission at individual hippocampal synapses. Neuron. 2014;82:848–858. doi: 10.1016/j.neuron.2014.03.031. [DOI] [PubMed] [Google Scholar]
- 57.Shanks NF, Cais O, Maruo T, Savas JN, Zaika EI, Azumaya CM, et al. Molecular dissection of the interaction between the AMPA receptor and cornichon homolog-3. J Neurosci. 2014;34:12104–12120. doi: 10.1523/JNEUROSCI.0595-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.GTEx Consortium. Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duffy DL, Iles MM, Glass D, Zhu G, Barrett JH, Höiom V, et al. IRF4 variants have age-specific effects on nevus count and predispose to melanoma. Am J Hum Genet. 2010;87:6–16. doi: 10.1016/j.ajhg.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gunduz-Cinar O, MacPherson KP, Cinar R, Gamble-George J, Sugden K, Williams B, et al. Convergent translational evidence of a role for anandamide in amygdala-mediated fear extinction, threat processing and stress-reactivity. Mol Psychiatry. 2013;18:813–823. doi: 10.1038/mp.2012.72. Epub 2012 Jun 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Day CA, JR, Dietze P, Dolan K. Initiation to heroin injecting among heroin users in Sydney, Australia: cross sectional survey. Harm Reduct J. 2005;2:2. doi: 10.1186/1477-7517-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Des Jarlais DC, Arasteh K, Perlis T, Hagan H, Heckathorn DD, Mcknight C, et al. The transition from injection to non-injection drug use: long-term outcomes among heroin and cocaine users in New York City. Addiction. 2007;102:778–785. doi: 10.1111/j.1360-0443.2007.01764.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.