Abstract
Identification of novel drug targets and affordable therapeutic agents remains a high priority in the fight against chronic hepatitis C virus (HCV) infection. Here, we report that the cellular proteins prohibitin 1 (PHB1) and 2 (PHB2) are pan-genotypic HCV entry factors functioning at a post-binding step. While predominantly found in mitochondria, PHBs localize to the plasma membrane of hepatocytes through their transmembrane domains and interact with both EGFR and CRaf. Targeting PHB by rocaglamide (Roc-A), a natural product that binds PHB1 and 2, reduced cell surface PHB1 and 2, disrupted PHB-CRaf interaction, and inhibited HCV entry at low nanomolar concentrations. A structure-activity analysis of 32 synthetic Roc-A analogs indicated that the chiral, racemic version of aglaroxin C, a natural product biosynthetically related to Roc-A, displayed improved potency and therapeutic index against HCV infection. This study reveals a new class of HCV entry inhibitors that target the PHB1/2-CRaf pathway.
Abbreviations: HCV, hepatitis C virus; PHB, prohibitin; HCVcc, cell culture grown HCV; HCVpp, lentiviral particles pseudotyped with HCV envelope proteins; VSV-Gpp, HIV particles pseudotyped with vesicular stomatitis virus envelope protein G; MOI, multiplicity of infection; CHIKVpp, HIV particles pseudotyped with Chikungunya virus envelope proteins
Keywords: HCV, Entry factors, Entry inhibitors, Rocaglates
Graphical abstract
Highlights
-
•
Cellular proteins prohibitins 1 and 2 are essential HCV entry factors that function at a post-binding step.
-
•
The natural compound Roc-A potently blocks HCV infection by disrupting prohibitins-CRaf interaction
-
•
The Roc-A derivative, aglaroxin C, displays improved potency and therapeutic index towards HCV infection
Current FDA-approved HCV drugs all target viral proteins. We now demonstrate that a group of small molecules, the rocaglates, potently block HCV entry at low nanomolar concentrations. Roc-A inhibits HCV entry by disrupting the important interaction between two pan-genomic HCV entry factors, PHB1 and 2, and the signaling molecule CRaf. Overall, Roc-A and related rocaglates represent a new class of compounds that hold significant therapeutic promise in treating HCV infection.
1. Introduction
Hepatitis C virus (HCV) is an important human pathogen that primarily infects human hepatocytes and causes chronic liver diseases (Kontorinis et al., 2004). It is undeniable that recently approved direct-acting antiviral (DAA)-containing regimens have radically changed the paradigm of chronic HCV treatment. However, it remains unclear whether DAAs fully prevent pathology or restore normal immunity. Moreover, with continuous and expanded usage of DAAs, HCV is expected to become progressively more drug resistant, thereby eroding the efficacy of DAAs (Pawlotsky et al., 2015). Lastly, most DAAs are hardly affordable to patients in resource-limited countries. For these reasons, druggable host targets and new lead compounds are highly desirable.
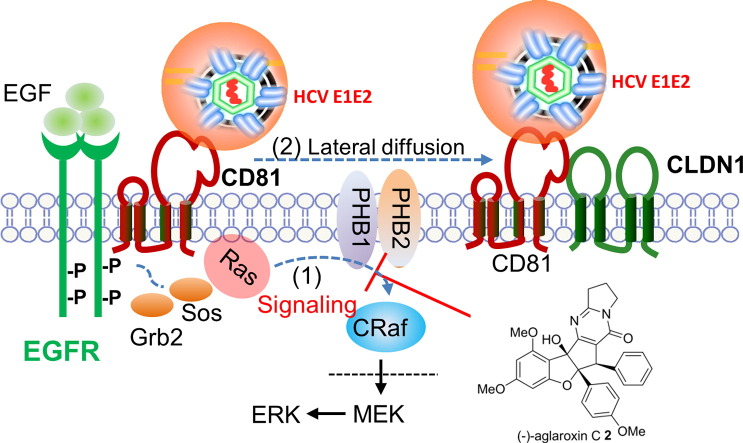
HCV entry is a multifaceted target for intervention. HCV encodes ten viral proteins to complete its life cycle. Viral glycoproteins E1 and E2 together form spikes on the viral envelope, which then engage with cell surface molecules (Lindenbach et al., 2005, Kato et al., 2005, Zhang et al., 2004, Cormier et al., 2004, Bartosch et al., 2003, Petracca et al., 2000) and trigger the endocytosis of the viral particle (Meertens et al., 2006, Hsu et al., 2003). In addition, E2 interacts with HCV nonstructural protein 2 (NS2) and plays an important role in virus morphogenesis (Selby et al., 1994). Recent advances have suggested that HCV enters hepatocytes in a step-wise fashion by utilizing multiple cellular membrane proteins, including CD81 (Pileri et al., 1998), scavenger receptor BI (SR-BI) (Scarselli et al., 2002), claudin-1 (CLDN1) (Evans et al., 2007), occludin (OCLN) (Liu et al., 2009, Ploss et al., 2009), epidermal growth factor receptor (EGFR) (Lupberger et al., 2011), and cholesterol-uptake receptor Niemann-Pick C1-like 1 (NPC1L1) (Sainz et al., 2012). Zona and coworkers further reported that the GTPase HRas acts as a signal transducer for EGFR-mediated HCV entry by regulating lateral membrane diffusion of CD81 which then enables tetraspanin receptor complex assembly (Zona et al., 2013).
In an attempt to identify novel cellular targets for antiviral development, we found that the cellular proteins prohibitin (PHB) 1 and 2 associate with HCV E2 in infected human hepatoma cells. We further demonstrated that PHBs mediate HCV entry at a post-binding step through their interaction with the signaling molecule CRaf. Blocking PHB-CRaf interactions with the natural product rocaglamide (Roc-A) potently inhibited HCV entry, and the biosynthetically-related rocaglate natural product aglaroxin C displayed both improved potency and therapeutic index.
2. Materials and Methods
Full experimental methods are provided in detail in the Supplemental Information.
2.1. Immunofluorescence Staining and Confocal Microscopy
Detailed procedures have been published (Yang et al., 2008). In brief, cells were stained with mitotracker red (1:10,000) at 37 °C for 15 min and then fixed in − 20 °C cold methanol for 5 min. Antibody dilutions are: anti-PHB1 (Santa Cruz, E-5 clone, 1:200), anti-PHB2 (Millipore, Cat No. 07–234, 1:200). Secondary antibodies were Alexa Fluor 488 or 568 Goat Anti-Mouse or Rabbit IgG (Life technologies 1:1000). Images were captured by a Zeiss LSM 700 laser scanning microscope.
2.2. HCV E1E2-Mediated Cell–Cell Fusion Assay
Detailed protocols have been published (Si et al., 2012). In brief, Huh7.5.1 cells were transfected with si-CTRL (control siRNA), or si-PHB1, or si-PHB2, and Stop-Luc construct which contains a firefly luciferase reporter gene whose transcription is prevented by a Stop cassette flanked by LoxP sites. 48 h post-transfection, these recipient cells were mixed at a 1:1 ratio with 293T-CLDN1 cells expressing Cre and HCV E1E2 (H77, genotype 1a) (donor cells) to initiate cell–cell fusion. Luciferase activity was measured 24 h thereafter.
2.3. Cell Surface Biotinylation Assay
Huh7.5.1 cells from four 150 mm plates were treated with DMSO, Roc-A (20 nM), or infected by HCVcc. 48 h post-transfection, cells were washed three times with ice-cold PBS and resuspended in PBS at a density of 25 × 106 cells/ml. Freshly prepared Sulfo-NHS-SS-biotin (Pierce) was added to the cells (final concentration 0.5 μg/ml) and allowed to incubate at 4 °C for 30 min. Cells were then washed three times with ice-cold PBS. 25 mM Tris (pH 8.0) was added in the initial wash to quench any non-reacted biotin reagent. Following cell lysis in RIPA buffer (50 mM Tris–HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 1 mM EDTA, 2 mM Na3VO4 and Pierce protease inhibitor cocktail), lysates were cleared by centrifugation at 13,000 × g for 15 min at 4 °C. The cleared lysates were used for immunoprecipitation using a 1:1 mixture of Streptavidin beads (Pierce). Beads were washed three times with RIPA buffer, and bound proteins were eluted by boiling the samples in SDS-PAGE sample buffer and then resolved on 9% SDS-PAGE. Biotinylated proteins were detected by anti-PHB1 and anti-PHB2 antibodies.
2.4. Cytotoxicity/Cell Viability Assay
PHHs (105 per well) were treated with Roc-A or DMSO at various concentrations for 48 h in 48-well plates. The numbers of viable cells in culture were determined using the CellTiter-Glo Cell Viability Luminescent Assay kit according to the manufacturer's instruction (Promega).
2.5. Statistical Analysis
Bar graphs were plotted to show mean ± standard deviation (SD) of at least two independent experiments. Statistical analyses were performed using Graphpad Prism 5. A p value of < 0.05 in the Student's test was considered statistically significant.
2.6. Chemical Synthesis
Synthetic rocaglates and derivatives were obtained from the chemical collection at the BU Center for Molecular Discovery (BU-CMD). Chiral, racemic rocaglates (Roche et al., 2010a, Roche et al., 2010b) and rocaglate hydroxamates (Rodrigo et al., 2012) were synthesized using the reported procedures. Chiral, non-racemic (−)-aglaroxin C and (+)-aglaroxin C were synthesized using biomimetic kinetic resolution of chiral, racemic aglain ketone precursors according to our published protocol (Stone et al., 2015) followed by further chemical transformations. (−)-Roc-A, and (+)-Roc-A were synthesized using the same protocol followed by amide formation (Gerard et al., 2006).
3. Results
3.1. PHB1 and 2 Interact with HCV E2
We have previously conducted a comparative proteomics analysis of the HCV-infected human hepatoma cell line Huh7.5.1 in order to identify HCV E2-interacting proteins. PHB1 and 2 were found to be the most abundant proteins in the E2 complex as detected by mass spectrometry. To validate the result, we performed immunoprecipitation using lysates from cells infected with the Flag-E2 JFH1 virus and confirmed that PHB1 and 2 co-precipitated with HCV E2 (Fig. S1A). The PHB-E2 association does not require the presence of other viral components as demonstrated in co-immunoprecipitation (Co-IP) studies (Fig. S1B).
3.2. PHB1 and 2 are Required for HCV Entry
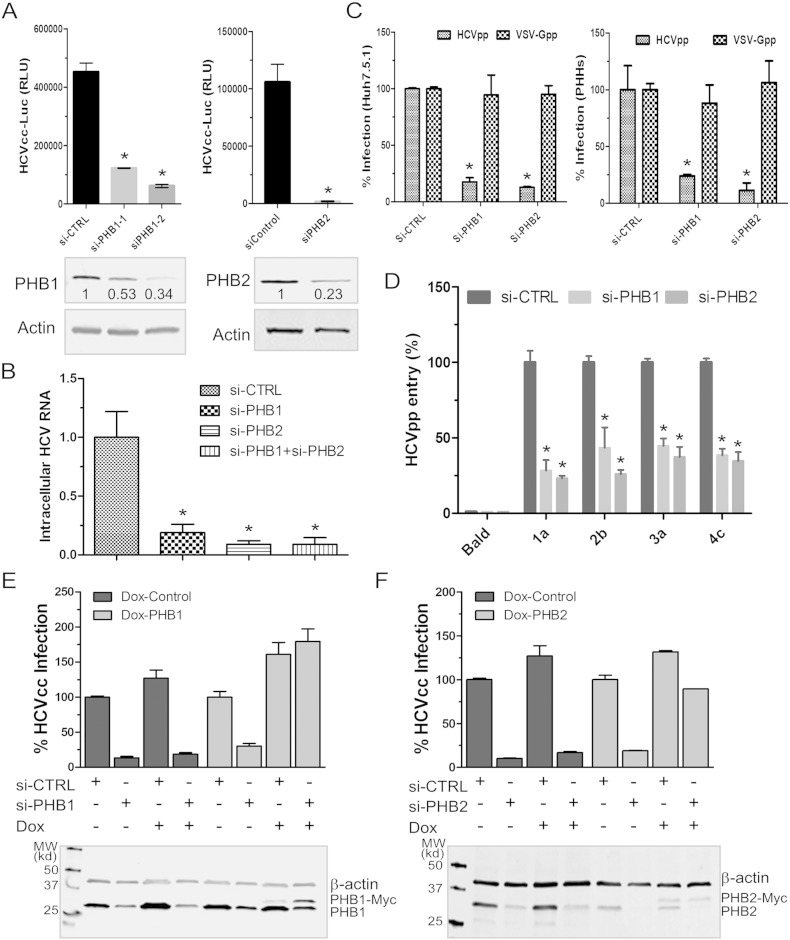
PHB1 is a ubiquitously expressed protein displaying antiproliferative activity (McClung et al., 1989). PHB2, also named repressor of estrogen receptor activity (REA), suppresses estrogen receptor (ER)-dependent gene activation (Montano et al., 1999). Interestingly, PHB has been implicated in the entry process of dengue and chikungunya virus (CHIKV) and also binds to HIV-1 glycoprotein and envelope proteins of white spot syndrome virus (Lan et al., 2013, Wintachai et al., 2012, Kuadkitkan et al., 2010, Emerson et al., 2010). To explore the role of PHB in modulating HCV infection, we transfected Huh7.5.1 cells with siRNA targeting PHB1 and PHB2, respectively. Reduction of endogenous PHB1 or 2 significantly inhibited cell culture grown HCV (HCVcc) as measured by either luciferase assays or real-time PCR quantification of viral RNA (Fig. 1A and B). By contrast, PHB knockdown had no effect at viral RNA levels if the infection took place first (Fig. S1C), suggesting that PHBs are required at an early stage of HCV infection. Notably, PHB1 and PHB2 knockdown also decreased the protein levels of each other (Fig. S1D).
Fig. 1.
Endogenous PHB1 and PHB2 are required for HCV infection.
(A–B) Endogenous PHB1 and 2 were knocked down by siRNA transfection followed by HCVcc-Luc infection (MOI ~ 0.3). Numbers shown below Western blot gel images indicate the relative expression levels quantified by Odyssey imaging system (LI-COR Biosciences). Luciferase activity was determined 72 h post-infection (A), intracellular viral RNA was quantified by RT-qPCR using protocols described in Experimental Procedures (B). Data are shown as mean ± SD, *p < 0.05.
(C) Knockdown of PHB1 and 2 in Huh7.5.1 (left) or PHHs (right) were achieved by transfecting cells with relevant siRNA for 48 h. Cells were infected by HCVpp (H77) or VSV-Gpp (MOI ~ 0.5). The percent of infection in cells transfected with si-CTRL (control) was arbitrarily set to 100% (mean ± SD, *p < 0.05).
(D) Huh7.5.1 were first transfected with siRNA targeting PHB1 and 2 and then infected by HCVpp bearing glycoproteins derived from genotypes 1a, 2b, 3a, and 4c. (mean ± SD, *p < 0.05).
(E & F) Restoration of HCV entry in PHBs knockdown cells by exogenously expressing PHB1 & 2. Silencing PHB1 (E) or 2 (F) by siRNA in Huh7.5.1 stable clones containing a control vector (Dox-CTRL) or a siRNA-resistant PHB-Myc (Dox-PHB1 & 2) expressing plasmid. Doxycycline was added to induce the expression of PHB-Myc. Cells were then infected by HCVcc-Luc for luciferase assay. The Western blot images were shown at the bottom of each panel to confirm the specific induction of PHB-Myc.
We then found that PHB1 and 2 are required for HCV entry, as silencing PHB1/2 in Huh7.5.1 cells or primary human hepatocytes (PHHs) ablated HIV–HCV pseudotype (HCVpp) infection while having no effect on pseudotyped virus displaying vesicular stomatitis virus G protein (VSV-Gpp) (Fig. 1C). Furthermore, PHB knockdown significantly reduced the infection of HCVpp-bearing glycoproteins from various HCV genotypes (Fig. 1D). When tested in a HCV replicon cell line, however, knockdown of endogenous PHB1 and 2 did not decrease viral RNA replication or protein translation (Fig. S1E & F).
To strengthen the above findings and to exclude the off-target effects of siRNAs, we generated Huh7.5.1 stable clones in which the PHB1 and 2 expressions are regulated by doxycycline. PHB1 or 2 silencing rendered these cells less susceptible to HCV infection. However, induction of siRNA-resistant PHB1 or PHB2 restored cell susceptibility to HCVcc infection (Fig. 1E and F). Altogether, our results indicate that PHB1 and 2 are pan-genomic HCV entry factors.
3.3. PHB1 and 2 can be Found on the Cell Surface and are Required for a Post-Binding Step in HCV Infection
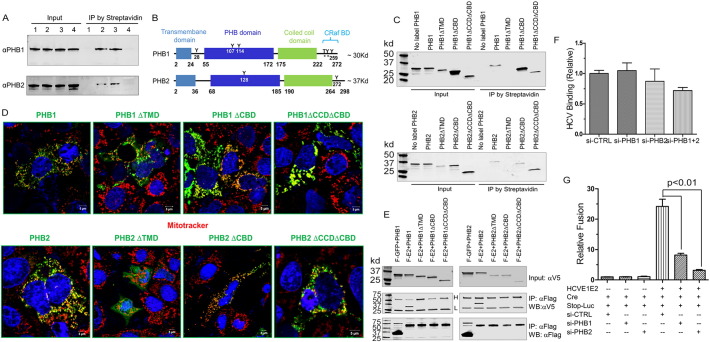
To gain insights into the roles of PHBs in HCV entry, we first examined the cellular distribution of PHB1/2. Typically found in the inner membrane of mitochondria, PHB1 and 2 form a large multimeric complex to stabilize newly synthesized mitochondrial proteins (Back et al., 2002). In addition, PHB1 and 2 are reportedly present on the plasma membrane, cytosol, and nucleus (Mishra et al., 2005, Theiss and Sitaraman, 2011, Kim Do et al., 2013, Kolonin et al., 2004). Confocal microscopy study of endogenous PHB1 and 2 in Huh7.5 cells revealed a typical mitochondria localization pattern, although a small portion of PHB2 appeared to traffic to cell surface (Fig. S2A). To definitively examine if PHB1 and 2 traffic to plasma membrane, cell surface proteins were biotinylated and then purified using streptavidin agarose beads. Both PHB1 and 2 were detected in the pull-down product, confirming their presence on the plasma membrane (Fig. 2A). Of note, HCV infection did not change the amount of PHB1 and 2 that were found in the precipitates (Fig. 2A, lane 3).
Fig. 2.
PHB1 and 2 mediate HCV entry via a novel mechanism.
(A) Huh7.5.1 cells were surface biotinylated as described in the Materials and Methods section. IP was done using streptavidin agarose beads. The presence of PHB1 and PHB2 was detected by Western blotting. Lane 1, no biotin labeling; lane 2, biotin labeled; lane 3, HCVcc infected cells labeled with biotin; lane 4, RocA (20 nM) treated cells labeled with biotin. Of note, because Roc-A treatment decreased the total cellular PHB1 and 2 (Shown in Fig. 3B), we intentionally used twice as many cells as starting materials for the Roc-A treated group (Lane 4) in order to achieve similar amount of prohibitins in the input in comparison to the levels in other lanes.
(B) Domain organization of PHBs.
(C) Surface biotinylation assay on individual PHB deletion mutants.
(D) Confocal images of PHB deletion mutants. Green, PHB mutants; Red, mitotracker. (E), co-IP studies of Flag-E2 (F-E2) and V5-tagged PHB deletion mutants.
(F) Real-time quantification of HCV RNA bound to cells in which PHB1 or PHB2 were knocked down. Results were calculated as relative RNA copies with numbers obtained from si-CTRL transfected cells set to 1 (mean ± SD, *p < 0.05).
(G) 293 T-CLDN1 cells expressing HCV E1E2 and Cre were fused to Huh7.5.1 cells that were transfected with siRNA and Stop-Luc expressing plasmid. Luciferase activity was determined 24 h after mixing (mean ± SD, *p < 0.05).
PHBs contain several functional domains, as illustrated in Fig. 2B. Mutational studies revealed that only the removal of the transmembrane domain (TMD) of PHBs led to the loss of cell surface localization (Fig. 2C). Interestingly, upon removal of their C-terminal domains, PHBs preserved both their cell surface and mitochondrial location, yet were unable to interact with HCV E2 (Fig. 2D & E).
To further investigate the mechanistic action of PHB, we directly evaluated the role of PHB on binding of HCV to the cell surface. To this end, HCV virions were incubated with PHB knockdown cells at 4 °C for 2 h to allow binding but not penetration. After extensive wash, surface-bound virions were quantified by measuring the abundance of viral RNA. Shown in Fig. 2F, the amount of viral RNA bound to PHB-knockdown cells was comparable to that of si-CTRL cells, indicating that PHB silencing has no effect on binding of HCV to cells. Similarly, PHB knockdown did not have significant effect on CD81, SR-BI, CLDN1, or OCLN expression (Fig. S2B), nor did PHBs interact with any of these known entry factors (Fig. S2B). Lastly, we measured the HCV E1E2-dependent fusion and found that PHB knockdown cells were impaired in fusing with 293T cells expressing HCV E1E2 (Fig. 2G). Altogether, these results suggest that PHB1 and 2 are involved in a late stage during virus entry.
3.4. PHB1 and 2 Interact with CRaf
Co-IP studies showed that PHB1 and 2 interact with EGFR and CRaf (Fig. S2C & D). Removal of the entire C-terminal domains of PHB1 and 2 abolished the interaction between PHB and CRaf. In contrast to a recent publication which reported that a Raf inhibitor (10 μM) failed to block HCV infection (Diao et al., 2012), silencing CRaf expression in Huh7.5.1 cells specifically suppressed HCV entry (Fig. S2E). Correctively, our data suggest that the PHB-CRaf pathway is critical for HCV infection.
3.5. A PHB Inhibitor, Rocaglamide A (Roc-A), Inhibits HCV Entry
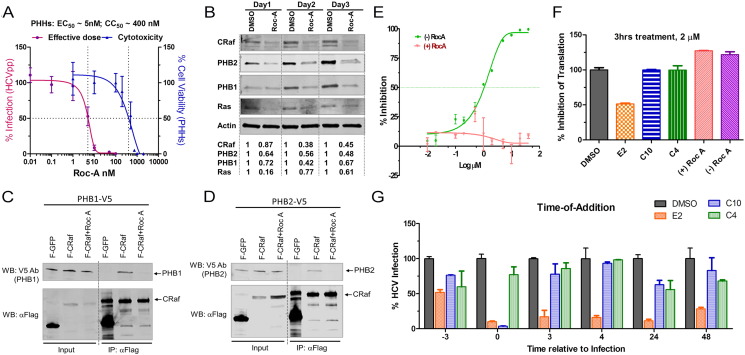
Next, we asked whether targeting the PHB-CRaf pathway would be a viable approach to block HCV infection. The natural product rocaglamide A (Roc-A) has been shown to directly bind to PHB1 and 2 and blocks PHB-CRaf-MEK-ERK signaling (Polier et al., 2012). Addition of Roc-A at 20 nM did not inhibit HCV RNA replication or protein translation (Fig. S3A & B). Rather, it significantly reduced HCVcc, HCVpp, CHIKVpp, and dengue virus infection (Fig. S3C–G) at low nanomolar concentrations. Moreover, pretreatment of PHHs with Roc-A at non-cytotoxic concentrations also suppressed HCVpp entry (Fig. 3A). The therapeutic index (TI) of Roc-A is greater than 80 when tested on primary human hepatocytes. Notably, Roc-A treatment decreased the cell surface expression of PHB1 and 2 (Fig. 2A) and also decreased PHB1, 2, Ras, and CRaf at the total protein level (Fig. 3B). Roc-A treatment disrupted the PHB-CRaf interaction in a co-IP experiment (Fig. 3C and D). Finally, Roc-A displayed a half-life of 37 min in a human liver microsomal stability assay (Fig. S4), suggesting that the compound has good metabolic stability.
Fig. 3.
Roc-A potently inhibits HCV infection.
(A) PHHs seeded in a 48-well plate were treated with Roc-A or DMSO and then infected by HCVpp. EC50 and CC50 (50% cytotoxicity dose) were calculated based on the fitted Sigmoid curves.
(B) Huh7.5.1 cells were treated with Roc-A (20 nM) or DMSO for 1, 2, and 3 days. Cellular PHB1, PHB2, CRaf, and Ras were detected by Western blotting. Numbers shown below Western blot gel images indicate the relative protein levels quantified by Odyssey imaging system (LI-COR Biosciences).
(C–D) Flag-CRaf (F-CRaf) and V5-tagged PHB1 & 2 were co-transfected into 293T cells and then left untreated or treated with Roc-A (20 nM) before immunoprecipitation with anti-Flag antibody. The pulled-down PHB1 & 2 as well as CRaf were detected by immunoblotting.
(E) Different inhibitory profiles of the (+)- and (−)-Roc-A enantiomers on HCVcc infection.
(F) A CMV-luciferase reporter construct was transfected into Huh7.5.1 cells. 24 h post-transfection each compound was added to the cells for 3 h at 2 μM. After removal of the compound, the cells were further incubated for 24 h followed by luciferase assay.
(G) C10 (racemic aglaroxin C) inhibited HCV infection when added together with the virus. HCVcc-Luc was added to Huh7.5.1 cells at 37 °C and incubated for 3 h. At the indicated time points, 2 μM of each compound or DMSO was added into the media and incubated for 3 h prior to removal. Infected cells were incubated at 37 °C for an additional 48 h prior to luciferase assay (mean of n = 3; error bars, s.d.). Compound E2 (−)-NH hydroxamate, is a potent translation inhibitor (Rodrigo et al., 2012) and hence appeared to inhibit HCV no matter when added; compound C4 (±)-β-lactone (Lajkiewicz et al., 2014), was added as a negative control as it exerted negligible effect.
3.6. Roc-A Derivatives Display Improved Therapeutic Index
To obtain compounds that display more favorable therapeutic index towards HCV, we evaluated a set of 32 additional rocaglate derivatives to ascertain structure-activity relationships (SAR) (Table S1). Interestingly, the natural (−) enantiomer of Roc-A displayed stronger inhibition on HCV entry than the (+) enantiomer (Fig. 3E). Moreover, the chiral, racemic version of the natural product aglaroxin C (Udom Kokpol, 1994) exhibited picomolar half-maximum effective concentrations (EC50) towards the JFH-1 genotype 2a infectious virus in cell culture and a therapeutic index of over 100 (Table 1). Further analysis indicated that (−)-aglaroxin C is the active enantiomer for HCV viral entry inhibition (Table 1). Moreover, these compounds, even when incubated at 2 μM for 3 h, did not inhibit protein translation (Fig. 3F). Results from a time-of-addition experiment further showed that chiral, racemic aglaroxin C displayed maximal anti-HCV activity when added together with the virus but lost its activity when added 3 h after the infection was initiated (Fig. 3G). This finding bolstered the notion that aglaroxin C is specifically inhibiting viral entry.
Table 1.
Evaluation of rocaglate derivatives as HCV viral entry inhibitors.a
| Compound | Structure | EC50 | CC50 | TI (CC50/EC50) | Translation inhibition at 2 μMc |
|---|---|---|---|---|---|
| (+) Roc-A | 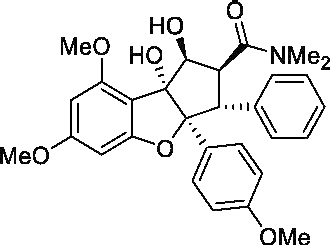 |
> 200 μMa | 300 μMa | N/A | No |
| (−) Roc-A | 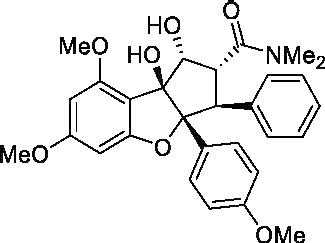 |
1 μMa 4 nMb |
50 μMa 300 nMb |
50a 75b |
No |
| (±) Aglaroxin C (C10)d |
 |
100 nMa 40 pMb |
10 μMa 20 nMb |
100a 500b |
No |
| (+)-Aglaroxin C | 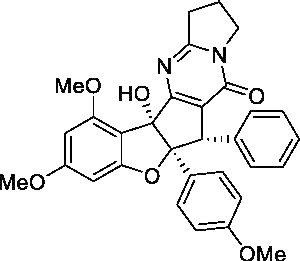 |
20 μMa | 100 μMa | 5a | No |
| (−)-Aglaroxin C | 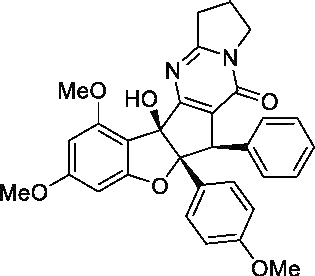 |
200 nMa | 12 μMa | 60a | No |
Compounds were incubated with cells for a total of 3 h on Huh7.5.1 cells.
Compounds were incubated with cells for a total of 48 h on Huh7.5.1 cells.
Measured using a luciferase reporter gene that was transfected into Huh7.5.1 cells. The compound was added for 3 h.
Name appearing in Fig. 3G.
4. Discussion
The identification of PHB1 and 2 as HCV entry factors is somewhat surprising because these two proteins are typically recognized as molecular chaperones that stabilize other mitochondrial proteins. A fraction of PHB1 and 2 can be found on the surface of Huh7.5.1 cells, although they are not required for the initial binding of virions. Given that PHB-CRaf interaction is necessary for CRaf activation by Ras (Rajalingam and Rudel, 2005, Polier et al., 2012), and that HRas is a key host signal transducer for EGFR-mediated HCV entry (Zona et al., 2013), a plausible role of PHBs in HCV entry is to link HRas to CRaf-mediated signaling. In support of this notion, we found that CRaf knockdown reduced HCV infection, as did disruption of PHB-CRaf interaction by Roc-A. PHBs have been implicated in facilitating signal transduction. For example, a recent study showed that PHB1, normally stored in mast cell granules, translocates to plasma membrane lipid rafts upon antigen stimulation in order to activate the tyrosine kinase Syk-dependent signaling that stimulates mast cell degranulation and the secretion of cytokines (28).
Exactly when and how HCV activates the PHBs-mediated signaling remains to be investigated. PHB1/2 do not precipitate with CD81, SR-BI, CLDN1, or OCLN, but are associated with HCV E2 in both infected and co-transfected cells. PHBs are anchored to the plasma membrane or mitochondrial inner membrane by their transmembrane domains with carboxyl termini facing cytoplasm or the intermembrane space of mitochondria. The C-termini of PHBs recruit CRaf to the inner plasma membrane (Mishra et al., 2005). Given that HCV E2 does not traffic to the intermembrane space of mitochondria, the interaction between PHBs and E2 likely takes place in close proximity to the inner plasma membrane although evidence for direct PHB–HCV E2 interaction is still lacking. Since the removal of either the transmembrane domain or C-terminal domain of PHBs abolishes PHB–E2 interaction, HCV E2 may form a signaling complex with membrane-bound PHB-CRaf at some point during entry. The PHBs–HCV E2 associations are not mediated by cell membranes, as C-terminal deletion of PHBs did not alter their membrane localization. C-terminal deletion did, however, abolish PHBs–E2 association. Plasma membrane-bound PHB1 is indispensable for the activation of CRaf by Ras (Rajalingam and Rudel, 2005, Rajalingam et al., 2005), and interaction between PHB1 and CRaf requires phosphorylation of PHB1 at Thr 258 and Tyr 259 (Chiu et al., 2013). Further investigation is needed to understand the topology, the phosphorylation status of plasma membrane-bound PHBs, and the likely signaling pathways that PHBs mediate during HCV entry.
An exciting finding of our study is that Roc-A, which binds PHB and inhibits its interaction with CRaf (Polier et al., 2012), potently inhibited HCV entry. Roc-A was first reported as an immunosuppressant and inhibitor of NF-kappa B activity (31). Roc-A and related rocaglates are also recognized as potent anticancer compounds (Kim et al., 2006, Ebada et al., 2011) by inhibiting translation initiation through inhibition of the RNA helicase eIF4a (Roche et al., 2010a, Roche et al., 2010b, Rodrigo et al., 2012, Chowdhury et al., 2014, Cencic et al., 2009). Roc-A has also been shown to indirectly target heat shock factor 1 (HSF1), a multifaceted transcriptional regulator of the heat-shock response and numerous other cellular processes essential for anabolic metabolism, cellular proliferation, and tumorigenesis (Santagata et al., 2013). In the current study, Roc-A treatment significantly reduced the protein levels of cell surface-bound PHB1 and PHB2 and disrupted PHB-CRaf interaction, indicating that it blocks HCV entry by targeting this pathway. The observation that a racemic, synthetic sample of the natural product aglaroxin C displays improved therapeutic index relative to enantiopure Roc-A suggests the possibility for synergy of enantiomers (Zhuang et al., 2014, Danielsson et al., 2011). In our studies, racemic aglaroxin C did not inhibit protein translation even at 2 μM over 3 h in a translation inhibition assay (Fig. 3F). Future investigations will be needed to understand whether it is possible to selectively target the PHB-CRaf pathway using appropriately functionalized rocaglates (flavaglines) and to what extent there may be synergy between translation inhibition or other mechanisms and HCV viral entry effects via PHB's (Cencic et al., 2010, Rozelle et al., 2014).
In conclusion, the identification of PHB1 and 2 adds additional targets to the repertoire of HCV entry factors. In contrast to most small molecule inhibitors that have advanced to the clinic targeting viral components, Roc-A, a PHB inhibitor, represents a promising drug lead that targets a host factor and hence reduces the likelihood that resistance will be developed. By virtue of its distinct mechanism of inhibition, Roc-A and its derivatives may also be used in combination with other anti-HCV drugs for potential synergistic effects in treating HCV infections, especially in settings where liver cancer is present. The observation that rocaglates (flavaglines) block CHIKVpp entry (Wintachai et al., 2015) (Fig. S3F) and dengue virus infection (Fig. S3G) also raises the hope that Roc-A or an optimized congener may be developed into a drug curbing infections by the two viruses.
Author contributions
S.L., T.Z., J. A.P., Jr., and T.W. designed the overall project and analyzed data. S.L., C.Q., J.Z., and T.W. performed most cellular and molecular experiments. W.W., N.L. and L.E.B. synthesized and curated rocaglate derivatives. S.L., J. A.P., Jr., and T.W. wrote the manuscript.
The following are the supplementary data related to this article.
Supplementary material.
Representative Rocaglate Derivatives Evaluated, Related to Table 1
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
This work was supported by grant from National Institute of Health (NIH R01DK088787 grant to T.W.), by NIH 5P30CA047904-25: HIV-Associated Malignancies Research (AMC) Supplement (to T.W.), and by an NIH R01 grant (GM073855, J.A.P, Jr.). Research in the BU-CMD is supported by NIH grant GM111625. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Health. We thank Ms. Madeline Weber (Boston University) for assistance with the compound library.
Contributor Information
Shufeng Liu, Email: shufeng.liu@sri.com.
John A. Porco, Jr, Email: porco@bu.edu.
Tony T. Wang, Email: tony.wang@sri.com.
References
- Back J.W., Sanz M.A., De Jong L., De Koning L.J., Nijtmans L.G., De Koster C.G., Grivell L.A., Van Der Spek H., Muijsers A.O. A structure for the yeast prohibitin complex: structure prediction and evidence from chemical crosslinking and mass spectrometry. Protein Sci. 2002;11:2471–2478. doi: 10.1110/ps.0212602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosch B., Dubuisson J., Cosset F.L. Infectious hepatitis C virus pseudo-particles containing functional E1–E2 envelope protein complexes. J. Exp. Med. 2003;197:633–642. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R., Carrier M., Galicia-Vazquez G., Bordeleau M.E., Sukarieh R., Bourdeau A., Brem B., Teodoro J.G., Greger H., Tremblay M.L., Porco J.A., Jr., Pelletier J. Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R., Carrier M., Trnkus A., Porco J.A., Jr., Minden M., Pelletier J. Synergistic effect of inhibiting translation initiation in combination with cytotoxic agents in acute myelogenous leukemia cells. Leuk. Res. 2010;34:535–541. doi: 10.1016/j.leukres.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C.F., Ho M.Y., Peng J.M., Hung S.W., Lee W.H., Liang C.M., Liang S.M. Raf activation by ras and promotion of cellular metastasis require phosphorylation of prohibitin in the raft domain of the plasma membrane. Oncogene. 2013;32:777–787. doi: 10.1038/onc.2012.86. [DOI] [PubMed] [Google Scholar]
- Chowdhury I., Thompson W.E., Thomas K. Prohibitins role in cellular survival through Ras-Raf-MEK-ERK pathway. J. Cell. Physiol. 2014;229:998–1004. doi: 10.1002/jcp.24531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier E.G., Tsamis F., Kajumo F., Durso R.J., Gardner J.P., Dragic T. CD81 is an entry coreceptor for hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 2004;101:7270–7274. doi: 10.1073/pnas.0402253101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsson M., Rosenthal-Aizman K., Bergsson G., Unden A. Pharmacodynamic synergy strictly dependent on the co-operative aggregation of enantiomers of cyclic peptides in the bacterial cell membrane. Bioorg. Med. Chem. Lett. 2011;21:5262–5265. doi: 10.1016/j.bmcl.2011.07.032. [DOI] [PubMed] [Google Scholar]
- Diao J., Pantua H., Ngu H., Komuves L., Diehl L., Schaefer G., Kapadia S.B. Hepatitis C virus induces epidermal growth factor receptor activation via CD81 binding for viral internalization and entry. J. Virol. 2012;86:10935–10949. doi: 10.1128/JVI.00750-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebada S.S., Lajkiewicz N., Porco J.A., Jr., Li-Weber M., Proksch P. Chemistry and biology of rocaglamides (= flavaglines) and related derivatives from aglaia species (meliaceae) Prog. Chem. Org. Nat. Prod. 2011;94:1–58. doi: 10.1007/978-3-7091-0748-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson V., Holtkotte D., Pfeiffer T., Wang I.H., Schnolzer M., Kempf T., Bosch V. Identification of the cellular prohibitin 1/prohibitin 2 heterodimer as an interaction partner of the C-terminal cytoplasmic domain of the HIV-1 glycoprotein. J. Virol. 2010;84:1355–1365. doi: 10.1128/JVI.01641-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M.J., Von Hahn T., Tscherne D.M., Syder A.J., Panis M., Wolk B., Hatziioannou T., Mckeating J.A., Bieniasz P.D., Rice C.M. Claudin-1 is a hepatitis c virus co-receptor required for a late step in entry. Nature. 2007;446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- Gerard B., Sangji S., O'leary D.J., Porco J.A., Jr. Enantioselective photocycloaddition mediated by chiral bronsted acids: asymmetric synthesis of the rocaglamides. J. Am. Chem. Soc. 2006;128:7754–7755. doi: 10.1021/ja062621j. [DOI] [PubMed] [Google Scholar]
- Hsu M., Zhang J., Flint M., Logvinoff C., Cheng-Mayer C., Rice C.M., Mckeating J.A. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. U. S. A. 2003;100:7271–7276. doi: 10.1073/pnas.0832180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T., Date T., Miyamoto M., Zhao Z., Mizokami M., Wakita T. Nonhepatic cell lines hela and 293 support efficient replication of the hepatitis C virus genotype 2a subgenomic replicon. J. Virol. 2005;79:592–596. doi: 10.1128/JVI.79.1.592-596.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Do K., Kim H.S., Kim A.R., Jang G.H., Kim H.W., Park Y.H., Kim B., Park Y.M., Beaven M.A., Kim Y.M., Choi W.S. The scaffold protein prohibitin is required for antigen-stimulated signaling in mast cells. Sci. Signal. 2013;6:ra80. doi: 10.1126/scisignal.2004098. [DOI] [PubMed] [Google Scholar]
- Kim S., Salim A.A., Swanson S.M., Kinghorn A.D. Potential of cyclopenta[b]benzofurans from aglaia species in cancer chemotherapy. Anti Cancer Agents Med. Chem. 2006;6:319–345. doi: 10.2174/187152006777698123. [DOI] [PubMed] [Google Scholar]
- Kolonin M.G., Saha P.K., Chan L., Pasqualini R., Arap W. Reversal of obesity by targeted ablation of adipose tissue. Nat. Med. 2004;10:625–632. doi: 10.1038/nm1048. [DOI] [PubMed] [Google Scholar]
- Kontorinis N., Agarwal K., Dieterich D.T. Current status of the use of growth factors and other adjuvant medications in patients receiving peginterferon and ribavirin. Rev. Gastroenterol. Disord. 2004;4(Suppl. 1):S39–S47. [PubMed] [Google Scholar]
- Kuadkitkan A., Wikan N., Fongsaran C., Smith D.R. Identification and characterization of prohibitin as a receptor protein mediating DENV-2 entry into insect cells. Virology. 2010;406:149–161. doi: 10.1016/j.virol.2010.07.015. [DOI] [PubMed] [Google Scholar]
- Lajkiewicz N.J., Cognetta A.B., 3rd, Niphakis M.J., Cravatt B.F., Porco J.A. JR, Remodeling natural products: chemistry and serine hydrolase activity of a rocaglate-derived beta-lactone. J. Am. Chem. Soc. 2014;136:2659–2664. doi: 10.1021/ja412431g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J.F., Li X.C., Sun J.J., Gong J., Wang X.W., Shi X.Z., Shi L.J., Weng Y.D., Zhao X.F., Wang J.X. Prohibitin Interacts with envelope proteins of white spot syndrome virus and prevents infection in the red swamp crayfish, procambarus clarkii. J. Virol. 2013;87:12756–12765. doi: 10.1128/JVI.02198-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach B.D., Evans M.J., Syder A.J., Wolk B., Tellinghuisen T.L., Liu C.C., Maruyama T., Hynes R.O., Burton D.R., Mckeating J.A., Rice C.M. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- Liu S., Yang W., Shen L., Turner J.R., Coyne C.B., Wang T. Tight junction proteins claudin-1 and occludin control hepatitis C Virus entry and are downregulated during infection to prevent superinfection. J. Virol. 2009;83:2011–2014. doi: 10.1128/JVI.01888-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupberger J., Zeisel M.B., Xiao F., Thumann C., Fofana I., Zona L., Davis C., Mee C.J., Turek M., Gorke S., Royer C., Fischer B., Zahid M.N., Lavillette D., Fresquet J., Cosset F.L., Rothenberg S.M., Pietschmann T., Patel A.H., Pessaux P., Doffoel M., Raffelsberger W., Poch O., Mckeating J.A., Brino L., Baumert T.F. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat. Med. 2011;17:589–595. doi: 10.1038/nm.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcclung J.K., Danner D.B., Stewart D.A., Smith J.R., Schneider E.L., Lumpkin C.K., Dell'orco R.T., Nuell M.J. Isolation of a cDNA that hybrid selects antiproliferative mRNA from rat liver. Biochem. Biophys. Res. Commun. 1989;164:1316–1322. doi: 10.1016/0006-291x(89)91813-5. [DOI] [PubMed] [Google Scholar]
- Meertens L., Bertaux C., Dragic T. Hepatitis C virus entry requires a critical postinternalization step and delivery to early endosomes via clathrin-coated vesicles. J. Virol. 2006;80:11571–11578. doi: 10.1128/JVI.01717-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S., Murphy L.C., Nyomba B.L., Murphy L.J. Prohibitin: a potential target for new therapeutics. Trends Mol. Med. 2005;11:192–197. doi: 10.1016/j.molmed.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Montano M.M., Ekena K., Delage-Mourroux R., Chang W., Martini P., Katzenellenbogen B.S. An estrogen receptor-selective coregulator that potentiates the effectiveness of antiestrogens and represses the activity of estrogens. Proc. Natl. Acad. Sci. U. S. A. 1999;96:6947–6952. doi: 10.1073/pnas.96.12.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlotsky J.M., Feld J.J., Zeuzem S., Hoofnagle J.H. From non-A, non-B hepatitis to hepatitis C virus cure. J. Hepatol. 2015;62:S87–S99. doi: 10.1016/j.jhep.2015.02.006. [DOI] [PubMed] [Google Scholar]
- Petracca R., Falugi F., Galli G., Norais N., Rosa D., Campagnoli S., Burgio V., Di Stasio E., Giardina B., Houghton M., Abrignani S., Grandi G. Structure-function analysis of hepatitis C virus envelope-CD81 binding. J. Virol. 2000;74:4824–4830. doi: 10.1128/jvi.74.10.4824-4830.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pileri P., Uematsu Y., Campagnoli S., Galli G., Falugi F., Petracca R., Weiner A.J., Houghton M., Rosa D., Grandi G., Abrignani S. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- Ploss A., Evans M.J., Gaysinskaya V.A., Panis M., You H., De Jong Y.P., Rice C.M. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polier G., Neumann J., Thuaud F., Ribeiro N., Gelhaus C., Schmidt H., Giaisi M., Kohler R., Muller W.W., Proksch P., Leippe M., Janssen O., Desaubry L., Krammer P.H., Li-Weber M. The natural anticancer compounds rocaglamides inhibit the Raf-MEK-ERK pathway by targeting prohibitin 1 and 2. Chem. Biol. 2012;19:1093–1104. doi: 10.1016/j.chembiol.2012.07.012. [DOI] [PubMed] [Google Scholar]
- Rajalingam K., Rudel T. Ras–Raf signaling needs prohibitin. Cell Cycle. 2005;4:1503–1505. doi: 10.4161/cc.4.11.2142. [DOI] [PubMed] [Google Scholar]
- Rajalingam K., Wunder C., Brinkmann V., Churin Y., Hekman M., Sievers C., Rapp U.R., Rudel T. Prohibitin is required for Ras-induced Raf-MEK-ERK activation and epithelial cell migration. Nat. Cell Biol. 2005;7:837–843. doi: 10.1038/ncb1283. [DOI] [PubMed] [Google Scholar]
- Roche S.P., C.,R., Pelletier J., Porco J.A., Jr. Biomimetic photocycloaddition of 3-hydroxyflavones: synthesis and evaluation of rocaglate derivatives as inhibitors of eukaryotic translation. Angew. Chem. Int. Ed. 2010;49:6338–‐6533. doi: 10.1002/anie.201003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche S.P., Cencic R., Pelletier J., Porco J.A., Jr. Biomimetic photocycloaddition of 3-hydroxyflavones: synthesis and evaluation of rocaglate derivatives as inhibitors of eukaryotic translation. Angew. Chem. 2010;49:6533–6538. doi: 10.1002/anie.201003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo C.M., Cencic R., Roche S.P., Pelletier J., Porco J.A. Synthesis of rocaglamide hydroxamates and related compounds as eukaryotic translation inhibitors: synthetic and biological studies. J. Med. Chem. 2012;55:558–562. doi: 10.1021/jm201263k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozelle D.K., Filone C.M., Kedersha N., Connor J.H. Activation of stress response pathways promotes formation of antiviral granules and restricts virus replication. Mol. Cell. Biol. 2014;34:2003–2016. doi: 10.1128/MCB.01630-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz B., Jr., Barretto N., Martin D.N., Hiraga N., Imamura M., Hussain S., Marsh K.A., Yu X., Chayama K., Alrefai W.A., Uprichard S.L. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat. Med. 2012;18:281–285. doi: 10.1038/nm.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata S., Mendillo M.L., Tang Y.C., Subramanian A., Perley C.C., Roche S.P., Wong B., Narayan R., Kwon H., Koeva M., Amon A., Golub T.R., Porco J.A., Jr., Whitesell L., Lindquist S. Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state. Science. 2013;341:1238303. doi: 10.1126/science.1238303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarselli E., Ansuini H., Cerino R., Roccasecca R.M., Acali S., Filocamo G., Traboni C., Nicosia A., Cortese R., Vitelli A. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002;21:5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby M.J., Glazer E., Masiarz F., Houghton M. Complex processing and protein:protein interactions in the E2:NS2 region of HCV. Virology. 1994;204:114–122. doi: 10.1006/viro.1994.1515. [DOI] [PubMed] [Google Scholar]
- Si Y., Liu S., Liu X., Jacobs J.L., Cheng M., Niu Y., Jin Q., Wang T., Yang W. A human claudin-1-derived peptide inhibits hepatitis C virus entry. Hepatology. 2012;56:507–515. doi: 10.1002/hep.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.D., Lajkiewicz N.J., Whitesell L., Hilmy A., Porco J.A., Jr. Biomimetic kinetic resolution: highly enantio- and diastereoselective transfer hydrogenation of aglain ketones to access flavagline natural products. J. Am. Chem. Soc. 2015;137:525–530. doi: 10.1021/ja511728b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theiss A.L., Sitaraman S.V. The role and therapeutic potential of prohibitin in disease. Biochim. Biophys. Acta. 2011;1813:1137–1143. doi: 10.1016/j.bbamcr.2011.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udom Kokpol B.V., Simpson J., Weavers R.T. Isolation and X-ray structure determination of a novel pyrimidinone from Aglaia odorata. J. Chem. Soc. Chem. Commun. 1994:773–774. [Google Scholar]
- Wintachai P., Thuaud F., Basmadjian C., Roytrakul S., Ubol S., Desaubry L., Smith D.R. Assessment of flavaglines as potential chikungunya virus entry inhibitors. Microbiol. Immunol. 2015;59:129–141. doi: 10.1111/1348-0421.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintachai P., Wikan N., Kuadkitkan A., Jaimipuk T., Ubol S., Pulmanausahakul R., Auewarakul P., Kasinrerk W., Weng W.Y., Panyasrivanit M., Paemanee A., Kittisenachai S., Roytrakul S., Smith D.R. Identification of prohibitin as a chikungunya virus receptor protein. J. Med. Virol. 2012;84:1757–1770. doi: 10.1002/jmv.23403. [DOI] [PubMed] [Google Scholar]
- Yang W., Qiu C., Biswas N., Jin J., Watkins S.C., Montelaro R.C., Coyne C.B., Wang T. Correlation of the tight junction-like distribution of claudin-1 to the cellular tropism of HCV. J. Biol. Chem. 2008;283(13):8643–8653. doi: 10.1074/jbc.M709824200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Randall G., Higginbottom A., Monk P., Rice C.M., Mckeating J.A. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 2004;78:1448–1455. doi: 10.1128/JVI.78.3.1448-1455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang C., Sheng C., Shin W.S., Wu Y., Li J., Yao J., Dong G., Zhang W., Sham Y.Y., Miao Z. A novel drug discovery strategy: mechanistic investigation of an enantiomeric antitumor agent targeting dual p53 and NF-kappaB pathways. Oncotarget. 2014;5:10830–10839. doi: 10.18632/oncotarget.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zona L., Lupberger J., Sidahmed-Adrar N., Thumann C., Harris H.J., Barnes A., Florentin J., Tawar R.G., Xiao F., Turek M., Durand S.C., Duong F.H., Heim M.H., Cosset F.L., Hirsch I., Samuel D., Brino L., Zeisel M.B., Le Naour F., Mckeating J.A., Baumert T.F. HRas signal transduction promotes hepatitis C virus cell entry by triggering assembly of the host tetraspanin receptor complex. Cell Host Microbe. 2013;13:302–313. doi: 10.1016/j.chom.2013.02.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Representative Rocaglate Derivatives Evaluated, Related to Table 1