Abstract
Risperidone is a second-generation antipsychotic that causes weight gain. We hypothesized that risperidone-induced shifts in the gut microbiome are mechanistically involved in its metabolic consequences. Wild-type female C57BL/6J mice treated with risperidone (80 μg/day) exhibited significant excess weight gain, due to reduced energy expenditure, which correlated with an altered gut microbiome. Fecal transplant from risperidone-treated mice caused a 16% reduction in total resting metabolic rate in naïve recipients, attributable to suppression of non-aerobic metabolism. Risperidone inhibited growth of cultured fecal bacteria grown anaerobically more than those grown aerobically. Finally, transplant of the fecal phage fraction from risperidone-treated mice was sufficient to cause excess weight gain in naïve recipients, again through reduced energy expenditure. Collectively, these data highlight a major role for the gut microbiome in weight gain following chronic use of risperidone, and specifically implicates the modulation of non-aerobic resting metabolism in this mechanism.
Keywords: Risperidone induced weight gain, Gut microbiome, Suppression of energy expenditure; Fecal transfer; Phage transfer; Non-aerobic resting metabolic rate
Highlights
-
•
Risperidone-induced weight gain correlates with an altered gut microbiome.
-
•
Risperidone-induced weight gain occurs due to suppressed energy expenditure.
-
•
Transfer of risperidone-treated microbiota or phage suppresses energy expenditure.
-
•
Reduction in energy expenditure is attributable to non-aerobic resting metabolism.
-
•
Transfer of risperidone-treated microbiota suppresses non-aerobic resting metabolism.
Risperidone is increasingly used for psychiatric disorders and is known to cause robust weight gain in humans. This study demonstrates that risperidone-induced weight gain correlates with alterations in the bacterial composition of the gut in mice. The observed weight gain is mediated through suppression of energy expenditure, specifically by reducing non-aerobic resting metabolic rate. The effect can be reproduced by transferring the gut microbiota or associated bacteriophage (bacterial viruses) from risperidone-treated animals to naïve animals. Thus, gut bacteria and their associated viruses can affect changes in resting metabolic rates leading to weight gain.
1. Introduction
Recent studies associate the gut microbiome, the bacterial ecosystem that resides in the human gut, with the modulation of weight gain and metabolic diseases. It has been shown that the microbiome contributes to host metabolism and physiology by several mechanisms including increased energy harvest from the diet (Turnbaugh et al., 2006, Turnbaugh et al., 2008), modulation of lipid metabolism (Bäckhed et al., 2004, Velagapudi et al., 2010), altered endocrine function (Dumas et al., 2006, Swann et al., 2011, Wang et al., 2011), and inflammatory stability (Elinav et al., 2011, Hall et al., 2011, Henao-Mejia et al., 2012, Vandanmagsar et al., 2011). Thus the gut microbiota is influential in modulating obesity and other metabolic diseases.
Contributing to the obesity epidemic over the last two decades, the prescribing rate of second-generation antipsychotics for children has increased nearly eight-fold due to their efficacy (Findling et al., 2005). Second generation antipsychotics (SGAs) are used to treat a variety of psychiatric illnesses, including autism, bipolar disorder and schizophrenia. It has also been well established that the most commonly prescribed SGA, risperidone (a benzylamino-piperidine derivative), causes significant weight gain, insulin resistance, and metabolic syndrome (Möller et al., 2015, Calarge et al., 2012, Correll et al., 2009, De Hert et al., 2011). Numerous observational studies of risperidone-induced weight gain have led to significant advancements in treatment guidelines. However, the multifactorial nature of cardiometabolic effects is not completely understood, and patients taking risperidone still suffer from the side effects associated with weight gain. In humans, risperidone may promote weight gain through appetite stimulation, although many animal studies comparing SGAs challenge this hypothesis (Baptista et al., 2004, Li et al., 2013a, Pouzet et al., 2003, Smith et al., 2012). Risperidone-induced weight gain is thought to be multifaceted, involving genetic, metabolic, and environmental contributors (Correll et al., 2011, Lett et al., 2011). Recent data implicates alterations in the gut microbiome as the mechanism by which SGAs impact metabolism and weight gain (Davey et al., 2013, Morgan et al., 2014). We have evidence that alterations in the gut microbiome of children chronically treated with risperidone are associated with an increase in BMI compared to antipsychotic-naïve children (Bahr et al., 2015).
In this study, we examined the hypothesis that shifts in the gut microbiome are mechanistically linked to weight gain that occurs in response to risperidone treatment. We examined energy balance and weight gain in wild type mice in response to treatment with risperidone alone or in combination with various xenobiotics, transfer of risperidone-altered microbiota, or transfer of the phage associated with risperidone-treated microbiota. We found that risperidone alters the gut microbiota resulting in weight gain via suppressed energy expenditure. Furthermore, transfer of risperidone-treated fecal material, including the phage fraction alone, was sufficient to induce similar effects in naïve mice. Identification of the gut microbiome as a critical mediator of energy homeostasis may identify novel therapeutic targets and approaches for risperidone-induced weight gain and obesity.
2. Materials and methods
2.1. Animal husbandry
Six to seven-week old C57BL/6J female mice from the Jackson Laboratory were housed in a standard 12:12 dark–light cycle with ad libitum access to standard chow (Teklad 7013; 18% kcal from fat) and water (pH 3.7 with acetic acid to match the pH of oral risperidone) or water with risperidone (20 μg/ml, pH 3.7). Antibiotics were administered for ten days in drinking water at 62.5 mg/l and 142.5 mg/l of ciprofloxacin and ampicillin respectively. All procedures were approved by the University of Iowa Animal Care and Use Committee in compliance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
2.2. Nuclear magnetic resonance
Body composition was assessed using nuclear magnetic resonance (NMR; Brucker LF90II). Mice were restrained (< 1 min) for analysis and then returned to home cages.
2.3. Energy intake
Mice were individually housed in metabolic cages (Nalgene) to monitor fluid and food intake daily. Additionally, urine and fecal outputs were measured and collected. Fecal material was utilized to determine digestive efficiency by bomb calorimetry.
2.4. Bomb calorimetry
Caloric densities of desiccated food and fecal samples were determined using a 50 mg semi-micro bomb calorimeter (Parr). Energy absorption was calculated as:
Energy Consumed is the product of the dry mass of food consumed and the caloric density of dry food, and Fecal Energy is the product of the dry mass of feces produced and the caloric density of the dried feces. Digestive efficiency was then calculated as:
Energy efficiency was calculated at various time points after the initiation of risperidone as:
2.5. Physical activity & core temperature
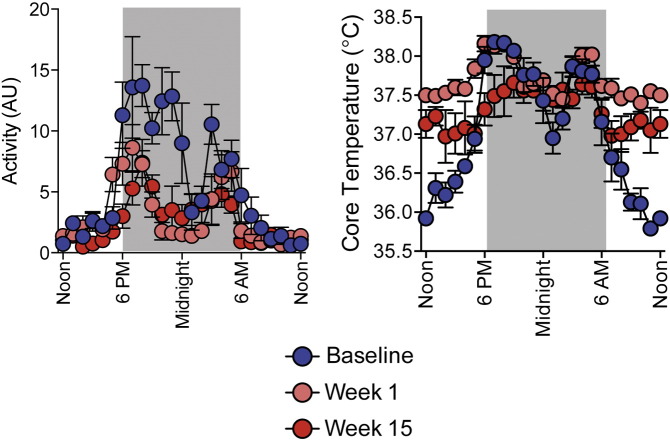
Physical activity and core temperature were determined using radiotelemetric probes (DSI), as previously described (Burnett and Grobe, 2014, Grobe et al., 2010). Mice were anesthetized using isoflurane and probes were inserted inside the abdominal wall. Following 1–2 weeks of recovery, activity and core temperature were recorded for 30 s every 5 min throughout the light–dark cycle (Fig. S1).
Supplemental Fig. S1.
Core body temperature and physical activity measured through radiotelemetry before and after 1 and 15 weeks of risperidone treatment. Data are representative of mean ± s.e.m.
2.6. Combined calorimetry
Resting metabolic rate (RMR) was measured simultaneously by direct calorimetry and respirometry as previously described (Burnett and Grobe, 2013, Burnett and Grobe, 2014). Briefly, rates of total heat dissipation, heat retention, and O2 and CO2 exchange of mice were assessed during sleep at thermoneutrality (30 °C) respectively, using a custom-built gradient-layer direct calorimeter, core temperature telemeters (DSI) and S-3A/II O2 and CD-3A CO2 analyzers (AEI). Mass flow of air was measured (Sensiron) and STP-corrected. Total RMR determined by direct calorimetry represents the sum of all heat dissipation (sensible and insensible) plus heat retained, which is determined by core temperature change and body composition as determined using NMR. Aerobic RMR determined by respirometry represents the estimated heat production using the formula derived from Lusk (1924):
VO2 represents STP-corrected rate of oxygen consumption (in ml/min), and the respiratory exchange ratio, RER, represents the ratio of carbon dioxide production to oxygen consumption. Non-aerobic RMR represents the difference between measured total heat production (from direct calorimetry) and the estimated rate of aerobic RMR (from respirometry):
It is important to note that throughout the current study, we report “RMR” data from mice, assessed while the animals are sleeping. The metabolic rate during sleep is more accurately referred to as “sleeping metabolic rate, SMR” but given the complexities of dissociating resting vs. sleeping metabolic rates in mice, few reports in the literature use this terminology in studies examining metabolic function in mice. In contrast, in humans SMR and RMR are clearly distinct. Thus although less technically accurate, we have chosen to report data herein as “RMR” instead of “SMR” as a reflection of the current vernacular of the field.
2.7. Bacterial DNA extraction and sequencing
16S rRNA gene sequencing methods were adapted from the methods developed for the NIH-Human Microbiome Project (Consortium, 2012). Briefly, bacterial genomic DNA was extracted using MO BIO PowerSoil DNA Isolation Kit (MO BIO Laboratories). The 16S rDNA V4 region was amplified by PCR and sequenced on the MiSeq platform (Illumina) using the 2 × 250 bp paired-end protocol yielding pair-end reads. The primers used for amplification contain adapters for MiSeq sequencing and dual-index barcodes so that the PCR products may be pooled and sequenced directly (Caporaso et al., 2012) targeting at least 6000 reads per sample.
2.8. Sequencing analysis
16S data was analyzed using QIIME v.1.9 software. Barcodes were matched to fastq files and then removed (Caporaso et al., 2010b). Similar sequences (cutoff 97%) were combined into operational taxonomic units (OTU) using sumaclust v1.1.00 and sortmerna 2.0. Representative sequences for each OTU were aligned using PyNAST (Caporaso et al., 2010a). The lanemaskPH was used to screen out the hypervariable regions and OTUs were classified with the greengenes database (DeSantis et al., 2006). Observed species, Chao1, and phylogenetic diversity metrics were assessed with alpha_diversity.py and compare_alpha_diversity.py in QIIME. Samples were rarefied to a depth of 6000 sequences/sample, which was shown to be sufficient by goods coverage. Beta diversity was assessed using UniFrac in QIIME (Lozupone et al., 2010). LEfSe was used with default parameters on species level OTU tables to determine taxa that best characterize each population (Segata et al., 2011). Only features with LDA scores > 2.0 were kept.
2.9. Fecal transfer
Fecal material (0.3 g) from respective donor cohorts was collected and pooled, ground by sterile mortar and pestle and suspended in sterile water (1 ml/0.1 g stool). Samples were centrifuged for 2 min at 3000 × g to remove large particulates. Individual mice received 100 μl of respective fecal supernatant by oral gavage daily for 14 consecutive days.
2.10. Risperidone measurement by HPLC–MS
Risperidone and 9-hydroxyrisperidone were dissolved in a mixture of water and methanol (1:1) to a concentration of 1 mg/ml and subsequently diluted with the same diluents to 10 mcg/ml, 1 mcg/ml and 100 ng/ml in order to prepare standards and controls for the calibration curve. Calibration standards were prepared by adding between 5 and 25 μl of the appropriate working standard to 0.5 ml water to make nine standards between 1 and 500 ng/ml. The concentrations of control samples were 3, 75 and 300 ng/ml. Internal standard solution was prepared by dissolving clozapine in methanol:water 1:1 to a concentration of 1 mcg/ml. The homogenized fecal samples, standards, blanks and controls were spiked with 50 μl of the internal standard and diluted with 500 μl of 4% H3PO4 in water. All samples were vortexed and subjected to solid phase extraction (SPE) prior to instrumental analysis. SPE was conducted using Oasis MCX cartridges (Waters Inc.). The SPE cartridges were conditioned with 1 ml of methanol and 1 ml of nanopure water. The samples were loaded into SPE cartridges and rinsed with 1 ml of 2% formic acid in water, followed by 1 ml of methanol. Then the samples were eluted with 1 ml of 5% ammonium hydroxide in methanol and then dried under nitrogen flow. The samples were reconstituted with 200 μl of 0.1% formic acid in water and acetonitrile mixture (1:1) and injected (15 μl) into an HPLC–MS (Shimadzu 2010A LC–MS platform in APCI ion mode operating under LCMS Solution software Version 2.04H3, Shimadzu, Columbia, MD, USA). A Phenomenex Synergi Polar RP analytical column (4 μm, 2 × 250 mm, Phenomenex, Torrance, CA) preceded by a Phenomenex Hydro PR 80A column was used for chromatographic separation of risperidone, 9-hydroxyrisperidone and clozapine (internal standard). Isocratic analysis was performed at 0.2 ml/min flow rate. Mobile phase consisted of 0.1% formic acid in water and acetonitrile (38:65). The mass spectrometer was tuned using a polyethylene glycol solution following the manufacturer's protocol. The scan interval was 0.3 s, microscan — 0.1 amu, APCI temperature was 400 °C, the CDL temperature was 200 °C, and the heat block temperature was 200 °C. Nitrogen flow rate was 2.5 L/min; detector voltage was 1.5 kV. The monitored mass-to-charge ratio was 427.4 for risperidone, 411.4 for 9-hydroxyrisperidone and 327.1 for clozapine. The assay was linear from 1 to 500 ng/ml and the coefficient of variation was < 15% for all control samples.
2.11. Phage isolation and transfer
Bacteriophage were isolated from stool as previously described (Thurber et al., 2009). Briefly, approximately 5 g of mouse stool was pooled from five treated or control mice over a 24 h time course. Stool was homogenized in PBS using a mortar and pestle and was centrifuged at 3000 × g for 3 min. Supernatant was collected and the pellet was homogenized in PBS to collect supernatant twice more. Total collected supernatant was filtered through 0.45 μm and 0.22 μm filters and then added to a gradient containing 1.75 g/ml, 1.55 g/ml, and 1.35 g/ml densities of CsCl. Gradients were spun at 60,000 × g for 2 h at 4 °C. Approximately 2 ml of bacteriophage were generated from each 50 ml gradient by inserting a 20-gauge needle into the 1.75 g/ml layer and drawing out the phage fraction. Pooled phage fractions were dialyzed to remove residual CsCl. Phage particles were assessed using an epiflourescent microscope after viral particles had been stained with SYBR-gold. This allowed us to document the presence of bacteriophage and absence of bacterial and/or eukaryotic cells. Pooled phage were aliquoted into 350 μl fractions for daily use to be combined in a 1:1 ratio by volume with rehydrated milk. Individual mice received 50 μl of respective phage fraction with 50 μl of milk by oral gavage daily for 4 weeks.
2.12. Growth of aerobic and anaerobic organisms
A freshly collected mouse fecal sample was homogenized with 1 × PBS for 10 min followed by centrifugation at 3000 × g for 3 min to remove debris. Organisms were grown aerobically and anaerobically overnight at 37 °C in BPRM broth. Suppressive effects of risperidone upon aerobic and anaerobic culture growth rates were determined after 5 h of treatment. Optical densities (OD600) of cultures were normalized against the growth exhibited by vehicle-treated cultures in matched aerobic/anaerobic conditions, and IC50 concentrations were calculated in SigmaPlot 13 by performing four-parameter logistic curve fitting using the following formula:
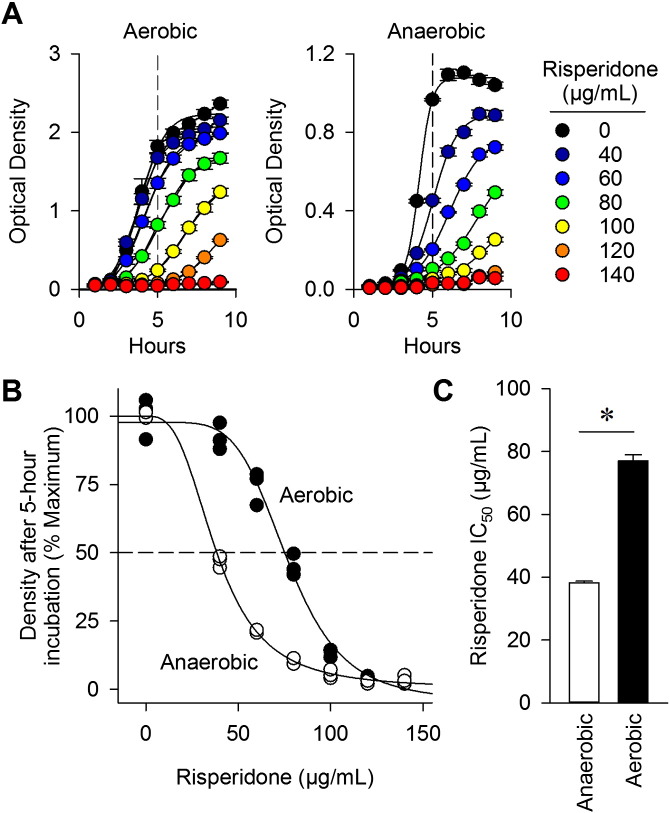
Regressions for aerobic and anaerobic cultures were reached with 11 and 8 iterations, with R2 values of 0.9876 and 0.9985, resulting in IC50 values of 77 ± 2 and 38 ± 1 μg/ml risperidone, and Hillslopes of − 5.13 ± 0.64 and − 2.99 ± 0.17, respectively.
2.13. Statistics
Analyses of phyla level relative abundances were performed by multiple t-test comparison. Analyses of genus relative abundance over time, and behavioral and physiological endpoints were performed using ANOVA with repeated measures as appropriate, followed by Tukey multiple-comparisons procedures. Significance was assigned as p < 0.05, and data are reported as mean ± sem throughout.
3. Results
3.1. Risperidone treatment results in increased weight gain that correlates with an altered gut microbiome
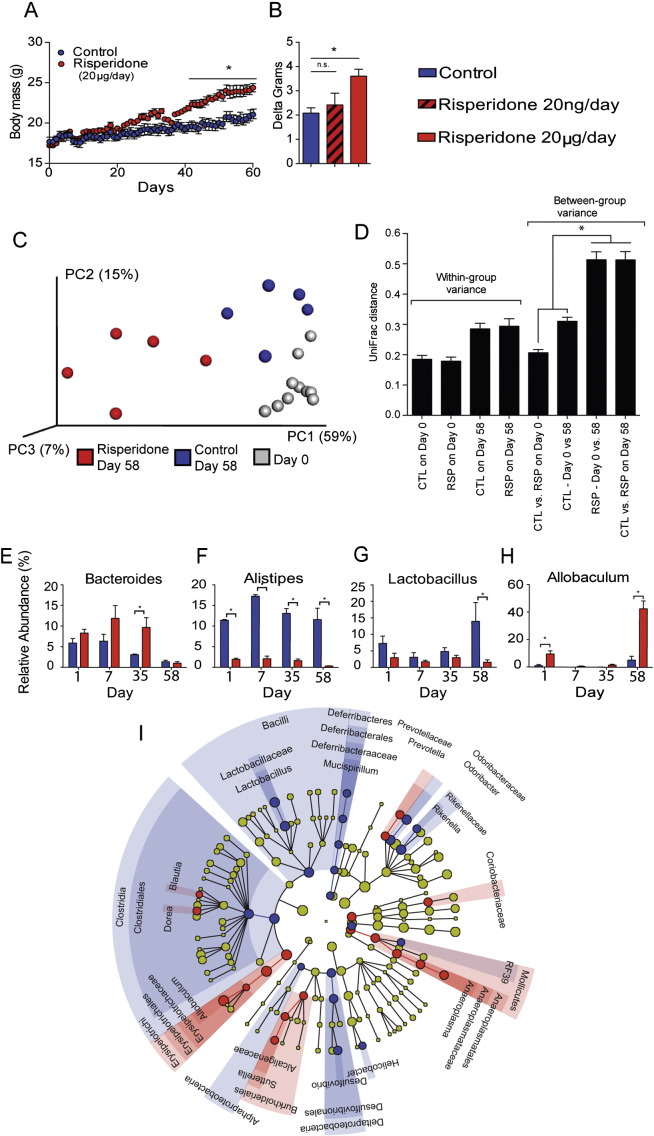
To better understand the role of the gut microbiota in the development of risperidone-induced weight gain, we conducted a prospective study on individually housed C57BL/6J mice over two months. Risperidone was provided in drinking water at 20 μg/ml. On average, mice consumed 4.1 ± 0.3 ml/day, which thereby resulted in an average 80 μg/day risperidone treatment. During risperidone treatment (n = 5) or treatment with acidified water to match the acidity of oral risperidone (pH = 3.5, n = 5), body masses were assessed daily, and fecal samples were collected weekly. By the sixth week, a significant increase in weight was observed for those mice exposed to risperidone, resulting in a 2.8-g difference relative to the control mice. In contrast, mice were offered a 1000-fold lower dose of risperidone (20 ng/ml in drinking water resulting in consumption of 80 ng/day) and exhibited normal rates of growth (Fig. 1A–B). This broadly establishes a dose-dependence for risperidone to cause weight gain and thus provides a rationale to treat mice with 20 μg/ml in subsequent studies.
Fig. 1.
Chronic risperidone treatment results in increased weight gain and an altered microbiome. For all panels, color indicates risperidone treatment (red) vs. control (blue). A) Body masses were assessed daily in 10 C57BL/6 female mice maintained on standard chow with ad libitum access to water (n = 5) or water with risperidone (20 μg/ml, n = 5). B) Body masses were assessed during risperidone treatment at a 1000-fold lower dose of risperidone (20 ng/ml, n = 5). C) PCoA of UniFrac distances illustrating the association between gut bacterial community structure after treatment with risperidone. Day 0 (Gray), Day 58 risperidone (red), and Day 58 control (blue). Risperidone Day 58 samples were compared to all other samples by ANOSIM (R = 0.895, p = 0.002). D) Mean pairwise intra- and intergroup UniFrac distances of control and chronic RSP treatment at days 0 and 58. p < 0.05 based on 1000 permutations. E–H) Genus-level percent relative abundances of the microbiome during treatment with risperidone vs. controls over time. Data are representative of mean ± s.e.m. *p < 0.05. I) Discriminatory OTUs among treatment populations as determined by LEfSe analysis.
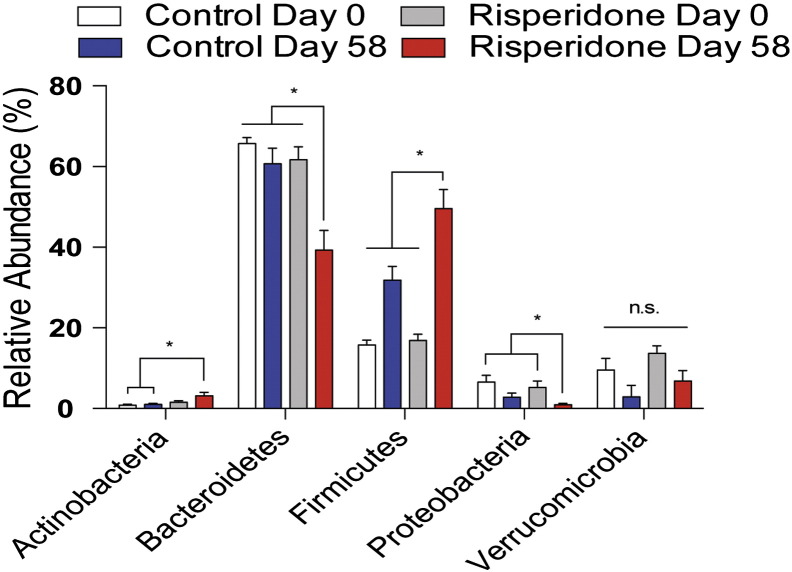
Next, we analyzed the composition of the gut microbiota by 16S rDNA sequencing. The composition of the gut microbiome of mice treated with risperidone (20 μg/ml) for 58 days was significantly different than the microbiome for control mice (ANOSIM R = 0.8954, p = 0.002) (Fig. 1C–D). Risperidone treated mice had a 32.6% increase in the relative abundance of Firmicutes with a reciprocal 22.4% decrease in the relative abundance of Bacteroidetes, two major phyla found in the gut microbiome (p < 0.05, Fig. S2) (Consortium, 2012). Within the phylum Bacteroidetes, the most abundant genera represented in either treatment group were the Bacteroides and Alistipes. Bacteroides spp. were more abundant following risperidone treatment compared to control mice (p < 0.05, Fig. 1E). Conversely, Alistipes spp. were consistently more abundant in controls compared to risperidone treated mice (p < 0.05, Fig. 1F). Within the phylum Firmicutes, Lactobacillus spp. were more abundant in controls compared to risperidone treated mice (p < 0.05, Fig. 1G). The most abundant genera found within risperidone treated mice were Allobaculum spp., with a very large increase, 36.5%, relative to controls (p < 0.05, Fig. 1H). Additional organisms were identified that differentiate the risperidone treated microbiota relative to controls as determined by LDA effect size analysis with LEfSe (Fig. 1I). Collectively, these organisms represent statistically significant discriminatory operational taxonomic units (OTUs) for risperidone treatment in mice.
Supplemental Fig. S2.
Relative abundance of the major phyla within the gut microbiome are shown before and after risperidone treatment. Data are representative of mean ± s.e.m.
3.2. Body mass gains in mice treated with risperidone are due to suppressed energy expenditure
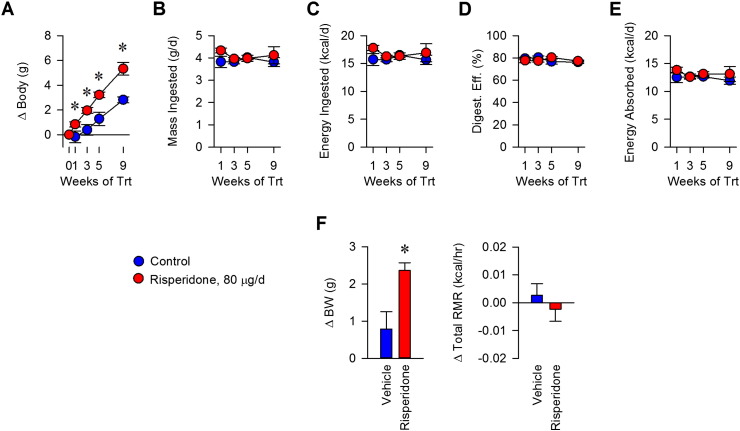
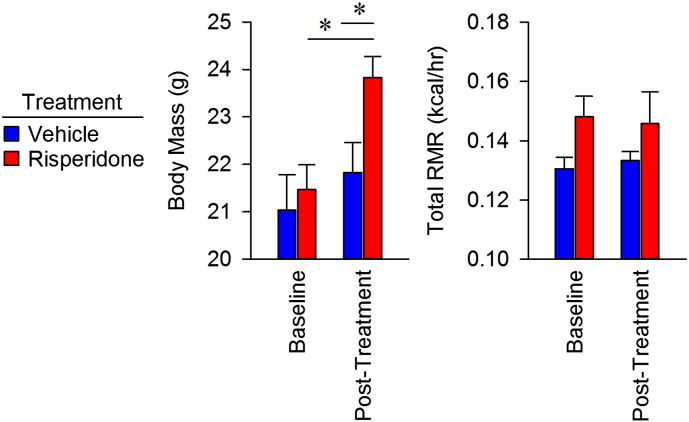
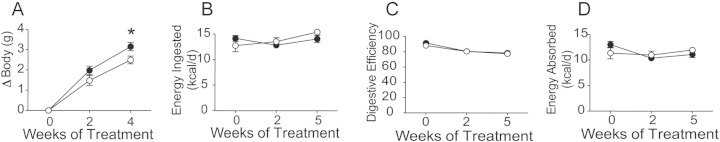
To understand the mechanism of weight gain in mice induced by risperidone, we assessed total energy flux using bomb calorimetry (Fink et al., 2014). Nine weeks of risperidone treatment led to a 2.51 g increase in body weight (Fig. 2A, p < 0.05). Food intake behavior for C57BL/6J mice was found to be similar between groups, resulting in normal daily caloric ingestion, as reported previously for risperidone (Fig. 2B) (Baptista et al., 2004, Li et al., 2013b, Pouzet et al., 2003, Smith et al., 2012). Similarly, risperidone treatment had no effect on digestive efficiency (Fig. 2C). Thus, with similar intake and efficiency, total daily caloric absorption was similar between groups (Fig. 2D). Energy efficiency, the rate of weight gain per unit energy absorbed, was significantly greater with risperidone treatment for the first few weeks of treatment, reflecting a reduction in energy expenditure compared to controls (Fig. 2E). We then determined that total resting metabolic rate did not increase despite gains in body mass following risperidone treatment (Figs. 2F, S3). Thus, we conclude that excess weight gain in mice following risperidone treatment occurs through suppressed energy expenditure, attributable to an altered resting metabolic rate.
Fig. 2.
Body mass gains in mice treated with risperidone result from suppression of energy efficiency via resting metabolic rate. A) Body mass gains were assessed daily in C57BL/6 female mice maintained on standard chow with ad libitum access to water (n = 5) or water with risperidone (20 μg/ml, n = 5) for nine weeks. B) Food intake behavior was measured daily during housing in metabolic cages. C) Digestive efficiency was determined by bomb calorimetry, and performed on desiccated food and feces. D) Energy absorbed was calculated as the subtraction of calculated fecal energy from energy consumed. E) Energy efficiency was calculated as the rate of weight gain per unit energy absorbed. F) Total resting metabolic rate, measured by direct calorimetry, was assessed in mice after treatment with water (n = 4) or water with risperidone (20 μg/ml, n = 4) for two weeks. Data are representative of mean ± s.e.m. *p < 0.05.
Supplemental Fig. S3.
Body mass, total and aerobic RMR in mice treated with risperidone for 2 weeks. Data are plotted as change from baseline, in Fig. 2F. Data are representative of mean ± s.e.m.
3.3. Other xenobiotics do not prevent risperidone-induced weight gain
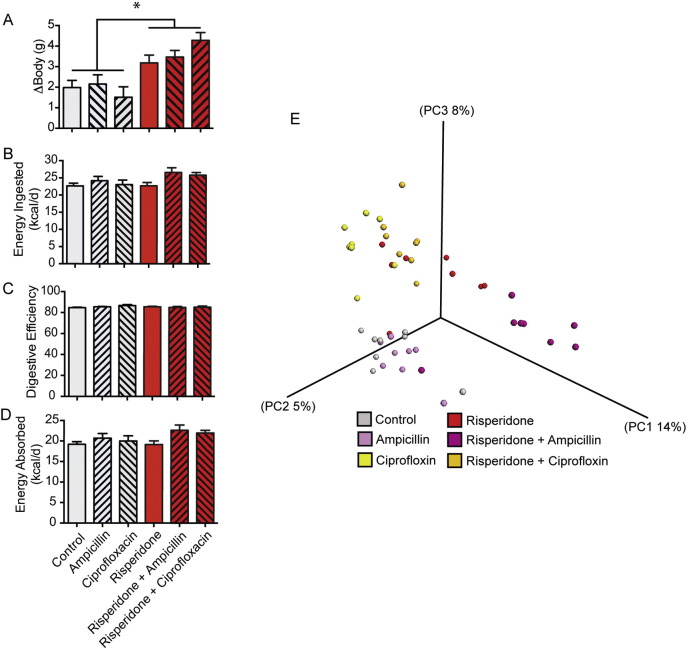
We hypothesized that synergistic or opposing shifts in the microbiome initiated through administering xenobiotics in addition to risperidone would alter energy expenditure and resulting weight gain. To test this possibility, we treated unique cohorts of mice with ampicillin, ciprofloxacin, and risperidone, alone or in combination, resulting in consumption of 0.54 mg, 0.24 mg, and 80 μg per day respectively. All mice were treated with either risperidone or acidified water for 48 days. Antibiotic administration began on the 10th day and continued for 10 days. Risperidone treatment led to a 2.80 g weight increase by day 48, while ten-day courses of antibiotics did not significantly change the outcome of weight gain (Fig. 3A). Food intake behavior and digestive efficiency were found to be similar among all treatment groups (Fig. 3B–C). Additionally, energy absorption was unchanged following antibiotic perturbation (Fig. 3D). We conclude that while other xenobiotics cause unique shifts in the gut microbiome which can synergize with risperidone (Fig. 3E), risperidone alone is sufficient to reduce energy expenditure and induce weight gain. These results provide additional support for the conclusion reached above, namely that risperidone-induced weight gain occurs specifically through suppressed energy expenditure.
Fig. 3.
Additional xenobiotic-induced shifts in the microbiome do not prevent risperidone-induced weight gain through suppressed energy expenditure. 6-Week old female C57Bl/6 mice were maintained on standard chow with ad libitum access to water (n = 8) or risperidone (n = 8, 20 μg/ml) for 7 weeks. During the second week of treatment, all mice (n = 48) were treated for 10 days with ampicillin (142.5 mg/l) or ciprofloxacin (62.5 mg/l). A) Body mass gains were assessed daily. B) Food intake behavior was measured during housing in metabolic cages during the seventh week of treatment. C) Digestive efficiency was determined through bomb calorimetry during the seventh week of treatment. D) Energy absorbed was calculated as the subtraction of calculated fecal energy from energy consumed. E) PCoA of UniFrac distances illustrating the change in the microbiome due to risperidone treatment and the changes in the microbiome due to antibiotic perturbation during the seventh week of treatment. Data are representative of mean ± s.e.m. *p < 0.05.
3.4. Risperidone differentially suppresses anaerobic growth of cultured gut microorganisms
Based on its molecular structure and effects on the microbiome (Figs. 1I and 3E), we hypothesized that risperidone may directly affect the growth of organisms within the gut. To test this possibility, microorganisms collected from fresh stool were grown overnight under either aerobic or anaerobic conditions. Increasing concentrations of risperidone were shown to inhibit the growth of organisms (Fig. 4A). The concentration of risperidone required to inhibit growth of aerobes was greater than the dose needed to suppress the growth of anaerobes (Fig. 4B). After 5 h of growth with risperidone, the IC50 for aerobes was 77 ± 2 μg/ml while the IC50 for anaerobes was 38 ± 0.4 μg/ml (Fig. 4C, p < 0.05). These data demonstrate that risperidone is capable of suppressing the growth of microorganisms cultured from the gut and that anaerobically growing bacteria display increased sensitivity to risperidone. These data suggest that risperidone could act directly on gut microbes to produce shifts in the microbiome described above.
Fig. 4.
Risperidone differentially suppresses growth of cultured gut microorganisms. Microorganisms were collected from stool and cultured either aerobically or anaerobically overnight at 37 °C in BPRM broth. A) Growth curves of aerobic and anaerobic organisms were determined in BPRM supplemented with risperidone at various concentrations. B) Differential growth due to risperidone treatment is observed at the five hour timepoint. C) The IC50 of risperidone for anaerobic and aerobic growth was determined. Data are representative of mean ± s.e.m. *p < 0.05.
3.5. Fecal transfer of risperidone-treated microbiota into naïve recipients suppresses non-aerobic resting metabolic rate
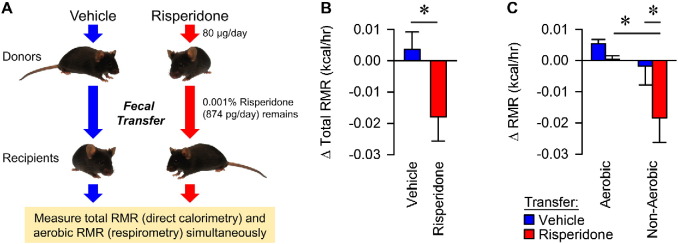
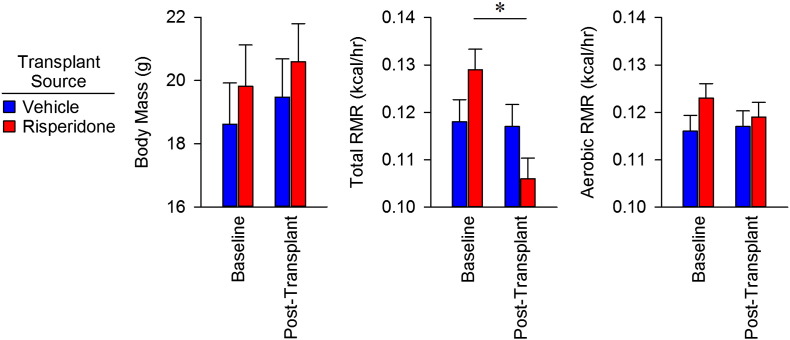
The results above led to the hypothesis that risperidone-altered microbiota are responsible for the associated weight gain in treated animals. To establish Koch's third postulate and attribute risperidone-induced weight gain to the gut microflora and consequent energy expenditure, we performed a fecal transfer from either vehicle- (n = 3) or risperidone-treated mice (n = 3) to a second cohort of naïve mice (n = 6 vehicle recipients and n = 7 risperidone recipients) (Fig. 5A). Transfer of the gut microbiota from vehicle-treated animals had no effect on total resting metabolic rate (RMR), but transfer from risperidone-treated microbiota caused a 16% reduction in total RMR for recipients (Fig. 5B). Importantly, the reduction in total RMR in mice receiving the transplant from a risperidone-treated donor was due entirely to a reduction in non-aerobic RMR, while transfer from risperidone-treated mice had no effect on aerobic RMR (Figs. 5C, S4). Together, these results establish that the shift in the microbiota caused by chronic exposure to risperidone is sufficient to cause the reduced energy expenditure described above. Lastly, a 16% reduction in RMR is sufficient to account for the observed weight gain (see Discussion).
Fig. 5.
Fecal transfer via gavage results in suppression of non-aerobic resting metabolic rates. A) C57BL/6 female donor mice were maintained on standard chow with ad libitum access to water (n = 4) or water with risperidone (20 mg/ml, n = 5) for nine weeks. Fecal material from donor mice was transferred by gavage to naive recipients (n = 7 for risperidone and n = 6 for control) daily for 14 days. Risperidone measured by HPLC in donor stool indicated that recipient mice received only 8.7 ± 1 ng/ml risperidone/day. Following fecal transfer, total and aerobic RMR was measured in recipient mice. B) Total RMR in fecal transfer recipients. C) Combined calorimetry determined the aerobic and non-aerobic components of total resting metabolic rate. Data are representative of mean ± s.e.m. *p < 0.05.
Supplemental Fig. S4.
Body mass, total and aerobic RMR in naïve mice receiving fecal transplants from vehicle- or risperidone-treated donors. Data plotted as change from baseline, in Fig. 5B–C. Data are representative of mean ± s.e.m.
Based on our experimental setup and procedures, we considered the possibility that risperidone present in the stool may have been transferred directly to the recipient mice via gavage. Thus, we performed HPLC to measure the amount of risperidone in stool from a donor prior to fecal transfer by gavage. The HPLC analysis indicated that the concentration of risperidone found in fecal samples was only 8.7 ± 1 ng/ml, corresponding to 0.87 ng delivered via gavage per day or 0.001% of the original dose received daily from drinking water. Thus, the total amount of risperidone found in donor stool is 10-fold less that that used to establish a dose–response curve above which produced no observable effects (Fig. 1B). Thus, we conclude that the microbiota itself is the obesigenic factor present within the stool of risperidone-treated mice.
3.6. Risperidone-treated phage fractions are capable of suppressing host energy expenditure
In addition to bacteria, phage are also present within the gut (Minot et al., 2012, Minot et al., 2013, Reyes et al., 2010). Phage genomes are known to encode traits such as antibiotic resistance, which can be mobilized by exposure to antibiotics and subsequently transferred to other organisms (Modi et al., 2013). Thus, we considered the possibility that risperidone treatment might alter the phageome in the mouse gut and could ultimately affect the energy expenditure and subsequent weight gain for the host.
To determine if bacteriophage play a contributing role in body mass gain, we isolated phage from the stool of risperidone and vehicle treated mice using methods described previously (Thurber et al., 2009) (Fig. S5). Two unique cohorts of mice received a dose of 7 × 109 phage particles (as estimated by microscopy) by gavage each day for 24 consecutive days either from risperidone (n = 7) or control (n = 6) donors. Transfer of phage isolated from risperidone-treated microbiota resulted in significant weight gain relative to vehicle (p < 0.05) (Fig. 6A). Furthermore, food intake behavior and digestive efficiency was unchanged with phage treatment (Fig. 6B–C). Energy absorbed was therefore also unchanged (Fig. 6D). Thus, we conclude that the phage fraction induces changes in the gut microbiota resulting in decreased energy expenditure, similar to that generated by risperidone treatment alone.
Supplemental Fig. S5.
Epifluorescent microscopy determined viral-particle concentration and purity. All images were taken at 600 × magnification with an oil immersion objective. A representative image is shown which shows the characteristic milky appearance of a filter loaded with virus particles.
Fig. 6.
Phage transfer via gavage results in increased body mass without affecting digestive efficiency. Naïve mice were treated daily with bacteriophage isolated from risperidone-treated mice (n = 7) or control (n = 6). A) Body mass gains were measured daily. B) Food intake behavior was measured during housing in metabolic cages. C) Digestive efficiency was determined through bomb calorimetry. D) Energy absorbed was calculated as the subtraction of calculated fecal energy from energy consumed. Data are representative of mean ± s.e.m. *p < 0.05.
4. Discussion
In this study, we demonstrate that risperidone causes major changes in the gut microbiome and that those changes are mechanistically linked to weight gain through the suppression of energy expenditure. We demonstrate that suppression of energy expenditure is recapitulated via transfer of fecal material or the phage fraction from a risperidone-treated donor to a naïve recipient. Risperidone-induced suppression of energy expenditure was not modified through addition of other microbiome-altering xenobiotics. In parallel, we found that cultured microorganisms from the gut were more sensitive to risperidone when grown anaerobically than aerobically. The mechanism for suppression of energy expenditure in vivo was found to be a reduction in non-aerobic RMR following fecal transfer of the gut microbiota. Therefore, host non-aerobic RMR can be suppressed by shifts in the gut microbiome in response to treatment with risperidone. Together, these findings highlight the critical role of the gut microbiota in energy homeostasis and provide a potent example of the physiological significance of non-aerobic RMR in whole-animal energy balance.
We used total calorimetry to assess the RMR of the subject, which includes simultaneous measurement of total RMR via direct calorimetry, and aerobic RMR via respirometry (Kaiyala and Ramsay, 2011). Simultaneous assessments of RMR in various species via direct calorimetry and respirometry by our group and others have uncovered large discrepancies (Walsberg and Hoffman, 2005, Walsberg and Hoffman, 2006, Burnett and Grobe, 2013, Burnett and Grobe, 2014), which we interpret as an index of non-aerobic processes. For example, we recently described a large contribution (7 to 12%) of non-aerobic RMR in both C57BL/6J and FVB/NCrl strains of mice (Burnett and Grobe, 2013, Burnett and Grobe, 2014). It is worth noting that contributions of non-aerobic RMR are variable even within a species. We also determined that one week of a 45% high fat diet in C57BL/6J mice specifically suppresses non-aerobic RMR (Burnett and Grobe, 2014). Because non-aerobic resting metabolism exists in humans (Pittet et al., 1974, Pittet et al., 1976), we are only beginning to appreciate its physiological significance (Burnett and Grobe, 2014).
We also demonstrated that risperidone treatment led to an overall reduction in observed taxa (OTUs) found within the gut microbiome (Fig. S6) as well as selectively suppressing growth of organisms under anaerobic conditions, similar to the environment found within the mammalian intestinal tract. The observed suppression of non-aerobic RMR was found to be 16% of the total RMR for the animal. The magnitude of this difference is substantial. For example, for a 2000 kcal/day human, roughly 80% of total energy expenditure (1600 kcal/day) occurs in the form of RMR. A 16% reduction in total RMR would therefore equate to increasing energy retention by ~ 260 kcal/day, nearly equivalent to consuming an extra standard McDonald's cheeseburger (290 kcal) (McDonald's Nutrition, 2015) each day — a decidedly obesigenic maneuver.
Supplemental Fig. S6.
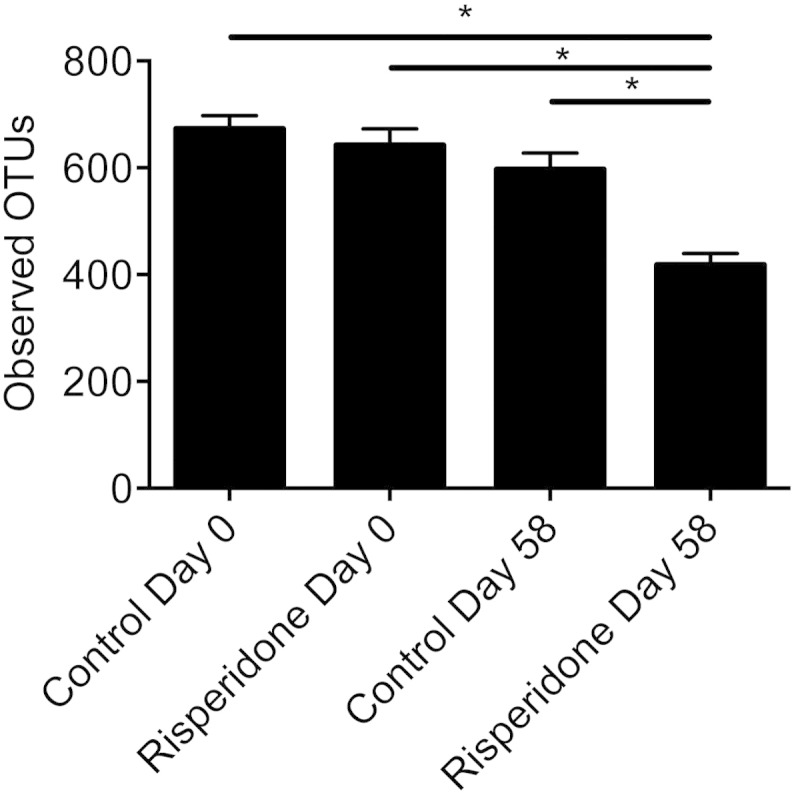
Observed OTUs in control and risperidone treated mice, before and after treatment (mean ± s.e.m.), calculated at a uniform depth of 6000 sequences/sample.
Chronic treatment with risperidone led to specific alterations in the microbiota. We found a significant enrichment in Allobaculum and Turicibacter spp., both members of the Firmicute family Erysipelotrichaceae, which has been associated with diet-induced obesity in mice (Ravussin et al., 2011). Interestingly, members of this family have been shown in several independent studies to change in abundance in response to dietary fat intake (Cani et al., 2008, Fleissner et al., 2010, Turnbaugh et al., 2008). Aneroplasma spp., a member of the Mollicutes class, was also found to be more abundant following risperidone treatment. Previously, Gordon et al. performed metagenomic sequencing and transcriptomics on diet-induced obesity cecal samples of the microbiome, revealing that Mollicutes are enriched for pathways involved in the import and fermentation of simple sugars and host glycans (Turnbaugh et al., 2008) thereby providing a mechanism for increased energy harvest from an obesigenic microbiota. We also found a depletion of Alistipes spp. and Akkermansia spp. in risperidone treated animals compared to controls, both of which have been associated with a lean microbiota (Ridaura et al., 2013). Therefore, our findings offer evidence that risperidone treatment alters the composition of the microbiome which suppresses resting metabolic rate and leads to weight gain. Because suppression of energy expenditure was conferred by the transfer of either the whole microbiota or the phage fraction from risperidone treated mice, we propose that non-aerobic RMR is regulated in part by the gut microbiota. Therefore, manipulation of resting metabolic rate, specifically via the gut microbiome, represents a largely untapped approach to treating obesity. Preventing alteration of the microbiome or the effects of the altered microbiome on RMR, may prove beneficial for patients undergoing risperidone treatment.
The following are the supplementary data related to this article.
NMR data for recipient animals before and after fecal transfer from vehicle- or risperidone-treated donors. Data are representative of mean ± s.e.m.
Conflict of interest
The authors declare no conflicts of interest.
Funding
This work was supported by grants from the NIH (HL098276 and HL084207 to JLG, and R01AI108255 to JK), the American Heart Association (15SFRN23730000 to JLG), the American Diabetes Association (1-14-BS-079 to JLG), the National Science Foundation (MCB-1244021 to JK), and the University of Iowa Fraternal Order of Eagles' Diabetes Research Center (to JK & JLG). Additional support was provided by the University of Iowa Department of Microbiology. BJW was supported by an undergraduate research fellowship from the American Physiological Society. CMLB was supported by the University of Iowa Medical Student Research Program. ANC was supported by the University of Iowa Dean's Graduate Research Fellowship and an Alfred P. Sloan Foundation Scholarship. OD was supported by an Alfred P. Sloan Foundation Scholarship.
Author contributions
The following aspects of this work were performed by the following authors: conceptualization, S.M.B., J.L.G. and J.R.K.; software, S.M.B. and C.M.L.B.; investigation, S.M.B., B.J.W., A.N.C., J.W.W., C.M.L.B., N.A.P., and D.J.M.; writing, review & editing, S.M.B., J.L.G., and J.R.K; and project administration, J.R.K.
Acknowledgements
The authors gratefully acknowledge the assistance of the University of Iowa Office of Animal Resources for assistance in the project. We also thank Samantha Atkinson for assistance with sequence deposition.
References
- Bäckhed F., Ding H., Wang T., Hooper L.V., Koh G.Y., Nagy A., Semenkovich C.F., Gordon J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahr S.M., Tyler B.C., Wooldridge N., Butcher B.D., Burns T.L., Teesch L.M., Oltman C.L., Azcarate-Peril M.A., Calarge C.A., Kirby J.R. Use of the Second generation antipsychotic, risperidone, and secondary-weight gain are associated with an altered gut microbiota in children. Transl. Psychiatry. 2015 doi: 10.1038/tp.2015.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista T., de Baptista E.A., Lalonde J., Plamondon J., Kin N.M.K.N.Y., Beaulieu S., Joober R., Richard D. Comparative effects of the antipsychotics sulpiride and risperidone in female rats on energy balance, body composition, fat morphology and macronutrient selection. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2004;28:1305–1311. doi: 10.1016/j.pnpbp.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Burnett C.M.L., Grobe J.L. Direct calorimetry identifies deficiencies in respirometry for the determination of resting metabolic rate in C57Bl/6 and FVB mice. AJP Endocrinol. Metab. 2013;305:E916–E924. doi: 10.1152/ajpendo.00387.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett C.M.L., Grobe J.L. Dietary effects on resting metabolic rate in C57BL/6 mice are differentially detected by indirect (O2/CO2 respirometry) and direct calorimetry. Mol. Metab. 2014;3:460–464. doi: 10.1016/j.molmet.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarge C.A., Xie D., Fiedorowicz J.G., Burns T.L., Haynes W.G. Rate of weight gain and cardiometabolic abnormalities in children and adolescents. J. Pediatr. 2012;161:1010–1015.e1011. doi: 10.1016/j.jpeds.2012.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani P.D., Bibiloni R., Knauf C., Waget A., Neyrinck A.M., Delzenne N.M., Burcelin R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- Caporaso J.G., Bittinger K., Bushman F.D., Desantis T.Z., Andersen G.L., Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Huntley J., Fierer N., Owens S.M., Betley J., Fraser L., Bauer M. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, T.H.M.P. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correll C.U., Manu P., Olshanskiy V., Napolitano B., Kane J.M., Malhotra A.K. Cardiometabolic risk of second-generation antipsychotic medications during first-time use in children and adolescents. JAMA. 2009;302:1765–1773. doi: 10.1001/jama.2009.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correll C.U., Lencz T., Malhotra A.K. Antipsychotic drugs and obesity. Trends Mol. Med. 2011;17:97–107. doi: 10.1016/j.molmed.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey K.J., Cotter P.D., Sullivan O.O.A., Crispie F., Dinan T.G., Cryan J.F., Mahony S.M.O.A. Antipsychotics and the gut microbiome: olanzapine-induced metabolic dysfunction is attenuated by antibiotic administration in the rat. Transl. Psychiatry. 2013;3:e309. doi: 10.1038/tp.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Hert M., Dobbelaere M., Sheridan E.M., Cohen D., Correll C.U. Metabolic and endocrine adverse effects of second-generation antipsychotics in children and adolescents: a systematic review of randomized, placebo controlled trials and guidelines for clinical practice. Eur. Psychiatry. 2011;26:144–158. doi: 10.1016/j.eurpsy.2010.09.011. [DOI] [PubMed] [Google Scholar]
- DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas M.-E., Barton R.H., Toye A., Cloarec O., Blancher C., Rothwell A., Fearnside J., Tatoud R., Blanc V., Lindon J.C. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12511–12516. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E., Strowig T., Kau A.L., Henao-Mejia J., Thaiss C.A., Booth C.J., Peaper D.R., Bertin J., Eisenbarth S.C., Gordon J.I. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findling R.L., Steiner H., Weller E.B. Use of antipsychotics in children and adolescents. J. Clin. Psychiatry. 2005;66(Supplement 7):29–40. [PubMed] [Google Scholar]
- Fink B.D., Herlein J.A., Guo D.F., Kulkarni C., Weidemann B.J., Yu L., Grobe J.L., Rahmouni K., Kerns R.J., Sivitz W.I. A mitochondrial-targeted coenzyme q analog prevents weight gain and ameliorates hepatic dysfunction in high-fat-fed mice. J. Pharmacol. Exp. Ther. 2014;351:699–708. doi: 10.1124/jpet.114.219329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleissner C.K., Huebel N., Abd El-Bary M.M., Loh G., Klaus S., Blaut M. Absence of intestinal microbiota does not protect mice from diet-induced obesity. Br. J. Nutr. 2010;104:919–929. doi: 10.1017/S0007114510001303. [DOI] [PubMed] [Google Scholar]
- Grobe J.L., Grobe C.L., Beltz T.G., Westphal S.G., Morgan D.A., Xu D., de Lange W.J., Li H., Sakai K., Thedens D.R. The brain renin–angiotensin system controls divergent efferent mechanisms to regulate fluid and energy balance. Cell Metab. 2010;12:431–442. doi: 10.1016/j.cmet.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall K.D., Sacks G., Chandramohan D., Chow C.C., Wang Y.C., Gortmaker S.L., Swinburn B.A. Quantification of the effect of energy imbalance on bodyweight. Lancet. 2011;378:826–837. doi: 10.1016/S0140-6736(11)60812-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henao-Mejia J., Elinav E., Jin C., Hao L., Mehal W.Z., Strowig T., Thaiss C.A., Kau A.L., Eisenbarth S.C., Jurczak M.J. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.2001. http://nutrition.mcdonalds.com/getnutrition/nutritionfacts.pdf
- Kaiyala K.J., Ramsay D.S. Direct animal calorimetry, the underused gold standard for quantifying the fire of life. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2011;158(3):252–264. doi: 10.1016/j.cbpa.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lett T.A.P., Wallace T.J.M., Chowdhury N.I., Tiwari A.K., Kennedy J.L., Müller D.J. Pharmacogenetics of antipsychotic-induced weight gain: review and clinical implications. Mol. Psychiatry. 2011;17:242–266. doi: 10.1038/mp.2011.109. [DOI] [PubMed] [Google Scholar]
- Li F., Jiang C., Krausz K.W., Li Y., Albert I., Hao H., Fabre K.M., Mitchell J.B., Patterson A.D., Gonzalez F.J. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat. Commun. 2013;4:1–10. doi: 10.1038/ncomms3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Johnson M.S., Smith D.L., Jr., Li Y., Kesterson R.A., Allison D.B., Nagy T.R. Effects of risperidone on energy balance in female C57BL/6J mice. Obesity. 2013;21(9):1850–1857. doi: 10.1002/oby.20350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C., Lladser M.E., Knights D., Stombaugh J., Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2010;5:169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk G. Animal calorimetry: twenty-fourth paper. Analysis of the oxidation of mixtures of carbohydrate and fat. J. Biol. Chem. 1924;59(1):41–42. [Google Scholar]
- Minot S., Grunberg S., Wu G.D., Lewis J.D., Bushman F.D. Hypervariable loci in the human gut virome. Proc. Natl. Acad. Sci. 2012;109:3962–3966. doi: 10.1073/pnas.1119061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minot S., Bryson A., Chehoud C., Wu G.D., Lewis J.D., Bushman F.D. Rapid evolution of the human gut virome. Proc. Natl. Acad. Sci. U. S. A. 2013;110:12450–12455. doi: 10.1073/pnas.1300833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi S.R., Lee H.H., Spina C.S., Collins J.J. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature. 2013;499:219–222. doi: 10.1038/nature12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller D., Salama I., Kling R.C., Hübner H., Gmeiner P. 1,4-Disubstituted aromatic piperazines with high 5-HT2A/D2 selectivity: quantitative structure-selectivity investigations, docking, synthesis and biological evaluation. Bioorg. Med. Chem. 2015;23(18):6195–6209. doi: 10.1016/j.bmc.2015.07.050. [DOI] [PubMed] [Google Scholar]
- Morgan A.P., Crowley J.J., Nonneman R.J., Quackenbush C.R., Miller C.N., Ryan A.K., Bogue M.A., Paredes S.H., Yourstone S., Carroll I.M. The antipsychotic olanzapine interacts with the gut microbiome to cause weight gain in mouse. PLoS One. 2014;9:e115225. doi: 10.1371/journal.pone.0115225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittet P., Gygax P.H., Jéquier E. Thermic effect of glucose and amino acids in man studied by direct and indirect calorimetry. Br. J. Nutr. 1974;31:343–349. doi: 10.1079/bjn19740042. [DOI] [PubMed] [Google Scholar]
- Pittet P., Chappuis P., Acheson K., De Techtermann F., Jéquier E. Thermic effect of glucose in obese subjects studied by direct and indirect calorimetry. Br. J. Nutr. 1976;35:281–292. doi: 10.1079/bjn19760033. [DOI] [PubMed] [Google Scholar]
- Pouzet B., Mow T., Kreilgård M., Velschow S. Chronic treatment with antipsychotics in rats as a model for antipsychotic-induced weight gain in human. 2003;75:133–140. doi: 10.1016/s0091-3057(03)00042-x. [DOI] [PubMed] [Google Scholar]
- Ravussin Y., Koren O., Spor A., LeDuc C., Gutman R., Stombaugh J., Knight R., Ley R.E., Leibel R.L. Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity. 2011;20:738–747. doi: 10.1038/oby.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A., Haynes M., Hanson N., Angly F.E., Heath A.C., Rohwer F., Gordon J.I. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura V.K., Faith J.J., Rey F.E., Cheng J., Duncan A.E., Kau A.L., Griffin N.W., Lombard V., Henrissat B., Bain J.R. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341 doi: 10.1126/science.1241214. 1241214-1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.C., Rachakonda S., Dwivedi S., Davis J.M. Olanzapine and risperidone effects on appetite and ghrelin in chronic schizophrenic patients. Psychiatry Res. 2012;199:159–163. doi: 10.1016/j.psychres.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Swann J.R., Want E.J., Geier F.M., Spagou K., Wilson I.D., Sidaway J.E., Nicholson J.K., Holmes E. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc. Natl. Acad. Sci. U. S. A. 2011;108(Suppl. 1):4523–4530. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber R.V., Haynes M., Breitbart M., Wegley L., Rohwer F. Laboratory procedures to generate viral metagenomes. Nat. Protoc. 2009;4:470–483. doi: 10.1038/nprot.2009.10. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P.J., Bäckhed F., Fulton L., Gordon J.I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. cell host microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandanmagsar B., Youm Y.-H., Ravussin A., Galgani J.E., Stadler K., Mynatt R.L., Ravussin E., Stephens J.M., Dixit V.D. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat. Med. 2011;17:179–188. doi: 10.1038/nm.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi V.R., Hezaveh R., Reigstad C.S., Gopalacharyulu P., Yetukuri L., Islam S., Felin J., Perkins R., Boren J., Oresic M. The gut microbiota modulates host energy and lipid metabolism in mice. J. Lipid Res. 2010;51:1101–1112. doi: 10.1194/jlr.M002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsberg G.E., Hoffman T.C. Direct calorimetry reveals large errors in respirometric estimates of energy expenditure. J. Exp. Biol. 2005;208:1035–1043. doi: 10.1242/jeb.01477. [DOI] [PubMed] [Google Scholar]
- Walsberg G.E., Hoffman T.C. Using direct calorimetry to test the accuracy of indirect calorimetry in an ectotherm. Physiol. Biochem. Zool. 2006;79(4):830–835. doi: 10.1086/505514. [DOI] [PubMed] [Google Scholar]
- Wang Z., Klipfell E., Bennett B.J., Koeth R., Levison B.S., DuGar B., Feldstein A.E., Britt E.B., Fu X., Chung Y.-M. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
NMR data for recipient animals before and after fecal transfer from vehicle- or risperidone-treated donors. Data are representative of mean ± s.e.m.