Abstract
Background
Anthrax is a rare disease in humans but elicits great public fear because of its past use as an agent of bioterrorism. Injectional anthrax has been occurring sporadically for more than ten years in heroin consumers across multiple European countries and this outbreak has been difficult to trace back to a source.
Methods
We took a molecular epidemiological approach in understanding this disease outbreak, including whole genome sequencing of Bacillus anthracis isolates from the anthrax victims. We also screened two large strain repositories for closely related strains to provide context to the outbreak.
Findings
Analyzing 60 Bacillus anthracis isolates associated with injectional anthrax cases and closely related reference strains, we identified 1071 Single Nucleotide Polymorphisms (SNPs). The synapomorphic SNPs (350) were used to reconstruct phylogenetic relationships, infer likely epidemiological sources and explore the dynamics of evolving pathogen populations. Injectional anthrax genomes separated into two tight clusters: one group was exclusively associated with the 2009–10 outbreak and located primarily in Scotland, whereas the second comprised more recent (2012–13) cases but also a single Norwegian case from 2000.
Interpretation
Genome-based differentiation of injectional anthrax isolates argues for at least two separate disease events spanning > 12 years. The genomic similarity of the two clusters makes it likely that they are caused by separate contamination events originating from the same geographic region and perhaps the same site of drug manufacturing or processing. Pathogen diversity within single patients challenges assumptions concerning population dynamics of infecting B. anthracis and host defensive barriers for injectional anthrax.
Funding
This work was supported by the United States Department of Homeland Security grant no. HSHQDC-10-C-00,139 and via a binational cooperative agreement between the United States Government and the Government of Germany. This work was supported by funds from the German Ministry of Defense (Sonderforschungsprojekt 25Z1-S-431,214). Support for sequencing was also obtained from Illumina, Inc. These sources had no role in the data generation or interpretation, and had not role in the manuscript preparation.
Panel 1: Research in Context Systematic Review
We searched PubMed for any article published before Jun. 17, 2015, with the terms “Bacillus anthracis” and “heroin”, or “injectional anthrax”. Other than our previously published work (Price et al., 2012), we found no other relevant studies on elucidating the global phylogenetic relationships of B. anthracis strains associated with injectional anthrax caused by recreational heroin consumption of spore-contaminated drug. There were, however, publically available genome sequences of two strains involved (Price et al., 2012, Grunow et al., 2013) and the draft genome sequence of Bacillus anthracis UR-1, isolated from a German heroin user (Ruckert et al., 2012) with only limited information on the genotyping of closely related strains (Price et al., 2012, Grunow et al., 2013).
Lay Person Interpretation
Injectional anthrax has been plaguing heroin drug users across Europe for more than 10 years. In order to better understand this outbreak, we assessed genomic relationships of all available injectional anthrax strains from four countries spanning a > 12 year period. Very few differences were identified using genome-based analysis, but these differentiated the isolates into two distinct clusters. This strongly supports a hypothesis of at least two separate anthrax spore contamination events perhaps during the drug production processes. Identification of two events would not have been possible from standard epidemiological analysis. These comprehensive data will be invaluable for classifying future injectional anthrax isolates and for future geographic attribution.
Keywords: Heroin, Whole Genome Sequencing, SNP, Phylogeny, Injectional Anthrax, Bacillus anthracis
Highlights
-
•
Whole genome sequences of injectional anthrax B. anthracis isolates fall in two tight but distinct genomic clusters.
-
•
The distinct genomic clusters are consistent with two or more disease events that overlap in time and space.
-
•
Defining pathogen clusters will lead to better public health responses to difficult to track disease outbreaks.
1. Introduction
Anthrax is a common disease in livestock and usually only infects humans that come into contact with infected animals or their products (Mock & Fouet, 2001). High hygienic standards, knowledge of the disease, and effective surveillance make human anthrax infections extremely rare in industrialized countries. Most commonly, human anthrax presents as a cutaneous lesion due to spore deposition from handling contaminated animal hair, meat, or skin. The three other recognized anthrax types are gastrointestinal, inhalational, and injectional anthrax, which have unique clinical presentations dependent on the infective route. Injectional anthrax (IA), which was only recently recognized, is associated with recreational drug use when Bacillus anthracis spores have contaminated heroin (Berger et al., 2014). Injectional drug consumers may have preexisting health issues that exacerbate their sensitivity to anthrax and victims may be reluctant to seek medical attention. The presentation of IA can be particularly gruesome with extensive tissue necrosis emanating from the injection site (Booth et al., 2010, Russell et al., 2013).
While the first case of IA was described in 2000 from Norway (Ringertz et al., 2000), it is remarkable that no additional cases of IA were documented over the following 10 years. The largest subsequent cluster of IA cases occurred in 2009–10 and predominantly centered in Scotland (Team, 2011). Other more sporadic cases have been observed in England, Wales, Scotland, Germany, Denmark, and France in 2012–13 (Hanczaruk et al., 2014). It is clear that not all drug users consuming a particular batch of heroin have contracted anthrax and attempts to directly detect or isolate B. anthracis from heroin have failed (Team, 2011). How and why heroin was contaminated with B. anthracis spores has been difficult to address due to the illicit nature of the drug, but likely occurred during its distribution.
B. anthracis evolved as a clonal lineage from a B. cereus-like ancestor with increased pathogenicity due to the acquisition of two virulence plasmids and chromosomal mutations (Mock and Fouet, 2001, Okinaka et al., 1999). The clonal nature of B. anthracis and, perhaps, slow evolutionary rates have resulted in a highly homogeneous species with little strain-to-strain diversity. Nevertheless, Single Nucleotide Polymorphism (SNP) variation has been readily discovered through the use of whole genome sequencing and the population structure has been defined to a high level at both a local and global scale (Pearson et al., 2004, Van Ert et al., 2007, Read et al., 2002).
In a previous analysis of the Scottish IA outbreak, a single whole genome sequence was used to identify clade-specific SNPs (Price et al., 2012). Diagnostic assays targeting some of these SNPs clustered all IA specimens, including those from 2012 (Price et al., 2012, Grunow et al., 2013), into a single subtype (defined by SNP1173928 and SNP1053700) nested within a clade that contained two closely affiliated strains from infected humans (Turkey) (Price et al., 2012). Given a suspected origin in Afghanistan, a major producer of heroin destined for Western Europe, these results led to the conclusion that the heroin may have been contaminated along transportation routes through the Middle East and not at the production source. Importantly, it was discovered that European IA cases could be differentiated by a VNTR allelic difference, raising questions about the epidemiological significance of the differentiation. However, as only IA strains appeared to be in a common clade within the comparison group, a single continuing contamination event and persistence of contaminated heroin circulating in Europe provided the most parsimonious description of this outbreak (Hanczaruk et al., 2014). Detailed knowledge of heroin production practices, transportation, processing, and potential sources of spore contamination is lacking, which makes understanding and controlling IA challenging. Here, we greatly expand upon our knowledge on the genetic make-up and relationships of IA-associated B. anthracis by analyzing whole genome sequences (WGS) from 58 newly sequenced isolates and two published isolates from 36 different IA patients and seven WGS from phylogenetically affiliated strains. We find that IA cases from the years 2000 to 2013 can be clustered into two distinct genetic groups that overlap in time and space, suggesting distinct contamination events, albeit with very similar but unique strains of B. anthracis that likely originated from a common geographic region.
2. Methods
B. anthracis strains. We analyzed the genomes of 67 B. anthracis isolates, including 60 associated with IA cases. Seven additional strains were not from IA cases but were phylogenetically affiliated. The more distantly related Ames Ancestor is a very high quality finished genome and was used as a reference. These strains are listed in Table S1 along with their original sources and NCBI accession numbers for each WGS. All isolates represented residual material from clinical diagnostic procedures and were de-identified. As such, they are exempt from the human subjects requirements for patient enrollment.
2.1. DNA Isolation and WGS Generation
B. anthracis isolates from IA cases and the reference strain collections were cultured on blood agar plates (bioMérieux) and incubated for 16–24 h at 37 °C and then used for DNA purification. DNA was harvested from single colonies, either directly or following liquid media culturing, using the QIAamp UCP Pathogen Mini Kit or Blood and Tissue kit (Qiagen, Valencia, CA) with an enzymatic (lysozyme) or mechanical pre-lysis step. DNA library preparations for WGS were performed using standard methods and performed on an Illumina MiSeq instrument (see Supplemental Methods for details).
2.2. SNP Analysis of WGS
Reference genome assemblies were downloaded from GenBank (Benson et al., 2012). Paired-end reads were aligned against the Ames Ancestor genome (NC_007530) using BWA-MEM (Li, 2013) and SNPs were called with the UnifiedGenotyper method in GATK (DePristo et al., 2011, McKenna et al., 2010); any SNPs that had a coverage less than 6X depth and majority allele frequencies < 90% were discarded. The reference (Ames Ancestor) was aligned against itself with NUCmer (Kurtz et al., 2004) and SNPs that fell into regions that mapped more than once were removed from the analysis. External genome assemblies were aligned against the Ames Ancestor genome with nucmer and a direct one-to-one mapping was generated to identify variant as well as invariant sites (Table S2). All of these methods are wrapped by the Northern Arizona SNP Pipeline (NASP; https://github.com/TGenNorth/NASP). Phylogenetic analysis was performed by maximum parsimony analysis using MEGA 6.0 (Tamura et al., 2013).
2.3. Identifying Close Relatives to the IA Strains
In order to identify close relatives of the B. anthracis IA strains, we screened two genetically diverse global collections comprising 1293 B. anthracis isolates. This included the historical CDC B. anthracis archives (Marston et al., 2005) and the NAU strain collection. We tested these strains using three published canSNP PCR assays (SNP1530761, SNP3836105, and SNP1053700), which define the heroin-associated strains and their close relatives (Price et al., 2012).
2.4. Molecular Confirmation of Phylogenetic Structure
In order to confirm branches identified by WGS analysis within the IA-subclade and close relatives, we developed 31 SNP assays using methods described by Birdsell et al. (Birdsell et al., 2012). A subset of these assays was used to screen 60 IA and close relatives strains (Fig. 1) (Supplemental Table S3). For one additional IA clinical tissue specimen (P35 A4569) collected from a victim for which no isolate was available, trace amounts of B. anthracis DNA were detected with these new SNP-PCR genotyping assays allowing this sample to be phylogenetically placed. We also used PCR on a subset of samples to confirm the WGS-based allele analysis of pXO1- and pXO2-VNTR loci allelic states (Hanczaruk et al., 2014, Van Ert et al., 2007, Keim et al., 2000). The pXO2 alleles were assigned as a code based on the copy number of VNTR repeats as described in Grunow et al. (Grunow et al., 2013).
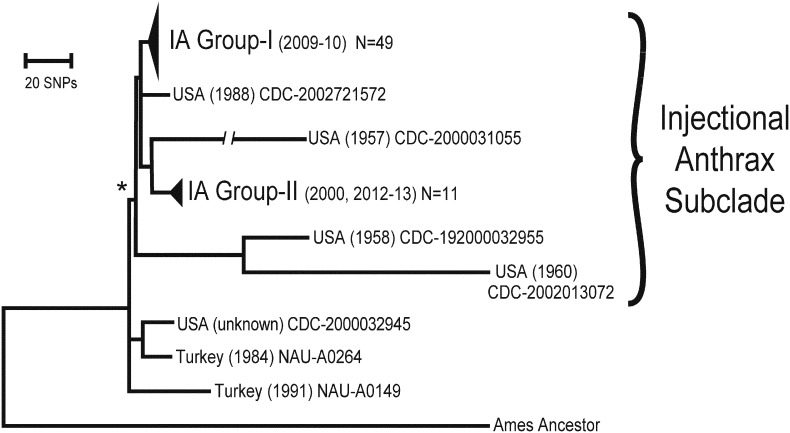
Fig. 1.
Whole Genome Phylogenetic Reconstruction of Injectional Anthrax (IA) B. anthracis Isolates. SNPs discovered from whole genome sequences of 60 B. anthracis isolates (isolation dates in parentheses) sampled from 36 IA patients, seven closely related B. anthracis isolates, and one outgroup genome were used to reconstruct phylogenetic relationships (68 OTUs). Of the 1079 SNPs, 350 synapomorphic SNPs were phylogenetically informative for determining the tree topology. Sixteen SNPs separated the two IA groups. There was only a single homoplastic SNP in this phylogenetic model (CI = 0.997), which did not affect the topology. The Injectional Anthrax Subclade is labeled and consistent with the Price et al. (Price et al., 2012) definition, which was based upon two branch-specific SNPs (SNP1173928 and SNP1053700) marked here with an asterisk (*). Variation within IA-Group I and IA-Group II is presented in Fig 2 and Fig S1.
3. Results
3.1. WGS Defined Phylogenetic Relationships
We performed WGS analyses on 58 new and two published isolates from 36 IA victims to understand how the different cases are epidemiologically related. These genomes were generated to a high quality draft level and compared to seven other closely related strain genomes, as well as the Ames Ancestor genome as a reference (Fig. 1). The seven closely related strains were identified by pre-screening clade specific SNPs across the global strain collections (Price et al., 2012). Notably, four of the non-IA strains also possessed the two SNPs (SNP1173928 and SNP1053700), which had been thought to be specific for the heroin-associated strains (Price et al., 2012). Thus, members of the IA clade themselves are relatively rare as only < 0.5% of the 1293 strains screened were identified as such.
The IA WGS phylogenetic relationships identify two distinct clusters (Fig. 1). Whereas 266 SNPs from 1071 total SNPs used in this analysis were discovered separating the IA subclade from the Ames Ancestor reference, only 77 parsimony informative (synapomorphic) SNPs were found within the IA subclade and six informative SNPs from the near-neighbor clade. An additional 721 SNPs were on terminal branches and unique for the corresponding isolate (autapomorphic). Excluding the long branch to CDC-2000031055, which is likely a mutator strain due to mutations in the yycJ gene (data not shown), there are 236 autapomorphic SNPs. Sixteen SNPs separate the two IA clusters as well as an allelic difference at the ‘AT’ repeat sequence of the VNTR locus on pXO2 (Table S4). The phylogenetic model is highly robust (Pearson et al., 2009) with only a single homoplasious SNP (4398276) that does not impact the phylogenetic topology of the IA subclade and results in an overall tree with a consistency index of 0.997. Price et al. (Price et al., 2012) reported the close relationship of two Turkish isolates to the Scottish IA outbreak strains, but we have now identified additional five close phylogenetic relatives that were isolated in the USA; unfortunately the original global sources of these strains are unknown as they were likely isolated from goods imported into the USA and were never ecologically established. Three of these seven closely related strains, including the two Turkish isolates, are basal to the IA subclade but four others are within this subclade. While the origins of the two most closely related non-IA strains (CDC-2002721572 and CDC-2000031055) would provide further details on the potential geographic source of contamination, these isolates are important for showing that the IA outbreak is indeed the result of more than one contamination event.
3.2. Phylogenetic Diversity Within Injectional Anthrax Groups
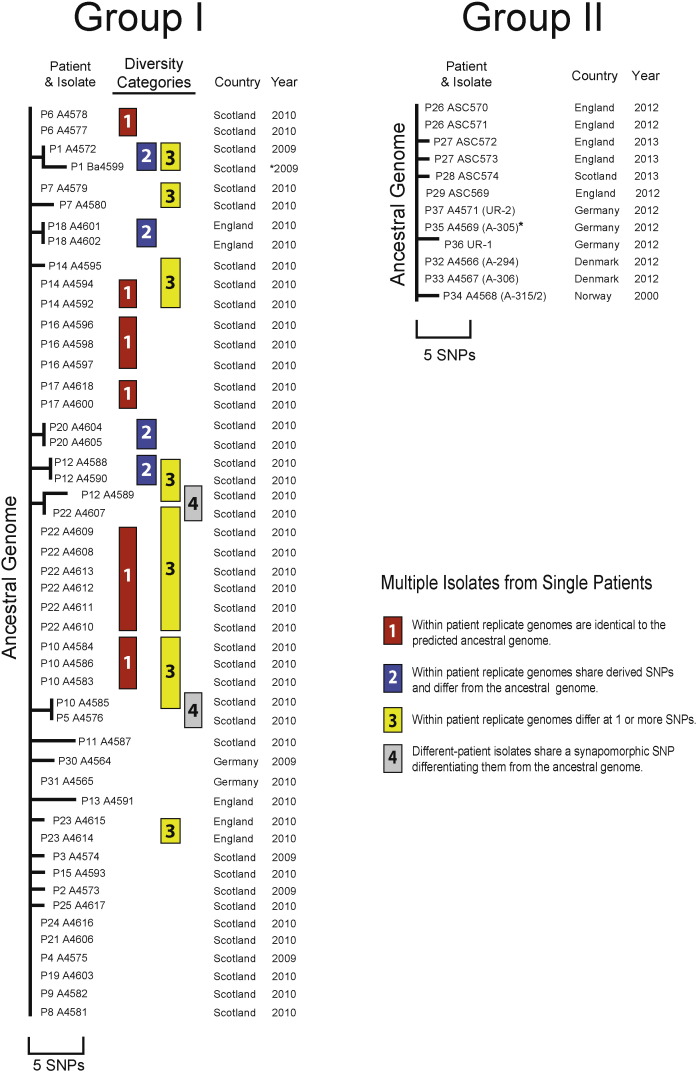
Although the two IA Groups are each dominated by an archetypal ancestral genome, there are minor variants observed in both (Fig. 2 and S1). Of the 58 new and two published genomes, 49 of these were members of G-I with 27 matching the predicted ancestral genome and 22 differentiated with derived SNP alleles (1 to 4 SNP differences). Six pairs of differentiated genotypes were observed among the 22 derived genomes, resulting in sixteen derived subgroups. The remaining 11 IA isolates formed G-II, with 6 genomes identical to the ancestral genotype and 5 genomes differentiated by derived alleles at 1 or 2 SNPs. The single B. anthracis positive complex clinical sample (P35 A4569) was phylogenetically placed in G-II by 11 new canSNP assays (Table S3). Due to a lack of WGS for A4569, it remains collapsed in the ancestral genotype. In G-II, all genomes with derived alleles were singletons (Fig. 2).
Fig. 2.
Phylogenetic Diversity Within the Two Injectional Anthrax Groups. The diversity within each of the two IA groups is represented as dendrograms where the predicted ancestral genomes are the left vertical lines. Genomes perfectly matching the ancestral allele states are flush with these lines, whereas genomes differing by one or more SNPs have a horizontal line of the appropriate length. Group I (N = 49) contains 27 strains with genomes identical to the ancestor and 22 strains with genomes that differ from the ancestor, including 16 unique derived types. Shared SNPs (synapomorphic) identify 6 derived pairs. For Group I, independent isolates from the same patient (replicate genomes) are labeled into four different categories of within single patient diversity. Group II (N = 11) has six ancestral genome types and five genomes with derived allele states. The single B. anthracis clinical tissue specimen (P35 A4569) is labeled with an asterisk (*) and was not WGS but rather analyzed phylogenetically by the 11 new canSNP assays (Table S3) and, thus, is collapsed into the ancestral genome. There is only one incidence of allelic variation within patient replicate isolates for Group II.
In G-I, multiple isolates were obtained from 12 patients and genome replicates from the same patient showed heterogeneity in seven of the 12 patients (Fig. 2). The within-patient diversity patterns can be captured by four categories, including: 1) multiple replicates from a single patient that are identical to each other as well as the ancestral genotype, 2) multiple replicates from a single patient that are identical to each other but differ from the ancestral genotype, 3) multiple replicates from a single patient that differ from each other as well as from the ancestral genotype, and 4) isolates from different patients that differ from the ancestral genotype but share rare SNP(s) and, for one, are even identical to each other. Identical replicate genomes commonly matched the ancestral genome, but there were three patients where duplicate isolates were identical but differed by 1 SNP from the ancestral genotype (P12, P18, P20). The replicate pair from one other patient (P1) differed from the ancestral genome by 2 shared SNPs, but also from each other by 2 additional SNP differences. Three other patients (P7, P14, P23) had genome pairs with replicates both matching and differing from the ancestral genotype (differing by 1 or 2 SNPs). All these minor variants could have been generated within single hosts during the infections or during subsequent laboratory culturing, but they also could have preexisted in the contaminated heroin. This scenario of the presence of pre-infection diversity is supported by the two category 4 examples that are most parsimoniously explained by SNP differences existing among the heroin spores themselves. These two derived genome pairs differed from the G-I ancestral genotype by a shared SNP but were isolated from different patients (P5/P10 and P12/P22). This argues for heterogeneity among the infective spores, as well as for infective doses sufficient to be detected in small end-point sample sizes and for biological diversity to be maintained within a single diseased host despite the potential for genetic drift.
3.3. Spatial and Temporal Distribution of the Phylogenetic Groups
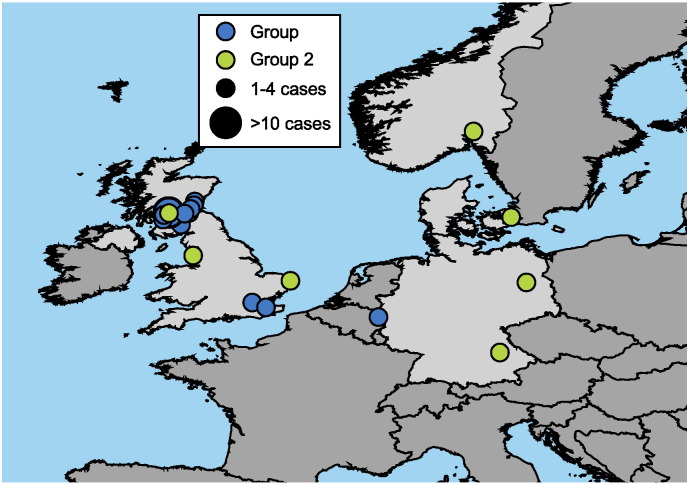
The two phylogenetic groups were found in both continental Europe and the UK (Fig. 3). The large IA outbreak of 2009–10 was focused in Scotland and most of G-I isolates were located there, with the exception of five isolates from England and two from Germany. G-II isolates were distributed across a larger region, including Denmark, England, Germany, and Scotland. The IA temporal occurrence is exclusively in the 2009–10 timeframe for the G-I and primarily in 2012–13 for G-II, with the single exception of the Norwegian 2000 isolate (Grunow et al., 2013). These spatial patterns must, at least partially, reflect the illicit drug distribution networks. The temporal patterns probably reflect the timing of spore contamination in drug production and subsequent drug distribution activities. We attempted to calibrate the phylogenetic branch lengths to epidemiological dates of isolate, but observed no correlation between the two (Fig S3).
Fig. 3.
Distribution of Heroin-Associated Anthrax Cases in this Study. The location of the 37 IA cases (from which a total of 61 patient isolates were retrieved) is indicated along with their membership in Group I or II.
4. Discussion
Understanding the IA outbreak has been difficult because of the mostly sporadic nature of cases that are restricted to a subset of the population with hidden activities (Berger et al., 2014). Furthermore, efforts to directly monitor heroin for anthrax spore contamination by culture or PCR have failed (Booth et al., 2010, Team, 2011). Epidemiological analyses to identify IA trends using fewer strains and with lower resolution molecular subtyping (clade specific canSNPs and MLVA-31) have been informative, yet incomplete. In this report, we have increased our discriminatory power and phylogenetic resolution by increased sampling of IA cases, screening two large strain collections to identify other phylogenetically relevant strains, and employing WGS to all relevant isolates.
The assumption that European heroin is produced in Afghanistan and then smuggled through intervening countries is well founded and has restricted hypotheses for the source of the IA anthrax spores to these regions. One use of genome databases is for source attribution, and in Price et al. (Price et al., 2012), the affiliation with Turkish isolates fueled speculation that the spore contamination event occurred while the heroin was transiting through this country. The identification of other isolates in the IA subclade, possibly from other parts of the world, illustrates the strengths and weakness of this argument and the necessity for extensive sampling in the suspected geographic region. Detailed sampling of B. anthracis in Afghanistan and other potential regional smuggling routes within the Middle East is greatly needed for eliminating and pinpointing likely sources. For most recent molecular epidemiological investigations, the current databases have been sufficient to identify possible sources, but additional contemporary sampling is needed to differentiate alternative source hypotheses. In this report, the linkage to the Middle East and Turkey still exists, but new strains utilized in this study emphasize the uncertainty in attribution as discussed by Price et al. (Price et al., 2012). These strains are more closely related to the IA subclade than the Turkish ones, and while their exact origins are unknown, they emphasize the need to consider and sample neighboring geographical regions as possible sources.
The phylogenetic and temporal patterns reported here indicate that at least two spore contamination events occurred. The core genome SNP analysis strongly supports the presence of two IA clusters, each of which is monophyletic and well supported by multiple synapomorphic SNPs. The phylogenetic presence of other isolates that are spatially and temporally unrelated to IA cases provides further evidence that these are distinct groups. Even though these non-IA strains probably originated from the same region as the IA strains, the dates of collection (1957 and 1988) are key to establishing that these isolates are not directly epidemiologically related to the IA outbreak and thus break apart the otherwise monophyletic clade of IA strains. The early isolation dates also demonstrate that strains representing these two clades could have been affecting human and animal health for decades before the IA was detected. Because of the lack of temporal calibration for B. anthracis evolution, we are limited to collection dates to make inferences about the duration of this outbreak. These two IA groups therefore must have arisen independently. There is also diversity within each group, suggesting diversity at the source and the possibility of even more contamination events. It is possible that variation is generated in a single host infection but some of the phylogenetic patterns shown here are best explained by diversity generated from several infection cycles and thus contamination of B. anthracis from several hosts. Although distinct, the two subclades are closely related with only 16 SNPs separating them. This level of variation is frequently seen within a single country (Khmaladze et al., 2014, Simonson et al., 2009, Kenefic et al., 2009) and could be indicative of a close geographic relationship for the two sources.
Whether each group represents a single contamination or multiple contaminations from a single source is hard to determine, but a single contamination event for each group is the simplest explanation. The temporal pattern of G-I in 2009–1010 and G-II mostly in 2012–13 is also mostly consistent with separate contamination events aligned with these outbreaks. The time point associated with the Norwegian 2000 isolate in G-II is most difficult to explain. The distribution of G-II across > 13 years is astounding and difficult to reconcile with a single contamination event. It has been argued that heroin supply and inventories would not last for 13 years, especially given the apparent 10-year lapse in G-II IA cases. We therefore suggest that there is a single contaminating source for the G-II cases but that there have been two or more (unobserved) contamination events.
Acknowledgments
We are very grateful to individuals and institutions that provided strains for this work. This includes Centers of Disease Control (Atlanta Georgia USA), Dr. Mehmet Doganay (Erciyes University, Kayseri, Turkey), Dr. Udo Reischl, and Dr. Thomas Holzmann (University Hospital Regensburg, Germany), Dr. Mandy Elschner (Friedrich-Loeffler-Institut, Institute of Bacterial Infections and Zoonoses, Jena, Germany), Dr. Anne Kjerulf and Dr. Jorgen Skov Jensen (both Statens Serum Institut, National Institute for Health Data and Disease Control, Copenhagen, Denmark) and Dr. Per Sandven (Norwegian Institute of Public Health, Department of Bacteriology and Immunology, Oslo, Norway).
The following are the supplementary data related to this article.
Strains Table.
SNP Genotype Matrix.
PCR Primer Table.
VNTR Genotype Matrix.
Supplementary material.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2015.10.004.
References
- Benson D.A., Karsch-Mizrachi I., Clark K., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucleic Acids Res. 2012;40(Database issue):D48–D53. doi: 10.1093/nar/gkr1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger T., Kassirer M., Aran A.A. Euro surveillance: Bulletin Europeen sur les maladies transmissibles = European Communicable Disease Bulletin. Vol. 19. 2014. Injectional anthrax — new presentation of an old disease. [DOI] [PubMed] [Google Scholar]
- Birdsell D.N., Pearson T., Price E.P. Melt analysis of mismatch amplification mutation assays (Melt-MAMA): a functional study of a cost-effective SNP genotyping assay in bacterial models. PLoS ONE. 2012;7(3):e32866. doi: 10.1371/journal.pone.0032866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth M.G., Hood J., Brooks T.J., Hart A. Health Protection Scotland Anthrax Clinical N. Anthrax infection in drug users. Lancet. 2010;375(9723):1345–1346. doi: 10.1016/S0140-6736(10)60573-9. [DOI] [PubMed] [Google Scholar]
- DePristo M.A., Banks E., Poplin R. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunow R., Klee S.R., Beyer W. Euro surveillance: Bulletin Europeen sur les maladies transmissibles = European Communicable Disease Bulletin. Vol. 18. 2013. Anthrax among heroin users in Europe possibly caused by same Bacillus anthracis strain since 2000. [PubMed] [Google Scholar]
- Hanczaruk M., Reischl U., Holzmann T. Injectional anthrax in heroin users, Europe, 2000–2012. Emerg. Infect. Dis. 2014;20(2):322–323. doi: 10.3201/eid2002.120921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim P., Price L.B., Klevytska A.M. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 2000;182(10):2928–2936. doi: 10.1128/jb.182.10.2928-2936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenefic L.J., Pearson T., Okinaka R.T. Pre-Columbian origins for North American anthrax. PLoS ONE. 2009;4(3) doi: 10.1371/journal.pone.0004813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmaladze E., Birdsell D.N., Naumann A.A. Phylogeography of Bacillus anthracis in the country of Georgia shows evidence of population structuring and is dissimilar to other regional genotypes. PLoS ONE. 2014;9(7):e102651. doi: 10.1371/journal.pone.0102651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Phillippy A., Delcher A.L. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning Sequence Reads, Clone Sequences and Assembly Contigs With BWA-MEM. (arXivorg). (arXiv:1303.3997 [q-bio.GN]) [Google Scholar]
- Marston C.K., Hoffmaster A.R., Wilson K.E. Effects of long-term storage on plasmid stability in Bacillus anthracis. Appl. Environ. Microbiol. 2005;71(12):7778–7780. doi: 10.1128/AEM.71.12.7778-7780.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock M., Fouet A. Anthrax. Annu. Rev. Microbiol. 2001;55:647–671. doi: 10.1146/annurev.micro.55.1.647. [DOI] [PubMed] [Google Scholar]
- Okinaka R.T., Cloud K., Hampton O. Sequence and organization of pXO1, the large Bacillus anthracis plasmid harboring the anthrax toxin genes. J. Bacteriol. 1999;181(20):6509–6515. doi: 10.1128/jb.181.20.6509-6515.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson T., Busch J.D., Ravel J. Phylogenetic discovery bias in Bacillus anthracis using single-nucleotide polymorphisms from whole-genome sequencing. Proc. Natl. Acad. Sci. U. S. A. 2004;101(37):13536–13541. doi: 10.1073/pnas.0403844101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson T., Okinaka R.T., Foster J.T., Keim P. Phylogenetic understanding of clonal populations in an era of whole genome sequencing. Infect. Gen. Evol. J. Mol. Epidemiol.Evol. Gen. Infect. Dis. 2009;9(5):1010–1019. doi: 10.1016/j.meegid.2009.05.014. [DOI] [PubMed] [Google Scholar]
- Price E.P., Seymour M.L., Sarovich D.S. Molecular epidemiologic investigation of an anthrax outbreak among heroin users, Europe. Emerg. Infect. Dis. 2012;18(8):1307–1313. doi: 10.3201/eid1808.111343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read T.D., Salzberg S.L., Pop M. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science. 2002;296(5575):2028–2033. doi: 10.1126/science.1071837. [DOI] [PubMed] [Google Scholar]
- Ringertz S.H., Hoiby E.A., Jensenius M. Injectional anthrax in a heroin skin-popper. Lancet. 2000;356(9241):1574–1575. doi: 10.1016/s0140-6736(00)03133-0. [DOI] [PubMed] [Google Scholar]
- Ruckert C., Licht K., Kalinowski J. Draft genome sequence of Bacillus anthracis UR-1, isolated from a German heroin user. J. Bacteriol. 2012;194(21):5997–5998. doi: 10.1128/JB.01410-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell L., Pedersen M., Jensen A.V., Soes L.M., Hansen A.B. Two anthrax cases with soft tissue infection, severe oedema and sepsis in Danish heroin users. BMC Infect. Dis. 2013;13:408. doi: 10.1186/1471-2334-13-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonson T.S., Okinaka R.T., Wang B. Bacillus anthracis in China and its relationship to worldwide lineages. BMC Microbiol. 2009;9:71. doi: 10.1186/1471-2180-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team N.A.O.C. Health Protection Scotland; Glasgow, Scotland: 2011. An Outbreak of Anthrax Among Drug Users in Scotland, December 2009 to December 2010. [Google Scholar]
- Van Ert M.N., Easterday W.R., Huynh L.Y. Global genetic population structure of Bacillus anthracis. PLoS ONE. 2007;2(5):e461. doi: 10.1371/journal.pone.0000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Strains Table.
SNP Genotype Matrix.
PCR Primer Table.
VNTR Genotype Matrix.
Supplementary material.