Abstract
Thyroglobulin (Tg) is a vertebrate secretory protein synthesized in the thyrocyte endoplasmic reticulum (ER), where it acquires N-linked glycosylation and conformational maturation (including formation of many disulfide bonds), leading to homodimerization. Its primary functions include iodide storage and thyroid hormonogenesis. Tg consists largely of repeating domains, and many tyrosyl residues in these domains become iodinated to form monoiodo- and diiodotyrosine, whereas only a small portion of Tg structure is dedicated to hormone formation. Interestingly, evolutionary ancestors, dependent upon thyroid hormone for development, synthesize thyroid hormones without the complete Tg protein architecture. Nevertheless, in all vertebrates, Tg follows a strict pattern of region I, II-III, and the cholinesterase-like (ChEL) domain. In vertebrates, Tg first undergoes intracellular transport through the secretory pathway, which requires the assistance of thyrocyte ER chaperones and oxidoreductases, as well as coordination of distinct regions of Tg, to achieve a native conformation. Curiously, regions II-III and ChEL behave as fully independent folding units that could function as successful secretory proteins by themselves. However, the large Tg region I (bearing the primary T4-forming site) is incompetent by itself for intracellular transport, requiring the downstream regions II-III and ChEL to complete its folding. A combination of nonsense mutations, frameshift mutations, splice site mutations, and missense mutations in Tg occurs spontaneously to cause congenital hypothyroidism and thyroidal ER stress. These Tg mutants are unable to achieve a native conformation within the ER, interfering with the efficiency of Tg maturation and export to the thyroid follicle lumen for iodide storage and hormonogenesis.
Introduction to Thyroglobulin and Its Role in Formation of Thyroid Hormones
The Tg Polypeptide and Thyroid Hormones in Evolution
-
Tg Primary Structure
Tg cysteine-rich repetitive units
The ChEL domain
Tg Iodination and Hormonogenesis
-
Tg Domain Foldability and Interdomain Interactions
Intramolecular chaperone and molecular courier functions
Dimerization function
The challenges of folding Tg region I
-
Chaperones and Oxidoreductases That Play a Role in Tg Maturation
The CRT/CNX cycle
Substrate recognition by CRT/CNX
Glycan-dependent oxidoreductase activity
Glycoprotein ERAD
Tg as a model substrate for CRT/CNX/ERp57 in secretory protein folding
Other oxidoreductases of the ER
Evidence of ER-oxidoreductase involvement in Tg folding
Alternatives Within the Tg Folding Pathway
-
Understanding Mutations Causing Congenital Hypothyroidism
Nonsense mutations and single nucleotide insertion/deletion
Acceptor and donor splice site mutations
Missense mutations
Conclusion
I. Introduction to Thyroglobulin and Its Role in Formation of Thyroid Hormones
The thyroid is the endocrine gland that synthesizes and secretes the thyroid hormones (THs) T3 and T4. Synthesis of T4, the primary form of TH released by the thyroid gland, consists of two sequential steps: iodination of selected tyrosines of thyroglobulin (Tg; a large glycoprotein), and coupling of two doubly iodinated tyrosines within Tg to produce T4. Thus, TH synthesis relies on iodide availability. Iodide abundance is not only limited in the terrestrial environment, but its intake is also variable, and this strikingly contrasts with the fact that TH is required continuously throughout human life. During fetal life and childhood, TH is critical for brain development, controlling myelination, somatogenesis, neuronal differentiation, and formation of neural processes, and the requirement continues in adult life as a regulator of intermediary metabolism (1). Thus, whereas insufficient iodide intake results in hypothyroidism and can promote goiter development in patients of all ages, in critical windows of development during fetal life and childhood it causes irremediable neurological manifestations.
To minimize the deleterious effects of iodide deficiency, a unique strategy for the structure of the thyroid gland evolved: specifically, iodinated TH precursor protein is stored extracellularly. This arrangement permits a massive storage that is even greater than the extensive intracellular storage capacity of the secretory granules of other endocrine cell types. Indeed, the anatomical unit of the thyroid follicle is the functional unit for TH synthesis within the mature thyroid gland. Thyroid follicles comprise a monolayer of polarized thyrocytes with the basolateral surface facing the bloodstream and the apical surface delimiting a central, spherical, follicle lumen (Figure 1). The lumen is filled with “colloid” (mainly highly concentrated Tg in different states of oligomerization) (2). Tg itself evolved under pressure to maximize iodide storage, as well as being an efficient molecular scaffold for TH synthesis, and the need to serve as the body's primary reservoir for iodide storage and TH synthesis amounts to two distinct evolutionary pressures that are simultaneously fulfilled by Tg within the thyroid follicular structure.
Figure 1.
TH synthesis and secretion. The thyroid gland is comprised of follicles and surrounding blood vessels. Follicles are the functional unit for TH synthesis and secretion. Thyroid follicles are a spherical monolayer of polarized thyrocytes with the basolateral surface facing the bloodstream and the apical surface delimiting a central follicle lumen. Iodine is taken up by thyrocytes by the action of the NIS exploiting the Na+ electrochemical gradient generated by the Na+/K+ ATPase. I− crosses the apical membrane and reaches the follicular lumen (I− efflux) by the action of the anion exchanger Pendrin and/or other transporters. I− is oxidized by TPO in the presence of H2O2 generated by the Duox2. Reactive iodide is covalently linked to selected tyrosyl residues of Tg to generate MIT and/or DIT. Under these oxidizing conditions, MIT and DIT are coupled to form THs T3 and T4. Newly synthesized Tg, TPO, and Duox2 are made in the ER and fold with the aid of general (CNX, CRT, etc) and dedicated (Duox maturation factor 2 for Duox2) chaperones before transport to the apical membrane. Newly secreted and iodinated Tg is endocytosed and degraded in lysosomes, and THs undergo transport to the bloodstream. The iodide from uncoupled MIT and DIT is recycled by tyrosine dehalogenase (Dehal1). Intact Tg is normally found in serum at low levels, but may exhibit increased concentrations in serum under conditions of increased thyroid cell mass (279), TSH stimulation (279), Graves' disease (280), subacute thyroiditis (281), and thyroid carcinoma. In the latter condition, post-thyroidectomy and ablation of residual thyroid tissue, serum Tg levels are used to monitor residual disease (282).
Remarkably, even when the huge extracellular colloid deposit is not considered, Tg is still the predominant protein of the thyroid gland. Tg has a monomeric molecular mass of approximately 330 kDa, and at approximately 2750 amino acid residues (after cleavage of the signal peptide), Tg falls in the largest 1% of proteins in the vertebrate proteome. The N-terminal two-thirds of the protein, known as Tg region I-II-III (3), contains multiple cysteine-rich repeat motifs, mainly the Tg type-1 domains (4), whereas the carboxyl-terminal region of Tg shows homology with the acetylcholinesterase (AChE; α-β hydrolase fold) superfamily (5) and is known as the cholinesterase-like (ChEL) domain. Roughly 10% of the molecular mass of Tg is carbohydrate, mostly N-linked oligosaccharides acquired as Tg (like other secretory and membrane glycoproteins) passes through the secretory pathway (6, 7). Tg possesses about 120 cysteine residues, and during its conformational maturation about 60 intramolecular disulfide bonds are formed (8). Thus, the achievement of the native three-dimensional structure of Tg is an elaborate and slow process (9, 10). Tg contains at least 66 tyrosine residues (in human Tg, and varying slightly between species). The percentage of tyrosine residues that become iodinated normally varies with dietary iodide intake, but under iodide-sufficient conditions, there may average 10–15 monoiodotyrosine (MIT) plus diiodotyrosine (DIT) residues, of which three or four may be recovered as TH (11, 12). At first glance, TH formation may appear as a wasteful process in which the thyrocyte must synthesize a protein of 330 kDa that can give rise to only three or four TH molecules. However, Tg is able to form THs at extremely low levels of iodination (even 4 mol of iodide per mole of Tg) (13), with the major TH-forming sites at the extreme N terminus (T4) and C terminus (T4 and T3), whereas other iodotyrosines constitute the body's primary reservoir of iodide for recycling within the thyroid gland (Figure 2).
Figure 2.
Cartoon view of Tg structure and function. Tg can be considered to have two main regions: the N-terminal, three-fourths are composed of cysteine-rich repeated motifs; and the carboxyl-terminal, one-fourth is composed of the ChEL domain. Thyroid hormonogenesis in Tg of most vertebrates favors T4 at a conserved site near the N terminus, T3 at a conserved site near the C terminus, and iodide storage via iodotyrosines located at various sites along the polypeptide chain.
II. The Tg Polypeptide and Thyroid Hormones in Evolution
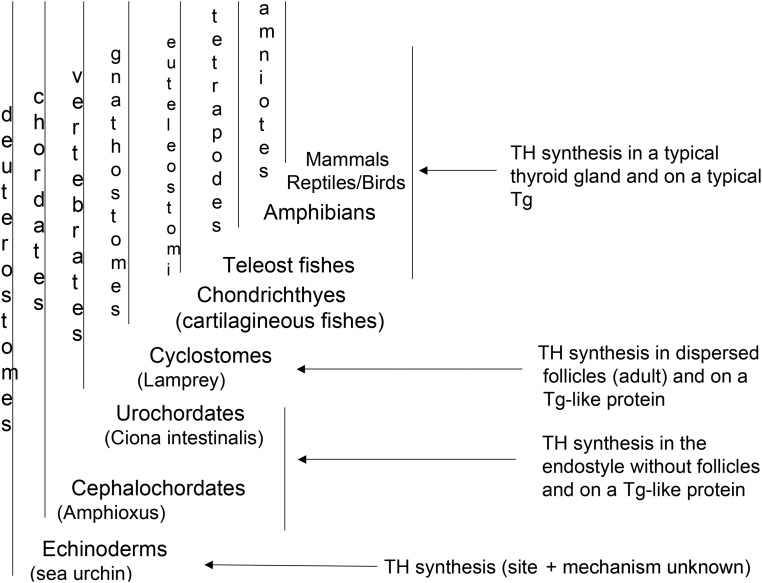
The appearance of a molecular mechanism of TH synthesis based on Tg-like and thyroperoxidase (TPO)-like proteins appears to have preceded the evolution of the thyroid follicle. Indeed, TH synthesis occurs in the most basal nonvertebrate cephalochordates such as amphioxus and urochordates such as the sea squirt Ciona intestinalis (Figure 3). In these organisms, the endostyle (contained within the hypobranchial groove of the pharynx) accumulates iodide and synthesizes TH despite the absence of a thyroid follicular structure (14). In amphioxus (Branchiostoma floridae), a large (still unidentified) protein that incorporates iodide via peroxidase activity (15) produces T3 and T4 (16). Recently, taking advantage of the complete amphioxus genome sequence (17, 18), orthologs of the main genes involved in TH synthesis and metabolism (sodium-iodide symporter [NIS], TPO, deiodinase enzymes) have been identified (19, 20). However, a typical Tg was not found, but many genes with homology to esterases or containing the Tg type-1 domain were noted. Among the latter predicted open reading frames is a protein of about 2400 amino acids (Bf_123169, estimated molecular mass of about 260 kDa), comprising numerous Tg type-1 domains (19). Thus, the hormonogenic function in an evolutionary ancestor of the Tg protein may have preceded the contiguity of N- and C-terminal regions found in modern vertebrate Tg. Moreover, these findings raise interesting questions as to whether the hormonogenic function associated with Tg appeared in evolution first in the region enriched in Tg type-1 repeat domains (the N-terminal region of modern vertebrate Tg) or in the ChEL-domain portion (the C-terminal region of modern vertebrate Tg). The answer to these questions may have to await cloning the TH precursor protein(s) of amphioxus and other nonvertebrate chordates. However, if the amphioxus TH precursor identified biochemically (16) does turn out to be the predicted 2400 residue protein (Bf_123169) (19), then this would imply that the capability to synthesize TH efficiently may have originated in proximity to Tg type-1 repeat domains.
Figure 3.
Evolutionary progression of TH synthesis (from left to right). TH synthesis preceded the appearance of thyroid follicles or modern vertebrate Tg. In simple chordates such as the nonvertebrate amphioxus and Ciona intestinalis, TH synthesis occurs in the endostyle, an exocrine gland located in the pharyngeal region. Typical thyroid follicles can be recognized in the vertebrate lamprey, but only at the time of metamorphosis. In these simpler organisms, many orthologs of genes involved in TH synthesis have been identified, but not a complete modern-day vertebrate Tg—only various gene products containing Tg type-1 repeat domains or others homologous to ChEL. A complete modern-day vertebrate Tg is detectable in all teleost fishes and above.
In species even more primitive than chordates, THs are nevertheless important inducers of metamorphosis, such as in some echinoderms (sea urchins, sea biscuits, and sand dollars) (21–23). In such species, several proteins containing Tg type-1 repeat domains and others homologous to ChEL have been found (4). Moreover, peroxidase activity-dependent T4 production has also been demonstrated (24). In the sea urchin genome, as in amphioxus, many orthologs of genes involved in TH synthesis and metabolism—but not Tg—have been identified (19). Once again, these results point strongly to an origin of TH synthesis that precedes evolutionary development of the modern vertebrate Tg protein.
As above, in nonvertebrate chordates and in lamprey (and amphibians), TH signaling is also required for metamorphosis. In the parasitic lamprey, a simple vertebrate, the endostyle is present during the larval stage (ammocoetes), but after metamorphosis, a thyroid follicular organization develops (25). When iodoproteins from adult lamprey thyroid tissue (Petromyzon fluviatilis L.) were analyzed by velocity gradient centrifugation, the major component had a sedimentation coefficient of 12S and a molecular mass of 331 000 (26) and a larger 17–18S species; thus, the lamprey iodoprotein appears—features characteristic of the modern vertebrate Tg monomer although no typical Tg gene is yet described in publicly available genomes (http://hgdownload.cse.ucsc.edu/downloads.html). Unfortunately, the low DNA coverage of publicly available genome sequences of cartilaginous fish (http://esharkgenome.imcb.a-star.edu.sg/) also does not yet reveal Tg (19), despite a follicular thyroid structure with extracellular colloidal protein that is highly likely to be enriched in stored THs (27) (Figure 4). From these analyses, it is still too early to say whether there is precise evolutionary coincidence of the follicular organization of the thyroid gland and a polypeptide exhibiting the typical structure of modern-day vertebrate Tg. However, the fact that vertebrate Tg is designed both for TH synthesis and iodide storage leaves open the possibility that these two functions might have derived from distinct gene products before the evolutionary need for iodide storage in sea creatures. Clearly, in teleost fishes and above, genetic rearrangements to include both Tg type-1 repeat domains and a contiguous carboxyl-terminal ChEL domain have already taken place.
Figure 4.
Thyroid morphology from thresher shark. This figure shows abundant extracellular colloidal protein, highly suggestive of Tg accumulation. [Reproduced from J. D. Borucinska and M. Tafur: Comparison of histological features, and description of histopathological lesions in thyroid glands from three species of free-ranging sharks from the northwestern Atlantic, the blue shark, Prionace glauca (L.), the shortfin mako, Isurus oxyrhinchus Rafinesque, and the thresher, Alopias vulpinus (Bonnaterre). J Fish Dis. 2009;32:785–793 (27), with permission. © John Wiley & Sons Ltd.]
III. Tg Primary Structure
The Tg gene structure, promoter region, splicing, and mRNA have been extensively described (28–33) and will not be reviewed again here. The primary structure of Tg was first deduced by reconstructing the predicted sequence from a series of overlapping bovine Tg cDNA clones (34). The encoded primary sequence of Tg (2749 amino acids in human Tg, after cleavage of the signal peptide) includes contiguous disulfide-rich regions I, II, and III (occupying the first ∼2170 residues) comprised of multiple repeat domains with cysteines at conserved positions, followed by the carboxyl-terminal ChEL region of approximately 520 residues. Beginning from the N terminus, the first 1200 residues of Tg are composed primarily of “Tg type-1 repeats” with an average size of roughly 60 amino acids, with three of the repeats bearing nonhomologous insertions as well as a “linker” between repeats 1–4 and 1–5, plus an approximately 235-residue “hinge” segment (all included within region I); three short type-2 repeats plus one additional type-1 repeat (altogether called region II); and finally, five type-3 repeats (about 520 residues, altogether called region III) (Figure 5). In sum, the first 2170 residues of Tg are described as regions I-II-III. The concluding ChEL domain contains only six of the many Cys residues of Tg.
Figure 5.
Schematic of the regional primary structure of Tg. The 270-kB homo sapiens Tgn gene (GenBank accession no. NT_008046 [older] or NG_015832 [current]) located on chromosome 8q24.2–8q24.3 (28–33) contains 48 exons separated by introns of varying sizes up to 64 kB, ultimately encoding an 8.5-kB mRNA (also see Figure 13). Within the encoded polypeptide shown in the figure, the various domains, insertions, and connecting sequences are not drawn to scale. The 11 Tg type-1 domains are in dark gray with four nonhomologous insertions designated as white vertical bars. The three type-2 repeats are in white, and five type-3 repeats are in light gray. The ChEL domain follows, and Tg concludes with a short stretch of nondomain sequence. The numbers above are approximate positions based on numbering the mature Tg peptide as position 1. The numbers below indicate the number of Cys residues in each domain, and numbers below the arrows specify Cys residues contained within linker and hinge and in a segment connecting type-2 and type-3 repeats.
A. Tg cysteine-rich repetitive units
1. Tg type-1 repeat: general structure and evolution
Tg type-1 repeats are classified as a domain superfamily recognized within the Structural Classification of Proteins database (35). The domain is notable for a central CWCV sequence and usually contains six cysteines in a conserved order (34). The Tg type-1 domain has been found as part of several functionally unrelated proteins. In the case of Tg itself, Tg type-1 repeats are subdivided into type-1A with six cysteines and type-1B with four cysteines (in the ninth and 11th repeats). Tg also contains an 11th type-1 repeat not contiguous with the others (residues 1492–1546) (36).
To begin to trace the evolution of the Tg type-1 domain, Novinec et al (4) collected a large dataset retrieved from GenBank or reconstructed from expressed sequence tag databases comprising genome sequences encoding 170 proteins containing 333 type-1 Tg domains. The Tg type-1 domains are specific modules that appeared early in the evolution of Metazoa, found mostly in extracellular proteins, and appear to be linked to the need to develop cell-cell or cell-matrix contacts. Many of the modern type-1 repeat-containing proteins appear to retain a function as a regulator of cysteine or cation-dependent protease activity (37, 38), and for some of them it has been shown that their inhibitory properties reside within the Tg type-1 domains (38–41)—including equistatin from sea anemone (40, 42), saxiphilin from bullfrog (38), and peptidase inhibitor from chum salmon egg (43).
Overall, in vertebrates, Tg type-1 repeats have been assembled into proteins organized in seven different modular architectures exemplified by: 1) testican; 2) secreted modular calcium binding protein (SMOC); 3) nidogens; 4) IGF binding proteins (IGFBPs); 5) major histocompatibility complex (MHC) class II-associated invariant chain; 6) Trop; and 7) Tg—with the last three being vertebrate-specific (Figure 6). These distinct architectures may allow type-1 domains to perform additional functions that are either superimposed or separated from the protease regulator function. As one example, human testican-1 (44) serves not only as a protease inhibitor but also as a substrate for certain cysteine cathepsin proteases (41). Early type-1 repeat-containing proteins evolved from their invertebrate homologs by incorporation of other vertebrate-specific domains. The oldest of the modular architectures, present throughout metazoans, are found in testican and the SMOCs, which are composed of Tg type-1 and SPARC-type extracellular calcium-binding (EC) domains with two different orders of presentation (EC-Tg type-1 in testican, Tg type-1-EC in SMOCs). Nidogen/entactin (a sulfated glycoprotein that is widely distributed in basement membranes and is tightly associated with laminin) (45) is already present in the urochordate Ciona intestinalis (together with IGFBP) and evolved by insertion of a Tg type-1 domain into a modular architecture composed of epidermal growth factor-like and nidogen domains. Three of the remaining four modular architectures (Trop, MHC class II-associated invariant chain, and Tg) appear to have been formed during vertebrate evolution (Figure 6). These unique modular architectures appear linked to specialized functions: the IGFBPs are involved IGF binding (46), the p41 splice variant of the MHC class II-associated invariant chain is involved in antigen processing (47), the Trops are involved in cell adhesion (48) and epithelial development (49), and of course Tg is involved in TH synthesis (50).
Figure 6.
Modular architecture of vertebrate Tg type-1 domain-containing proteins. The various domains and connecting sequences are not to scale. Tg type-1 domains are in gray, with other domains in white. AC, testican acidic region; CL, invariant chain class II-associated invariant chain peptide (CLIP) fragment; EC, SMOC, and Testican, extracellular calcium-binding domains; EGF, epidermal growth factor–like domain; FS, follistatin-like domain; G1, nidogen G1 domain; G2, nidogen G2 domain; IC, invariant chain intracellular domain; IGFBP, IGF binding protein domain; LY, low-density lipoprotein receptor-like YWTD repeat; SMOC, SMOC-unique domain; Tg1, Tg type-1 repeat; Tg2, Tg type-2 repeat; Tg3, Tg type-3 repeat; TM, transmembrane region; TST, testican-unique domain.
In teleost fishes, the Tg type-1 repeat domains can already be found to be organized in two clusters: an N-terminal cluster of four domains followed by a linker, with subsequent type-1 repeats thereafter. The N-terminal cluster of four domains may be linked to thyroid hormonogenesis—when the positions of iodinated tyrosyl residues are analyzed in teleost fishes, these map to iodination sites at Tyr-130 and Tyr-239 in the N-terminal cluster (51)—and certainly iodination at Tyr-130 has been suggested by several groups to be linked to T4 formation in mammals. Thus, there is a real possibility that at least a portion of thyroid hormonogenesis may be conserved within the N-terminal cluster of Tg type-1 repeat domains in all vertebrates. Many of the additional downstream iodination sites of Tg regions I-II-III of amniotes may have appeared as a consequence of the sea-to-land habitat change of tetrapods, favoring long-term iodide storage.
2. Sequence conservation and insertions in the Tg type-1 domains
Between different members of the Tg type-1 domain superfamily, there is high primary sequence variation. The residues that are most conserved include the central cysteine structure containing the CWCV motif and a Cys that tends to represent the final residue of each domain (36), plus Gly at positions 28 and 49, Gln at position 34, and Val at position 45 (Figure 7A). The relatively low number of conserved residues indicates the spectrum of Tg type-1 domain-containing protein variability (52).
Figure 7.
Targeting Tg type-1 domains 1–10 with protease sensitivity, glycosylation, and iodination: insertions in the basic domain. A, Tg domains are aligned according to Ref. 29. Conserved residues are indicated when present at least five times (in bold), with the exception of Y130 and Y685; otherwise they are generically indicated as X. Numbers within the sequence indicate the length of segments without similarities between domains. The four large insertions in domains 1.3 (two), 1.7, and 1.8, and the linker (248 residues between domains 1.4 and 1.5) appear to be regions of preferential proteolytic sensitivity, glycosylation, and iodination. B, Spacing between cysteine residues in proteins possessing type-1 domains. Disulfide linkages and spacing between the cysteines for GA733–2 and IGFBP-1,6 are from Refs. 54 and 53, respectively. In Tg, the spacings between linked cysteines in the basic domain common to GA733–2 and IGFBP-1,6 are variable as a result of four major (and some minor) insertions (see Figures 5 and 7A), whereas other spacings between adjacent disulfide linked pairs are highly conserved and equal to those of the GA733–2 and IGFBP-1,6.
The disulfide-bonding pattern of the Tg type-1 domain, although not determined experimentally in Tg, has been determined in IGFBP-6 and GA733-2 (53, 54), producing covalent bonds in the order Cys1-Cys2, Cys3-Cys4, and Cys5-Cys6. In Tg are four major sequences of variable length, unrelated to the repetitive unit but inserted into repeats 1–3, 1–7, and 1–8 (Figure 5). As a result of these insertions, the spacings between presumptive disulfide partners in some of these repeat domains are quite variable, including between Cys3-Cys4 (in repeats 1–3 and 1–7), Cys5-Cys6 (in repeat 1–3), and Cys1-Cys2 (in repeat 1–8). By contrast, Tg type-1 domains across other proteins (IGFBP-1 and -6 [Ref. 53], and GA733–2 [Ref. 54]) do not show such variations (Figure 7B).
In addition to the four inserted sequences are the linker region that falls cleanly between Tg repeats 1–4 and 1–5 (55) and the hinge region that follows immediately after Tg repeat 1–10 (56). The linker region is likely to be important for the ultimate proteolytic cleavage of Tg (see Section III.A.4), whereas the novel hinge region is likely to be critical to overall Tg folding (55).
3. Origin of type-1 insertions, linker, and hinge regions
Parma et al (29) sequenced the exon/intron junctions of the first 16 exons of the Tg gene, corresponding to the 10 Tg type-1 domains, and compared them with the protein sequence. There is often a correlation of “one exon-one domain” within modular proteins (57). Indeed, Tg repeat domains 1–2, 1–4, 1–7, and 1–10 are each encoded by a single exon, and domain 1–5 together with 1–6 is encoded by one exon. To the contrary, domains 1–1, 1–3, and 1–9 are interrupted by one intron, and domain 1–8 is interrupted by two introns. Of these “intradomain” interruptions, introns 5, 6, and 11 are located near the borders of the novel inserted sequences, intron 8 is near the border of the linker, and other introns appear to separate natural disulfide-linked cysteine partners (Figure 7A). This suggests the possibility that during evolution, there may have been exonization of previous intronic sequences to create the novel insertions (29).
4. Functional roles of the type-1 insertions, linker, and hinge regions
Preservation of possible exonization events during evolutionary development of the vertebrates suggests that they may provide selective advantage. The actual fold of the Tg type-1 domain has been resolved in the crystal structure of the p41 fragment–cathepsin L complex (52), and more recently, in solution, for the isolated p41 fragment (58) and the C-terminal part of IGFBP-6 (46). When the two conformations are superimposed, the three disulfide bonds create three loops, with the first containing an α-helix in its N-terminal part and the second connecting an antiparallel β-sheet, but the IGFBP-6 domain exhibits prolongation of the N-terminal α-helix and elaboration of the second loop because of insertions in these positions (Figure 8). As noted above, several type-1 domains within Tg contain insertions falling between disulfide-linked cysteines, plus there is the presence of the linker region between type 1–4 and type 1–5 repeats (Figure 7A). These features are also likely to extend loops within Tg region I (to a greater extent than in IGFBP-6) as well as to present unique peptide sequence for post-translational modifications and proteolytic cleavage (Table 1). Interestingly, except for the proximal linker region, these additional peptide sequences lack Cys residues (Table 1), indicating that they have high flexibility rather than participating in covalent scaffolding of the protein.
Figure 8.
The fold of Tg type-1 domain. Superimposition of three-dimensional structures of the Tg type-1 domain of IGFBP-6 (light gray) (46) and p41 fragment (dark gray) (58). The three disulfide bonds (shown as sticks) determine three loops, with the first containing an α-helix in its N-terminal part, and the second connecting an antiparallel β-sheet held in place by the second disulfide bond. Between various members of the family bearing this domain, including Tg itself, the domain structure tends to differ in the elaboration of sequences within loop I and loop II.
Table 1.
Features of Tg Type-1 Repeat Domains
| Repeat | C-C | aa | Cysteine Residues | Protease Sensitivity |
Glycosylation |
Iodination |
|||
|---|---|---|---|---|---|---|---|---|---|
| Human | Bovine | Human | Bovine | Human | Mouse | ||||
| Large insertions and linker | |||||||||
| 1.3 | C3-C4 | 40 | No | 179 | |||||
| 1.3 | C5-C6 | 43 | No | 240 | 239 | 239don | |||
| Linker | 248 | 2 | Cluster A | Cluster A | 465 | 464 | 363 | ||
| 532* | 510 | 476 | 410 | ||||||
| 551* | |||||||||
| 1.7 | C3-C4 | 123 | No | 795* | 797 | 834 | 766 | 765 | |
| 847don | |||||||||
| 864 | |||||||||
| 1.8 | C1-C2 | 99 | No | Cluster B | Cluster B | 928 | 926 | 973 | 973acc |
| Small insertions | |||||||||
| 1.2 | C1-C2 | 18 | No | 91 | |||||
| 1.5 | C1-C2 | 6 | No | 597 | |||||
| 1.7 | C1-C2 | 27 | No | 729 | |||||
| 1.10 | C1-C2 | 14 | No | 1142 | |||||
| 1.10 | C5-C6 | 3 | No | 1184 | |||||
| Outside insertions | 57 | 1121 | 130don | 130 | |||||
| 685acc | 672 | ||||||||
| 684 | |||||||||
| 1095 | |||||||||
Abbreviations: C-C, the pair of disulfide-linked cysteines in a given Tg type-1 repeat; a-a, number of amino acids inserted in the corresponding pair of disulfide-linked cysteines; acc, TH acceptor site; don, TH donor site. Cysteine residues indicates the number of cysteine residues by inspection of the sequence of human Tg (van de Graaf et al, 2001 [33]). Protease sensitivity sites, glycosylation sites, and iodinated tyrosines primarily cluster in the four major insertions and linker region of the Tg type-1 domains, secondarily in the small insertions, suggesting that they represent solvent-exposed segments available for modifications and cleavage.
Cathepsins.
When sites of proteolytic cleavage of human, bovine, and rat Tg were analyzed, four major clusters of cleavage sites were identified: cluster A falls around residue 500 in the linker region, cluster B is around residue 990 in an inserted sequence within the type 1–8 repeat, cluster C is located near residue 1800 (within the type-3 repeats), and cluster D is around residue 2515 (in the ChEL domain). Dunn et al (59) analyzed the sensitivity of Tg to cathepsins—the enzymes that initially hydrolyze Tg upon endocytosis to liberate TH—and described primary cleavage sites in these clusters, which are thought to enhance liberation of TH. Additional cleavages have been found to occur in the insertion within domain 1–3 (position 240; Figure 7A and Table 1), in small insertions in domain 1–10 of bovine Tg (positions 1142 and 1184; Figure 7A and Table 1), and in the small insertion in domain 1–5 of human Tg (position 597; Figure 7A and Table 1) (60). Thus, of those cleavage sites that reside within the Tg type-1 domains, all are located within disulfide loops or in the linker region, suggesting that these segments “present themselves” to proteases for processing. Moreover, when the locations of actual glycosylation sites have been analyzed, all but one (residue 57) fall in these loops or in the linker region of human Tg (61), and the same pattern is observed in bovine Tg (Figure 7A and Table 1). Altogether, these data strongly suggest that the insertion and linker regions represent solvent-exposed segments available for modifications and cleavage (59, 62, 63).
Because Tg is the molecular scaffold for TH synthesis and iodide storage, it is worthwhile to note the location of iodination sites within Tg type-1 domains. Of the 7–9 Tg type-1 tyrosines that have been found to be iodinated (51, 64) 55–70% are present in insertion and linker segments, with most being involved in iodide storage rather than hormonogenesis. Thus, we hypothesize that the insertions and linker segments have been preserved because of their evolutionary advantage in long-term iodide storage (which occurred upon sea-to-land migration of vertebrates), whereas the flexibility of these loops may not be optimal to meet the rigid structural requirements needed for iodotyrosine coupling for hormonogenesis.
5. Tg type-2 and -3 repeats
The middle part of Tg is constituted of three type-2 repeats, each of 14 to 17 residues, two of which are cysteines that are distant members of the GCC2/GCC3 domain superfamily (65). Additionally, there are five type-3 repeats, subdivided into 3a with eight cysteines each and 3b with six cysteines each, alternating with each other, and which exhibit only internal homology. Similar to the Tg type-1 repeats, it is thought that the Cys residues of type-2 and type-3 repeats are all consumed in internal disulfide bonds (34).
B. The ChEL domain
The carboxyl-terminal ChEL domain of Tg (about 520 amino acids) shows homology with the AChE (66) and other esterases (5). These enzymes belong to the class of α-β hydrolase fold superfamily, characterized by α–helices and β-strands that roughly alternate along the polypeptide chain (67). The crystal structure of several members of this superfamily has been solved; they are characterized by a common core: an α/β sheet of eight β-strands connected by α-helices (68, 69) (Figure 9, boxed). AChE, the prototype from this superfamily (70), is a homodimer held together by a four-helix bundle composed of helices α-7/8 and α-10 from each subunit (where 7/8 indicates the loop connecting β-strands 7 and 8, and 10 indicates the C-terminal segment following β-strand 10) (71–73). The four-helix bundle may be further stabilized by a interchain disulfide involving the C-terminal C536, but this is not required for homodimerization (74).
Figure 9.
The Cholinesterase family fold. The ChEL domain of Tg shows homology with members of the α-β hydrolase fold superfamily (68, 69) including AChE (66, 70), whose structure is shown. Like ChEL, AChE forms a homodimer held together by a four-helix bundle composed of helices α-7/8 and α-10 from each subunit (gray cylindrical segments) (71–73).
A comparison of the known three-dimensional structures of AChE and a neuroligin (a related ChEL protein) (75, 76) with that predicted for ChEL by ESyPred3D (77) supports a shared overall protein architecture between these AChE paralogs (78). Notably, all six Cys residues involved in AChE intrachain disulfide pairing are conserved in Tg, suggesting that they form the same intradomain disulfide bonds (79). However, whereas some members of the superfamily have enzymatic activity that depends upon a catalytic triad of three amino acids (Ser, Glu/Asp, and His) (80) (Figure 9) positioned in a β-sheet platform at the bottom of a narrow and deep gorge (81), the ChEL domain of Tg lacks the Ser residue of the catalytic triad as well as conserved flanking residues (Table 2) such that it does not encode an active esterase (82). Thus, the Tg ChEL domain has diverged away from catalytic functions in favor of a critical role in three-dimensional protein assembly (see Section V) as well as in TH synthesis.
Table 2.
Absence of the Esterase Catalytic Triad in Tg
| 200 | 326 | 440 | ||
|---|---|---|---|---|
| Torpedo | AChE | VTLFGE S AGGASV | NKD E GSFF | GVI H GY |
| Bovine | AChE | VTLFGE S AGAASV | AKD E GSYF | GVP H GY |
| Human | AChE | VTLFGE S AGAASV | VKD E GSYF | GVI H GY |
| Rat | Tg | VTLAAD R GGADVA | SQD D GLIN | G– H GS |
| Mouse | Tg | VTLAAD R SGADVA | SQD D GLIN | G– H GS |
| Bovine | Tg | VTLAAD R GGADIA | SQD D GLIN | S– H SS |
| 2368 | 2496 | 2601 |
Numbering is provided for the critical residues of Torpedo AChE (above) as well as for bovine Tg (below). Note that Tg in all species lacks the critical Ser residue of the catalytic triad of AChE.
IV. Tg Iodination and Hormonogenesis
The most specific post-translational modification of Tg is iodination leading to T4 and T3 synthesis. Indeed, through the combined actions of the NIS, dual-function oxidase (Duox), and TPO, iodide becomes covalently linked to tyrosyl residues on Tg to form MIT or DIT. Selected pairs of DIT residues serve as functional hormonogenic units; a coupling reaction permits a donor DIT to contribute its iodophenyl group via a quinol-ether linkage to a DIT acceptor that serves as the site of T4 formation. Analogous coupling of an MIT donor with a DIT acceptor leads to de novo formation of T3. (Coupling of noniodinated tyrosine donor with a DIT acceptor could result in formation of 3,5-T2, which is not an active TH.) Both T4 and T3 are formed while still residing within the Tg polypeptide backbone (83). Release of the iodophenyl moiety at the donor site during the coupling reaction leaves a dehydroalanine that also resides within Tg, and this is converted to pyruvic acid upon cleavage of the polypeptide chain (84–88). Interestingly, when Tg is iodinated (even nonenzymatically) in vitro, it leads to similar levels of iodide incorporation as well as TH synthesis (86), indicating that hormone formation is encoded primarily within the structure of the Tg substrate rather than via specificity of the TPO enzyme.
The three-dimensional structure of Tg is designed for efficient TH formation within the Tg molecule. Iodinated Tg yields much more T4 and T3 than iodination of other proteins, whereas denatured Tg is an incompetent substrate for hormonogenesis (89). Indeed, even limited structural modifications decrease the efficiency of hormone formation (90). The initial iodination of Tg tyrosyl residues tends to occur at preferred sites and in a preferred order (91). After incorporation of the first few iodide atoms into Tg, formation of DIT becomes favored over the formation of additional MIT residues (92), increasing the chances for hormone formation. Moreover, because DIT becomes deprotonated with a pKa of 6.5 (whereas MIT has a pKa around 8.5), physiological pH favors DIT as an acceptor site in the coupling reaction, creating the hormonogenic inner ring of T4 and T3 (93). In vivo, T4 is formed in Tg from human goiters even at 4 mol of iodide per mole of Tg (13). Thus, the tyrosine residues that are iodinated first are preferentially used for hormone formation, whereas preferential synthesis of T4 is not observed upon iodination of other proteins such as casein or fibrinogen or upon iodination of denatured Tg (94). At an average level of iodination, out of 134 tyrosyl residues per human Tg dimer, only 25–30 are iodinated, and only six to eight of these form iodothyronines (11, 12). Tyrosine 5 is the most efficient T4-forming residue in most species, and in rat Tg this has been shown to be the first tyrosine residue to become iodinated (95). In short, Tg encodes a “hierarchy of tyrosyl residues” that preferentially are mono-iodinated, di-iodinated, and coupled to form THs using donor and acceptor sites at specific positions in the three-dimensional Tg protein structure.
The identification of these sites has been performed by limited proteolysis of Tg followed by sequencing or mass spectrometry of peptides to find iodotyrosines, iodothyronines (acceptor sites), and dehydroalanine or pyruvic acid (donor sites). As reviewed elsewhere (33), four main hormonogenic acceptor DIT residues have been identified in Tg from several animal species, including position 5 (“site A”), 2554 (“site B”), 2747 (“site C”), and 1291 (“site D”). (These sites correspond to most reports in which Tg numbering begins with the first residue after signal peptide cleavage.) Other minor acceptor sites include 2568, 685, and 632 (not present in human Tg). As noted above, Tyr-5 is the most favored site for T4 formation (Tyr-2554 is second most efficient for T4 formation), whereas Tyr-2747 is the preferential site of T3 formation (51, 96). It has been noted that T3 is specifically increased within the Tg protein of patients with Graves' disease and iodide deficiency (11, 97).
One explanation for a relative increase of T3 synthesis in Tg at low iodine intake may be a higher MIT/DIT molar ratio at low levels of Tg iodination (12, 13, 98). In rabbit Tg, at an average level of iodination, the two major T4-forming sites (Tyr-5 and Tyr-2554) can be found to contain both T4 and T3, with a prevalence of T4 (96), indicating that donor tyrosines are fractionally distributed between a majority of DIT and a minority of MIT. Interestingly, in human Tg, the iodination status of the acceptor Tyr-5 is relatively insensitive to the level of iodination, being mostly DIT under all conditions (51), whereas the major T4 donor site (Tyr-130) shows a 1.57-fold increase in MIT/DIT ratio under low iodine conditions (51), which could increase the relative synthesis of T3. However, in Graves' patients, Tg also shows a higher T3 content that cannot be explained by diminished iodination (11). It has been reported that TSH increases the T3/T4 ratio in Tg by directly enhancing T3 synthesis (99). TSH receptor activation might enhance monoiodination of donor tyrosines or might even change donor iodotyrosines to different MIT residues, which would clearly imply structural changes in the Tg synthesized under high TSH conditions. Because TSH has known structural effects on Tg (eg, altered glycosylation) (100), the possibility exists that enhancement of T3 formation is the indirect result of TSH receptor activation affecting earlier post-translational modifications that subtly change Tg structure, which may occur in iodine deficiency as well as in Graves' disease (because both have TSH receptor stimulation). (As a caveat, it must be noted that the T3/T4 ratio embedded within the Tg protein can be further increased in thyroid secretion [Ref. 101] as a result of cytosolic T4 to T3 deiodination by thyroid-expressed iodothyronine deiodinases type-1 [D1] and type-2 [D2]) (102, 103). Indeed, increased TSH receptor activation in Graves' patients may lead to increased D1 mRNA and enzyme activity (104–106). However, because Tg itself is never free in the cytosol, to the best of our knowledge, Tg itself does not serve as a substrate for these deiodinases.
Several donor DIT or MIT residues have been described, including at positions 5, 926, 986 or 1008, and 1375 of bovine Tg (88); 2469 and/or 2522 of calf Tg (107); and 130, 847, and 1448 of human Tg (51).
Recently, using a shotgun-based mass spectrometry approach, the iodination sites have been directly analyzed on mouse Tg without purification, which may avoid inherent bias (64). In this case, 36 iodinated tyrosines, four hormonogenic sites, and two donor sites were identified. In the mouse Tg protein sequence, homonogenic sites were reported at positions 5 (T4 only), 973 (T3 only, and this position had not previously been reported as a hormonogenic site), and 1290 and 2552 (both T3 and T4). Dedieu et al (64) did not detect any dehydroalanine (donors) in mouse Tg, but they did detect pyruvic acid (donors) with peptide bond cleavage at Tyr-239 (a previously unreported donor site) and Tyr-2519 (which corresponds to the donor site identified in calf Tg at position 2522) (107). However, in these types of studies, the identified donors will not be matched with particular acceptors until we learn which donor-acceptor couples are brought into close proximity within the three-dimensional structure of Tg and in an orientation that permits formation of a charge transfer complex between the partners (108). As noted above, Tg polypeptide is comprised of four regions bearing intra-domain and intra-regional disulfide bonds; thus, it seems likely that the folding domains of Tg may coincide with at least some if not all of the hormone-forming sites because many donor-acceptor iodotyrosine pairs are localized in the same region of individual Tg monomers. With this in mind, it is not surprising that DIT at position 130 is a donor to DIT at position 5 for T4 formation (84, 109)—representing one of the most prominent early iodination sites of Tg. Position 1375 was suggested to be a donor for position 1291, both being in the middle part of Tg (87). Analogously, a segment comprising the 224 C-terminal amino acids of rat Tg was reported to be capable of hormone formation (110), suggesting that donor(s) site(s) may also be contained in this fragment (although they are neither positions 2469 nor 2522 reported in calf Tg [Ref. 107]—corresponding to positions 2468 and 2521 of rat Tg—because these residues are not contained within the 224 residue C-terminal fragment).
Besides the information encoded by the three-dimensional organization of Tg, the residues in the local environment flanking the iodination sites may also favor iodination and/or coupling. In vitro iodination of human Tg reveals three “consensus sequences” that, according to the prevailing view, favor iodination and coupling (50): Asp/Glu-Tyr, Ser/Thr-Tyr-Ser, and Glu-X-Tyr. The two major T4-forming sites at positions 5 and 2554 have the sequence Glu-Tyr and Asp-Tyr, respectively (51). The sequence Asp-Tyr is also found at minor hormone forming sites including 1291, 2568, and 973 in human Tg (and the corresponding positions in other species) (96). However, in human Tg, the Asp/Glu-Tyr “consensus sequence” is also found at four other sites that have not been reported to be iodinated. In mouse Tg, Dedieu et al (64) found Asp/Glu-Tyr at eight positions with hormone formation at positions 5, 2552, 1290, and 973 (position 5 carried only T4 and position 973 carried only T3), plus iodination without hormone formation at four remaining positions. Thus, in both humans and mice, the Asp/Glu-Tyr sequence appears associated with acceptor tyrosyl residues, although not all Asp/Glu-Tyr sites are associated with hormone formation.
The Ser/Thr-Tyr-Ser “consensus sequence” is associated with T3 synthesis at the carboxyl terminus of Tg in many species, including human, mouse, calf, guinea pig, and rabbit (50). In human Tg, the Ser/Thr-Tyr-Ser sequence also surrounds positions 864 and 1448, sites that have been found to be iodinated without hormone formation (51); but in mouse Tg, this sequence is present only at the carboxyl-terminal T3-forming site (and it has not yet been possible to determine whether this is invariably associated with T3 formation or also with T4 formation).
The Glu-X-Tyr “consensus sequence” appears to favor iodination over hormone formation. In human Tg, this sequence occurs seven times; all the tyrosyl residues are iodinated, with only one being a minor acceptor (at position 685), although two are potential donor sites (at positions 130 and 847) (51). In the mouse, six of eight such occurrences were found to be associated with iodination rather than hormone formation (64), and again, two potential donor sites were identified (at positions 130 and 239). On the basis of these results in both mice and humans, it appears that Glu-X-Tyr is most favorable for iodination, whereas the Asp/Glu-Tyr sequence does indeed tend to favor acceptor-site function in the hormonogenic coupling reaction. More work is still clearly needed to understand fully the chemical mechanism underlying the donor-acceptor pairing.
To liberate T4-forming as well as T3-forming sites from Tg, cathepsin proteases are likely to be involved (59). Upon incubation of endopeptidase-rich lysosomal extracts with in vivo-labeled Tg, it was shown that initial cleavages involve the liberation of hormone-containing iodopeptides that precede the release of free TH, and cathepsins B and L (more than cathepsin D) are important in this process (111). More recently, the presence in thyroid of cathepsins K (112) and S (113) has also been demonstrated. Early cathepsin-mediated proteolytic processing begins with attack near the major hormonogenic sites at both ends of the Tg molecule, resulting in a slightly smaller-than-normal dimeric Tg with diminished TH content, removed by limited proteolysis (114). Although cathepsins reside mostly in endo-lysosomes (59, 111), minor amounts of all cathepsins can be detected in the follicular lumen (113) and may be functional within the extracellular environment (115, 116). In the follicular lumen, especially in conjunction with iodination, higher-order oligomers of Tg are formed (2) via disulfide bonds (117) and dityrosine bridges (118). Such iodine-rich, multimeric Tg is not enriched in TH (119) possibly because hormone-rich iodopeptides have already been released by limited proteolysis. Indeed, follicular Tg analyzed under reducing conditions gives rise to multiple iodinated fragments, including several fragments that contain the main N-terminal hormonogenic site (120). These findings suggest the hypothesis that secreted cathepsins may be responsible for initial processing of follicular Tg, including the higher-order covalently-linked oligomers (112, 113, 117, 118)—although it must be noted that peptide bond cleavage may also occur during the oxidative process of iodination itself (121).
V. Tg Domain Foldability and Interdomain Interactions
A. Intramolecular chaperone and molecular courier functions
Because iodination and coupling reactions are catalyzed at the apical surface of the thyrocyte, Tg must be transported along the secretory pathway as a prerequisite for thyroid hormonogenesis. Interestingly, rather than causing defects in its ability to synthesize TH per se, most Tg mutations responsible for congenital hypothyroidism (see Section VIII) cause folding defects that result in a failure of Tg intracellular transport to the site of iodination. Tg folding engages assistance from multiple endoplasmic reticulum (ER) chaperones and oxidoreductases, but also requires internal regional interactions within Tg.
When Tg region I-II-III is expressed separately from ChEL (stop codon at position 2169) and ChEL is also expressed in isolation (by introducing an artificial N-terminal signal peptide), ChEL is rapidly folded and secreted, whereas region I-II-III remains incompletely folded and trapped within the ER (3) (Figure 10A). By contrast, simultaneous coexpression of the secretory ChEL with Tg region I-II-III rescues oxidative folding, stability, and secretion of this latter (Figure 10B), and the two separate proteins (I-II-III and ChEL) remain associated even after their secretion (3). Intriguingly, when an artificial N-terminal signal peptide is positioned at the 5′ end of II-III, this polypeptide also folds easily in the ER and is rapidly secreted (56) (Figure 10A). Thus, it seems likely that the intramolecular rescue by ChEL is directed mostly toward the improved folding stability of Tg region I. Curiously, other members of the family of ChEL proteins such as AChE and neuroligins (75, 76) can also bind to I-II-III, but they are not able to perform the intramolecular rescue of I-II-III folding, stability, or secretion (78).
Figure 10.
Folding and secretion of truncated Tg segments. A, Autonomous folding and secretion of distinct Tg truncation mutations. From this we propose that folding the segment shown at bottom, including both linker and hinge regions, may be rate-limiting for Tg folding and export. See Section V for details. B, Intramolecular chaperone function of ChEL. ChEL rescues secretion of Tg region I-II-III. However, isolated II-III easily folds in the ER and is also rapidly secreted. Thus, the intramolecular rescue by ChEL appears to be directed mostly toward the improved folding stability of a portion of Tg region I. However, ChEL rescue of region I folding/stability requires the presence of II-III. See Section V for details.
A true molecular chaperone shows preferential binding to early folding intermediates over more/fully mature forms. In contrast, secretory ChEL shows preference for mature forms of I-II-III, and as noted above, it remains associated with I-II-III along the entire secretory pathway (3). Moreover, when ChEL was engineered to be retained in the ER (by appending to it the retention signal KDEL), ChEL-KDEL facilitated the oxidative maturation of I-II-III, but secretion of I-II-III was no longer promoted (3). Thus, ChEL stabilizes a well-folded form of I-II-III and functions as a molecular courier for Tg transport along the secretory pathway.
B. Dimerization function
Follicular 19S Tg (660 kDa, the predominant form of Tg in the thyroid follicle lumen) is a homodimer of two 12S monomers (330 kDa) assembled into a compact ovoid structure (122). However, formation of 19S Tg does not take place in the follicular lumen, but occurs rather in the ER before intracellular transport to the Golgi complex (9, 123). Moreover, inhibiting dimerization decreases the efficiency of Tg export from the ER and its ensuing secretion (124). These findings are in accordance with the view that acquisition of a proper quaternary structure is required for efficient secretory protein export from the ER (125).
Like AChE (66) and its related family members, ChEL undergoes homodimerization via formation of a four-helix bundle at its C-terminal end (71–73). Using the PSIPRED program for prediction of protein secondary structure (126), it was noted that sequences in the carboxyl-terminal portion of the Tg ChEL domain are also predicted with high confidence to form helical segments that are positioned identically to the dimerization helices of AChE (74). Indeed, the isolated ChEL domain was found capable of dimerization with full-length Tg (whereas isolated I-II-III cannot), whereas insertion of an N-linked glycan into the upstream dimerization helix inhibits homodimerization of the isolated ChEL domain. Indeed, even AChE and neuroligins can cross-dimerize with the Tg ChEL domain (although the dimers are less stable than those formed by ChEL with Tg). These results indicate that the ChEL domain in general, and these helices in particular, plays a role in Tg dimerization. However, addition of the same glycosylation site in full-length Tg did not efficiently block Tg dimerization and secretion. Thus, additional interactions involving upstream Tg regions with ChEL domain further stabilize the Tg dimer complex in a way that can overcome a modest insult to the four-helix bundle (74, 78). It is currently unknown whether the stable Tg dimer-formation and molecular courier functions represent the same ChEL binding associations or are two discrete interactions within the Tg molecule.
Cross-dimerization of ChEL despite the presence of modest structural insult to one of the dimerization partners (78) has interesting pathophysiological implications. In congenital hypothyroidism with defective Tg, an autosomal recessive trait, most patients bearing a mutant Tgn allele are heterozygotes. Importantly, through cross-dimerization in such heterozygous patients, it is formally possible that the ChEL domain from the wild-type gene product may suppress in trans the intracellular transport defect of the mutant gene product. For example, missense mutations in the ChEL domain of Tg from rodent models of congenital hypothyroidism (rdw/rdw homozygous G2300R [Refs. 127 and 128] and cog/cog homozygous L2263P [Ref. 129]) have been introduced into the homologous sequence within AChE, and not surprisingly they inhibit enzyme activity (82). Moreover, these mutations also block Tg homodimerization (130) and interfere with the molecular courier function of ChEL (3). However, when coexpressed with wild-type Tg, the mutant rdw-Tg cross-dimerizes with the wild-type protein, and secretion of rdw-Tg is partially restored (78). Thus, in patients heterozygous for mutant Tg, the possibility of (partial or total) trans-suppression of the mutant phenotype exists, which may help to account for the recessive phenotype.
The phenomenon of suppression in trans may also imply that different naturally occurring mutations within the Tg ChEL domain may have different cross-dimerization capability and/or different molecular courier activity (and different degrees of suppression of the disease phenotype in heterozygotes). This possibility suggests that some Tgn mutant heterozygotes may exhibit predisposition to mild hypothyroidism and goiter.
C. The challenges of folding Tg region I
The final step(s) of Tg conformational maturation appear(s) to involve a covalent plication of the protein as detected by a sudden jump in band mobility by nonreducing SDS-PAGE (55). This process has been referred to as the “D-to-E transition,” implying formation of one or two disulfide bonds engaging Cys partners that are normally separated by a significant distance along the primary sequence. In contrast, the vast majority of Tg disulfide bridges employ cysteines that form much shorter disulfide loops (see Section III.A.3). The nine type-1A and two type-1B repeats are thought to consume most or all of their internal thiols (36), and this is thought to occur also in type-2 and type-3 repeats (34), as well as in the ChEL domain (66, 79). Thus, it is likely that the long-range disulfide(s) involved in the D-to-E transition employ cysteines outside the repeats or ChEL units. Region I is particularly implicated in the long-range disulfide(s) because the oxidative folding of isolated region I appears to reproduce the D-to-E transition when it is expressed in the presence of II-III and ChEL (55). Naturally occurring mutation of Tg-C1245R is associated with Tg misfolding, leading to hypothyroidism and goiter (131, 132), and indeed mutation of Cys1245 (and to a lesser extent, Cys1489) inhibits the D-to-E transition, strongly implicating the hinge region in the formation of at least one long-range disulfide bond within Tg region I (55).
Within the many repeats of Tg region I, a truncation Tg-A340X, comprising precisely the first through fourth type-1 repeats, has also been found to function as an independent folding unit that is rapidly and efficiently secreted without assistance from other Tg regions (55) (Figure 10A). The transport competence of this truncated segment of region I suggests an evolutionary significance of this cluster comprised of the first four Tg type-1 that, remarkably, also encompasses the main T4-forming site at Tyr-5 and the DIT donor at Tyr-130. Presumably, expansion of this cluster with more Tg type-1 domains and insertions provided evolutionary benefit in iodide storage, even as it rendered the larger Tg protein more difficult to fold.
The isolated region I, especially that ranging from the hinge segment to the linker segment, is the most difficult part of Tg to fold (Figure 10A) and is in part responsible for the strong binding of Tg to BiP and GRP94 (56). Interestingly, ChEL rescues the isolated region I only in cells that also coexpress secretory II-III (Figure 10B), although ChEL does not bind to isolated II-III (56). These data suggest that conformational maturation of region I—particularly the portion from the linker to the hinge segment—is facilitated by the presence of both II-III and ChEL, altogether suggesting a ternary complex involving regions I + II-III + ChEL.
VI. Chaperones and Oxidoreductases That Play a Role in Tg Maturation
A. The CRT/CNX cycle
About 20–30% of proteins synthesized in eukaryotic cells are translocated into the lumen of the ER en route to their final destination. Of these, about 80% are N-glycosylated by transfer en bloc of the Glc3Man9GlcNAc2 oligosaccharide by the action of the oligosaccharyl transferase complex (Figure 11) (133). Addition of N-glycans to the nascent polypeptide has two major consequences. First, by virtue of their hydrophilicity, glycans shield hydrophobic stretches on nascent proteins (134), increasing protein solubility and decreasing nonproductive interactions (135). Because the molecular chaperone BiP also binds hydrophobic stretches on nascent proteins (136, 137), N-linked glycosylation helps to limit BiP binding. Second, this modification shifts the selection of ER chaperones for assistance in protein folding. Specifically, N-linked glycans of nascent folding proteins attract the binding of calreticulin (CRT) and calnexin (CNX) that act as lectins for monoglucosylated-Man9GlcNAc2. Indeed, the distance from the N terminus to the first N-glycan can help determine whether the emerging polypeptide will interact first with these lectin-like chaperones or with BiP (138–140).
Figure 11.
Structure of N-linked core oligosaccharide added to the polypeptide chains.
The monoglucosylated N-glycan is initially generated by the action of glucosidase I (which produces doubly glucosylated-Man9GlcNAc2) and glucosidase II (which generates sequentially monoglucosylated-Man9GlcNAc2 and unglucosylated Man9GlcNAc2) (141–143). Thus, the action of glucosidase II first generates the monoglucosylation binding site for CRT/CNX but ultimately eliminates the binding site, preventing further CRT/ CNX association (Figure 11). Productive protein folding may occur either during the period of association with CRT/CNX (144–146) or immediately after dissociation from these lectin-like chaperones (133, 147). However, many proteins have domains that need more time to fold to the native state. Such non-native glycoproteins are recognized by UDP- Glc:glycoprotein glucosyltransferase (UGGT), which restores one glucose to the A-branch of Man9GlcNAc2, regenerating monoglucosylated oligosaccharides that can rebind to CRT/CNX (Figure 11) (141, 142). In vitro studies have demonstrated that UGGT recognizes exposed hydrophobic patches situated C-terminal to the glycan (148). Although the severity of misfolding is correlated with the degree of reglucosylation (149), UGGT is not thought to recognize completely disordered conformations (150–152) that tend to be preferred by BiP.
Deglucosylation-reglucosylation reactions, catalyzed by sequential activities of glucosidase II and UGGT, continue to cycle until the glycoprotein is no longer processed by UGGT (which may signify that it has reached its native fold) or until it becomes degraded by the ER-associated degradation (ERAD) pathway. Some glycoproteins may require only the single initial deglucosylation event and single round of binding to lectin-like chaperones en route to the native state; other substrates require multiple iterative rounds of reglucosylation and lectin-chaperone binding (146). For multidomain proteins like Tg, each round of reglucosylation allows that individual misfolded domain to selectively recruit CRT/CNX for folding assistance (152, 153), whereas other domains may engage other chaperones.
B. Substrate recognition by CRT/CNX
CRT and CNX possess identical oligosaccharide binding specificity (154–157), but their function is not identical because the newly synthesized protein substrates that they bind are not identical (154, 158, 159). Substrate selection by CRT and CNX may be related, at least in part, to protein folding context and topographic considerations (154, 158). CNX is a type-I membrane protein comprised of a globular domain that binds oligosaccharide and an extended P (proline-rich) domain (160), whereas CRT is a soluble protein containing an oligosaccharide-binding domain and a P domain (161). CNX is in close association with the translocon complex and may interact preferentially with membrane-associated glycans once they are exposed, whereas CRT tends to interact with glycans when they become more freely accessible in the ER lumen (162–164); thus CNX tends to interact first, whereas CRT interactions tend to peak thereafter (162). Consequently, CRT may have a preference for lumenal proteins, and CNX may have a preference for membrane proteins (154, 165). However, CNX substrates do not automatically associate with CRT upon selective CNX depletion; instead, they tend to associate with BiP (159). This finding suggests the possibility that substrate proteins not selected by CNX may tend to bypass the CRT/CNX cycle, instead utilizing distinct ER chaperones (138–140). In multidomain proteins, it is likely that some domains exhibit narrow specificity for selected ER chaperones, whereas others may be more promiscuous in chaperone binding.
C. Glycan-dependent oxidoreductase activity
The CRT/CNX chaperones can facilitate formation, isomerization, and reduction of disulfide bonds through the action of ERp57, a member of the protein disulfide isomerase (PDI) family of ER oxidoreductases (166–168). ERp57 associates noncovalently with the P domain of either CRT or CNX (169). In addition there is an ERp57 pool associated with tapasin—a protein involved in MHC class I peptide loading (170–172). Independently of CRT/CNX, ERp57 can interact with no more than a small subset of secretory protein substrates (170, 171, 173).
ERp57 deletion is lethal at the whole organism level but is not at the cellular level—similar to that seen for CRT and CNX (174, 175). This is explained by the fact that only a finite number of proteins in an organism are strictly dependent on the CRT/CNX/ERp57 system for folding (Table 3); thus, there are backup folding systems operative at the cellular level. Indeed, ERp57 deletion may be compensated by the function of other oxidoreductases, such as ERp72 (176, 177).
Table 3.
Classification of Cellular Proteins Based on UGGT/CRT/CNX/ERp57 Requirement
| ER Chaperone Requirement | Substrate | Folding in ERp57−/− Cells | Folding in Castanospermine-Treated Cells | Small Disulfide-Rich Domains |
|---|---|---|---|---|
| Do not prefer UGGT-CRT/CNX-ERp57 | Most cellular glycoproteins | Yes | Yes | No |
| Prefer (but do not require) UGGT-CRT/CNX-ERp57 | Clusterina | No | Yes | No |
| Require UGGT-CRT/CNX-ERp57 | β1 integrin (LDL-R) Tg | No | No | Yes |
Abbreviation: LDL-R, low-density lipoprotein receptor.
In ERp57−/− cells, clusterin is recruited to a nonfunctional cycle by the presence of CNX, whereas in castanospermine-treated cells, clusterin does not even enter the cycle and may adopt surrogate chaperones.
However, for a selected subset of glycoprotein substrates, there can be devastating consequences for the loss of ERp57 activity (176)—for example, β1 integrin does not fold in ERp57−/− cells or in cells treated with castanospermine so as to prevent formation of a monoglucosylated substrate (and thereby deny entry into the CRT/CNX cycle) (173, 177). A similar folding defect is observed upon expression of an ERp57-R282A mutant that does not bind CRT or CNX, indicating a requirement for these lectin-like chaperones (173). The “small disulfide-rich” domains of β1 integrin that ultimately engage all internal cysteines into intradomain disulfide bonds do have a high potential for disulfide mispairing during folding. Thus, the requirement for UGGT/CRT/CNX may be linked to ERp57-induced formation of proper disulfide bonds in small disulfide-rich domains (177). Another secretory glycoprotein, prosaposin (178), comprised of four small disulfide-rich domains and a single N-linked glycan (179), has similarly been found to be a prominent UGGT substrate that is acted upon by ERp57 (180). In UGGT−/− MEF cells, prosaposin secretion is impaired, and ER-retained prosaposin displays improper disulfide bond formation thought related to insufficient ERp57 engagement. Table 2 provides a list of cellular glycoproteins and their relative dependence upon UGGT/CRT/CNX/ERp57 for successful folding.
D. Glycoprotein ERAD
Trimming of mannose residues from N-linked glycans occurs as a part of successful transport of native-state glycoproteins and is a prerequisite for the formation of complex (Golgi-modified) glycans found on mature glycoproteins (181). Mannose trimming is also linked to processing of misfolded glycoproteins that are targeted for ERAD (182). There may be decreased efficiency of UGGT activity on lower mannose N-glycan forms (183) and decreased efficiency of ER glucosidase-II (184); both may limit further rounds of reglucosylation/deglucosylation, favoring release of misfolded glycoproteins from CRT/CNX and promoting ERAD (185).
For substrates entrapped in the ER, glycoproteins first tend to lose a “B-chain (middle branch) mannose” (Figure 11) (186)—indicative of ER mannosidase I (ER man-I) activity, which remains compatible with continued reglucosylation of the misfolded substrate by UGGT (187). ER man-I, which provides an α1,2-mannosidase activity, is implicated in ERAD based on evidence that the α1,2-mannosidase inhibitor, kifunensine, stabilizes glycoproteins that are normally degraded—including misfolded Tg (188). The mechanism of degradation includes recognition of the misfolded Man8 glycoprotein by EDEM-I (189, 190) that facilitates ERAD (189, 191). Additional components, including EDEM-3 and OS-9, may engage the remaining trimmed glycan (192) for subsequent delivery of the misfolded glycoprotein to the cytosol via retrotranslocation. Indeed, ER mannose trimming events appear to enhance ERAD efficiency (193). Conceivably, ER man-I could be responsible for N-glycan cleavage all the way down to the Man5GlcNAc2 form (Figure 11) (194), especially if the glycoprotein substrates are targeted to the pericentriolar “ER quality control compartment” where ER man-I is particularly concentrated (193). Ultimately, retrotranslocation initiated by OS-9 includes roles for SEL1 and HRD1 (189, 191). SEL1 together with HRD1 activates a transmembrane E3 ubiquitin ligase complex that promotes degradation of ubiquitylated ERAD substrates by cytosolic proteasomes (189).
E. Tg as a model substrate for CRT/CNX/ERp57 in secretory protein folding
Tg, with up to 60 disulfide bonds and between 10 and 15 N-linked oligosaccharides per Tg monomer (depending on the species) (61), is a good candidate to be a CRT/CNX/ERp57 client protein. Indeed, newly synthesized Tg interacts with CRT and CNX in a carbohydrate-dependent manner, with kinetics that are concomitant with the maturation of Tg intrachain disulfide bonds (10). Indeed, Tg is the first endogenous protein reported in ternary complexes with CRT and CNX in the ER of live cells (10).
Tg type-1 repeats share structural homology with the small disulfide-rich domains found in several proteins noted above (53, 54, 177) (see Section VI.C). These small disulfide-rich domains have a high potential to form non-native disulfide bonds and are substrates for the reductase activity of ERp57 (177); thus, Tg type-1 repeats could be a good target for CRT/CNX/ERp57. Consistent with this, when thyrocytes were treated with castanospermine to block Tg entry in the CRT/CNX cycle, the oxidative folding of Tg monomers and Tg dimerization was impaired, Tg association with BiP/GRP78 and GRP94 was increased, the ER stress response (including ERAD) was activated, and a partial Tg escape from ER quality control with increased secretion of free monomers yet decreased overall Tg secretion was evident (124).
F. Other oxidoreductases of the ER
Formation and isomerization of disulfide bonds (195) are catalyzed by multiple ER oxidoreductases that belong to the PDI family, comprising over 20 members (196–198). Formation and isomerization of disulfide bonds implies transient formation of a mixed disulfide between the enzyme and the substrate. Because it is believed that the time needed to complete one catalytic cycle of each oxidoreductase is quite short, the mixed disulfides are extremely short-lived. Recently, mutant oxidoreductases—so-called “trap mutants”—have been employed to prolong the half-life of these mixed disulfide species (177, 199).
G. Evidence of ER-oxidoreductase involvement in Tg folding
Even without the use of trap mutants, it has been possible to isolate several mixed disulfides between wild-type Tg and endogenous oxidoreductases, represented by “adduct bands A, B, and C” (124, 200). Because Tg adducts appear during the time course when Tg oxidative folding is ongoing (200), and because their resolution is not blocked by inhibition of ERAD (124), they most likely represent folding intermediates of Tg. One subcomponent within adduct B involves Tg-ERp57 complexes, and another subcomponent includes Tg-PDI complexes. Adduct A is enriched in Tg-ERp72 complexes, and adduct C is enriched in Tg-CaBP1 (the human P5) complexes. Interestingly, each adduct band contains not only the primary-associated ER oxidoreductase, but also lower levels of additional co-associated ER oxidoreductases. Thus, the same Tg molecule can simultaneously bind to more than one ER oxidoreductase, each working on its respective preferred binding site in a kinetically overlapping way (200).
In recent years, the concept of different “chaperone networks” has been raised. In this concept, a subset of ER chaperones constitutes a preformed network that can bind protein substrates as a unit (201). Different chaperone networks may be preassembled with specific oxidoreductases, as is the case for ERp57 with CRT/CNX (168, 176, 173) or for P5/CaBP1 with BiP (199). PDI, on the other hand, is thought to bind substrates directly through its b' domain (202). Indeed, for some oxidoreductases, substrate specificity may be linked to an interaction with a specific chaperone network. Thus, the simultaneous binding of different oxidoreductases to the same Tg molecule may also imply the simultaneous binding of different chaperone networks that can act on different Tg domains. Importantly, oxidative maturation of Tg appears to continue—seemingly within the adducts—in parallel with the oxidative maturation of Tg monomers. Presumably, this is because the adducts represent a small fraction of Tg molecules “caught in the act” of formation of a disulfide bond. If adducts A, B, and C formed along a linear folding pathway, then it would be expected that, as oxidation proceeds, Tg molecules would successively pass from A to B to C. However, adducts A, B, and C do not follow a precursor-product relationship. Thus it appears that, when engaged in an adduct enriched in a given chaperone/oxidoreductase complex, the Tg molecules are undergoing oxidative folding with assistance of that chaperone/oxidoreductase network (200), resulting in a finite number of parallel tracks of oxidative Tg folding leading to the native (transport-competent) state.
VII. Alternatives Within the Tg Folding Pathway
Large multidomain proteins are commonly thought to fold in the ER of eukaryotic cells in a sequential or vectorial manner (203, 204), although the low-density lipoprotein receptor is an exception to this view (205). During folding of the low-density lipoprotein receptor in the ER, non-native disulfide bonds between cysteines of different small disulfide-rich domains may form (205, 206), which are then isomerized via the function of ERp57 (177) and other oxidoreductases of the PDI family, as well as ERdj5 (207) that works in conjunction with BiP to play a profolding role distinct from its recognized role in misfolded glycoprotein degradation (208–211). For Tg, the difficult to fold region I is indeed comprised of small disulfide-rich domains (177). Thus, Tg type-1 repeats may form non-native disulfides that require the isomerase function of ERp57 in conjunction with CRT/CNX rebinding as directed by UGGT. Indeed, the association of ERp57 with Tg I-II-III can mostly be attributed to its binding to Tg region I (200). Interestingly, as noted above, ChEL must ultimately serve to stabilize one or more folded segments within Tg region I. Because ChEL also binds well to ERp57 (200), ChEL might possibly assist in part by recruiting CRT/CNX/ERp57 to region I to execute its intramolecular chaperone function. Moreover, in addition to ERp57, Tg folding is simultaneously promoted by a different chaperone/oxidoreductase network that operates in a parallel track (200). These features tend to support a nonsequential model of Tg folding involving several predominant intermediates, all of which are en route to the native state.
Experimental evidence suggests that the amino terminus of Tg starts to fold with assistance of the CRT/CNX cycle (124). The unfolded N-terminal type-1 repeats are likely to form non-native mixed disulfides, including with ER oxidoreductases (PDI, ERp72, CaBP1), thereby becoming favorable substrates of UGGT and subjected to numerous cycles of folding. Because other parts of Tg cotranslationally appear in the ER lumen, part II-III readily folds, and lastly, the ChEL domain emerges. This last domain may also be captured in the CRT/CNX/ERp57 cycle (the ChEL domain of human Tg has the first glycan at position 40) (61) and is brought into proximity of Tg region I to which it binds. The ChEL domain may complete its own folding in just one or few cycles, but once associated with region I, ChEL can assist in stabilizing Tg region I.
VIII. Understanding Mutations Causing Congenital Hypothyroidism
Congenital TH insufficiency affects 1 in 2000–4000 newborns (212). Early diagnosis and TH replacement are critical to promote normal growth and neurological development. Most countries have adopted primary newborn screening of TSH or TH levels. Causes of congenital hypothyroidism include: 1) thyroid dysgenesis and resistance to TSH, characterized by deficient thyroid gland development (for review, see Ref. 213); and 2) dyshormonogenesis, including defects in the NIS (214), pendrin (SLC26A4) (215), TPO (216), Duox 2 (217), Duox maturation factor 2 (217), iodotyrosine dehalogenase 1 (218), and Tg (219). Dyshormonogenesis comprises a heterogeneous spectrum of hypothyroidism with a typically enlarged thyroid (goiter) because of chronic stimulation by TSH that is elevated in response to the reduced circulating levels of TH. The various forms of congenital hypothyroidism are traditionally diagnosed on the basis of the presence of goiter, iodide uptake, serum Tg, and perchlorate discharge test (212). In particular, patients with Tg defects present with hypothyroidism and very low levels of serum Tg (220). However, all congenital hypothyroidism patients are ultimately treated with TH supplementation.
Tg deficiency is generally inherited in an autosomal recessive manner; thus, affected patients are either homozygotes or compound heterozygotes. Up to now, about 70 mutations have been described in the human Tg gene, including nonsense mutations (Table 4), single nucleotide insertion/deletion (Table 4), acceptor and donor splice site mutations (Table 5), and missense (many of these are summarized in Table 6). This review does not intend to be exhaustive of all described mutations, but rather to describe the molecular and cellular defects inferred from the considerations described in preceding sections, thereby offering a molecular explanation for the clinical phenotype.
Table 4.
Tg Gene Nonsense Mutations and Single Nucleotide Insertion/Deletion
| Exon/Intron Position | Nucleotide Position | Amino Acid Position | Tg Region Position | First Author, Year (Ref.) |
|---|---|---|---|---|
| Nonsense mutationsa | ||||
| Exon 4 | c.378C>A | p.Y107X | 1.2; stop between the 2nd and 3rd cys | Citterio, 2013 (234) |
| Exon 7 | c.886C>T | p.R277X | 1.3; disrupts the last disulfide bond of the 1.3 | van de Graaf, 1999 (228); Gutnisky, 2004 (243); Rivolta, 2006 (244); Pardo, 2009 (229); Machiavelli, 2010 (237) |
| Exon 9 | c.1351C>T | p.R432X | Linker; stop downstream the 2 cys of the linker | Niu, 2009 (232) |
| Exon 9 | c.2131C>T | p.Q692X | 1.6; disrupts the last disulfide of 1.6 | Hishinuma, 2006 (132) |
| Exon 10 | c.2206C>T | p.Q717X | 1.7; disrupts the first disulfide of 1.7 | Citterio, 2011 (233) |
| Exon 10 | c.2359C>T | p.R768X | 1.7; disrupts the second disulfide of 1.7 | Citterio, 2013 (234) |
| Exon 20 | c.4310G>A | p.W1418X | Hinge; stop between the 10th and 11th cys of the hinge | Hishinuma, 2006 (132) |
| Exon 22 | c.4588C>T | p.R1511X | Skipping of exon 22 corresponding to repeat 1.11 plus deletion of cys-1491 | Targovnik, 1993 (235), 2010 (219); Gutnisky, 2004 (243); Mendive, 2005 (236) |
| Exon 27 | c.5350C>T | p.Q1765X | 3b.1; stop between the 7th and 8th cys | Niu, 2009 (232) |
| Exon 27 | c.5386C>T | p.Q1777X | 3b.1; stop between the 7th and 8th cys | Targovnik, 2010 (219) |
| Exon 37 | c.6481C>T | p.Q2142X | 3a.3; stop between the 4th and 5th cys | Pardo, 2009 (229) |
| Exon 40 | c.7006C>T | p.R2317X | ChEL; stop between the 2nd and 3rd cys | Machiavelli, 2010 (237) |
| Exon 46 | c.7969C>T | p.Q2638X | ChEL; stop between the 5th and 6th cys, disrupts the third disulfide | Hishinuma, 2006 (132) |
| Single nucleotide deletions/insertionsb | ||||
| Exon 7 | c.759–760insA | p.L234fsX237 | 1.3; stop between the 4th and 5th cys | Targovnik, 2010 (219) |
| Exon 9 | c.1143delC | p.G362fsX382 | Linker; fs between the 1st and 2nd cys | Targovnik, 2010 (219) |
| Exon 9 | c.1348delT | p.S431fsX459 | Linker; fs after the 2nd cys | Niu, 2009 (232) |
| Exon 9 | c.1712delT | p.L552fsX576 | Linker; fs after the 2nd cys | Niu, 2009 (232) |
| Exon 10 | c.2736delG | pR893fsX946 | 1.7; fs between the 5th and 6th cys | Citterio, 2013 (234) |
| Exon 17 | c.3788–3789insT | p.I1244fsX1246 | Hinge; stop between the 4th and 5th cys | Targovnik, 2012 (283) |
| Exon 22 | c.4537delG | p.D1494fsX1547 | 1.11; fs between the 1st and 2nd cys | Hishinuma, 2006 (132) |
| Exon 28 | c.5466delA | p.K1803fsX1832 | 3b.1; fs between the 7th and 8th cys | Citterio, 2013a (234) |
| Exon 33 | c.6047delA | p.Q1997fsX1998 | 3b.2; stop between the 4th and 5th cys | Niu, 2009 (232) |
The nucleotide position is designated according to Tg mRNA reference sequences (GenBank accession number: NM 003235). The A of the ATG of the initiator codon is considered nucleotide + 1. The amino acid positions are numbered after subtracting the 19-amino acid of the signal peptide.
“X” indicates the new stop codon generated by the mutation.
Frame shifting mutations are indicated by “fs” after the first amino acid affected by the change (insertion or deletion) and followed by “X,” indicating the stop codon of the new reading frame.
Table 5.
Acceptor and Donor Splice Site Mutations
| Exon/Intron Position | Nucleotide Position | Amino Acid Position | Tg Region Position | First Author, Year (Ref.) |
|---|---|---|---|---|
| Intron 3 | g.IVS3 + 2T>G | Skipping of exon 3 and stop codon in the exon 4 | Disrupts 2nd and 3rd disulfide of 1.1; fs; stop in domain 1.2 | Niu, 2009 (232) |
| Intron 3 | g.IVS3 − 3 C<G | Skipping of exon 4 | Eliminates domain 1.2; domain 1.3 in frame | Ieiri, 1991 (238) |
| Intron 5 | g.IVS5 + 1G>A | Skipping of exon 5 and stop codon in the exon 6 | Disrupt 2nd disulfide of 1.3; fs; stop in 1.3 | Targovnik, 2010 (219) |
| Intron 6 | g.IVS6 + 1G>A | Skipping of exon 6 and stop codon in exon 7 or cryptic 5'ss activation in exon 6 and stop codon in exon 9 | Disrupt 2nd and 3rd disulfide of 1.3; fs and stop in 1.3 or disrupt 2nd and 3rd disulfide of 1.3; fs and stop after 153 residues | Citterio, 2015 (284) |
| Intron 10 | g.IVS10–1G<A | Skipping of exon 11 | Disrupt 1st disulfide of 1.8; remaining of 1.8 in frame | Hishinuma, 2006 (132) |
| Intron 19 | g.IVS19 + 3__ + 4delAT | Skipping of exon 19 and stop codon in exon 20 or cryptic 5'ss activation in exon 19 and stop codon in exon 20 | Deletions and stops in the C-terminal of hinge | Targovnik, 2012 (283) |
| Intron 24 | g.IVS24 + 1G>C | Skipping of exon 24 and stop codon in the exon 26 | Deletion of part of 3a1; fs; stop | Hishinuma, 2006 (132) |
| Intron 30 | g.IVS30 + 1G>T | Skipping of exon 30 | Deletion of C-t part of 3b1 and N-t part of 3a2; rest of 3a2 in frame | Targovnik, 1995 (239), 2001 (240); Pardo, 2008 (241), 2009 (229) |
| Intron 30 | g.IVS30 + 1G>A | Skipping of exon 30 | As above | Hishinuma, 2006 (132) |
| Intron 30 | g.IVS30 + 1G>C | Skipping of exon 30 | As above | Hermanns, 2013 (242) |
| Intron 34 | g.IVS34–1G<C | Skipping of exon 35 | Deletion in the middle of 3b2; rest of 3b2 in frame | Gutnisky, 2004 (243) |
| Intron 35 | g.IVS35 + 1delG | Skipping of exon 35 | As above | Peteiro-Gonzalez, 2010 (230) |
| Intron 40 | g.IVS40 + 2T>A | Skipping of exon 40 and stop codon in exon 41 | Delete 2274–2327 of ChEL; fs; stop | Citterio, 2015 (284) |
| Intron 45 | g.IVS45 + 2T>A | Skipping of exon 45 | Delete 2567–2602; rest of ChEL in frame | Hishinuma, 2006 (132) |
| Intron 46 | g.IVS46–1G<A | Skipping of exon 47 | Delete 2603–2647; rest of ChEL in frame | Pardo, 2009 (229) |
The nucleotide position is designated according to Tg mRNA reference sequences (GenBank accession number: NM 003235). The A of the ATG of the initiator codon is considered nucleotide + 1. The amino acid positions are numbered after subtracting the 19-amino acid of the signal peptide. Intronic nucleotides of donor splice site, located at the 5′ of the intron, have positive numbering and may produce skipping or alteration (cryptic donor site activation) of the upstream exon, whereas intronic nucleotides of acceptor splice site, located at the 3′ of the intron, have negative numbering and may produce skipping or alteration (cryptic acceptor site activation) of the downstream exon. Both exon skipping and cryptic splice site activation may result in either a frameshift (causing downstream generation of a premature stop codon with probable nonsense-mediated mRNA decay) or preservation of the reading frame (with normal mRNA translation of a protein carrying an internal deletion).
Table 6.
Missense Mutations
| Exon/Intron Position | Nucleotide Position | Amino Acid Position | Tg Region Position | First Author, Year (Ref.) |
|---|---|---|---|---|
| Exon 2 | c.113G>A | p.R19K | 1.1 4 residues C-t C1 | Kim, 2008 (275) |
| Exon 5 | c.548G>A | p.C164Y | 1.3 hits C2 | Targovnik, 2010 (219) |
| Exon 5 | c.580T>G | p.C175G | 1.3 hits C3 | Hishinuma, 2006 (132) |
| Exon 8 | c.986A>C | p.Q310P | 1.4 adjacent N-t C3 | Hishinuma, 2006 (132) |
| Exon 10 | c.2610G>T | p.Q851H | 1.7 insert C3-C4 | Corral, 1993 (276); Pérez-Centeno, 1996 (277) |
| Exon 11 | c.2969G>A | p.S971I | 1.8 insert C1-C2 | Hishinuma, 2006 (132) |
| Exon 12 | c.3022C>T | p.R989C | 1.8 insert C1-C2 introduces a cys | Hishinuma, 2006 (132) |
| Exon 12 | c.3035C>T | p.P993 liter | 1.8 insert C1-C2 | Hishinuma, 2006 (132) |
| Exon 14 | c.3229T>C | p.C1058R | 1.9 hits C1 | Hishinuma, 2006 (132) |
| Exon 17 | c.3790T>C | p.C1245R | Hinge hits a cys making a long-range S-S | Hishinuma, 1999 (131), 2006 (132); Baryshev, 2004 (274) |
| Exon 17 | c.3842G>A | p.C1262Y | Hinge Hits a cys | Citterio, 2013 (234) |
| Exon 21 | c.4397G>A | p.S1447N | 2.1 | Hishinuma, 2006 (132) |
| Exon 24 | c.4820G>T | p.C1588F | 3a1 hits C2 | Hishinuma, 2006 (132) |
| Exon 31 | c.5690G>A | p.C1878Y | 3a2 hits C2 | Hishinuma, 2006 (132) |
| Exon 31 | c.5791A>G | p.I1912V | 3a2 between C5-C6 | Hishinuma, 2006 (132) |
| Exon 33 | c.5986T>A | p.C1977S | 3b2 hits C1 | Hishinuma, 1999 (131), 2006 (132); Baryshev, 2004 (274) |
| Exon 33 | c.6000C>G | p.C1981W | 3b2 hits C2 | Citterio, 2013 (234) |
| Exon 33 | c.6017G>A | p.C1987Y | 3b2 hits C4 | Hishinuma, 2006 (132) |
| Exon 37 | c.6461G>A | p.C2135Y | 3a3 hits C4 | Hishinuma, 2006 (132) |
| Exon 38 | c.6605C>G | p.P2183R | Segment between 3a3 and ChEL | Citterio, 2013 (234) |
| Exon 38 | c.6701C>A | p.A2215D | ChEL before C1 | Pardo, 2008 (241), 2009 (229); Machiavelli, 2010 (237) |
| Exon 38 | c.6725G>A | p.R2223H | ChEL before C1 | Machiavelli, 2010 (237) |
| Exon 40 | c.6956G>A | p.G2300D | ChEL after C2 | Hishinuma, 2006 (132) |
| Exon 40 | c.7007G>A | p.R2317Q | ChEL after C2 | Hishinuma, 2006 (132) |
| Exon 41 | c.7121G>T | p.G2355V | ChEL after C2 | Hishinuma, 2006 (132) |
| Exon 41 | c.7123G>A | p.G2356R | ChEL after C2 | Hishinuma, 2006 (132) |
The nucleotide position is designated according to Tg mRNA reference sequences (GenBank accession number: NM 003235). The A of the ATG of the initiator codon is considered nucleotide + 1. The amino acid positions are numbered after subtracting the 19-amino acid of the signal peptide.
Nonsense mutations and single nucleotide insertions/deletions tend to generate premature stop codons (either directly or after frameshift), and such messages are often eliminated by nonsense-mediated mRNA decay (221), especially when an intron is located more than 50 nucleotides downstream of the stop codon (222, 223). Such messages will often produce lower levels of translated protein, and this may be less harmful to cells (224). In other cases, the premature stop codon can be eliminated by skipping the affected exon (225).
Acceptor and donor splice site mutations also produce exon skipping when alternative natural splice sites are used or altered exons when cryptic splice sites are used. Such altered exon mechanisms may exclude part of a normal exon or result in exonization of intronic sequence (Figure 12). Exon skipping and cryptic splice site activation may result in frameshift or preservation of reading frame, the latter of which results in translation of a Tg polypeptide with a small internal deletion.
Figure 12.
Acceptor and donor splice site mutations. Acceptor and donor splice site mutations produce exon skipping, when alternative natural splice sites are used (A); or altered exons, when cryptic splice sites are used (B). In exon skipping, if a donor splice site is mutated, skipping of the upstream exon may occur (A.i); or if an acceptor splice site is mutated, skipping of the downstream exon may occur (A.ii). In altered exons, if the cryptic splice site is located in an exon, part of a normal exon may be excluded (B.i); or, if the cryptic splice site is located in an intron, it may result in exonization of intronic sequences (B.ii). Both exon skipping and cryptic splice site activation may result in either a frameshift and downstream generation of a premature stop codon (the mRNA is then often subjected to nonsense-mediated mRNA decay), or preservation of the reading frame (the mRNA is normally translated, and a protein carrying an internal deletion is synthesized).
Missense mutations give rise to single amino acid substitutions, in which case the translated Tg protein is full-length. Other types of mutations include a large deletion in the 5′-region of the Tgn gene that involves the promoter region and the first 11 exons (226) as well as a large DNA inversion of 16 962 bp at the 3′-end of the Tgn gene (227). Figure 13 depicts a map of Tgn exons and Tg domains to locate the various Tg mutations.
Figure 13.
Alignment of Tgn exons with Tg repeat domains. Amino acid numbering is indicated both above the Tg protein structure and below the Tgn exons. The sequences are not drawn to scale. At the exon/exon boundary, a single number indicates that the nucleotide coding triplet is split by the boundary, whereas two numbers indicate that the adjacent nucleotides coding triplets are not split by the exon/exon boundary. A general exon/domain correspondence is notable in most type-1 repeats involving one exon for one repeat domain (repeats 1–2, 1–4, 1–7, and 1–10) or one exon for two repeats (1–5 plus 1–6). However, even when a repeat is interrupted by introns, the repeat boundaries correspond to exon boundaries, with the exception of repeat 1–1.
A. Nonsense mutations and single nucleotide insertion/deletion
The rare p.Y107X mutant has a stop codon downstream of the first disulfide of domain 1–2, and it lacks the minimal requirement for TH formation (acceptor Tyr-5 and donor Tyr-130). By contrast, the p.R277X mutation is the most frequently reported Tg mutation. In homozygous patients, p.R277X mutation causes severe to moderate hypothyroidism (228, 229). In a homozygous patient, RT-PCR amplification of a Tg fragment from total thyroidal RNA suggested similar Tg mRNA abundance between patient and control (228). The p.R277X truncation disrupts the last disulfide bond of the 1–3 repeat—leaving exposed novel inserted sequence of the 1–3 repeat—and the recombinant p.R277X expressed in vitro is completely retained in cells, indicating misfolding incompatible with ER export (55). Synthesis of the truncated Tg protein has been confirmed in the heterozygous p.R277X/ /g.IVS35+1delG, but the truncated protein is highly unstable (230). Perhaps surprisingly, constructs with a complete third repeat (Tg-P279X) and even including several amino acids of the fourth type-1 repeat (Tg-G293X) also are hardly secreted (55), explaining the phenotype caused by the naturally occurring Tg-Y296X mutation in hypothyroid Dutch goats (231). Curiously, recombinant Tg-G175X that leaves intact the first two cysteines of region 1–3, and Tg-A340X comprising precisely the first through fourth type-1 domain are both readily secreted (55). It thus appears that the first four type-1 domains, together, constitute an independent folding unit.
For additional region I stop codons (p.R432X, p.Q692X, p.Q717X, p.768X, p.W1418X), the presence of mRNA/protein in the thyroid gland of patients has not been studied (132, 232–234). However, these mutations may disrupt internal domain disulfides and potentially introduce sequences of the linker and hinge segments that are very difficult to fold, accounting for failed ER export.
The p.R1511X mutation in exon 22 produces the phenomenon of exon skipping, joining exon 21 with 23 in frame, allowing translation to reach the normal stop codon (235). The alternative transcript is present at a level similar to the full-length transcript in control tissue (236) and produces a Tg with a deletion of 57 amino acids, comprising the whole 1–11 domain plus the immediately preceding Cys-1491. Cys-1491 is a solitary Cys in a short segment between type-2 and the 11th type-1 repeat (see Figure 5), which may be engaged in a long-range disulfide bond (55). Indeed missense mutation of this residue in mouse Tg (C1489S) produces a protein that is inefficiently secreted (55), at least partially accounting for the congenital hypothyroidism of patients expressing p.R1511X.
p.Q1765X, p.Q1777X, and p.Q2142X introduce stop codons in region III (219, 229, 232). Finally, p.R2317X and p.Q2638X introduce stop codons within the ChEL domain (132, 237). In the eventually synthesized proteins, p.R2317X deletes the two α-helices α7/8 and α10, and p.Q2638X deletes the α10 helix involved in Tg homodimerization plus loss of the third disulfide bond of ChEL. In the absence of a functional ChEL domain, it is highly likely that region I-II-III is secretion incompetent, which explains the molecular defect seen in all of these patients.
The single nucleotide deletions/insertions cause a frameshift that generates a stop codon either very close downstream or at a distance. Presumably, in most these cases, the mRNA is degraded and the protein translated at low levels, but in the absence of nonsense-mediated mRNA decay, the produced polypeptides exhibit truncation and a changed stretch of downstream amino acids. p.L234fsX237 is similar to p.R277X, disrupting the last disulfide bond in the critical 1–3 repeat. p.G362fsX382, p.S431fsX459, p.L552fsX576, pR893fsX946, and p.I1244fsX1246 cause frameshifts that introduce stop codons in the critical stretch from linker to hinge, and they may introduce novel amino acids. In addition, p.I1244fsX1246 disrupts Cys1245, which normally participates in a critical long-range disulfide bond (55). Thus, this subset of frame shift mutations phenotypically resembles the nonsense mutations described above.
p.D1494fsX1547 introduces a frameshift leading to a long novel sequence that encodes almost complete replacement of domain 1–11 before terminating in a truncation close to a site (Tg-Q1509X) already known to be incompetent for secretion. p.K1803fsX1832 and p.Q1997fsX1998, either with or without a frameshift, respectively, present stop codons in region III that certainly cannot be secreted because they are similar to the isolated I-II-III.
B. Acceptor and donor splice site mutations
Acceptor and donor splice site mutations are shown in Table 5. It may be anticipated that an internally deleted Tg is produced by g.IVS3–3C<G, g.IVS10–1G<A, g.IVS30+1G>T, g.IVS30+1G>A, g.IVS30+1G>C, g.IVS34–1G<C, g.IVS35+1delG, g.IVS45+2T>A, and g.IVS46-1G<A.
g.IVS3–3C<G produces skipping of exon 4, which neatly corresponds to domain 1–4. The homozygous patient studied by Ieiri et al (238) was affected by severe hypothyroidism. Normal amounts of the altered Tg mRNA were observed in the goiter, but immunoreactive Tg protein was not detected in the surgical sample, and serum Tg was not increased after TSH stimulation. Rather than hypothyroidism based on loss of the donor Tyr130 as hypothesized by Ieiri et al (238), the above results and severe clinical phenotype suggest the synthesis of a misfolded, highly unstable Tg as the primary cause of disease.
A homozygous patient bearing g.IVS10–1G<A produces a Tg skipping exon 11 (132), which should cause loss of the N-terminal part of domain 1–8, including its first disulfide bond.
g.IVS30+1G>T, g.IVS30+1G>A, and g.IVS30+1G>C represent different mutations of the same donor splice site and produce skipping of exon 30. These mutations have been reported in homozygosity (239–242), and Tg protein expression in the goitrous tissue is severely decreased (239), yet one patient was able to maintain normal serum T3 levels with relatively low serum total T4 levels (240). The predicted Tg protein is expected to carry a deletion eliminating the last Cys of repeat 3b1 and the first Cys of repeat 3a2, disrupting one disulfide bond in each of these repeats and thereby destabilizing the Tg protein structure.
g.IVS34–1G<C and g.IVS35+1delG both produce skipping of exon 35. These mutations have been reported to occur in heterozygosity with p.R277X (230, 243). Tg protein expression was studied in a patient bearing p.R277X/ g.IVS35+1delG (230)—both Tg mutants were detected in negligible amounts, and ultrastructural analyses of thyroid surgical specimens showed dilated ER (230). The fate of a recombinant exon 35-deleted Tg was also studied in HEK293 cells where the mutant Tg was not secreted (230). The exon 35-deleted Tg has a deletion of the middle of repeat 3b2, with loss of the last Cys causing disruption of one disulfide bond and producing Tg misfolding, ER retention, and ERAD.
Tg mutants g.IVS45+2T>A and g.IVS46–1G<A (132) are thought to skip exons 45 and 47, respectively (244). These exons encode sequences in the ChEL domain, with exon 45 amino acids immediately preceding helix α7/8, whereas exon 47 amino acids include helix α10. Both sets of helices are involved in homodimerization of Tg; thus, these Tg mutants are likely dimerization-defective, retained in the ER and subjected to ERAD.
C. Missense mutations
The pathogenic potential of missense mutations is likely to involve perturbation of local folding caused by nonconservative substitutions. These replacements primarily affect quality control within the ER, preventing trafficking to the follicular lumen; there is currently no evidence that such mutations limit TH synthesis via defective iodination or coupling.
1. The cellular response to protein misfolding: ER stress
Unfolded/misfolded proteins that accumulate in the ER can cause an imbalance between protein load and folding capacity, a condition known as ER stress, which may be either physiological or pathological (245, 246). Under such conditions, cells attempt to increase protein folding capacity and to decrease protein load through activation of the ER stress response. Three major classes of ER resident transmembrane proteins, inositol-requiring protein-1 (IRE1), activating transcription factor-6, and protein kinase RNA-like ER kinase, sense the ER protein misfolding with their intralumenal domains, and each can then generate a signal using their cytoplasmic domain (247). In mammalian cells, the ER stress response has several functional outcomes, including attenuation of protein synthesis (248), activation of ERAD (249) and autophagy (250), cotranslational degradation of secretory proteins (251), and IRE1-mediated degradation of ER-localized mRNAs (regulated IRE1-dependent decay) (252, 253). Additionally, there is activation of a complex transcriptional program, including genes encoding components of the ER protein-folding machinery and ERAD (247). These responses represent a cellular attempt to reestablish equilibrium between protein load and folding capability. If these attempts fail, programmed cell death becomes activated, eliminating overwhelmed cells (247).
Until recently, two possible outcomes of ER stress response so far considered have been life or death, with efforts devoted to understand the life/death switch mechanisms (254–257). However, recent data suggest that what appears to be a pure binary decision may in fact be more complex, and survival may be achieved at the expense of nonessential cell functions, such as expression of differentiation genes (258, 259). Indeed, reprogramming gene expression to a less differentiated state after ER stress has been shown, among others, in primary and immortalized chondrocytes after drug-induced ER stress (260), in hypertrophic chondrocytes of transgenic mice models expressing mutant secretory protein (261–263), in lens fiber cells expressing misfolded collagens (264), and in thyroid differentiated cells subjected to drug-induced ER stress (265).
2. ER storage diseases
Missense mutations and other mutations allowing translation of a misfolded mutant Tg polypeptide causing congenital hypothyroidism are a prototypic example of a larger group of ER storage diseases (266). Other examples in this group of disorders are cystic fibrosis (mutant CFTR), juvenile pulmonary emphysema (mutant α-1-antitrypsin), osteogenesis imperfecta (mutant type-I procollagen), metaphyseal chondrodysplasia type Schmid (mutant collagen X), juvenile diabetes insipidus (mutant vasopressin precursor), familial hypercholesterolemia (mutant low-density lipoprotein receptor), and others (267). The pathogenic mechanism common to these disorders is misfolding of a secretory or membrane protein caused by a mutation with failure of protein export out of ER. Generally in these disorders, the ER stress response is activated, which may in part alleviate the cellular defect. However, in general, these disorders lead to loss-of-function in which the membrane or secreted protein retained in the ER is disposed by the above-mentioned mechanisms (primarily ERAD and regulated IRE1-dependent decay) so that insufficient levels of the protein reach the destination at which its physiological function normally takes place. However, there are also examples of gain-of-toxic function—specifically, if disposal is not efficient, aberrant proteins may accumulate inside cells and may initiate cell responses that eventually lead to both dedifferentiation and cell death.
Two rodent models of congenital hypothyroidism highlight these different outcomes of ER stress responses triggered by Tg missense mutations. The L2263P mutation in the ChEL domain of Tg causes congenital hypothyroid goiter in homozygous cog/cog mice (129). A markedly distended thyrocyte ER results from accumulation of the mutant Tg (268) with induction of the ER stress response that up-regulates molecular chaperones of the ER lumen (130). However, the adult animals gradually achieve normal serum T4 levels, which requires thyroid cell survival despite chronic ER stress, as well as chronic TSH stimulation that leads to a grossly enlarged thyroid gland (269). The G2320R mutation in the ChEL domain of Tg is responsible for congenital hypothyroidism in rdw/rdw rats that also display dilated thyrocyte ER and very low levels of Tg in the colloid lumen (270) with marked elevation of ER molecular chaperones (271)—a hallmark of activated ER stress response. Surprisingly, despite elevated circulating levels of TSH, these rats have a hypoplastic thyroid gland (272), which may derive from incomplete ERAD of mutant Tg accumulated in the ER that may trigger a toxic gain-of-function triggering apoptotic cell death (273).
3. Human missense mutations
About half of the missense mutations directly hit cysteine residues (Table 6), 11 substituting a cysteine and one introducing a Cys (p.R989C) that perturbs the normal Cys1-Cys2 disulfide pattern within type-1 repeat 1–8. Of these, nine (p.C164Y, p.C175G, p.C1058R, p.C1588F, p.C1878Y, p.C1977S, p.C1981W, p.C1987Y, and p.C2135Y) substitute Cys residues within the type-1 and type-3 repeats, thereby perturbing their internal disulfide bonding. The remaining two (p.C1245R and p.C1262Y) substitute a Cys residue in the hinge region, with p.C1245R disrupting a long-range disulfide bond (55). In addition to these mutations is p.Q310P, which replaces a Gln with Pro near the Cys3-Cys4 disulfide bond within repeat 1–4.
There are likely to be a range of conformational alterations introduced by these mutations, which may differentially impact on clinical phenotype. For instance, the p.C1977S is reported to cause a mild defect in a homozygous patient with adenomatous goiter, whereas homozygous p.C1245R causes a more severe congenital hypothyroidism (131, 132). Upon analysis of goiter tissue, both p.C1977S and p.C1245R activate the unfolded protein response, with the Tg-C1245R mutant inducing more ER molecular chaperones than Tg-C1977S (274). Moreover, whereas Tg-C1245R was completely entrapped within the ER (completely endo-H sensitive), a fraction of Tg-C1977S could be transported to the Golgi complex (partially endo-H resistant) (131). This demonstrates a degree of permissiveness of ER quality control, indicating that it is possible for some Tg to escape the ER despite the absence of a single small disulfide loop within repeat 3b2, whereas loss of the long-range disulfide involving C1245 creates a more severe defect (55).
Of the six other missense mutations localized in region I-II-III, (p.Q851H, p.S971I, p.R19K, p.P993L, p.S1447N, and p.I1912V), three (p.Q851H, p.S971I, and p.P993L) are clustered in a small region of Tg comprising the insert Cys3-Cys4 of repeat 1–7 and Cys1-Cys2 of 1–8. These latter three mutations are not far from donor Tyr847 (51), acceptor Tyr973 (64), and donor Tyr986 (88), respectively, raising the theoretical possibility of their local perturbation of TH formation. However, until proven otherwise, we posit that the main molecular defects triggered by these mutations involve partial deficiency in intracellular transport from the ER as the primary cause of congenital hypothyroidism. Indeed, secretion of recombinant Tg-R19K and Tg-Q851H has been directly examined in COS-7 cells (275), and both are defective—although Tg-Q851H has a milder secretory phenotype that seems to correlate with a milder clinical phenotype (goiter without biochemical hypothyroidism) (276, 277), whereas Tg-R19K is associated with clinically severe congenital goiter (278). Due to its location, Arg19 (conserved between human and mouse Tg) may impair the normal Cys1–Cys2 partnership in Tg repeat 1–1.
Six missense mutations occur in the ChEL domain and one in the short segment between the last type-3 repeat and ChEL; all of these mutations occur within the first 160 out of a total of 520 amino acids, and none of them directly substitutes any of the six essential Cys residues of ChEL (although the mouse cog mutation is separated by only two intervening residues from Cys2263 engaged in the first disulfide bond of ChEL) (129). Three of these mutations (p.P2183R, p.A2215D, and p.R2223H) fall before the first disulfide bond of ChEL, whereas the remainder fall between the first and second disulfide bonds. This finding suggests that the structure of the segments preceding and following the first disulfide loop is critical for ChEL folding. This region is well apart from the two predicted α-helices involved in Tg dimerization (helix α7/8 and helix α10) that are directly affected by in-frame skipping of exon 45 (g.IVS45+2T>A) or exon 47 (g.IVS46–1G<A). Nevertheless, the ChEL missense mutations probably alter global ChEL folding, changing the spatial orientation of these α-helices and their ability to partner in the 4-helix bundle and in this way are likely to inhibit Tg dimerization and export from the ER (130). In the case of the homozygous p.A2215D mutant, Tg protein was decreased disproportionately to Tg mRNA (229), and Tg protein was entrapped intracellularly with electron microscopy, revealing a dilated ER.
IX. Conclusion
Tg is a complex protein that has developed under competing evolutionary pressures. A Tg-G175X mutant can be successfully secreted from cells and theoretically would be sufficient to preserve much of the T4 formation needed for animal development and metabolism—yet natural selection of this simple precursor would not: 1) match pre-existing exon-intron boundaries; or 2) allow for long-term iodide storage in the organism. We postulate that long-term iodide storage in vertebrates is a primary driving force preserving the complex structure of Tg—especially including the difficult to fold portion of region I from the linker to hinge segments. Evolutionary insertions (into otherwise conserved Tg type-1 repeats) and ancestral gene fusions have allowed for thyroid-specific specialization including presentation of glycosylation sites, iodination to include T3 formation in the C-terminal portion of the molecule, as well as protease cleavage sites to help liberate iodotyrosines and iodothyronines. At the same time, the complexity of vertebrate Tg has created new challenges in protein folding, not only for the conformational maturation of multiple independent folding units within the Tg polypeptide, but also how these units fit together into a singular native tertiary and quaternary structure. The fact that the thyroid must synthesize much more Tg than any other protein creates considerable physiological stress on the ER, but vertebrate design copes with this better in thyrocytes (that use TSH-driven cell T4 proliferation to expand thyroid mass) than, say, in pancreatic islets that may face similar physiological ER stress but have exceedingly limited proliferative capacity to expand β-cell mass. However, because of its complex structure, Tg is highly susceptible to small mutations that further compromise the difficult protein folding pathway. As a result, most if not all pathogenic Tg mutations result from misfolded Tg accumulation in the ER—a hallmark of ER storage disease, requiring ultimate disposal by ERAD—as a central feature to the thyroid pathology of human patients with congenital hypothyroidism due to deficient Tg.
Acknowledgments
This work was supported by a grant from “Fondazione Cassa di Risparmio di Puglia” (to B.D.J.) and by National Institutes of Health Grant R01 DK40344 (to P.A.).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- AChE
- acetylcholinesterase
- ChEL
- cholinesterase-like
- CNX
- calnexin
- CRT
- calreticulin
- DIT
- diiodotyrosine
- Duox
- dual-function oxidase
- EC
- extracellular calcium-binding
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- ER man-I
- ER mannosidase I
- IGFBP
- IGF binding protein
- IRE1
- inositol-requiring protein-1
- MHC
- major histocompatibility complex
- MIT
- monoiodotyrosine
- NIS
- sodium-iodide symporter
- P
- proline-rich
- PDI
- protein disulfide isomerase
- SMOC
- secreted modular calcium binding protein
- Tg
- thyroglobulin
- TH
- thyroid hormone
- TPO
- thyroperoxidase
- UGGT
- UDP-Glc:glycoprotein glucosyltransferase.
References
- 1. Pharoah PO, Buttfield IH, Hetzel BS. Neurological damage to the fetus resulting from severe iodine deficiency during pregnancy. Lancet. 1971;1:308–310. [DOI] [PubMed] [Google Scholar]
- 2. Frati L, Bilstad J, Edelhoch H, Rall JE, Salvatore G. Biosynthesis of the 27 S thyroid iodoprotein. Arch Biochem Biophys. 1974;162:126–134. [DOI] [PubMed] [Google Scholar]
- 3. Lee J, Di Jeso B, Arvan P. The cholinesterase-like domain of thyroglobulin functions as an intramolecular chaperone. J Clin Invest. 2008;118:2950–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Novinec M, Kordis D, Turk V, Lenarcic B. Diversity and evolution of the thyroglobulin type-1 domain superfamily. Mol Biol Evol. 2006;23:744–755. [DOI] [PubMed] [Google Scholar]
- 5. Takagi Y, Omura T, Go M. Evolutionary origin of thyroglobulin by duplication of esterase gene. FEBS Lett. 1991;282:17–22. [DOI] [PubMed] [Google Scholar]
- 6. Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. [DOI] [PubMed] [Google Scholar]
- 7. Hebert DN, Garman SC, Molinari M. The glycan code of the endoplasmic reticulum: asparagine-linked carbohydrates as protein maturation and quality-control tags. Trends Cell Biol. 2005;15:364–370. [DOI] [PubMed] [Google Scholar]
- 8. Malthièry Y, Marriq C, Bergé-Lefranc JL, et al. Thyroglobulin structure and function: recent advances. Biochimie. 1989;71:195–209. [DOI] [PubMed] [Google Scholar]
- 9. Kim PS, Arvan P. Folding and assembly of newly synthesized thyroglobulin occurs in a pre-Golgi compartment. J Biol Chem. 1991;266:12412–12418. [PubMed] [Google Scholar]
- 10. Di Jeso B, Ulianich L, Pacifico F, et al. Folding of thyroglobulin in the calnexin/calreticulin pathway and its alteration by loss of Ca2+ from the endoplasmic reticulum. Biochem J. 2003;370:449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Izumi M, Larsen PR. Triiodothyronine, thyroxine, and iodine in purified thyroglobulin from patients with Graves' disease. J Clin Invest. 1977;59:1105–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ogawara H, Bilstad JM, Cahnmann HJ. Iodoamino acid distribution in thyroglobulin iodinated in vivo and in vitro. Biochim Biophys Acta. 1972;257:339–349. [DOI] [PubMed] [Google Scholar]
- 13. Rolland M, Montfort MF, Valenta L, Lissitzky S. Iodoamino acid composition of the thyroglobulin of normal and diseased thyroid glands. Comparison with in vitro iodinated thyroglobulin. Clin Chim Acta. 1972;39:95–108. [DOI] [PubMed] [Google Scholar]
- 14. Paris M, Laudet V. The history of a developmental stage: metamorphosis in chordates. Genesis. 2008;46:657–672. [DOI] [PubMed] [Google Scholar]
- 15. Fredriksson G, Öfverholm T, Ericson LE. Electron-microscopic studies of iodine-binding and peroxidase activity in the endostyle of the larval amphioxus (Branchiostoma lanceolatum). Cell Tissue Res. 1985;241:257–266. [Google Scholar]
- 16. Monaco F, Dominici R, Andreoli M, De Pirro R, Roche J. Thyroid hormone formation in thyroglobulin synthesized in the amphioxus (Branchiostoma lanceolatum pallas). Comp Biochem Physiol B. 1981;70:341–343. [Google Scholar]
- 17. Holland LZ, Albalat R, Azumi K, et al. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 2008;18:1100–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Putnam NH, Butts T, Ferrier DE, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. [DOI] [PubMed] [Google Scholar]
- 19. Paris M, Brunet F, Markov GV, Schubert M, Laudet V. The amphioxus genome enlightens the evolution of the thyroid hormone signaling pathway. Dev Genes Evol. 2008;218:667–680. [DOI] [PubMed] [Google Scholar]
- 20. Paris M, Hillenweck A, Bertrand S. Active metabolism of thyroid hormone during metamorphosis of amphioxus. Integr Comp Biol. 2010;50:63–74. [DOI] [PubMed] [Google Scholar]
- 21. Hodin J. Expanding networks: Signaling components in and a hypothesis for the evolution of metamorphosis. Integr Comp Biol. 2006;46:719–742. [DOI] [PubMed] [Google Scholar]
- 22. Heyland A, Reitzel AM, Hodin J. Thyroid hormones determine developmental mode in sand dollars (Echinodermata: Echinoidea). Evol Dev. 2004;6:382–392. [DOI] [PubMed] [Google Scholar]
- 23. Saito M, Seki M, Amemiya S, Yamasu K, Suyemitsu T, Ishihara K. Induction of metamorphosis in the sand dollar Peronella japonica by thyroid hormones. Dev Growth Differ. 1998;40:307–312. [DOI] [PubMed] [Google Scholar]
- 24. Heyland A, Reitzel AM, Price DA, Moroz LL. Endogenous thyroid hormone synthesis in facultative planktotrophic larvae of the sand dollar Clypeaster rosaceus: implications for the evolutionary loss of larval feeding. Evol Dev. 2006;8:568–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eales JG. Iodine metabolism and thyroid-related functions in organisms lacking thyroid follicles: are thyroid hormones also vitamins? Proc Soc Exp Biol Med. 1997;214:302–317. [DOI] [PubMed] [Google Scholar]
- 26. Aloj S, Salvatore G, Roche J. Isolation and properties of a native subunit of lamprey thyroglobulin. J Biol Chem. 1967;242:3810–3814. [PubMed] [Google Scholar]
- 27. Borucinska JD, Tafur M. Comparison of histological features, and description of histopathological lesions in thyroid glands from three species of free-ranging sharks from the northwestern Atlantic, the blue shark, Prionace glauca (L.), the shortfin mako, Isurus oxyrhinchus Rafinesque, and the thresher, Alopias vulpinus (Bonnaterre). J Fish Dis. 2009;32:785–793. [DOI] [PubMed] [Google Scholar]
- 28. Baas F, van Ommen GJ, Bikker H, Arnberg AC, de Vijlder JJ. The human thyroglobulin gene is over 300 kb long and contains introns of up to 64 kb. Nucleic Acids Res. 1986;14:5171–5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Parma J, Christophe D, Pohl V, Vassart G. Structural organization of the 5′ region of the thyroglobulin gene. Evidence for intron loss and “exonization” during evolution. J Mol Biol. 1987;196:769–779. [DOI] [PubMed] [Google Scholar]
- 30. Mendive FM, Rivolta CM, Moya CM, Vassart G, Targovnik HM. Genomic organization of the human thyroglobulin gene: the complete intron-exon structure. Eur J Endocrinol. 2001;145:485–496. [DOI] [PubMed] [Google Scholar]
- 31. Brocas H, Christophe D, Pohl V, Vassart G. Cloning of human thyroglobulin complementary DNA. FEBS Lett. 1982;137:189–192. [DOI] [PubMed] [Google Scholar]
- 32. Malthiéry Y, Lissitzky S. Primary structure of human thyroglobulin deduced from the sequence of its 8448-base complementary DNA. Eur J Biochem. 1987;165:491–498. [DOI] [PubMed] [Google Scholar]
- 33. van de Graaf SA, Ris-Stalpers C, Pauws E, Mendive FM, Targovnik HM, de Vijlder JJ. Up to date with human thyroglobulin. J Endocrinol. 2001;170:307–321. [DOI] [PubMed] [Google Scholar]
- 34. Mercken L, Simons MJ, Swillens S, Massaer M, Vassart G. Primary structure of bovine thyroglobulin deduced from the sequence of its 8,431-base complementary DNA. Nature. 1985;316:647–651. [DOI] [PubMed] [Google Scholar]
- 35. Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. [DOI] [PubMed] [Google Scholar]
- 36. Molina F, Bouanani M, Pau B, Granier C. Characterization of the type-1 repeat from thyroglobulin, a cysteine-rich module found in proteins from different families. Eur J Biochem. 1996;240:125–133. [DOI] [PubMed] [Google Scholar]
- 37. Lenarcic B, Bevec T. Thyropins–new structurally related proteinase inhibitors. Biol Chem. 1998;379:105–111. [PubMed] [Google Scholar]
- 38. Lenarcic B, Krishnan G, Borukhovich R, Ruck B, Turk V, Moczydlowski E. Saxiphilin, a saxitoxin-binding protein with two thyroglobulin type 1 domains, is an inhibitor of papain-like cysteine proteinases. J Biol Chem. 2000;275:15572–15577. [DOI] [PubMed] [Google Scholar]
- 39. Bevec T, Stoka V, Pungercic G, Dolenc I, Turk V. Major histocompatibility complex class II-associated p41 invariant chain fragment is a strong inhibitor of lysosomal cathepsin L. J Exp Med. 1996;183:1331–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lenarcic B, Turk V. Thyroglobulin type-1 domains in equistatin inhibit both papain-like cysteine proteinases and cathepsin D. J Biol Chem. 1999;274:563–566. [DOI] [PubMed] [Google Scholar]
- 41. Meh P, Pavsic M, Turk V, Baici A, Lenarcic B. Dual concentration-dependent activity of thyroglobulin type-1 domain of testican: specific inhibitor and substrate of cathepsin L. Biol Chem. 2005;386:75–83. [DOI] [PubMed] [Google Scholar]
- 42. Galesa K, Pain R, Jongsma MA, Turk V, Lenarcic B. Structural characterization of thyroglobulin type-1 domains of equistatin. FEBS Lett. 2003;539:120–124. [DOI] [PubMed] [Google Scholar]
- 43. Yamashita M, Konagaya S. A novel cysteine protease inhibitor of the egg of chum salmon, containing a cysteine-rich thyroglobulin-like motif. J Biol Chem. 1996;271:1282–1284. [DOI] [PubMed] [Google Scholar]
- 44. Bocock JP, Edgell CJ, Marr HS, Erickson AH. Human proteoglycan testican-1 inhibits the lysosomal cysteine protease cathepsin L. Eur J Biochem. 2003;270:4008–4015. [DOI] [PubMed] [Google Scholar]
- 45. Durkin ME, Chakravarti S, Bartos BB, Liu SH, Friedman RL, Chung AE. Amino acid sequence and domain structure of entactin. Homology with epidermal growth factor precursor and low density lipoprotein receptor. J Cell Biol. 1988;107:2749–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Headey SJ, Keizer DW, Yao S, et al. C-terminal domain of insulin-like growth factor (IGF) binding protein-6: structure and interaction with IGF-II. Mol Endocrinol. 2004;18:2740–2750. [DOI] [PubMed] [Google Scholar]
- 47. Stumptner-Cuvelette P, Benaroch P. Multiple roles of the invariant chain in MHC class II function. Biochim Biophys Acta. 2002;1542:1–13. [DOI] [PubMed] [Google Scholar]
- 48. Litvinov SV, Balzar M, Winter MJ, et al. Epithelial cell adhesion molecule (Ep-CAM) modulates cell-cell interactions mediated by classic cadherins. J Cell Biol. 1997;139:1337–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guillemot JC, Naspetti M, Malergue F, Montcourrier P, Galland F, Naquet P. Ep-CAM transfection in thymic epithelial cell lines triggers the formation of dynamic actin-rich protrusions involved in the organization of epithelial cell layers. Histochem Cell Biol. 2001;116:371–378. [DOI] [PubMed] [Google Scholar]
- 50. Dunn JT, Dunn AD. The importance of thyroglobulin structure for thyroid hormone biosynthesis. Biochimie. 1999;81:505–509. [DOI] [PubMed] [Google Scholar]
- 51. Lamas L, Anderson PC, Fox JW, Dunn JT. Consensus sequences for early iodination and hormonogenesis in human thyroglobulin. J Biol Chem. 1989;264:13541–13545. [PubMed] [Google Scholar]
- 52. Guncar G, Pungercic G, Klemencic I, Turk V, Turk D. Crystal structure of MHC class II-associated p41 Ii fragment bound to cathepsin L reveals the structural basis for differentiation between cathepsins L and S. EMBO J. 1999;18:793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Neumann GM, Bach LA. The N-terminal disulfide linkages of human insulin-like growth factor-binding protein-6 (hIGFBP-6) and hIGFBP-1 are different as determined by mass spectrometry. J Biol Chem. 1999;274:14587–14594. [DOI] [PubMed] [Google Scholar]
- 54. Chong JM, Speicher DW. Determination of disulfide bond assignments and N-glycosylation sites of the human gastrointestinal carcinoma antigen GA733–2 (CO17–1A, EGP, KS1–4, KSA, and Ep-CAM). J Biol Chem. 2001;276:5804–5813. [DOI] [PubMed] [Google Scholar]
- 55. Lee J, Di Jeso B, Arvan P. Maturation of thyroglobulin protein region I. J Biol Chem. 2011;286:33045–33052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee J, Arvan P. Repeat motif-containing regions within thyroglobulin. J Biol Chem. 2011;286:26327–26333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Südhof TC, Goldstein JL, Brown MS, Russell DW. The LDL receptor gene: a mosaic of exons shared with different proteins. Science. 1985;228:815–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chiva C, Barthe P, Codina A, et al. Synthesis and NMR structure of p41icf, a potent inhibitor of human cathepsin L. J Am Chem Soc. 2003;125:1508–1517. [DOI] [PubMed] [Google Scholar]
- 59. Dunn AD, Crutchfield HE, Dunn JT. Thyroglobulin processing by thyroidal proteases. Major sites of cleavage by cathepsins B, D, and L. J Biol Chem. 1991;266:20198–20204. [PubMed] [Google Scholar]
- 60. Gentile F, Salvatore G. Preferential sites of proteolytic cleavage of bovine, human and rat thyroglobulin. The use of limited proteolysis to detect solvent-exposed regions of the primary structure. Eur J Biochem. 1993;218:603–621. [DOI] [PubMed] [Google Scholar]
- 61. Yang SX, Pollock HG, Rawitch AB. Glycosylation in human thyroglobulin: location of the N-linked oligosaccharide units and comparison with bovine thyroglobulin. Arch Biochem Biophys. 1996;327:61–70. [DOI] [PubMed] [Google Scholar]
- 62. Conte M, Arcaro A, D'Angelo D, et al. A single chondroitin 6-sulfate oligosaccharide unit at Ser-2730 of human thyroglobulin enhances hormone formation and limits proteolytic accessibility at the carboxyl terminus: potential insights into thyroid homeostasis and autoimmunity. J Biol Chem. 2006;281:22200–22211. [DOI] [PubMed] [Google Scholar]
- 63. Pungercic G, Dolenc I, Dolinar M, et al. Individual recombinant thyroglobulin type-1 domains are substrates for lysosomal cysteine proteinases. Biol Chem. 2002;383:1809–1812. [DOI] [PubMed] [Google Scholar]
- 64. Dedieu A, Gaillard JC, Pourcher T, Darrouzet E, Armengaud J. Revisiting iodination sites in thyroglobulin with an organ-oriented shotgun strategy. J Biol Chem. 2011;286:259–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hall N, Pain A, Berriman M, et al. Sequence of Plasmodium falciparum chromosomes 1, 3–9 and 13. Nature. 2002;419:527–531. [DOI] [PubMed] [Google Scholar]
- 66. Schumacher M, Camp S, Maulet Y, et al. Primary structure of Torpedo californica acetylcholinesterase deduced from its cDNA sequence. Nature. 1986;319:407–409. [DOI] [PubMed] [Google Scholar]
- 67. Levitt M, Chothia C. Structural patterns in globular proteins. Nature. 1976;261:552–558. [DOI] [PubMed] [Google Scholar]
- 68. Ollis DL, Cheah E, Cygler M, et al. The α/β hydrolase fold. Protein Eng. 1992;5:197–211. [DOI] [PubMed] [Google Scholar]
- 69. Cygler M, Schrag JD, Sussman JL, et al. Relationship between sequence conservation and three-dimensional structure in a large family of esterases, lipases, and related proteins. Protein Sci. 1993;2:366–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dvir H, Silman I, Harel M, Rosenberry TL, Sussman JL. Acetylcholinesterase: from 3D structure to function. Chem Biol Interact. 2010;187:10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sussman JL, Harel M, Frolow F, et al. Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science. 1991;253:872–879. [DOI] [PubMed] [Google Scholar]
- 72. Bourne Y, Taylor P, Marchot P. Acetylcholinesterase inhibition by fasciculin: crystal structure of the complex. Cell. 1995;83:503–512. [DOI] [PubMed] [Google Scholar]
- 73. Morel N, Leroy J, Ayon A, Massoulié J, Bon S. Acetylcholinesterase H and T dimers are associated through the same contact. Mutations at this interface interfere with the C-terminal T peptide, inducing degradation rather than secretion. J Biol Chem. 2001;276:37379–37389. [DOI] [PubMed] [Google Scholar]
- 74. Lee J, Wang X, Di Jeso B, Arvan P. The cholinesterase-like domain, essential in thyroglobulin trafficking for thyroid hormone synthesis, is required for protein dimerization. J Biol Chem. 2009;284:12752–12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gilbert MM, Auld VJ. Evolution of clams (cholinesterase-like adhesion molecules): structure and function during development. Front Biosci. 2005;10:2177–2192. [DOI] [PubMed] [Google Scholar]
- 76. De Jaco A, Comoletti D, King CC, Taylor P. Trafficking of cholinesterases and neuroligins mutant proteins. An association with autism. Chem Biol Interact. 2008;175:349–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lambert C, Léonard N, De Bolle X, Depiereux E. ESyPred3D: prediction of proteins 3D structures. Bioinformatics. 2002;18:1250–1256. [DOI] [PubMed] [Google Scholar]
- 78. Wang X, Lee J, Di Jeso B, et al. Cis and trans actions of the Cholinesterase-Like domain within the thyroglobulin dimer. J Biol Chem. 2010;285:17564–17573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. MacPhee-Quigley K, Vedvick TS, Taylor P, Taylor SS. Profile of the disulfide bonds in acetylcholinesterase. J Biol Chem. 1986;261:13565–13570. [PubMed] [Google Scholar]
- 80. Richardson JS. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. [DOI] [PubMed] [Google Scholar]
- 81. Silman I, Sussman JL. Acetylcholinesterase: how is structure related to function? Chem Biol Interact. 2008;175:3–10. [DOI] [PubMed] [Google Scholar]
- 82. Park YN, Arvan P. The acetylcholinesterase homology region is essential for normal conformational maturation and secretion of thyroglobulin. J Biol Chem. 2004;279:17085–17089. [DOI] [PubMed] [Google Scholar]
- 83. Taurog A, Dorris ML, Doerge DR. Mechanism of simultaneous iodination and coupling catalyzed by thyroid peroxidase. Arch Biochem Biophys. 1996;330:24–32. [DOI] [PubMed] [Google Scholar]
- 84. Dunn AD, Corsi CM, Myers HE, Dunn JT. Tyrosine 130 is an important outer ring donor for thyroxine formation in thyroglobulin. J Biol Chem. 1998;273:25223–25229. [DOI] [PubMed] [Google Scholar]
- 85. Gavaret JM, Nunez J, Cahnmann HJ. Formation of dehydroalanine residues during thyroid hormone synthesis in thyroglobulin. J Biol Chem. 1980;255:5281–5285. [PubMed] [Google Scholar]
- 86. Gavaret JM, Cahnmann HJ, Nunez J. Thyroid hormone synthesis in thyroglobulin. The mechanism of the coupling reaction. J Biol Chem. 1981;256:9167–9173. [PubMed] [Google Scholar]
- 87. Gentile F, Ferranti P, Mamone G, Malorni A, Salvatore G. Identification of hormonogenic tyrosines in fragment 1218–1591 of bovine thyroglobulin by mass spectrometry. J Biol Chem. 1997;272:639–646. [DOI] [PubMed] [Google Scholar]
- 88. Ohmiya Y, Hayashi H, Kondo T, Kondo Y. Location of dehydroalanine residues in the amino acid sequence of bovine thyroglobulin. Identification of “donor” tyrosine sites for hormonogenesis in thyroglobulin. J Biol Chem. 1990;265:9066–9071. [PubMed] [Google Scholar]
- 89. Rolland M, Montfort MF, Lissitzky S. Efficiency of thyroglobulin as a thyroid hormone-forming protein. Biochim Biophys Acta. 1973;303:338–347. [DOI] [PubMed] [Google Scholar]
- 90. Maurizis JC, Marriq C, Michelot J, Rolland M, Lissitzky S. Thyroid peroxidase-induced thyroid hormone synthesis in relation to thyroglobulin structure. FEBS Lett. 1979;102:82–86. [DOI] [PubMed] [Google Scholar]
- 91. Gavaret JM, Dème D, Nunez J, Salvatore G. Sequential reactivity of tyrosyl residues of thyroglobulin upon iodination catalyzed by thyroid peroxidase. J Biol Chem. 1977;252:3281–3285. [PubMed] [Google Scholar]
- 92. Van Zyl A, Edelhoch H. The properties of thyroglobulin. XV. The function of the protein in the control of diiodotyrosine synthesis. J Biol Chem. 1967;242:2423–2427. [PubMed] [Google Scholar]
- 93. de Vijlder JJ, den Hartog MT. Anionic iodotyrosine residues are required for iodothyronine synthesis. Eur J Endocrinol. 1998;138:227–231. [DOI] [PubMed] [Google Scholar]
- 94. Lamas L, Taurog A, Salvatore G, Edelhoch H. Preferential synthesis of thyroxine from early iodinated tyrosyl residues in thyroglobulin. J Biol Chem. 1974;249:2732–2737. [PubMed] [Google Scholar]
- 95. Palumbo G, Gentile F, Condorelli GL, Salvatore G. The earliest site of iodination in thyroglobulin is residue number 5. J Biol Chem. 1990;265:8887–8892. [PubMed] [Google Scholar]
- 96. Dunn JT, Anderson PC, Fox JW, et al. The sites of thyroid hormone formation in rabbit thyroglobulin. J Biol Chem. 1987;262:16948–16952. [PubMed] [Google Scholar]
- 97. Cetrangolo GP, Arcaro A, Lepore A, et al. Hormonogenic donor Tyr2522 of bovine thyroglobulin. Insight into preferential T3 formation at thyroglobulin carboxyl terminus at low iodination level. Biochem Biophys Res Commun. 2014;450:488–493. [DOI] [PubMed] [Google Scholar]
- 98. Sorimachi K, Ui N. Comparison of the iodoamino acid distribution in various preparations of hog thyroglobulin with different iodine content and subunit structure. Biochim Biophys Acta. 1974;342:30–40. [DOI] [PubMed] [Google Scholar]
- 99. Fassler CA, Dunn JT, Anderson PC, et al. Thyrotropin alters the utilization of thyroglobulin's hormonogenic sites. J Biol Chem. 1988;263:17366–17371. [PubMed] [Google Scholar]
- 100. Di Jeso B, Liguoro D, Ferranti P, et al. Modulation of the carbohydrate moiety of thyroglobulin by thyrotropin and calcium in Fisher rat thyroid line-5 cells. J Biol Chem. 1992;267:1938–1944. [PubMed] [Google Scholar]
- 101. Larsen PR, Zavacki AM. The role of the iodothyronine deiodinases in the physiology and pathophysiology of thyroid hormone action. Eur Thyroid J. 2012;1:232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Salvatore D, Tu H, Harney JW, Larsen PR. Type 2 iodothyronine deiodinase is highly expressed in human thyroid. J Clin Invest. 1996;98:962–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Bianco AC, Salvatore D, Gereben B, Berry MJ, Larsen PR. Biochemistry, cellular and molecular biology, and physiological roles of the iodothyronine selenodeiodinases. Endocr Rev. 2002;23:38–89. [DOI] [PubMed] [Google Scholar]
- 104. Nishikawa M, Toyoda N, Yonemoto T, et al. Quantitative measurements for type 1 deiodinase messenger ribonucleic acid in human peripheral blood mononuclear cells: mechanism of the preferential increase of T3 in hyperthyroid Graves' disease. Biochem Biophys Res Commun. 1998;250:642–646. [DOI] [PubMed] [Google Scholar]
- 105. Ishii H, Inada M, Tanaka K, et al. Triiodothyronine generation from thyroxine in human thyroid: enhanced conversion in Graves' thyroid tissue. J Clin Endocrinol Metab. 1981;52:1211–1217. [DOI] [PubMed] [Google Scholar]
- 106. Toyoda N, Nishikawa M, Horimoto M, et al. Graves' immunoglobulin G stimulates iodothyronine 5′-deiodinating activity in FRTL-5 rat thyroid cells. J Clin Endocrinol Metab. 1990;70:1506–1511. [DOI] [PubMed] [Google Scholar]
- 107. Palumbo G. Thyroid hormonogenesis. Identification of a sequence containing iodophenyl donor site(s) in calf thyroglobulin. J Biol Chem. 1987;262:17182–17188. [PubMed] [Google Scholar]
- 108. Cahnmann HJ, Pommier J, Nunez J. Spatial requirement for coupling of iodotyrosine residues to form thyroid hormones. Proc Natl Acad Sci USA. 1977;74:5333–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Marriq C, Lejeune PJ, Venot N, Vinet L. Hormone formation in the isolated fragment 1–171 of human thyroglobulin involves the couple tyrosine 5 and tyrosine 130. Mol Cell Endocrinol. 1991;81:155–164. [DOI] [PubMed] [Google Scholar]
- 110. Asunción M, Ingrassia R, Escribano J, et al. Efficient thyroid hormone formation by in vitro iodination of a segment of rat thyroglobulin fused to Staphylococcal protein A. FEBS Lett. 1992;297:266–270. [DOI] [PubMed] [Google Scholar]
- 111. Dunn AD, Crutchfield HE, Dunn JT. Proteolytic processing of thyroglobulin by extracts of thyroid lysosomes. Endocrinology. 1991;128:3073–3080. [DOI] [PubMed] [Google Scholar]
- 112. Tepel C, Brömme D, Herzog V, Brix K. Cathepsin K in thyroid epithelial cells: sequence, localization and possible function in extracellular proteolysis of thyroglobulin. J Cell Sci. 2000;113:4487–4498. [DOI] [PubMed] [Google Scholar]
- 113. Jordans S, Jenko-Kokalj S, Kühl NM, et al. Monitoring compartment-specific substrate cleavage by cathepsins B, K, L, and S at physiological pH and redox conditions. BMC Biochem. 2009;10:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Rousset B, Selmi S, Bornet H, Bourgeat P, Rabilloud R, Munari-Silem Y. Thyroid hormone residues are released from thyroglobulin with only limited alteration of the thyroglobulin structure. J Biol Chem. 1989;264:12620–12626. [PubMed] [Google Scholar]
- 115. Reddy A, Caler EV, Andrews NW. Plasma membrane repair is mediated by Ca(2+)-regulated exocytosis of lysosomes. Cell. 2001;106:157–169. [DOI] [PubMed] [Google Scholar]
- 116. Jaiswal JK, Andrews NW, Simon SM. Membrane proximal lysosomes are the major vesicles responsible for calcium-dependent exocytosis in nonsecretory cells. J Cell Biol. 2002;159:625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Berndorfer U, Wilms H, Herzog V. Multimerization of thyroglobulin (TG) during extracellular storage: isolation of highly cross-linked TG from human thyroids. J Clin Endocrinol Metab. 1996;81:1918–1926. [DOI] [PubMed] [Google Scholar]
- 118. Herzog V, Berndorfer U, Saber Y. Isolation of insoluble secretory product from bovine thyroid: extracellular storage of thyroglobulin in covalently cross-linked form. J Cell Biol. 1992;118:1071–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Baudry N, Lejeune PJ, Delom F, Vinet L, Carayon P, Mallet B. Role of multimerized porcine thyroglobulin in iodine storage. Biochem Biophys Res Commun. 1998;242:292–296. [DOI] [PubMed] [Google Scholar]
- 120. Dunn JT, Kim PS, Dunn AD. Favored sites for thyroid hormone formation on the peptide chains of human thyroglobulin. J Biol Chem. 1982;257:88–94. [PubMed] [Google Scholar]
- 121. Dunn JT, Kim PS, Dunn AD, Heppner DG, Jr, Moore RC. The role of iodination in the formation of hormone-rich peptides from thyroglobulin. J Biol Chem. 1983;258:9093–9099. [PubMed] [Google Scholar]
- 122. Berg G, Björkman U, Ekholm R. The structure of newly synthesized intracellular thyroglobulin molecules. Mol Cell Endocrinol. 1980;20:87–98. [DOI] [PubMed] [Google Scholar]
- 123. Di Jeso B, Pereira R, Consiglio E, Formisano S, Satrustegui J, Sandoval IV. Demonstration of a Ca2+ requirement for thyroglobulin dimerization and export to the golgi complex. Eur J Biochem. 1998;252:583–590. [DOI] [PubMed] [Google Scholar]
- 124. Di Jeso B, Park YN, Ulianich L, et al. Mixed-disulfide folding intermediates between thyroglobulin and endoplasmic reticulum resident oxidoreductases ERp57 and protein disulfide isomerase. Mol Cell Biol. 2005;25:9793–9805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Reddy PS, Corley RB. Assembly, sorting, and exit of oligomeric proteins from the endoplasmic reticulum. BioEssays. 1998;20:546–554. [DOI] [PubMed] [Google Scholar]
- 126. Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–W38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Kim PS, Ding M, Menon S, et al. A missense mutation G2320R in the thyroglobulin gene causes non-goitrous congenital primary hypothyroidism in the WIC-rdw rat. Mol Endocrinol. 2000;14:1944–1953. [DOI] [PubMed] [Google Scholar]
- 128. Hishinuma A, Furudate S, Oh-Ishi M, Nagakubo N, Namatame T, Ieiri T. A novel missense mutation (G2320R) in thyroglobulin causes hypothyroidism in rdw rats. Endocrinology. 2000;141:4050–4055. [DOI] [PubMed] [Google Scholar]
- 129. Kim PS, Hossain SA, Park YN, Lee I, Yoo SE, Arvan P. A single amino acid change in the acetylcholinesterase-like domain of thyroglobulin causes congenital goiter with hypothyroidism in the cog/cog mouse: a model of human endoplasmic reticulum storage diseases. Proc Natl Acad Sci USA. 1998;95:9909–9913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Kim PS, Kwon OY, Arvan P. An endoplasmic reticulum storage disease causing congenital goiter with hypothyroidism. J Cell Biol. 1996;133:517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Hishinuma A, Takamatsu J, Ohyama Y, et al. Two novel cysteine substitutions (C1263R and C1995S) of thyroglobulin cause a defect in intracellular transport of thyroglobulin in patients with congenital goiter and the variant type of adenomatous goiter. J Clin Endocrinol Metab. 1999;84:1438–1444. [DOI] [PubMed] [Google Scholar]
- 132. Hishinuma A, Fukata S, Nishiyama S, et al. Haplotype analysis reveals founder effects of thyroglobulin gene mutations C1058R and C1977S in Japan. J Clin Endocrinol Metab. 2006;91:3100–3104. [DOI] [PubMed] [Google Scholar]
- 133. D'Alessio C, Caramelo JJ, Parodi AJ. UDP-GlC:glycoprotein glucosyltransferase-glucosidase II, the ying-yang of the ER quality control. Semin Cell Dev Biol. 2010;21:491–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Imperiali B, O'Connor SE. Effect of N-linked glycosylation on glycopeptide and glycoprotein structure. Curr Opin Chem Biol. 1999;3:643–649. [DOI] [PubMed] [Google Scholar]
- 135. Ferris SP, Jaber NS, Molinari M, Arvan P, Kaufman RJ. UDP-glucose:glycoprotein glucosyltransferase (UGGT1) promotes substrate solubility in the endoplasmic reticulum. Mol Biol Cell. 2013;24:2597–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Flynn GC, Pohl J, Flocco MT, Rothman JE. Peptide-binding specificity of the molecular chaperone BiP. Nature. 1991;353:726–730. [DOI] [PubMed] [Google Scholar]
- 137. Blond-Elguindi S, Cwirla SE, Dower WJ, et al. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993;75:717–728. [DOI] [PubMed] [Google Scholar]
- 138. Wang N, Daniels R, Hebert DN. The cotranslational maturation of the type I membrane glycoprotein tyrosinase: the heat shock protein 70 system hands off to the lectin-based chaperone system. Mol Biol Cell. 2005;16:3740–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Molinari M, Helenius A. Chaperone selection during glycoprotein translocation into the endoplasmic reticulum. Science. 2000;288:331–333. [DOI] [PubMed] [Google Scholar]
- 140. Hammond C, Helenius A. Folding of VSV G protein: sequential interaction with BiP and calnexin. Science. 1994;266:456–458. [DOI] [PubMed] [Google Scholar]
- 141. Hebert DN, Foellmer B, Helenius A. Glucose trimming and reglucosylation determine glycoprotein association with calnexin in the endoplasmic reticulum. Cell. 1995;81:425–433. [DOI] [PubMed] [Google Scholar]
- 142. Hebert DN, Foellmer B, Helenius A. Calnexin and calreticulin promote folding, delay oligomerization and suppress degradation of influenza hemagglutinin in microsomes. EMBO J. 1996;15:2961–2968. [PMC free article] [PubMed] [Google Scholar]
- 143. Ou WJ, Cameron PH, Thomas DY, Bergeron JJ. Association of folding intermediates of glycoproteins with calnexin during protein maturation. Nature. 1993;364:771–776. [DOI] [PubMed] [Google Scholar]
- 144. Gidalevitz T, Stevens F, Argon Y. Orchestration of secretory protein folding by ER chaperones. Biochim Biophys Acta. 2013;1833:2410–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Molinari M, Galli C, Vanoni O, Arnold SM, Kaufman RJ. Persistent glycoprotein misfolding activates the glucosidase II/UGT1-driven calnexin cycle to delay aggregation and loss of folding competence. Mol Cell. 2005;20:503–512. [DOI] [PubMed] [Google Scholar]
- 146. Soldà T, Galli C, Kaufman RJ, Molinari M. Substrate-specific requirements for UGT1-dependent release from calnexin. Mol Cell. 2007;27:238–249. [DOI] [PubMed] [Google Scholar]
- 147. Pearse BR, Hebert DN. Lectin chaperones help direct the maturation of glycoproteins in the endoplasmic reticulum. Biochim Biophys Acta. 2010;1803:684–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Taylor SC, Thibault P, Tessier DC, Bergeron JJ, Thomas DY. Glycopeptide specificity of the secretory protein folding sensor UDP-glucose glycoprotein:glucosyltransferase. EMBO Rep. 2003;4:405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Pearse BR, Gabriel L, Wang N, Hebert DN. A cell-based reglucosylation assay demonstrates the role of GT1 in the quality control of a maturing glycoprotein. J Cell Biol. 2008;181:309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Caramelo JJ, Castro OA, Alonso LG, De Prat-Gay G, Parodi AJ. UDP-Glc:glycoprotein glucosyltransferase recognizes structured and solvent accessible hydrophobic patches in molten globule-like folding intermediates. Proc Natl Acad Sci USA. 2003;100:86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Caramelo JJ, Castro OA, de Prat-Gay G, Parodi AJ. The endoplasmic reticulum glucosyltransferase recognizes nearly native glycoprotein folding intermediates. J Biol Chem. 2004;279:46280–46285. [DOI] [PubMed] [Google Scholar]
- 152. Ritter C, Quirin K, Kowarik M, Helenius A. Minor folding defects trigger local modification of glycoproteins by the ER folding sensor GT. EMBO J. 2005;24:1730–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Ritter C, Helenius A. Recognition of local glycoprotein misfolding by the ER folding sensor UDP-glucose:glycoprotein glucosyltransferase. Nat Struct Biol. 2000;7:278–280. [DOI] [PubMed] [Google Scholar]
- 154. Peterson JR, Ora A, Van PN, Helenius A. Transient, lectin-like association of calreticulin with folding intermediates of cellular and viral glycoproteins. Mol Biol Cell. 1995;6:1173–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Spiro RG, Zhu Q, Bhoyroo V, Söling HD. Definition of the lectin-like properties of the molecular chaperone, calreticulin, and demonstration of its copurification with endomannosidase from rat liver Golgi. J Biol Chem. 1996;271:11588–11594. [DOI] [PubMed] [Google Scholar]
- 156. Vassilakos A, Michalak M, Lehrman MA, Williams DB. Oligosaccharide binding characteristics of the molecular chaperones calnexin and calreticulin. Biochemistry. 1998;37:3480–3490. [DOI] [PubMed] [Google Scholar]
- 157. Ware FE, Vassilakos A, Peterson PA, Jackson MR, Lehrman MA, Williams DB. The molecular chaperone calnexin binds Glc1Man9GlcNAc2 oligosaccharide as an initial step in recognizing unfolded glycoproteins. J Biol Chem. 1995;270:4697–4704. [DOI] [PubMed] [Google Scholar]
- 158. Wada I, Imai S, Kai M, Sakane F, Kanoh H. Chaperone function of calreticulin when expressed in the endoplasmic reticulum as the membrane-anchored and soluble forms. J Biol Chem. 1995;270:20298–20304. [DOI] [PubMed] [Google Scholar]
- 159. Pieren M, Galli C, Denzel A, Molinari M. The use of calnexin and calreticulin by cellular and viral glycoproteins. J Biol Chem. 2005;280:28265–28271. [DOI] [PubMed] [Google Scholar]
- 160. Schrag JD, Bergeron JJ, Li Y, et al. The structure of calnexin, an ER chaperone involved in quality control of protein folding. Mol Cell. 2001;8:633–644. [DOI] [PubMed] [Google Scholar]
- 161. Baksh S, Michalak M. Expression of calreticulin in Escherichia coli and identification of its Ca2+ binding domains. J Biol Chem. 1991;266:21458–21465. [PubMed] [Google Scholar]
- 162. Daniels R, Kurowski B, Johnson AE, Hebert DN. N-linked glycans direct the cotranslational folding pathway of influenza hemagglutinin. Mol Cell. 2003;11:79–90. [DOI] [PubMed] [Google Scholar]
- 163. Hebert DN, Zhang JX, Chen W, Foellmer B, Helenius A. The number and location of glycans on influenza hemagglutinin determine folding and association with calnexin and calreticulin. J Cell Biol. 1997;139:613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Danilczyk UG, Cohen-Doyle MF, Williams DB. Functional relationship between calreticulin, calnexin, and the endoplasmic reticulum luminal domain of calnexin. J Biol Chem. 2000;275:13089–13097. [DOI] [PubMed] [Google Scholar]
- 165. Molinari M, Eriksson KK, Calanca V, et al. Contrasting functions of calreticulin and calnexin in glycoprotein folding and ER quality control. Mol Cell. 2004;13:125–135. [DOI] [PubMed] [Google Scholar]
- 166. Oliver JD, van der Wal FJ, Bulleid NJ, High S. Interaction of the thiol-dependent reductase ERp57 with nascent glycoproteins. Science. 1997;275:86–88. [DOI] [PubMed] [Google Scholar]
- 167. Oliver JD, Roderick HL, Llewellyn DH, High S. ERp57 functions as a subunit of specific complexes formed with the ER lectins calreticulin and calnexin. Mol Biol Cell. 1999;10:2573–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Zapun A, Darby NJ, Tessier DC, Michalak M, Bergeron JJ, Thomas DY. Enhanced catalysis of ribonuclease B folding by the interaction of calnexin or calreticulin with ERp57. J Biol Chem. 1998;273:6009–6012. [DOI] [PubMed] [Google Scholar]
- 169. Frickel EM, Riek R, Jelesarov I, Helenius A, Wuthrich K, Ellgaard L. TROSY-NMR reveals interaction between ERp57 and the tip of the calreticulin P-domain. Proc Natl Acad Sci USA. 2002;99:1954–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Peaper DR, Wearsch PA, Cresswell P. Tapasin and ERp57 form a stable disulfide-linked dimer within the MHC class I peptide-loading complex. EMBO J. 2005;24:3613–3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Peaper DR, Cresswell P. The redox activity of ERp57 is not essential for its functions in MHC class I peptide loading. Proc Natl Acad Sci USA. 2008;105:10477–10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Peaper DR, Cresswell P. Regulation of MHC class I assembly and peptide binding. Annu Rev Cell Dev Biol. 2008;24:343–368. [DOI] [PubMed] [Google Scholar]
- 173. Jessop CE, Tavender TJ, Watkins RH, Chambers JE, Bulleid NJ. Substrate specificity of the oxidoreductase ERp57 is determined primarily by its interaction with calnexin and calreticulin. J Biol Chem. 2009;284:2194–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Denzel A, Molinari M, Trigueros C, et al. Early postnatal death and motor disorders in mice congenitally deficient in calnexin expression. Mol Cell Biol. 2002;22:7398–7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Mesaeli N, Nakamura K, Zvaritch E, et al. Calreticulin is essential for cardiac development. J Cell Biol. 1999;144:857–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Soldà T, Garbi N, Hämmerling GJ, Molinari M. Consequences of ERp57 deletion on oxidative folding of obligate and facultative clients of the calnexin cycle. J Biol Chem. 2006;281:6219–6226. [DOI] [PubMed] [Google Scholar]
- 177. Jessop CE, Chakravarthi S, Garbi N, Hämmerling GJ, Lovell S, Bulleid NJ. ERp57 is essential for efficient folding of glycoproteins sharing common structural domains. EMBO J. 2007;26:28–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Tadano-Aritomi K, Matsuda J, Fujimoto H, Suzuki K, Ishizuka I. Seminolipid and its precursor/degradative product, galactosylalkylacylglycerol, in the testis of saposin A- and prosaposin-deficient mice. J Lipid Res. 2003;44:1737–1743. [DOI] [PubMed] [Google Scholar]
- 179. Kishimoto Y, Hiraiwa M, O'Brien JS. Saposins: structure, function, distribution, and molecular genetics. J Lipid Res. 1992;33:1255–1267. [PubMed] [Google Scholar]
- 180. Pearse BR, Tamura T, Sunryd JC, Grabowski GA, Kaufman RJ, Hebert DN. The role of UDP-Glc:glycoprotein glucosyltransferase 1 in the maturation of an obligate substrate prosaposin. J Cell Biol. 2010;189:829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Moremen KW. Golgi alpha-mannosidase II deficiency in vertebrate systems: implications for asparagine-linked oligosaccharide processing in mammals. Biochim Biophys Acta. 2002;1573:225–235. [DOI] [PubMed] [Google Scholar]
- 182. Ruddock LW, Molinari M. N-Glycan processing in ER quality control. J Cell Sci. 2006;119:4373–4380. [DOI] [PubMed] [Google Scholar]
- 183. Sousa MC, Ferrero-Garcia MA, Parodi AJ. Recognition of the oligosaccharide and protein moieties of glycoproteins by the UDP-Glc:glycoprotein glucosyltransferase. Biochemistry. 1992;31:97–105. [DOI] [PubMed] [Google Scholar]
- 184. Stigliano ID, Alculumbre SG, Labriola CA, Parodi AJ, D'Alessio C. Glucosidase II and N-glycan mannose content regulate the half-lives of monoglucosylated species in vivo. Mol Biol Cell. 2011;22:1810–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Fagioli C, Sitia R. Glycoprotein quality control in the endoplasmic reticulum. Mannose trimming by endoplasmic reticulum mannosidase I times the proteasomal degradation of unassembled immunoglobulin subunits. J Biol Chem. 2001;276:12885–12892. [DOI] [PubMed] [Google Scholar]
- 186. Suh K, Bergmann JE, Gabel CA. Selective retention of monoglucosylated high mannose oligosaccharides by a class of mutant vesicular stomatitis virus G proteins. J Cell Biol. 1989;108:811–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Trombetta ES, Parodi AJ. Quality control and protein folding in the secretory pathway. Annu Rev Cell Dev Biol. 2003;19:649–676. [DOI] [PubMed] [Google Scholar]
- 188. Tokunaga F, Brostrom C, Koide T, Arvan P. Endoplasmic reticulum (ER)-associated degradation of misfolded N-linked glycoproteins is suppressed upon inhibition of ER mannosidase I. J Biol Chem. 2000;275:40757–40764. [DOI] [PubMed] [Google Scholar]
- 189. Hoseki J, Ushioda R, Nagata K. Mechanism and components of endoplasmic reticulum-associated degradation. J Biochem. 2010;147:19–25. [DOI] [PubMed] [Google Scholar]
- 190. Molinari M, Calanca V, Galli C, Lucca P, Paganetti P. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science. 2003;299:1397–1400. [DOI] [PubMed] [Google Scholar]
- 191. Christianson JC, Shaler TA, Tyler RE, Kopito RR. OS-9 and GRP94 deliver mutant α1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 2008;10:272–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Hosokawa N, Kamiya Y, Kamiya D, Kato K, Nagata K. Human OS-9, a lectin required for glycoprotein endoplasmic reticulum-associated degradation, recognizes mannose-trimmed N-glycans. J Biol Chem. 2009;284:17061–17068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Avezov E, Frenkel Z, Ehrlich M, Herscovics A, Lederkremer GZ. Endoplasmic reticulum (ER) mannosidase I is compartmentalized and required for N-glycan trimming to Man5–6GlcNAc2 in glycoprotein ER-associated degradation. Mol Biol Cell. 2008;19:216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Herscovics A, Romero PA, Tremblay LO. The specificity of the yeast and human class I ER α 1,2-mannosidases involved in ER quality control is not as strict previously reported. Glycobiology. 2002;12:14G–15G. [PubMed] [Google Scholar]
- 195. Sevier CS, Kaiser CA. Formation and transfer of disulphide bonds in living cells. Nat Rev Mol Cell Biol. 2002;3:836–847. [DOI] [PubMed] [Google Scholar]
- 196. Hatahet F, Ruddock LW. Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid Redox Signal. 2009;11:2807–2850. [DOI] [PubMed] [Google Scholar]
- 197. Appenzeller-Herzog C, Ellgaard L. The human PDI family: versatility packed into a single fold. Biochim Biophys Acta. 2008;1783:535–548. [DOI] [PubMed] [Google Scholar]
- 198. Feige MJ, Hendershot LM. Disulfide bonds in ER protein folding and homeostasis. Curr Opin Cell Biol. 2011;23:167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Jessop CE, Watkins RH, Simmons JJ, Tasab M, Bulleid NJ. Protein disulphide isomerase family members show distinct substrate specificity: P5 is targeted to BiP client proteins. J Cell Sci. 2009;122:4287–4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Di Jeso B, Morishita Y, Treglia AS, et al. Transient covalent interactions of newly synthesized thyroglobulin with oxidoreductases of the endoplasmic reticulum. J Biol Chem. 2014;289:11488–11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Meunier L, Usherwood YK, Chung KT, Hendershot LM. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell. 2002;13:4456–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Pirneskoski A, Klappa P, Lobell M, et al. Molecular characterization of the principal substrate binding site of the ubiquitous folding catalyst protein disulfide isomerase. J Biol Chem. 2004;279:10374–10381. [DOI] [PubMed] [Google Scholar]
- 203. Netzer WJ, Hartl FU. Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature. 1997;388:343–349. [DOI] [PubMed] [Google Scholar]
- 204. Jaenicke R. Stability and folding of domain proteins. Prog Biophys Mol Biol. 1999;71:155–241. [DOI] [PubMed] [Google Scholar]
- 205. Jansens A, van Duijn E, Braakman I. Coordinated nonvectorial folding in a newly synthesized multidomain protein. Science. 2002;2401–2403. [DOI] [PubMed] [Google Scholar]
- 206. Kurniawan ND, Aliabadizadeh K, Brereton IM, Kroon PA, Smith R. NMR structure and backbone dynamics of a concatemer of epidermal growth factor homology modules of the human low-density lipoprotein receptor. J Mol Biol. 2001;311:341–356. [DOI] [PubMed] [Google Scholar]
- 207. Oka OB, Pringle MA, Schopp IM, Braakman I, Bulleid NJ. ERdj5 is the ER reductase that catalyzes the removal of non-native disulfides and correct folding of the LDL receptor. Mol Cell. 2013;50:793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Dong M, Bridges JP, Apsley K, Xu Y, Weaver TE. ERdj4 and ERdj5 are required for endoplasmic reticulum-associated protein degradation of misfolded surfactant protein C. Mol Biol Cell. 2008;19:2620–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321:569–572. [DOI] [PubMed] [Google Scholar]
- 210. Hagiwara M, Maegawa K, Suzuki M, et al. Structural basis of an ERAD pathway mediated by the ER-resident protein disulfide reductase ERdj5. Mol Cell. 2011;41:432–444. [DOI] [PubMed] [Google Scholar]
- 211. Riemer J, Appenzeller-Herzog C, Johansson L, Bodenmiller B, Hartmann-Petersen R, Ellgaard L. A luminal flavoprotein in endoplasmic reticulum-associated degradation. Proc Natl Acad Sci USA. 2009;106:14831–14836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Rastogi MV, LaFranchi SH. Congenital hypothyroidism. Orphanet J Rare Dis. 2010;5:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. De Felice M, Di Lauro R. Minireview: intrinsic and extrinsic factors in thyroid gland development: an update. Endocrinology. 2011;152:2948–2956. [DOI] [PubMed] [Google Scholar]
- 214. Portulano C, Paroder-Belenitsky M, Carrasco N. The Na+/I− symporter (NIS): mechanism and medical impact. Endocr Rev. 2014;35:106–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Bizhanova A, Kopp P. Genetics and phenomics of Pendred syndrome. Mol Cell Endocrinol. 2010;322:83–90. [DOI] [PubMed] [Google Scholar]
- 216. Ris-Stalpers C, Bikker H. Genetics and phenomics of hypothyroidism and goiter due to TPO mutations. Mol Cell Endocrinol. 2010;322:38–43. [DOI] [PubMed] [Google Scholar]
- 217. Grasberger H. Defects of thyroidal hydrogen peroxide generation in congenital hypothyroidism. Mol Cell Endocrinol. 2010;322:99–106. [DOI] [PubMed] [Google Scholar]
- 218. Moreno JC, Visser TJ. Genetics and phenomics of hypothyroidism and goiter due to iodotyrosine deiodinase (DEHAL1) gene mutations. Mol Cell Endocrinol. 2010;322:91–98. [DOI] [PubMed] [Google Scholar]
- 219. Targovnik HM, Esperante SA, Rivolta CM. Genetics and phenomics of hypothyroidism and goiter due to thyroglobulin mutations. Mol Cell Endocrinol. 2010;322:44–55. [DOI] [PubMed] [Google Scholar]
- 220. Targovnik HM, Citterio CE, Rivolta CM. Thyroglobulin gene mutations in congenital hypothyroidism. Horm Res Paediatr. 2011;75:311–321. [DOI] [PubMed] [Google Scholar]
- 221. Schell T, Kulozik AE, Hentze MW. Integration of splicing, transport and translation to achieve mRNA quality control by the nonsense-mediated decay pathway. Genome Biol. 2002;3:REVIEWS1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Thermann R, Neu-Yilik G, Deters A, et al. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17:3484–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Zhang J, Sun X, Qian Y, LaDuca JP, Maquat LE. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol Cell Biol. 1998;18:5272–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Bhuvanagiri M, Schlitter AM, Hentze MW, Kulozik AE. NMD: RNA biology meets human genetic medicine. Biochem J. 2010;430:365–377. [DOI] [PubMed] [Google Scholar]
- 225. Dietz HC, Valle D, Francomano CA, Kendzior RJ, Jr, Pyeritz RE, Cutting GR. The skipping of constitutive exons in vivo induced by nonsense mutations. Science. 1993;259:680–683. [DOI] [PubMed] [Google Scholar]
- 226. González-Sarmiento R, Corral J, Mories MT, Corrales JJ, Miguel-Velado E, Miralles-Garcia JM. Monoallelic deletion in the 5′ region of the thyroglobulin gene as a cause of sporadic nonendemic simple goiter. Thyroid. 2001;11:789–793. [DOI] [PubMed] [Google Scholar]
- 227. Citterio CE, Rossetti LC, Souchon PF, et al. Novel mutational mechanism in the thyroglobulin gene: imperfect DNA inversion as a cause for hereditary hypothyroidism. Mol Cell Endocrinol. 2013;381:220–229. [DOI] [PubMed] [Google Scholar]
- 228. van de Graaf SA, Ris-Stalpers C, Veenboer GJ, et al. A premature stopcodon in thyroglobulin messenger RNA results in familial goiter and moderate hypothyroidism. J Clin Endocrinol Metab. 1999;84:2537–2542. [DOI] [PubMed] [Google Scholar]
- 229. Pardo V, Vono-Toniolo J, Rubio IG, et al. The p.A2215D thyroglobulin gene mutation leads to deficient synthesis and secretion of the mutated protein and congenital hypothyroidism with wide phenotype variation. J Clin Endocrinol Metab. 2009;94:2938–2944. [DOI] [PubMed] [Google Scholar]
- 230. Peteiro-Gonzalez D, Lee J, Rodriguez-Fontan J, et al. New insights into thyroglobulin pathophysiology revealed by the study of a family with congenital goiter. J Clin Endocrinol Metab. 2010;95:3522–3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. de Vijlder JJ, van Voorthuizen WF, van Dijk JE, Rijnberk A, Tegelaers WH. Hereditary congenital goiter with thyroglobulin deficiency in a breed of goats. Endocrinology. 1978;102:1214–1222. [DOI] [PubMed] [Google Scholar]
- 232. Niu DM, Hsu JH, Chong KW, et al. Six new mutations of the thyroglobulin gene discovered in Taiwanese children presenting with thyroid dyshormonogenesis. J Clin Endocrinol Metab. 2009;94:5045–5052. [DOI] [PubMed] [Google Scholar]
- 233. Citterio CE, Coutant R, Rouleau S, et al. A new compound heterozygous for c.886C>T/c.2206C>T[p.R277X/p.Q717X] mutations in the thyroglobulin gene as a cause of foetal goitrous hypothyroidism. Clin Endocrinol (Oxf). 2011;74:533–535. [DOI] [PubMed] [Google Scholar]
- 234. Citterio CE, Machiavelli GA, Miras MB, et al. New insights into thyroglobulin gene: molecular analysis of seven novel mutations associated with goiter and hypothyroidism. Mol Cell Endocrinol. 2013;365:277–291. [DOI] [PubMed] [Google Scholar]
- 235. Targovnik HM, Medeiros-Neto G, Varela V, Cochaux P, Wajchenberg BL, Vassart G. A nonsense mutation causes human hereditary congenital goiter with preferential production of a 171-nucleotide-deleted thyroglobulin ribonucleic acid messenger. J Clin Endocrinol Metab. 1993;77:210–215. [DOI] [PubMed] [Google Scholar]
- 236. Mendive FM, Rivolta CM, González-Sarmiento R, Medeiros-Neto G, Targovnik HM. Nonsense-associated alternative splicing of the human thyroglobulin gene. Mol Diagn. 2005;9:143–149. [DOI] [PubMed] [Google Scholar]
- 237. Machiavelli GA, Caputo M, Rivolta CM, et al. Molecular analysis of congenital goitres with hypothyroidism caused by defective thyroglobulin synthesis. Identification of a novel c.7006C>T [p.R2317X] mutation and expression of minigenes containing nonsense mutations in exon 7. Clin Endocrinol (Oxf). 2010;72:112–121. [DOI] [PubMed] [Google Scholar]
- 238. Ieiri T, Cochaux P, Targovnik HM, et al. A 3′ splice site mutation in the thyroglobulin gene responsible for congenital goiter with hypothyroidism. J Clin Invest. 1991;88:1901–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Targovnik HM, Vono J, Billerbeck AE, et al. A 138-nucleotide deletion in the thyroglobulin ribonucleic acid messenger in a congenital goiter with defective thyroglobulin synthesis. J Clin Endocrinol Metab. 1995;80:3356–3360. [DOI] [PubMed] [Google Scholar]
- 240. Targovnik HM, Rivolta CM, Mendive FM, Moya CM, Vono J, Medeiros-Neto G. Congenital goiter with hypothyroidism caused by a 5′ splice site mutation in the thyroglobulin gene. Thyroid. 2001;11:685–690. [DOI] [PubMed] [Google Scholar]
- 241. Pardo V, Rubio IG, Knobel M, et al. Phenotypic variation among four family members with congenital hypothyroidism caused by two distinct thyroglobulin gene mutations. Thyroid. 2008;18:783–786. [DOI] [PubMed] [Google Scholar]
- 242. Hermanns P, Refetoff S, Sriphrapradang C, et al. A clinically euthyroid child with a large goiter due to a thyroglobulin gene defect: clinical features and genetic studies. J Pediatr Endocrinol Metab. 2013;26:119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Gutnisky VJ, Moya CM, Rivolta CM, et al. Two distinct compound heterozygous constellations (R277X/IVS34–1G>C and R277X/R1511X) in the thyroglobulin (TG) gene in affected individuals of a Brazilian kindred with congenital goiter and defective TG synthesis. J Clin Endocrinol Metab. 2004;89:646–657. [DOI] [PubMed] [Google Scholar]
- 244. Rivolta CM, Targovnik HM. Molecular advances in thyroglobulin disorders. Clin Chim Acta. 2006;374:8–24. [DOI] [PubMed] [Google Scholar]
- 245. Gass JN, Gifford NM, Brewer JW. Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J Biol Chem. 2002;277:49047–49054. [DOI] [PubMed] [Google Scholar]
- 246. Christis C, Fullaondo A, Schildknegt D, Mkrtchian S, Heck AJ, Braakman I. Regulated increase in folding capacity prevents unfolded protein stress in the ER. J Cell Sci. 2010;123:787–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. [DOI] [PubMed] [Google Scholar]
- 248. Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. [DOI] [PubMed] [Google Scholar]
- 249. Termine DJ, Moremen KW, Sifers RN. The mammalian UPR boosts glycoprotein ERAD by suppressing the proteolytic downregulation of ER mannosidase I. J Cell Sci. 2009;122:976–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Kouroku Y, Fujita E, Tanida I, et al. ER stress (PERK/eIF2α phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–239. [DOI] [PubMed] [Google Scholar]
- 251. Oyadomari S, Yun C, Fisher EA, et al. Cotranslocational degradation protects the stressed endoplasmic reticulum from protein overload. Cell. 2006;126:727–739. [DOI] [PubMed] [Google Scholar]
- 252. Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. [DOI] [PubMed] [Google Scholar]
- 253. Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Rutkowski DT, Arnold SM, Miller CN, et al. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 2006;4:e374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Lin JH, Li H, Yasumura D, et al. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Lin JH, Li H, Zhang Y, Ron D, Walter P. Divergent effects of PERK and IRE1 signaling on cell viability. PLoS One. 2009;4:e4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Han D, Lerner AG, Vande Walle L, et al. IRE1α kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Tsang KY, Chan D, Bateman JF, Cheah KS. In vivo cellular adaptation to ER stress: survival strategies with double-edged consequences. J Cell Sci. 2010;123:2145–2154. [DOI] [PubMed] [Google Scholar]
- 259. Treglia AS, Turco S, Ulianich L, et al. Cell fate following ER stress: just a matter of “quo ante” recovery or death? Histol Histopathol. 2012;27:1–12. [DOI] [PubMed] [Google Scholar]
- 260. Yang L, Carlson SG, McBurney D, Horton WE., Jr Multiple signals induce endoplasmic reticulum stress in both primary and immortalized chondrocytes resulting in loss of differentiation, impaired cell growth, and apoptosis. J Biol Chem. 2005;280:31156–31165. [DOI] [PubMed] [Google Scholar]
- 261. Ho MS, Tsang KY, Lo RL, et al. COL10A1 nonsense and frame-shift mutations have a gain-of-function effect on the growth plate in human and mouse metaphyseal chondrodysplasia type Schmid. Hum Mol Genet. 2007;16:1201–1215. [DOI] [PubMed] [Google Scholar]
- 262. Tsang KY, Chan D, Cheslett D, et al. Surviving endoplasmic reticulum stress is coupled to altered chondrocyte differentiation and function. PLoS Biol. 2007;5:e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Rajpar MH, McDermott B, Kung L, et al. Targeted induction of endoplasmic reticulum stress induces cartilage pathology. PLoS Genet. 2009;5:e1000691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Firtina Z, Danysh BP, Bai X, Gould DB, Kobayashi T, Duncan MK. Abnormal expression of collagen IV in lens activates unfolded protein response resulting in cataract. J Biol Chem. 2009;284:35872–35884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Ulianich L, Garbi C, Treglia AS, et al. ER stress is associated with dedifferentiation and an epithelial-to-mesenchymal transition-like phenotype in PC Cl3 thyroid cells. J Cell Sci. 2008;121:477–486. [DOI] [PubMed] [Google Scholar]
- 266. Kim PS, Arvan P. Endocrinopathies in the family of endoplasmic reticulum (ER) storage diseases: disorders of protein trafficking and the role of ER molecular chaperones. Endocr Rev. 1998;19:173–202. [DOI] [PubMed] [Google Scholar]
- 267. Hebert DN, Molinari M. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol Rev. 2007;87:1377–1408. [DOI] [PubMed] [Google Scholar]
- 268. Mayerhofer A, Amador AG, Beamer WG, Bartke A. Ultrastructural aspects of the goiter in cog/cog mice. J Hered. 1988;79:200–203. [DOI] [PubMed] [Google Scholar]
- 269. Adkison LR, Taylor S, Beamer WG. Mutant gene-induced disorders of structure, function and thyroglobulin synthesis in congenital goitre (cog/cog) in mice. J Endocrinol. 1990;126:51–58. [DOI] [PubMed] [Google Scholar]
- 270. Sakai Y, Yamashina S, Furudate SI. Missing secretory granules, dilated endoplasmic reticulum, and nuclear dislocation in the thyroid gland of rdw rats with hereditary dwarfism. Anat Rec. 2000;259:60–66. [DOI] [PubMed] [Google Scholar]
- 271. Oh-Ishi M, Omori A, Kwon JY, Agui T, Maeda T, Furudate S. Detection and identification of proteins related to the hereditary dwarfism of the rdw rat. Endocrinology. 1998;139:1288–1299. [DOI] [PubMed] [Google Scholar]
- 272. Umezu M, Fujimura T, Sugawara S, Kagabu S. Pituitary and serum levels of prolactin (PRL), thyroid stimulating hormone (TSH) and serum thyroxine (T4) in hereditary dwarf rats (rdw/rdw). Jikken Dobutsu. 1993;42:211–216. [DOI] [PubMed] [Google Scholar]
- 273. Menon S, Lee J, Abplanalp WA, et al. Oxidoreductase interactions include a role for ERp72 engagement with mutant thyroglobulin from the rdw/rdw rat dwarf. J Biol Chem. 2007;282:6183–6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Baryshev M, Sargsyan E, Wallin G, et al. Unfolded protein response is involved in the pathology of human congenital hypothyroid goiter and rat non-goitrous congenital hypothyroidism. J Mol Endocrinol. 2004;32:903–920. [DOI] [PubMed] [Google Scholar]
- 275. Kim PS, Lee J, Jongsamak P, et al. Defective protein folding and intracellular retention of thyroglobulin-R19K mutant as a cause of human congenital goiter. Mol Endocrinol. 2008;22:477–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Corral J, Martín C, Pérez R, et al. Thyroglobulin gene point mutation associated with non-endemic simple goitre. Lancet. 1993;341:462–464. [DOI] [PubMed] [Google Scholar]
- 277. Pérez-Centeno C, González-Sarmiento R, Mories MT, Corrales JJ, Miralles-García JM. Thyroglobulin exon 10 gene point mutation in a patient with endemic goiter. Thyroid. 1996;6:423–427. [DOI] [PubMed] [Google Scholar]
- 278. Vono-Toniolo J, Medeiros-Neto G, Kopp P. Three novel homozygous nucleotide substitutions in the TG gene in an inbred Brazilian kindred with congenital goiter and defective thyroglobulin synthesis. In: Proceedings from the Endocrine Society; June 19–22, 2002; San Francisco, CA; p 536 (Abstract P3-185). [Google Scholar]
- 279. Feldt-Rasmussen U, Hegedüs L, Perrild H, Rasmussen N, Hansen JM. Relationship between serum thyroglobulin, thyroid volume and serum TSH in healthy non-goitrous subjects and the relationship to seasonal variations in iodine intake. Thyroidology. 1989;1:115–118. [PubMed] [Google Scholar]
- 280. Izumi M, Larsen PR. Correlation of sequential changes in serum thyroglobulin, triiodothyronine, and thyroxine in patients with Graves' disease and subacute thyroiditis. Metabolism. 1978;27:449–460. [DOI] [PubMed] [Google Scholar]
- 281. Hidaka Y, Nishi I, Tamaki H, et al. Differentiation of postpartum thyrotoxicosis by serum thyroglobulin: usefulness of a new multisite immunoradiometric assay. Thyroid. 1994;4:275–278. [DOI] [PubMed] [Google Scholar]
- 282. Mazzaferri EL, Robbins RJ, Spencer CA, et al. A consensus report of the role of serum thyroglobulin as a monitoring method for low-risk patients with papillary thyroid carcinoma. J Clin Endocrinol Metab. 2003;88:1433–1441. [DOI] [PubMed] [Google Scholar]
- 283. Targovnik HM, Edouard T, Varela V, et al. Two novel mutations in the thyroglobulin gene as cause of congenital hypothyroidism: identification a cryptic donor splice site in the exon 19. Mol Cell Endocrinol. 2012;348:313–321. [DOI] [PubMed] [Google Scholar]
- 284. Citterio CE, Morales CM, Bouhours-Nouet N, et al. Novel compound heterozygous thyroglobulin mutations c.745+1G>A/c.7036+2T>A associated with congenital goiter and hypothyroidism in a Vietnamese family: identification of a new cryptic 5 splice site in the exon 6. Mol Cell Endocrinol. 2015;404:102–112. [DOI] [PubMed] [Google Scholar]