Abstract
Stimulus reconstruction or decoding methods provide an important tool for understanding how sensory and motor information is represented in neural activity. We discuss Bayesian decoding methods based on an encoding generalized linear model (GLM) that accurately describes how stimuli are transformed into the spike trains of a group of neurons. The form of the GLM likelihood ensures that the posterior distribution over the stimuli that caused an observed set of spike trains is log-concave so long as the prior is. This allows the maximum a posteriori (MAP) stimulus estimate to be obtained using efficient optimization algorithms. Unfortunately, the MAP estimate can have a relatively large average error when the posterior is highly non-Gaussian. Here we compare several Markov chain Monte Carlo (MCMC) algorithms that allow for the calculation of general Bayesian estimators involving posterior expectations (conditional on model parameters). An efficient version of the hybrid Monte Carlo (HMC) algorithm was significantly superior to other MCMC methods for Gaussian priors. When the prior distribution has sharp edges and corners, on the other hand, the “hit-and-run” algorithm performed better than other MCMC methods. Using these algorithms we show that for this latter class of priors the posterior mean estimate can have a considerably lower average error than MAP, whereas for Gaussian priors the two estimators have roughly equal efficiency. We also address the application of MCMC methods for extracting non-marginal properties of the posterior distribution. For example, by using MCMC to calculate the mutual information between the stimulus and response, we verify the validity of a computationally efficient Laplace approximation to this quantity for Gaussian priors in a wide range of model parameters; this makes direct model-based computation of the mutual information tractable even in the case of large observed neural populations, where methods based on binning the spike train fail. Finally, we consider the effect of uncertainty in the GLM parameters on the posterior estimators.
1 Introduction
Understanding the exact nature of the neural code is a central goal of theoretical neuroscience. Neural decoding provides an important method for comparing the fidelity and robustness of different codes (Rieke et al., 1997). The decoding problem, in its general form, is the problem of estimating the relevant stimulus, x, that elicited the observed spike trains, r, of a population of neurons over a course of time. Neural decoding is also of crucial importance in the design of neural prosthetic devices (Donoghue, 2002).
A large literature exists on developing and applying different decoding methods to spike train data, both in single cell and population decoding. Bayesian methods lie at the basis of a major group of these decoding algorithms (Sanger, 1994; Zhang et al., 1998; Brown et al., 1998; Maynard et al., 1999; Stanley and Boloori, 2001; Shoham et al., 2005; Barbieri et al., 2004; Wu et al., 2004; Brockwell et al., 2004; Kelly and Lee, 2004; Karmeier et al., 2005; Truccolo et al., 2005; Pillow et al., 2010; Jacobs et al., 2006; Yu et al., 2009; Gerwinn et al., 2009). In such methods the a priori distribution of the sensory signal, p(x), is combined, via Bayes’ rule, with an encoding model describing the probability, p(r|x), of different spike trains given the signal, to yield the posterior distribution, p(x|r), that carries all the information contained in the observed spike train responses about the stimulus. A Bayesian estimate is one that, given a definite cost function on the amount of error, minimizes the expected error cost under the posterior distribution. Assuming the prior distribution and the encoding model are appropriately chosen, the Bayes estimate is thus optimal by construction. Furthermore, since the Bayesian approach yields a distribution over the possible stimuli that could lead to the observed response, Bayes estimates naturally come equipped with measures of their reliability or posterior uncertainty.
In a fully Bayesian approach, one has to be able to evaluate any desired functional of the high dimensional posterior distribution. Unfortunately, calculating these can be computationally very expensive. For example, most Bayesian estimates involve integrations over the (often very high-dimensional) space of possible signals. Accordingly, most work on Bayesian decoding of spike trains has either focused on cases where the signal is low dimensional (Sanger, 1994; Maynard et al., 1999; Abbott and Dayan, 1999; Karmeier et al., 2005) or on situations where the joint distribution, p(x, r), has a certain Markov tree decomposition, so that computationally efficient recursive techniques may be applied (Zhang et al., 1998; Brown et al., 1998; Barbieri et al., 2004; Wu et al., 2004; Brockwell et al., 2004; Kelly and Lee, 2004; Shoham et al., 2005; Eden et al., 2004; Truccolo et al., 2005; Ergun et al., 2007; Yu et al., 2009; Paninski et al., 2010). The Markov setting is extremely useful; it lends itself naturally to many problems of interest in neuroscience and has thus been fruitfully exploited. In particular, this setting is very useful in an important class of decoding problems where stimulus estimation is performed online, i.e., the stimulus at some time, t, is estimated conditioned on the observation of the spike trains only up to that time, as opposed to the entire spike train.
However, some decoding problems can not be formulated in the online estimation framework. In such cases quantities of interest should naturally be conditioned on the entire history of the spike train. In this paper, we focus on this latter class of problems (although many of the methods we discuss can potentially be adopted to the online case as well). Furthermore, it is awkward to cast many decoding problems of interest in the Markov setting. A more general method that does not require such tree decomposition properties is to calculate the maximum a posteriori (MAP) estimate xMAP (Stanley and Boloori, 2001; Jacobs et al., 2006; Gerwinn et al., 2009) – see the companion paper (Pillow et al., 2010) for further review and discussion. The MAP estimate requires no integration, but only maximization of the posterior distribution, and can remain computationally tractable even when the stimulus space is very high-dimensional. This is the case for general log-concave posterior distributions; many problems in sensory and motor coding fall in this class (it should be noted, however, that in many cases of interest where this condition is not satisfied, e.g., when the distributions are inherently multi-modal, posterior maximization can become highly intractable). The MAP is a good estimator when the posterior is well-approximated by a Gaussian distribution centered at xMAP (Tierney and Kadane, 1986; Kass et al., 1991). As the mode and the mean of a Gaussian distribution are identical, in this case the MAP is approximately equal to the posterior mean as well. This Gaussian approximation is expected to be sufficiently accurate, e.g., when the prior distribution and the likelihood function (i.e., p(r|x) as function of x) are not very far from Gaussian, or when the likelihood is sharply concentrated around xMAP. However, in cases where the prior distribution has sharp boundaries and corners and the likelihood function does not constrain the estimate away from such non-Gaussian regions, the Gaussian approximation can fail, resulting in a large average error in the MAP estimate. In such cases, one expects the MAP to be inferior to the posterior mean E(x|r), which is the optimal estimate under squared error loss.
Accordingly, in Sec. 3 of this paper we develop efficient Markov chain Monte Carlo (MCMC) techniques for sampling from general log-concave posterior distributions, and compare their performance in situations relevant to our neural decoding setting (for comprehensive introductions to MCMC methods, including their application in Bayesian problems, see, e.g., Robert and Casella (2005) and Gelman (2004)). By providing a tool for approximating averages (integrals) over the exact posterior distribution, p(x|r, θ) (where θ are the parameters of the encoding forward model, in principle obtained by fitting to experimental data), these techniques allow us to calculate general Bayesian estimates such as E(x|r, θ), and provide estimates of their uncertainty. Although, in principle many of the MCMC methods we discuss in this paper are applicable even to posterior distributions that are not log-concave, they may lose their efficiency in such cases, and furthermore estimates based on them may not even converge to true posterior averages. In Sec. 4 we compare the MAP and the posterior mean stimulus estimates based on the simulated response of a population of retinal ganglion cells (RGC). In Sec. 5 we discuss the applications of MCMC for calculating more complicated properties of p(x|r, θ) beyond marginal statistics, such as the statistics of first-passage times. We also discuss an MCMC-based method known as “bridge sampling” (Bennett, 1976; Meng and Wong, 1996) that provides a tool for a direct calculation of the mutual information. Using this technique, we show that for Gaussian priors the estimates of (Pillow et al., 2010) for this quantity based on the Laplace approximation are robust and accurate. Finally, in Sec. 6 we discuss the effect of uncertainty in the parameters of the forward model, θ, on the MAP and posterior mean estimate. We proceed by first introducing the forward model used to calculate the likelihood p(r|x, θ), in the next section.
2 The encoding model, the MAP, and the stimulus ensembles
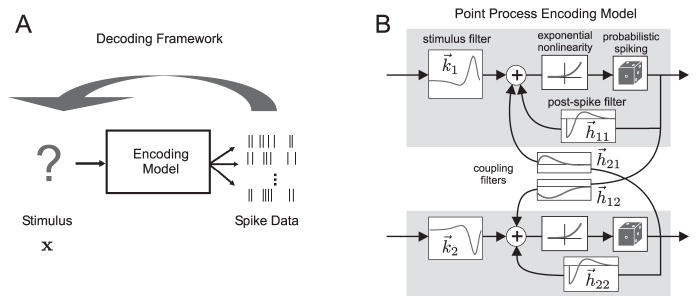
In this section we give an overview of neural encoding models based on generalized linear models (GLM) (Brillinger, 1988; McCullagh and Nelder, 1989; Paninski, 2004; Truccolo et al., 2005), and briefly review the treatment of (Pillow et al., 2010) for MAP based decoding. (Note that much of the material in this section was previously covered in (Pillow et al., 2010), but we include a brief review here to make this paper self-contained.) A neural encoding model is a model that assigns a conditional probability to the neural response given the stimulus. We take the stimulus to be an artificially discretized, possibly multi-component, function of time, x(t, n), which will be represented as a d-dimensional vector x.1
In response to x, the i-th neuron emits a spike train response
| (1) |
where ti,α is the time of the α-th spike of the i-th neuron. We represent this function by ri (we will use bold face symbols for both continuous time and discretized, finite-dimensional vectors), and the collection of response data of all cells by r.
The response, r, is not fully determined by x, and is subject to trial to trial variations. We model r as a point process whose instantaneous firing rate is the output of a generalized linear model (Brillinger, 1988; McCullagh and Nelder, 1989; Paninski, 2004). This class of models has been extensively discussed in the literature. Briefly, it is a generalization of the popular Linear-Nonlinear-Poisson model that includes feedback and interaction between neurons, with parameters that have natural neurophysiological interpretations (Simoncelli et al., 2004) and has been applied in a wide variety of experimental settings (Brillinger, 1992; Dayan and Abbott, 2001; Chichilnisky, 2001; Theunissen et al., 2001; Brown et al., 2003; Paninski et al., 2004; Truccolo et al., 2005; Pillow et al., 2008). The model gives the conditional (on the stimulus, as well as the history of the observed spike train) instantaneous firing rate of the i-th observed cell as
| (2) |
which we write more concisely as
| (3) |
Here, the linear operators (filters) Ki, and ℋij have causal,2 time translation invariant kernels ki(t, n) and hij(t) (we note that the causality condition for ki(t, n) is only true for sensory neurons). The kernel ki(t, n) represents the i-th cell’s linear ‘receptive field’, and hij(t) describe possible excitatory or inhibitory post-spike effect of the j-th observed neuron on the i-th. The diagonal components hii describe the post-spike feedback of the neuron to itself, and can account for refractoriness, adaptation and burstiness depending on their shape (see (Paninski, 2004) for details). The constant bi is the DC bias of the i-th cell, such that f(bi) may be considered as the i-th cell’s constant “baseline” firing rate. Finally, f(·) is a nonlinear, nonnegative, increasing function.3
Given the firing rate, Eq. (3), the forward probability, p(r|x, θ), can be written as (Snyder and Miller, 1991; Paninski, 2004; Truccolo et al., 2005)
| (4) |
where θ = {bi, ki, hij} is the set of GLM parameters. The constant term serves to normalize the probability and does not depend on x or θ. We will restrict ourselves to f(u) that are convex and log-concave (e.g., this is the case for f(u) = exp(u)). Then the log-likelihood function L(x, θ) is guaranteed to be a separately concave function of either the stimulus x or the model parameters,4 irrespective of the observed spike data r. The log-concavity with respect to the model parameters makes maximum likelihood fitting of this model very easy, as concave functions on convex parameter spaces have no nonglobal local maxima. Therefore simple gradient ascent algorithms can be used to find the maximum likelihood estimate.
The prior distribution describes the statistics of the stimulus in the natural world or that of an artificial stimulus ensemble used by the experimentalist. In this paper we only consider priors relevant for the latter case. Given a prior distribution, p(x), and having observed the spike trains, r, the posterior probability distribution over the stimulus is given by Bayes’ rule
| (5) |
where
| (6) |
The MAP estimate is by definition
| (7) |
(Except for in Sec. 6, in the following sections we will drop θ from the arguments of xMAP or the distributions, it being understood that they are conditioned on the specific θ obtained from the experimental fit). As discussed above, for the GLM nonlinearities that we consider, the likelihood, p(r|x, θ), is log-concave in x. If the prior, p(x), is also log-concave, then the posterior distribution is log-concave as a function of x, and its maximization (Eq. (7)) can also be achieved using simple gradient ascent techniques. The class of log-concave prior distributions is quite large, and it includes exponential, triangular, and general Gaussian distributions as well as uniform distributions with convex support.5
The MAP is a good, low-error estimate when Laplace’s method provides a good approximation for the posterior mean, which has the minimum mean square error. This method is a general asymptotic method for approximating integrals when the integrand peaks sharply at its global maximum and is exponentially suppressed away from it. In the Bayesian setting this corresponds to posterior integrals of interest (e.g., posterior averages, and so-called Bayes factors) receiving their dominant contribution from the vicinity of the main mode of p(x|r, θ), i.e., xMAP – for a comprehensive review of Laplace’s method in Bayesian applications see Kass et al. (1991), and books on Bayesian analysis, such as Berger (1993). In that case, we can Taylor expand the log-posterior to the first non-vanishing order around xMAP (i.e., the second order, since the derivative vanishes at the maximum), obtaining the Gaussian approximation (hereinafter also referred to as the Laplace approximation)
| (8) |
Here the matrix J is the Hessian of the negative log-posterior at xMAP
| (9) |
Normally, in the statistical setting the Laplace approximation is formally justified in the limit of large samples due to the central limit theorem, leading to a likelihood function with a very sharp peak (in neural decoding the meaning of “large samples” depends, in general, on the nature of the stimulus – we will discuss this point further in Sec. 4). However, this approximation often proves adequate even for moderately strong likelihoods, as long as the posterior is not grossly nonnormal. An obvious case where the approximation fails is for strongly multimodal distributions where no particular mode dominates. Here, we restrict our attention to the class of log-concave posteriors which as mentioned above are unimodal. For this class, and for a smooth enough GLM nonlinearity, f(·), we expect Eq. (8) to hold for prior distributions that are close to normal, even when the likelihood is not extremely sharp. However, for flatter priors with sharp boundaries or “corners” we expect it to fail unless the likelihood is narrowly concentrated away from such non-Gaussian regions.
In this paper, we set out to verify this intuition by studying two extreme cases within the class of log-concave priors, namely Gaussian and at distributions with convex support, given by
| (10) |
and
| (11) |
respectively.6 Here, 𝒞 is the d × d covariance matrix, and I𝒮 is the indicator function of a convex region, 𝒮, of Rd. In particular, for the white-noise stimuli we consider in Sec. 4, 𝒞 = c2Id×d in the Gaussian case, and 𝒮 is the d-dimensional cube , in the flat case (this choice for 𝒮 corresponds to a uniformly distributed white noise stimulus). Here, c is the standard deviation of the stimulus on a subinterval, and in the case where x(t) is the normalized light intensity (with the average luminosity removed), it is referred to as the contrast. We will compare the performance of the MAP and posterior mean estimates in each case, in Sec. 4. In Sec. 5.2 we will verify the adequacy of this approximation for the estimation of the mutual information in the case of Gaussian priors.
3 Monte Carlo techniques for Bayesian estimates
For the sake of completeness, we start this section by reviewing the basics of the Markov chain Monte Carlo (MCMC) method (for comprehensive textbooks on MCMC methods, see, e.g., Gelman (2004); Robert and Casella (2005)). However, the main point of this section is the discussion of the applications of this method to the neural case and ways of making the method more efficient, as well as a comparison of the efficiency of different MCMC algorithms, in this specific setting. As noted in the introduction, the posterior distribution, Eq. (5), represents the full information about the stimulus as encoded in the prior distribution and carried by the observed spike trains, r. However, a much simpler (and therefore less complete) representation of this information can be provided by a so-called Bayesian estimate for the stimulus, possibly accompanied by a corresponding estimate of its error. A commonly used Bayesian estimate is the posterior mean,
| (12) |
which is the optimal estimator with respect to average square error. The uncertainty of this estimator is in turn provided by the posterior covariance matrix. When the posterior distribution can be reasonably approximated as Gaussian, the posterior mean can be approximated by its mode, i.e. the MAP estimate, Eq. (7), and the inverse of the log-posterior Hessian, Eq. (9), can represent its uncertainty. In this paper we adopt the posterior mean, E(x|r), as a benchmark for comparing the performance of the two estimates, and we take the deviation of the MAP from the latter as a measure of the validity of the Gaussian approximation for the posterior distribution.
To calculate the posterior mean Eq. (12), we have to perform a high-dimensional integral over x. Computationally, this is quite costly. The Monte Carlo method is based on the idea that if one could generate N i.i.d. samples, xt (t = 1, …, N), from a probability distribution, π(x),7 then one could approximate integrals involved in expectations such as Eq. (12) by sample averages. This is because, by the law of large numbers, for any R g(x) (such that ∫|g(x)|π(x)dx < ∞)
| (13) |
Also, to decide how many samples are sufficient, we may estimate
| (14) |
when N is large enough that this variance is sufficiently small, we may stop sampling. However, it is often quite challenging to sample directly from a complex multi-dimensional distribution, and the efficiency of methods yielding i.i.d. samples often decreases exponentially with the number of dimensions.
Fortunately, Eq. (13) (the law of large numbers) still holds if the i.i.d. samples are replaced by an ergodic Markov chain, xt, whose equilibrium distribution is π(x). This is the idea behind the Markov chain Monte Carlo (MCMC) method based on the Metropolis-Hastings (MH) algorithm (Metropolis et al., 1953; Hastings, 1970). In the general form of this algorithm, the Markov transitions are constructed as follows. Starting at point x, we first sample a point y from some “proposal” density q(y|x), and then accept this point as the next point in the chain, with probability
| (15) |
If y is rejected, the chain stays at point x, so that the conditional Markov transition probability, T(y|x), is given by
| (16) |
where
| (17) |
is the rejection probability of proposals from x. The reason for accepting the proposals according to Eq. (15) is that doing so guarantees that π(x) is invariant under the Markov evolution (see, e.g., Robert and Casella (2005) for details). It is important to note that, from Eq. (15), to execute this algorithm we only need to know π(x) up to a constant, which is an advantage because often, particularly in Bayesian settings, normalizing the distribution itself requires the difficult integration for which we are using MCMC (we will discuss a method of calculating the normalization constant in Sec. 5.2).
The major drawback of the MCMC method is that the generated samples are dependent and thus it is harder to estimate how long we need to run the chain to get an accurate estimate, and in general we may need to run the chain much longer than the i.i.d. case. Thus, we would like to choose a proposal density, q(y|x), which gives rise to a chain that explores the support of π(x) (i.e., mixes) quickly, and has a small correlation time (roughly the number of steps separation to yield i.i.d samples), to reduce the number of steps the chain has to be iterated and hence the computational time (see Sec. 3.5 and (Gelman, 2004) and (Robert and Casella, 2005) for further details). In general, a good proposal density q(y|x) should allow for large jumps with higher probability for falling in regions of larger π(x) (so as to avoid a high MH rejection rate). A good rule of thumb is for the proposals q(.|x) to resemble the true density π(.) as well as possible. We review a few useful well-known proposals below, and explore different ways of boosting their efficiency in the GLM-based neural decoding setting. We note here, that these algorithms can be applied to general distributions, and do not require the log-concavity condition for π(x). However, some of the enhancements that we consider can only be implemented, or are only expected to boost up the performance of the chain, when the distribution π(x) is log-concave – see the discussion of non-isotropic proposals in Sec. 3.1 and Sec. 3.2, and that of adaptive rejection sampling in Sec. 3.4.
3.1 Non-isotropic random-walk Metropolis (RWM)
Perhaps the most common proposal is of the random walk type: q(x|y) = q(x − y), for some fixed density q(.). Centered isotropic Gaussian distributions are a simple choice, leading to proposals
| (18) |
where z is Gaussian of zero mean and identity covariance, and σ determines the proposal jump scale. (In this simple form, the RWM chain was used in a recent study to fit a hierarchical model of tuning curves of neurons in the primary visual cortex to experimental data (Cronin et al., 2009).) Of course, different choices of the proposal distribution will affect the mixing rate of the chain. To increase this rate, it is generally a good idea to align the axes of q(.) with the target density, if possible, so that the proposal jump scales in different directions are roughly proportional to the width of π(x) along those directions. Such proposals will reduce the rejection probability and increase the average jump size by biasing the chain to jump in more favorable directions. For Gaussian proposals, we can thus choose the covariance matrix of q(.) to be proportional to the covariance of π(x). Of course, calculating the latter covariance is often a difficult problem (which the MCMC method is intended to solve!), but we can exploit the Laplace approximation, Eq. (8), and take the inverse of the Hessian of the log-posterior at MAP, Eq. (9), as a first approximation for the covariance. This is equivalent to modifying the proposal rule (18) into
| (19) |
where A is the Cholesky decomposition of J−1
| (20) |
and J was defined in Eq. (9). We refer to chains with such jump proposals as non-isotropic Gaussian RWM. Figure 2 compares the isotropic and nonisotropic proposals. The modification Eq. (19) is equivalent to running a chain with isotropic proposals Eq. (18), but for the auxiliary distribution π̃ (x̃) = |A|π(Ax̃) (whose corresponding Laplace approximation corresponds to a standard Gaussian with identity covariance), and subsequently transforming the samples, x̃t, by the matrix A to obtain samples xt = Ax̃t from π(x). Implementing non-isotropic sampling using the transformed distribution π̃ (x̃), instead of modifying the proposals as in Eq. (19), is more readily extended to chains more complicated than RWM (see below) and therefore we used this latter method in our simulations using different chains.
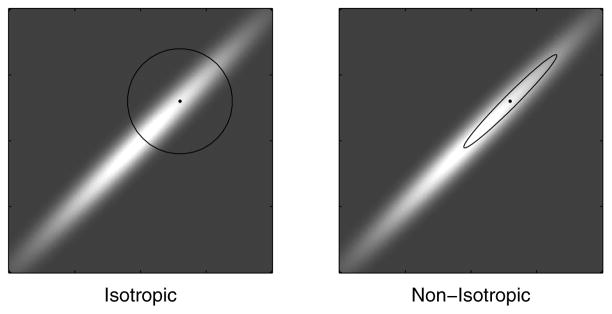
Figure 2.
Comparison of isotropic and non-isotropic Markov jumps for the Gaussian RWM and hit-and-run chains. In the RWM case, the circle and the ellipse are level sets of the Gaussian proposal distributions for jumping from the dot at their center. In isotropic (non-isotropic) hit-and-run, the jump direction n is generated by normalizing a vector sampled from an isotropic (non-isotropic) Gaussian distribution centered at the origin. The non-isotropic distributions were constructed using the Hessian, Eq. (9), in the Laplace approximation, so that the ellipse is described by xTJx = const. When the underlying distribution, π(x), is highly non-isotropic, it is disadvantageous to jump isotropically, as it reduces the average jump size and slows down the chain. In RWM, the proposal jump scale can not be much larger than the scale of the narrow “waist” of the underlying distribution, lest the rejection rate gets large (as most proposals will fall in the dark region of small π(x)) and the chain gets stuck. For hit-and-run, there is no jump scale to be set by the user, and the jump size in a given direction, n, is set by the scale of the “slice” distribution Eq. (25). Thus in the isotropic case the average jump size will effectively be a uniform average over the scales of π(x) along its principal axes. In the non-isotropic case, however, the jump size will be determined mainly by the scale of the “longer” dimensions, as the non-isotropic distribution gives more weight to these.
As we will see in the next section, in the flat prior case and for weak stimulus filters or a small number of identical cells, the Laplace approximation can be poor. In particular, the Hessian, Eq. (9), does not contain any information about the prior in the flat case, and therefore the approximate distribution, Eq. (8), can be significantly broader than the extent of the prior support in some directions. To take advantage of the Laplace approximation in this case, we regularized the Hessian by adding to it the inverse covariance matrix of the flat prior, obtaining a matrix that would be the Hessian if the flat prior was replaced by a Gaussian with the same mean and covariance. Even though the Gaussian with this regularized Hessian is still not a very good approximation for the posterior, we saw that in many cases it improved the mixing rate of the chain.
In general, the multiplication of a vector of dimensionality d by a matrix involves 𝒪(d2), and the inversion of a d×d matrix involves 𝒪(d3) basic operations. In the decoding examples we consider, the dimension of x is most often proportional to the temporal duration of the stimulus. Thus, naively, the one-time inversion of J and calculation of A takes 𝒪(T3) basic operations, where T is the duration of the stimulus, while the multiplication of x by A in each step of the MCMC algorithm takes 𝒪(T2) operations. This would make the decoding of stimuli with even moderate duration forbidding. Fortunately, the quasi-locality of the GLM model allows us to overcome this limitation. Since the filters Ki in the GLM have a finite temporal duration, Tk, the Hessian of the GLM log-likelihood Eqs. (4) is banded in time: the matrix element vanishes when |t1 − t2| ≥ 2Tk − 1. The Hessian of the log-posterior Eq. (9) is the sum of the Hessians of the log-prior and the log-likelihood, which in the Gaussian case is
| (21) |
where 𝒞 is the prior covariance (see Eq. (10)). Thus, if 𝒞−1 is also banded, J will be banded in time as well. As an example, Gaussian autoregressive processes of any finite order form a large class of priors which have banded C−1. In particular, for white-noise stimuli, C−1 is diagonal, and therefore J will have the same bandwidth as JLL. Efficient algorithms can find the Cholesky decomposition of a banded d×d matrix, with bandwidth nb, in a number of computations , instead of ∝ d3 (for example, the command chol in Matlab uses the 𝒪(d) method automatically if J is banded and is encoded as a sparse matrix). Likewise, if B is a banded matrix with bandwidth nb, the linear equation Bx = y can be solved for x in ∝ nbd computations. Therefore, to calculate x = Ax̃ from x̃ in each step of the Markov chain, we proceed as follows. Before starting the chain, we first calculate the Cholesky decomposition of J, such that J = BTB and x = Ax̃ = B−1x̃. Then, at each step of the MCMC, given x̃t, we find xt by solving the equation Bxt = x̃t. Since both of these procedures involve a number of computations that only scale with d (and thus with T), we can perform the whole MCMC decoding in 𝒪(T) computational time. This allows us to decode stimuli with durations on the order of many seconds. Similar methods with 𝒪(T) computational cost have been used previously in applications of MCMC to inference and estimation problems involving state-space models (Shephard and Pitt, 1997; Davis and Rodriguez-Yam, 2005; Jungbacker and Koopman, 2007), but these had not been generalized to non-state-space models (such as the GLM model we consider here) where the Hessian has a banded structure nevertheless. For a review of applications of state-space methods to neural data analysis see Paninski et al. (2010). That review also elucidates the close relationship between methods based on state-space models, and methods exploiting the bandedness of the Hessian matrix, as described here. Exploiting the bandedness of the Hessian matrix in the optimization problem of finding the MAP was discussed in Pillow et al. (2010).
3.2 Hybrid Monte Carlo and MALA
A more powerful method for constructing rapidly mixing chains is the so-called hybrid or Hamiltonian Monte Carlo (HMC) method. In a sense, HMC is at the opposite end of the spectrum with respect to RWM, in that it is designed to suppress the random walk nature of the chain by exploiting information about the local shape of π(x), via its gradient, to encourage steps towards regions of higher probability. This method was originally inspired by the equations of Hamiltonian dynamics for the molecules in a gas (Duane et al., 1987), but has since been used extensively in Bayesian settings (for its use in sampling from posteriors based on GLM see Ishwaran (1999); see also Neal (1996) for further applications and extensions).
This method starts with augmenting the vector x with an auxiliary vector of the same dimension z. Let us define the “potential energy” as ℰ(x) = −log π(x) up to a constant, and a “Hamiltonian function” by . Instead of sampling points, {xt}, from π(x), the HMC method constructs an MH chain that samples points, {(xt, zt)}, from the joint distribution . But since this distribution is factorized into the products of its marginals for x and z, the x-part of the obtained samples yield samples from π(x). On the other hand, sampling from the marginal over z is trivial, since z is normally distributed. In a generic step of the Markov chain, starting from (xt, zt), the HMC algorithm performs the following steps to generate (xt+1, zt+1). First, to construct the MH proposal.
Set x0:= xt, and sample z0 from the isotropic Gaussian distribution 𝒩d(0, 1).
-
Set (x, z) := (x0, z0), and evolve (x, z) according to the equations of Hamiltonian dynamics8 discretized based on the “leapfrog” method, by repeating the following steps, L times
x: = x + σz
Finally, to implement the MH acceptance step, Eq. (15),
with probability min {1, exp (−ΔH)}, where ΔH ≡ H(x, z) − H(x0, z0), accept the proposal x as xt+1. Otherwise reject it and set xt+1 = xt. (It can be shown that this is a bonafide Metropolis-Hastings rejection rule, ensuring that the resulting MCMC chain indeed has the desired equilibrium density (Duane et al., 1987).)
This chain has two parameters, L and σ, which can be chosen to maximize the mixing rate of the chain while minimizing the number of evaluations of ℰ(x) and its gradient. In practice, even a small L, requiring fewer gradient evaluations, often yields a rapidly mixing chain, and therefore in our simulations we used L ∈ {1, …, 5}. The special case of L = 1 corresponds to a chain that has proposals of the form
| (23) |
where z is normal with zero mean and identity covariance, and the proposal y is accepted according to the MH rule Eq. (15). In the limit σ → 0, this chain becomes a continuous Langevin process with the potential function ℰ(x) = −log π(x), whose stationary distribution is the Gibbs measure, π(x) = exp(−ℰ(x)), without the Metropolis-Hastings rejection step. For a finite σ, however, the Metropolis-Hastings acceptance step is necessary to guarantee that π(x) is the invariant distribution. The chain is thus refered to as the “Metropolis-adjusted Langevin” algorithm (MALA) (Roberts and Tweedie, 1996).
The scale parameter σ, which also needs to be adjusted for the RWM chain, sets the average size of the proposal jumps: we must typically choose this scale to be small enough to avoid jumping wildly into a region of low π(x), and therefore wasting the proposal, since it will be rejected with high probability. At the same time, we want to make the jumps as large as possible, on average, in order to improve the mixing time of the algorithm. See (Roberts and Rosenthal, 2001) and (Gelman, 2004) for some tips on how to find a good balance between these two competing desiderata for the RWM and MALA chains. For the HMC chains with L > 1, we chose σ, by trial and error, to obtain an MH acceptance rate of 60%–70%. We adopted this rule of thumb, based on a qualitative extrapolation of the results of (Roberts and Rosenthal, 1998) for the special cases of L = 0 and 1 (corresponding to the RWM and MALA chains, respectively), and their suggestion to tune the acceptance rate in those cases to ~25% and ~55%, respectively, for optimal mixing (for further discussion see Sec. 3.5; for a study on tuning the σ parameter for HMC with general L, see, e.g., (Kennedy et al., 1996)).
For highly non-isotropic distributions, the HMC chains can also be enhanced by exploiting the Laplace approximation (or its regularized version in the uniform prior case, as explained in the RWM case) by modifying the HMC proposals. Equivalently, as noted after Eq. (20), we can sample from the auxiliary distribution π̃(x̃) = |A|π (Ax̃) (where A is given in Eq. (20)) using the unmodified HMC chain, described above, and subsequently transforming the samples by A. As explained in the final paragraph of Sec. 3.1, we can perform this transformation efficiently in 𝒪(T) computational time, where T is the stimulus duration. Another practical advantage of this transformation by A is that the process of finding the appropriate scale parameter σ simplifies considerably, since π̃(x̃) may be approximated as a Gaussian distribution with identity covariance irrespective of the scaling of different dimensions in the original distribution π(x). To our knowledge, this 𝒪(T) enhancement of the HMC chain using the Laplace approximation is novel. This chain turned out to be the most efficient in most of the decoding examples we explored – we will discuss this in more detail in Sec. 3.5.
It is worth noting that when sampling from high-dimensional distributions with sharp gradients, the MALA, HMC, and RWM chains have a tendency to be trapped in “corners” where the log-posterior changes suddenly. This is because when the chain eventually ventures close to the corner, a jump proposal will very likely fall on the exterior side of the sharp high-dimensional corner (the probability of jumping to the interior side from the tip of a cone decreases exponentially with increasing dimensionality). Thus most proposals will be rejected, and the chain will effectively stop. As we will see below, the “hit-and-run” chain is known to have an advantage in escaping from such sharp corners (Lovasz and Vempala, 2004). We will discuss this point further in Sec. 3.4.
3.3 The Gibbs sampler
Gibbs sampling (Geman and Geman, 1984) is an important MCMC scheme. It is particularly efficient when, despite the complexity of the distribution π(x) = p(x|r, θ), its one-dimensional conditionals p(xm|x⊥m, r, θ) are easy to sample from. Here, xm is the m-th component of x, and x⊥m denotes the other components, i.e., the projection of x on the subspace orthogonal to the m-th axis. The Gibbs update is defined as follows: first choose the dimension m randomly or in order. Then update x along this dimension, i.e., sample xm from π(xm|x⊥m) (while leaving the other components fixed). This is equivalent to sampling a one-dimensional auxiliary variable, s, from
| (24) |
and setting y = x+sem, where em is the unit vector along the m-th axis (we will discuss how to sample from this one-dimensional distribution in Sec. 3.4). It is well-known that the Gibbs rule is indeed a special case of the MH algorithm where the proposals, Eq. (24), is always accepted. For applications of the Gibbs algorithm for sampling from posterior distributions involving GLM-like likelihoods see Chan and Ledolter (1995); Gamerman (1997, 1998); see also Smith et al. (2007) for some related applications in neural data analysis (discussed further below in section 5.1).
It is important to note that the Gibbs update rule can sometimes fail to lead to an ergodic chain, i.e., the chain can get “stuck” and not sample from π(x) properly (Robert and Casella, 2005). An extreme case of this is when the conditional distributions pm(xm|x⊥m, r, θ) are deterministic. then the Gibbs algorithm will never move, clearly breaking the ergodicity of the chain. More generally, in cases where strong correlations between the components of x lead to nearly deterministic conditionals, the mixing rate of the Gibbs method can be extremely low (panel (a) of Fig. 3, shows this phenomenon for a 2-dimensional distribution with strong correlation between the two components). Thus, it is a good idea to choose the parameterization of the model carefully before blindly applying the Gibbs algorithm. For example, we can change the basis, or more systematically, exploit the Laplace approximation, as described above, to sample from the auxiliary distribution π̃(x̃) instead.
Figure 3.
Comparison of different MCMC algorithms in sampling from a non-isotropic truncated Gaussian distribution. This distribution can arise as a posterior distribution resulting from a non-isotropic Gaussian likelihood and a uniform prior with square boundaries (at the frame borders). Panels (a–c) show 50-sample chains for a Gibbs, isotropic hit-and-run, and isotropic random walk Metropolis (RWM) samplers, respectively. The grayscale indicates the height of the probability density. As seen in panel (a), the narrow, non-isotropic likelihood can significantly hamper the mixing of the Gibbs chain as it chooses its jump directions unfavorably. The hit-and-run chain, on the other hand, mixes much faster as it samples the direction randomly and hence can move within the narrow high likelihood region with relative ease. The mixing of the RWM chain is relatively slower due to its rejections (note that there are fewer than 50 distinct dots in panel (c) due to rejections; the acceptance rate was about 0.4 here). For illustrative purposes, the hit-and-run direction and the RWM proposal distributions were taken to be isotropic here, which is disadvantageous, as explained in the text (also see Fig. 2).
3.4 The hit-and-run algorithm
The hit-and-run algorithm (Boneh and Golan, 1979; Smith, 1980; Lovasz and Vempala, 2004) can be thought of as “random-direction Gibbs”: in each step of the hit-and-run algorithm, instead of updating x along one of the coordinate axes, we update it along a random general direction not necessarily parallel to any coordinate axis. More precisely, the sampler is defined in two steps: first, choose a direction n from some positive density ρ(n) (with respect to the normalized Lebesgue measure) on the unit sphere nTn = 1. Then, similar to Gibbs, sample the new point on the line defined by n and x, with a density proportional to the underlying distribution. That is sample s from
| (25) |
and set y = x + sn.9 Even though the hit-and-run chain is well known in the statistics literature, it has not been used in neural decoding.
The main gain over RWM or HMC is that instead of taking small local steps (of size proportional to σ, in eq. 18 or 23)), we may take very large jumps in the n direction; the jump size is set by the underlying distribution itself, not an arbitrary scale, σ, which has to be tuned by the user to achieve optimal efficiency. there is no jump scale to be set by the user, and the jump size in a given direction, n, is set by the scale of the “slice” distribution Eq. (25).
This, together with the fact that all hit-and-run proposals are accepted, makes the chain better at escaping from sharp high-dimensional corners (see (Lovasz and Vempala, 2004) and the discussion at the end of Sec. 3.2 above). The advantage over Gibbs is in situations such as depicted in Fig. 2, where jumps parallel to coordinates lead to small steps but there are directions that allow long jumps to be made by hit-and-run. The price to pay for these possibly long nonlocal jumps, however, is that now (as well as in the Gibbs case) we need to sample from the one-dimensional density , which is in general non-trivial. Fortunately, as we mentioned above (see the discussion leading to Eqs. (5)–(7) and following it), in the case of neurons modeled by the GLM, the posterior distribution and thus all its “slices” are log-concave, and efficient methods such as adaptive rejection sampling (ARS) (Gilks, 1992; Gilks and Wild, 1992) can be used to sample from the one-dimensional slice in the hit-and-run step. Let us emphasize, however, that the hit-and-run algorithm, by itself, does not require the distribution π(x) to be log-concave. Given a method other than ARS for sampling from the one-dimensional conditional distributions, π(x + sn), hit-and-run can be applied to general distributions that are not log-concave, as well.
Regarding the direction density, ρ(n), the easiest choice is the isotropic ρ(n) = 1. More generally it is easy to sample from ellipses, by sampling from the appropriate Gaussian distribution and normalizing. Thus, again, a reasonable approach is to exploit the Laplace approximation: we sample n by sampling an auxiliary point x̃ from 𝒩(0, J−1), where J is the Hessian, Eq. (9), and setting n = x̃/||x̃|| (see Fig. 2). This prescription is equivalent to sampling n from the distribution , which is referred to as the angular central Gaussian distribution in the statistical literature (see e.g., (Tyler, 1987)). This adds to hit-and-run’s advantage over Gibbs by giving more weight to directions that allow larger jumps to be made.
3.5 Comparison of different MCMC chains
Above, we pointed out some qualitative reasons behind the strengths and weaknesses of the different MCMC algorithms, in terms of their mixing rates and computational costs. Here we give a more quantitative account, and also compare the different methods based on their performance in the neural decoding setting.
From a practical point of view, the most relevant notion of mixing is how fast the estimate of Eq. (13) converges to the true expectation of the quantity of interest, f. As one always has access to finitely many samples, N, even in the optimal case of i.i.d. samples from π, has a finite random error, Eq. (14). For the correlated samples of the MCMC chain, and for large N, the error is larger, and Eq. (14) generalizes to (see (Kipnis and Varadhan, 1986))
| (26) |
for N ≥ τcorr, independent of the starting point.10 Here, τcorr is the equilibrium autocorrelation time of the scalar process g(xi), based on the chain xi. It is defined by
| (27) |
where we refer to γt as the lag-t autocorrelation for g(x). Thus the smaller the τcorr, the more efficient is the MCMC algorithm, as one can run a shorter chain to achieve a desired estimated error.
Another measure of mixing speed which has the merit of being more amenable to analytical treatment is the mean squared jump size of the Markov chain
| (28) |
this has been termed the first-order efficiency (FOE) by (Roberts and Rosenthal, 1998). Let us define to be the lag-1 autocorrelation of the m-th component of x, xm. From the definition Eq. (28), it follows that the weighted average of over all components (with weights Var[xm]), is given by . Thus maximizing the FOE is roughly equivalent to minimizing correlations. One analytical result concerning the mixing performance of different MCMC chains was obtained in (Roberts and Rosenthal, 1998) for the FOE of RWM and MALA when sampling from the restricted class of product distributions , and asymptotically large dimension d = dim(x) (often a relevant limit in neural decoding). Based on their results, the authors also argue that in general, the jump scales of RWM and MALA proposals may be chosen such that their acceptance rates are roughly 0.25 and 0.55, respectively. For the special case of sampling from a d dimensional standard Gaussian distribution, π(x) ∝ exp (−||x||2/2), and for optimally chosen proposal jump scales they show that the FOE of Gaussian MALA and RWM are asymptotically equal to 1.6d2/3 and 1.33, respectively.
To enable a comparison with hit-and-run, we can calculate its FOE directly. Using y = x + sn, with s sampled as in Eq. (25), we see that
| (29) |
Now, from Eq. (25), , and using E (s2|n, x) = E (s|n, x)2 + Var (s|n, x), we obtain E (s2|n, x) = (n · x)2 + 1. Thus
| (30) |
| (31) |
where we used Eπ(xnxm) = δnm for the standard Gaussian distribution 𝒩d(0, 1), and n · n = 1. Therefore, while hit-and-run has higher FOE than RWM in this case, we see that for unimodal, nearly Gaussian distributions, MALA will mix much faster (by a factor ∝ d2/3) than both RWM and hit-and-run in large dimensions. Although we know of no such result for general HMC chains with higher-order leapfrog steps than the onestep MALA algorithm, we expect their mixing speed to increase even further for higher leapfrog steps. The superiority of HMC over the other chains is clearly visible in panel (a) of Fig. 4, which shows a plot of the estimated autocorrelation function γt for the sampling of the three chains from the GLM posterior with standard Gaussian priors, and a weak stimulus filter leading to a weak likelihood. More generally, in our simulations with Gaussian priors and smooth GLM nonlinearities, HMC (including MALA) had an order of magnitude advantage over the other chains for most of the relevant parameter ranges. Thus we used this chain in Sec. 5.2 for evaluating the mutual information with Gaussian priors.
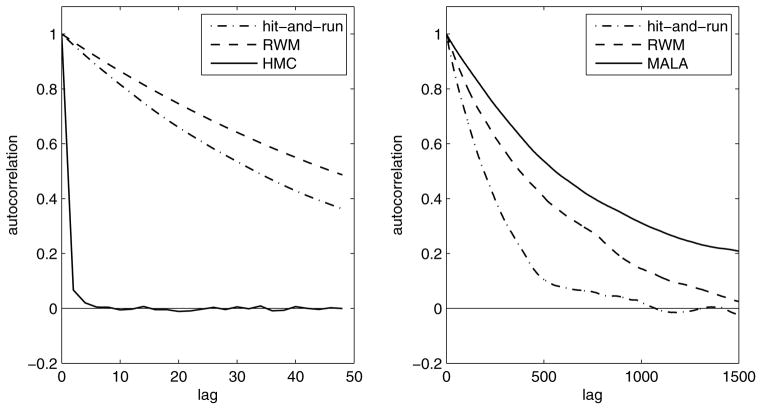
Figure 4.
The estimated autocorrelation function for the hit-and-run, Gaussian random-walk metropolis, and HMC chains, based on 7 separate chains in each case. The chains were sampling from a posterior distribution over a 50-dimensional stimulus (x) space with white noise Gaussian (a) and uniform (b) priors with contrast c = 1 (see Eqs. (10)–(11)), and with GLM likelihood (see Eqs. (3)–(4)) based on the response of two simulated ganglion cells. The GLM nonlinearity was exponential and the stimulus filters ki(t) were taken to be weak “delta functions” with heights ±0.1. For the HMC, we used L = 5 leapfrog steps in the Gaussian prior case, and L = 1 steps (corresponding to MALA) in the at prior case. The autocorrelation was calculated for a certain one-dimensional projection of x. In general, in the Gaussian prior case, HMC was superior by an order of magnitude. For uniform priors, however, hit-and-run was seen to mix faster than the other two chains over a wide range of parameters such as the stimulus filter strength (unless the filter was strong enough so that the likelihood determined the shape of the posterior, confining its effective support away from the edges of the at prior). This is mainly because hit-and-run is better in escaping from the sharp, high-dimensional corners of the prior support 𝒮. Here, MALA need not be slower than RWM, and its larger autocorrelation in the plot is because its jump size was chosen suboptimally, according to a rule (Roberts and Rosenthal, 1998) that is optimal only for smooth distributions. For both priors, using non-isotropic proposal or direction distributions improved the mixing of all three chains.
The situation can be very different, however, for highly non-Gaussian (but still log-concave) distributions, such as those with sharp boundaries. In our GLM setting this can be the case with flat priors on convex sets, Eq. (11), when the likelihood is broad and does not restrict the posterior support away from the boundaries and corners of the prior support 𝒮. In this case, HMC and MALA lose their advantage because they do not take advantage of the information in the prior distribution, which has zero gradient within its support. Furthermore, as mentioned in Secs. 3.2 and 3.4, when the convex body 𝒮 has sharp corners, hit-and-run will have an advantage over both RWM and HMC in avoiding getting trapped in those corners, which can otherwise considerably slow down the chain in large dimensionality (see the arguments in (Lovasz and Vempala, 2004)). Finally, we mention that the MALA or HMC proposals can in principle be inefficient in regions of sharp gradient changes; for example, in the GLM setting, if the nonlinearity f(.) is very sharp, then the log-likelihood might vary much more quickly than quadratic. In such cases the HMC proposal jumps can be too large, falling in regions where π(x) is very low and leading to high rejection rates. This can potentially reduce HMC’s advantage significantly even in case that the prior is Gaussian. However, in our experience, with f(.) = exp(.), this did not occur.
Figure 4, panel (b), shows the estimated autocorrelation function for different chains in sampling from the posterior distribution in GLM-based decoding with a flat stimulus prior distribution, Eq. (11), with cubic support.11 For this prior, the correlation time of the hit-and-run chain was consistently lower than those of the RWM, MALA, and Gibbs (not shown in the figure) chains, unless the likelihood was sharp and concentrated away from the boundaries of the prior cube. As we mentioned above (also see the next section), the Laplace approximation is adequate in this latter case. Thus we see that hit-and-run is the faster chain when this approximation fails, which is also the case where MCMC is more indispensable. We thus used the hit-and-run algorithm in our decoding examples for the flat prior case presented in the next section.
Finally, we note that other methods of diagnosing mixing and convergence, such as the so-called r-hat (R̂) statistic (Brooks and Gelman, 1998) gave consistent results with those based on the autocorrelation time, τcorr, presented here.
4 Comparison of MAP and Monte Carlo decoding
In this section we compare Bayesian stimulus decoding using the MAP and the posterior mean estimates, Eqs. (7) and (12), based on the response of a population of neurons modeled via the GLM introduced in section 2. We will show that in the flat prior case, Eq. (11), the MAP estimate, in terms of its mean squared error, is much less efficient than the posterior mean estimate. We contrast this with the Gaussian prior case, where the Laplace approximation is accurate in a large range of model parameters, and thus the two estimates are close. Furthermore, for both kinds of priors, in the limit of strong likelihoods (e.g., due to a strong stimulus filter or a large number of neurons) the posterior distribution will be sharply concentrated, the Laplace approximation becomes asymptotically more and more accurate, and both estimates will eventually converge to the true stimulus (more precisely the part of the stimulus that is not outside the receptive field of all the neurons; see footnote 13, below).
In the first two examples (Figs. 5–6), the stimulus estimates were computed given the simulated spike trains of a population of pairs of ON and OFF retinal ganglion cells (RGC), in response to a spatially uniform, full-field fluctuating light intensity signal. The stimuli were discretized white-noise with Gaussian and flat distributions (see the paragraph after Eq. (11)). Spike responses were generated by simulating the GLM point process encoding model, described by Eqs. (3)–(4), with exponential nonlinearity, f(u) = exp (u). The coupling between different cells (ℋij of Eq. (3) for i ≠ j) were set equal to zero, but the diagonal kernels, ℋii, representing the spike history feedback of each cell to itself were closely matched to those found with fits to macaque ON and OFF RGC’s reported in Pillow et al. (2008), and so were the DC biases, bi; the value of the DC biases were such that the baseline firing rate, exp (bi), in the absence of stimulus was approximately 7 Hz (see the appendix of Pillow et al. (2010) for a more detailed description of the fits for stimulus and spike history filters). However, for demonstration purposes, the stimulus filters, Ki, were set to positive and negative delta functions (for ON and OFF cells, respectively), resulting in Ki · x being proportional to the light stimulus, x(t), so that band-pass filtering of the stimulus did not result in information loss, and convergence of the estimates to the true stimulus could be observed more easily. For a fixed number of cells, the parameter of relevance here, which determines the signal to noise ratio of the RGCs’ spike trains, is the strength of the filtered stimulus input, Ki · x, to the GLM nonlinearity. The magnitude of this input is proportional to c||k||, where c is the stimulus contrast, and ||k|| is the norm of the receptive field filter (which we have taken to be the same for all cells in this example). Figure 5 shows the stimulus, the spike trains, and the two estimates for three different magnitudes of c||k||, based on the response of one pair of ON and OFF cells. Figure 6 shows the same based on the response of ten identical pairs of RGCs.
Figure 5.
Comparison of MAP and posterior mean estimates, for a pair of ON and OFF RGC’s (see the main text), for different values of the stimulus filter amplitude (||k|| = 0.5, 1, and 2.4 from left to right) and contrast c = 1 (defined after Eq. (11) – the product c||k|| represents the scale of the filtered stimulus input term to the GLM nonlinearity (see the main text for the full description of the GLM parameters used in this simulation). The stimulus (black traces in the first row panels and dotted traces in other rows) consists of a 500 ms interval of uniformly distributed white noise, refreshed every 10 ms. Thus the stimulus space is 50 dimensional. The dashed horizontal lines mark the boundaries of the flat prior distribution of the stimulus intensity on each 10 ms subinterval. They are set at , corresponding to intensity variance of 1 and zero mean. Dots on the top row show the spikes of the ON (gray) and the OFF (black) cell. The solid traces in the middle row are the MAP estimates, and the solid traces in the bottom rows show the posterior means estimated from 10000 samples of a hit-and-run chain (after burning 2500 samples). The shaded regions in the second and third rows are error bars showing the estimated marginal posterior uncertainties about the stimulus value. For the MAP (second rows), these are calculated as the square root of the diagonal of the inverse Hessian, J−1, but they have been cut-off where they would have encroached on the zero prior region beyond the horizontal dashed lines. For the posterior mean (third rows), the error bars represent one standard deviation about the mean, and are calculated as the square root of the diagonal of the covariance matrix, which is itself estimated from the MCMC chain (the standard error of the posterior mean estimate due to the finite sample size of the MCMC were much smaller than these error bars, and are not shown). Note that the errorbars of the mean are in general smaller than those for the MAP, and that all estimate uncertainties decrease as the stimulus filter amplitude grows.
Figure 6.
Comparison of MAP and posterior mean estimates, for 10 identical and independent pairs of ON and OFF RGC’s, for different values of the stimulus filter. The stimulus and all GLM parameters are the same as in Fig. 5, except for the number of pairs of RGC. The increase in the number of cells leads to the sharpening of the likelihood, leading to smaller error bars on the estimates, and a more accurate Laplace approximate and smaller disparity between the two estimates. Here a 20000 sample long MALA chain (after burning 5000 samples) was used to estimate the posterior mean.
Because the prior distribution here is flat on the 50-dimensional cube centered at the origin, the Laplace approximation, Eq. (8), will be justified only when the likelihood is sharp and supported away from the edges of the cube.12 Moreover, since the flat prior is only “felt” on the boundaries of the cube (the horizontal dashed lines in Figs. 5–6), the MAP will lie in the interior of the cube only if the likelihood has a maximum there. For filtered stimulus inputs with small magnitude, c||k||, the log-likelihood, Eqs. (3)–(4), becomes approximately linear in the components of x. With a flat prior, the almost linear log-posterior will very likely be maximized only on the boundaries of the cube (since linear functions on convex domains attain their maxima at the “corners” of the domain). Thus in the absence of a strong, confining likelihood, the MAP has a tendency to stick to the boundaries, as seen in the first two columns of Fig. 5; in other words, the MAP falls on a corner of the cube, where the Laplace approximation is worst and where MALA and RWM are least efficient. We note that the likelihood will be further weakened in fact, if we replace the delta function stimulus filters with more realistic filters, as the band-pass filtering will remove the dependence of the likelihood on the features of the stimulus that were filtered out – c.f. a similar discussion in our companion paper on MAP decoding (Pillow et al., 2010).
On the other hand, a sharp likelihood confines the posterior away from the boundaries of the prior support, and solely determines the position of both the MAP and the posterior mean. In this case the Gaussian approximation for the posterior distribution is valid and the two estimates will in fact be very close (as the mean and the mode of a Gaussian are one and the same). This can be seen in the right column of Fig. 5, where the large value of the stimulus filter has sharpened the likelihood. Also, as is generally true in statistical parameter estimation, when the number of data points becomes large the likelihood term gets very sharp, leading to accurate estimates.13 In our case this corresponds to increasing the number of cells with similar receptive fields, leading to the smaller error bars in Fig. 6 and the more accurate and closer MAP and mean estimates.
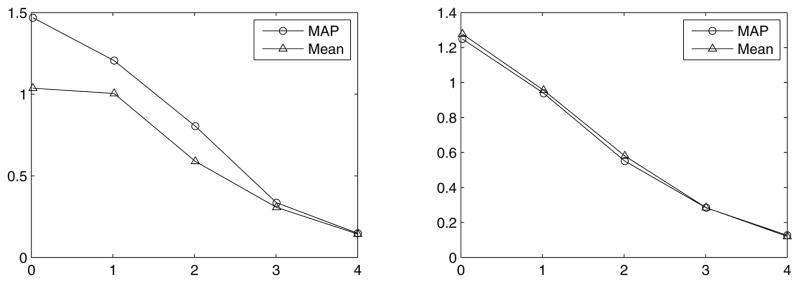
To compare the performance of the two estimates more quantitatively, in Fig. 7, we have plotted the average squared errors of the two estimates under the full stimulus-response distribution, p(x, r) (for the same type of stimulus and cell pair as in the Fig. 5 simulations), as function of the magnitude of the filtered stimulus input, c||k||. This was done by generating 5 samples of the stimulus in each case, and then simulating the GLM to generate the spike train response of the pair of ON and OFF cells to each stimulus, leading to sample pairs (xi, ri) for i = 1, · · ·, 5. For each of the responses, ri, the MAP and MCMC mean were computed based on the posteriors p(x|ri). The average (over p(x, r)) square error, 〈||x̂(r) − x||2〉, was then approximated by its sample mean, . The left and right panels in Fig. 7 show plots of the squared error per dimension, for MAP and mean estimates, as a function of the stimulus filter strength for the case of the flat and Gassian white-noise stimulus ensembles, respectively. As is evident from the plots, in the former ensemble, the MAP is inferior to the mean, due to its higher mean squared error, unless the filter strength is large. For the Gaussian ensemble, the plot shows that the error of the MAP and posterior mean estimates are very close, throughout the range of stimulus filter strength. Thus, due to its much lower computational cost, the MAP-based decoding method of (Pillow et al., 2010) is superior for this prior. Let us mention that the magnitude of the filtered stimulus, c||k||, in the experimental data reported in Pillow et al. (2008) (which is also the basis of the final example in this section – see Fig. 8) was in the range 3 ± 1, depending on the cell; smaller values of c||k|| can be achieved experimentally by lowering the contrast of the visual stimulus as needed. Thus the values of this parameter used in Fig. 7, as well as in Figs. 5–6, are on the same order of magnitude as those used in that experiment, and cover a range of values that is experimentally and biologically relevant.
Figure 7.
Comparison of mean squared error (〈||x̂ − x||2〉 /d) of MAP and posterior mean estimates for uniform (left panel) and Gaussian (right panel) white-noise stimulus distributions as a function of the stimulus filter strength times contrast. In the left panel, the data points at ||k|| = 0 were obtained for very small but non-zero ||k||. As seen here, for flat priors, MAP has a higher average squared error than the posterior mean, except for large values of the stimulus filter where both estimates converge to the true value. For Gaussian priors, on the other hand, the Laplace approximation is accurate and therefore the posterior mean and MAP are very close. Thus their efficiency (e.g., as measured by the inverse of their mean squared error) is very similar even for small values of the stimulus filter, and the fact that the computational cost of calculating MAP is much lower makes it the preferable estimate here.
Figure 8.
The top six panels show the posterior mean (solid curve) the MAP (dashed curve) estimates of the stimulus input to 3 pairs of ON and OFF RGC’s given their spike trains from multi-electrode array recordings. The GLM parameters used in this example were fit to data from the same recordings – see Pillow et al. (2008) for the full description of the fit GLM parameters. The jagged black traces are the actual inputs. The bottom panel shows the recorded spike trains. The posterior means were estimated using an HMC chain with 15000 samples (after an initial 3750 samples were burnt). The gray error-bars around the blue curve are represent its marginal standard deviations which were estimated using the MCMC itself (the error-bars for the MAP, e.g. based on the Hessian, would not be distinguishable in this figure, and are not shown). The closeness of the posterior mean to the MAP is an indication of the accuracy of the Laplace approximation. (This decoding example also appeared briefly in Paninski et al. (2010); see also Pillow et al. (2010))
Finally, we compared the MAP and posterior mean estimates in decoding of experimentally recorded spike trains. The spike trains were recorded from a group of 11 ON and 16 OFF RGCs (whose receptive fields fully cover a patch of the visual field) in response to the light signal of the optically reduced image of a cathode ray display which refreshes at 120 Hz, and is projected on the retina (Litke et al., 2004; Shlens et al., 2006). The stimlulus, x, in this case, is a spatiotemporally fluctuating binary white-noise, with x(t, n) representing the contrast of the pixel n at time t. In Pillow et al. (2008), 20 minutes of this data were used to fit the GLM model parameters including cross-couplings, hij, to these cells – see that reference for details about the recording and the fitting method, and a full description of the fit GLM parameters. Here, we took a 500 ms portion of the recorded spike trains of 6 neighboring RGCs (3 ON and 3 OFF), and using the fit GLM parameters for them, decoded the filtered inputs,
| (32) |
to these cells using the MAP and posterior mean (calculated using an HMC chain). The inputs are a priori correlated due to the overlaps between the cell’s receptive fields, and the covariance matrix of the yi is given by , where 𝒞x = c21 is the covariance of the white-noise visual stimulus. More explicitly
| (33) |
Notice that with the experimentally fit ki, which have a finite temporal duration Tk, the covariance matrix, 𝒞y is banded: it vanishes when |t1 − t2| ≥ 2Tk − 1. Since x is binary, yi is not a Gaussian vector. However, because the filters Ki(t, n) have a relatively large spatiotemporal dimension, yi(t) are weighted sums of many independent identically distributed binary random variables, and their prior marginal distributions can be well approximated by Gaussian distributions. For this reason, and because the likelihood was relatively strong for this data (and hence the dependence on the prior relatively weak), we replaced the true (highly non-Gaussian) joint prior distribution of yi with a Gaussian distribution with zero mean and covariance Eq. (33). This allowed us to implement the efficient non-isotropic HMC chain, described above, so that its computational cost scales only linearly with the stimulus duration T, allowing us to decode very long stimuli. However, in this case the details of the procedure explained in the final paragraph of Sec. 3.1 have to be modified as follows. The Hessian for y is given by
| (34) |
where the Hessian of the negative log-likelihood term, , is now diagonal, because yi(t) affects the conditional firing rate instantaneously (see Eq. (3)). Let , similar to Eq. (20). The non-isotropic chain requires the calculation of Aỹ for some vector ỹ at each step of the MCMC. In order to carry this out in 𝒪(T) computational time, we proceed as follows. First we calculate the Cholesky decomposition, L, of 𝒞y, satisfying LLT = 𝒞y. As mentioned in Sec. 3.1, since 𝒞y is banded this can be performed in 𝒪(T) operations. Then we can rewrite Eq. (34) as
| (35) |
Since L is banded (due to the bandedness of 𝒞y) and is diagonal, it follows that Q is also banded. Therefore its Cholesky decomposition, B, satisfying BTB = Q, can be calculated in 𝒪(T) time, and is also banded. Using this definition and inverting Eq. (35), we obtain , from which we deduce A = LB−1, or
| (36) |
The calculation of L and B can be performed before running the HMC chain. Then at each step we need to perform Eq. (36). As described in the final paragraph of Sec. 3.1, calculating B−1ỹ and the multiplication of the resulting vector by L, both require only 𝒪(T) elementary operations due to the bandedness of B and L.
Figure 8 shows the spike trains, as well as the corresponding true inputs and MAP and posterior mean estimates. The closeness of the posterior mean to the MAP (the L2 norm of their difference is only about 9% of the L2 norm of the MAP) is an indication of the accuracy of the Laplace approximation in this case.
5 Other applications: estimation of non-marginal quantitites
So far we focused on using the MCMC samples to estimate E(x|r) or the posterior covariance. Both of these quantities involve separate averaging over the marginal distribution of single components or pairs of components of x. However, since MCMC provides samples from the joint distribution p(x|r), we can also calculate quantities that cannot be reduced to averages over one or two dimensional marginal distributions, and involve the whole joint distribution p(x|r). We consider two examples below.
5.1 Posterior statistics of crossing times
One important example of these non-marginal computations involves the statistics (e.g., mean and variance) of some crossing time for the time series x, e.g., the time that xt first crosses some threshold value. (First-passage time computations are especially important, for example, in the context of integrate-and-fire-based neural encoding models Paninski et al. (2008).) In Smith et al. (2004), the authors proposed a hidden state-space model that provides a dynamical description for the learning process of an animal in a task learning experiment (with binary responses), and yields suitable statistical indicators for establishing the occurrence of learning or determining the “learning trial.” In the proposed model, the state variable, xt, evolves according to a Gaussian random walk from trial to trial (labeled by t), and the probability of a correct response on every trial, qt, is given by a logistic function of the corresponding state variable, xt. Given the observation of the responses in all trials, the hidden state variable trajectory can be inferred. In Smith et al. (2007), the authors carried out this inference in Bayesian fashion by using Gibbs sampling from the posterior distribution over the state variable time-series and the model parameters conditioned on the observed responses. There, the learning trial was defined as the first trial after which the ideal (Bayesian) observer can state with 95% confidence that the animal will perform better than chance. More mathematically, using the MCMC samples (using the winBUGS package), they obtained the sequence of the lower 95% confidence bounds for qt for all t’s (for each t, this bound depends only on the one-dimensional marginal distribution of qt). The authors defined the learning trial as the t for which the value of this lower confidence bound crosses the probability value corresponding to chance performance, and stays above it in all the following trials.
However, it is reasonable to consider several alternative definitions of the “learning trial” in this setting. One plausible approach is to define the learning trial, tL, in terms of certain passage times of qt, e.g., the trial in which qt first exceeds the chance level and does not become smaller than this value at later trials. In this definition, tL is a random variable whose value is not known by the ideal observer with certainty, and its statistics is determined by the full joint posterior distribution and can not be obtained from its marginals. The posterior mean of tL provides an estimate for this quantity, and its posterior variance, an estimate of its uncertainty. These quantities involve nonlinear expectations over the full joint posterior distribution of {xt}, and can be estimated by the MCMC samples from that distribution.
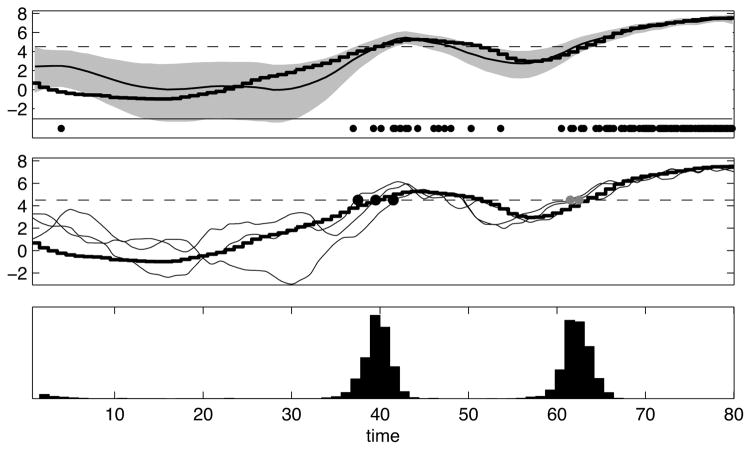
Figure 9 shows a simulated example in which we used our MCMC methods to decode the crossing times of the input to a Poisson neuron, based on the observation of its spike train. The neuron’s rate was given by λ(t) = exp(xt + b) and the threshold corresponded to a value xt = x0. The hidden process xt was assumed to evolve according to a Gaussian AR(1) process, as in Smith et al. (2007). Having observed a spike train, samples from the posterior distribution of xt were obtained by an HMC chain. To estimate the first and the last times that xt crosses x0 from below, we calculate these times for each MCMC sample, obtaining samples from the posterior distribution of these times. Then we calculate their sample mean to estimate when learning occurs. Fig. 9 shows the full histograms of these passage times, emphasizing that these statistics are not fully determined by a single observation of the spike train.
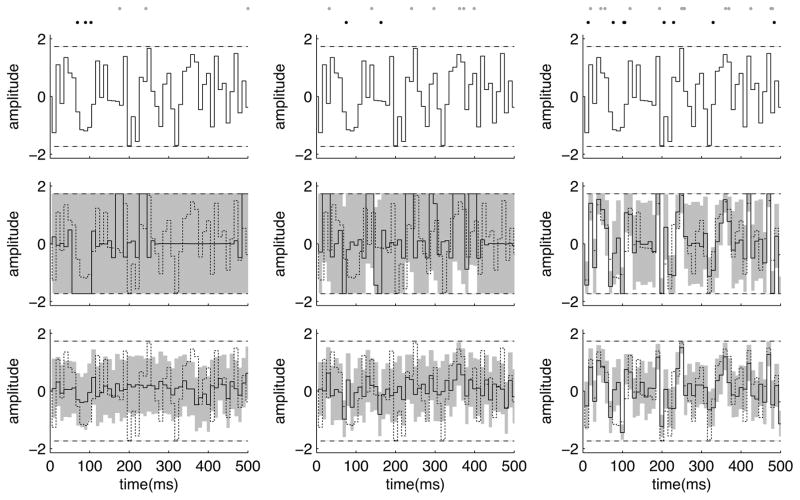
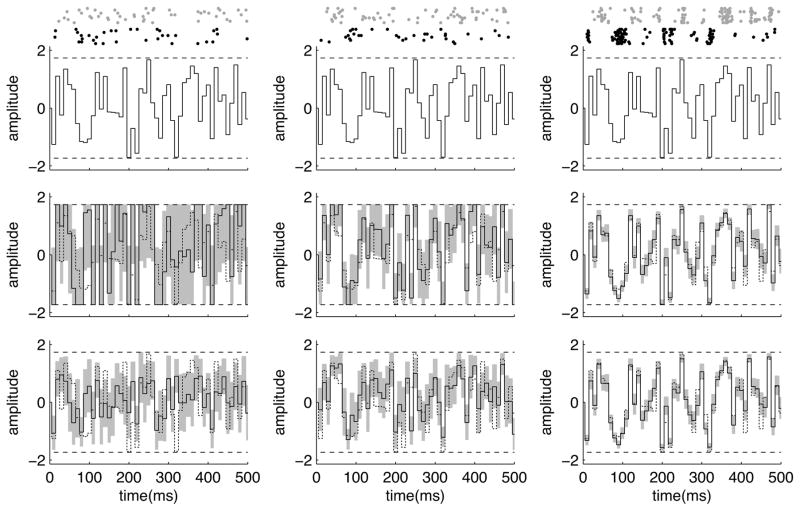
Figure 9.
Estimation of threshold crossing times using MCMC sampling. The spike trains are generated by an inhomogenous Poisson process with a rate λ(t) = exp(xt + b) that depends on a changing hidden variable xt (times are in arbitrary units). Having observed a particular spike train (bottom row of the top panel), the goal is to estimate the first or the last time that xt crosses a threshold from below. The top and the middle plots show the true xt (black jagged lines) and the threshold (the dashed horizontal lines). The top plot also shows the posterior marginal median for xt (curvy line) given the observed spike train, and its corresponding posterior marginal 90% confidence interval (shaded area). In Smith et al. (2007), these marginal statistics were used to estimate the crossing times. However, a more systematic way of estimating these times is to directly use their (non-marginal) posterior statistics. The middle plot also shows three posterior samples of xt (curvy lines) obtained using an HMC Markov chain. The first and last crossing times are well-defined for these three curves, and are marked by black and gray dots, respectively. For each MCMC sample curve, we calculated these crossing times, and then we tabulated the statistics of these times across all samples. The bottom panel shows the MCMC-based posterior histograms of these crossing times thus obtained. The two separated peaks corresponds to the first and the last crossing times. The posterior mean and variance of the crossing times can then be calculated from these histograms.
As a side note, to obtain a comparison between the performance of the Gibbs-based winBUGS package employed in Smith et al. (2007) versus the HMC chain used here, we simulated a Gibbs chain for y(t) on the same posterior distribution. The estimated correlation time of the Gibbs chain was ≈ 130 — i.e., Gibbs mixes a hundred times slower than the HMC chain here, due to the nonnegligible temporal correlations in xt (Fig. 9); recall Fig. 3. In addition, due to the state-space nature of the prior on xt here, the Hessian of the log-posterior on x is tridiagonal, and therefore the HMC update requires just 𝒪(T) time, just like a full Gibbs sweep.
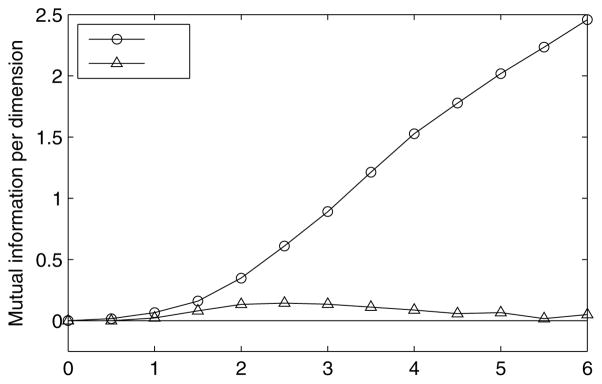
5.2 Mutual Information
Our second example is the calculation of the mutual information. Estimates of information transfer rates of neural systems, and the mutual information between the stimulus and response of some neural population, are essential in the study of the neural encoding and decoding problems (Bialek et al., 1991; Warland et al., 1997; Barbieri et al., 2004). Estimating this quantity is known to be often computationally quite difficult, particularly for high-dimensional stimuli and responses (Paninski, 2003). In (Pillow et al., 2010), the authors presented an easy and efficient method for calculating the mutual information for neurons modeled by the GLM, Eqs. (3)–(4), based on the Laplace approximation Eq. (8). As discussed above, this approximation is expected to hold in the case of Gaussian priors, in a broad region of the GLM parameter space. Our goal here is to verify this intuition, by comparing the Laplace approximation for the mutual information with an exact direct estimation using MCMC integration. As we will see, the main difficulty in using MCMC to estimate the mutual information lies in the fact that we can only calculate p(x|r) up to an unknown normalization constant. Estimating this unknown constant turns out to be tricky, in that naive methods for calculating it lead to large sampling errors. Below, we use an efficient, low error method, known as bridge sampling, for estimating this constant.
The mutual information is by definition equal to the average reduction in the uncertainty regarding the stimulus (i.e., the entropy, H, of the distribution over the stimulus) of an ideal observer having access to the spike trains of the RGC, from its prior state of knowledge about the stimulus:
| (37) |
Here, p(r) is given by Eq. (6), and the posterior probability p(x|r) is given by Bayes’ rule Eq. (5). The logarithms are assumed to be in base 2, so that information is measured in bits. We consider Gaussian priors given by Eq. (10), for which we can compute the entropy H[x] explicitly,
| (38) |
Thus the real problem is to evaluate the second term in Eq. (37). The integral involved in the definition of H[x|r] is in general hard to evaluate. One approach which is computationally very fast, is to use the Laplace approximation, Eq. (8), if it is justified – we took this approach in Pillow et al. (2010). In that case, from Eq. (8), we obtain
| (39) |
where J(r) is the Hessian Eq. (9).
More generally, we can use the MCMC method developed in Sec. 3 to estimate H[x|r] directly. The integral involved in H[x|r], Eq. (37), (before averaging over p(r)) has the form
| (40) |
i.e., one representing the posterior expectation of a function g(x). If we could evaluate g(x) for arbitrary x, we could evaluate this expectation by the MCMC method, via Eq. (13). As we mentioned above, however, the difficulty lies in that in general we can only evaluate an unnormalized version of the posterior distribution, and thus g(x) = −log p(x|r), only up to an additive constant. Suppose we can evaluate
| (41) |
for some Z(r) at any arbitrary x. Then H[x|r] can be rewritten as
| (42) |
From the normalization condition for p(x|r), Z(r) is given by
| (43) |
The main difficulty in calculating the mutual information lies in estimating Z(r); for a discussion of the difficulties involved in estimating normalization constants and marginal probabilities, see Meng and Wong (1996), the discussion of the paper by Newton and Raftery (1994), and Neal (2008). By contrast, the first term in Eq. (42) already has the form Eq. (40) (with q(x|r) replacing g(x)) and can be estimated using Eq. (13). In the following we introduce an efficient method for estimating Z(r) and I(r).
As noted above, if in Eqs. (42)–(43), we replace q(x|r) with the Laplace approximation
| (44) |
we obtain the result Eq. (39), as a first approximation to the mutual information. Here we defined
| (45) |
and from the normalization condition for pL(x|r) we must have
| (46) |
We included the constant ℒ0 in the exponent in Eq. (44) so that when the Laplace approximation is accurate we have log qL(x|r) ≈ log q(x|r), and log Z(r) ≈ log ZL(r).
We now write the exact mutual information as I[x; r] = 〈I(r)〉p(r), and write I(r) ≡ H[x] − H[x|r] as the Laplace approximation for it plus a difference
| (47) |
where
| (48) |
Using the general formula (42) both for the true distribution, Eq. (41), and its Gaussian approximation, Eq. (44), we obtain
| (49) |
with η ≡ Z(r)/ZL(r). Thus, after averaging over p(r), δI(r), calculated using Eq. (49), gives the correction to the Laplace approximation to the mutual information, Eq. (39). When the Laplace approximation is justified, this correction will be small (even before averaging over p(r)). Also, note that in that case η ≈ 1, and the last term in Eq. (49) is small on its own.
The second term in Eq. (49) is readily evaluated:
| (50) |
and the first term can be evaluated using the MCMC, via Eq. (13). To evaluate the third term, we use the following trick. For any well-behaved function α(x), we have
| (51) |
Using this formula we can estimate η by estimating the numerator and denominator on the right hand side according to Eq. (13) with samples drawn from p(x|r) and pL (x|r), respectively, e.g., by MCMC. However, as we only have access to finitely many samples from each distribution care must be taken in the choice of the function α to avoid large estimation errors. For example, if the support of p(x|r) and pL(x|r) have a small overlap, α(x) has to be chosen such that it amplifies the contribution from the region of overlap of the two distributions, thus acting like a bridge connecting the two supports. Otherwise (e.g., if α(x) is a constant), both the numerator and denominators in Eq. (51) can be very small in such a case, leading to an almost indeterminate ratio with large random error.14 A method of evaluating η using Eq. (51) by employing an optimal α (x), was originally developed by (Bennett, 1976) and was further refined by (Meng and Wong, 1996), and is referred to as “bridge sampling” for the above reason. These authors have shown that the asymptotically optimal (for large number of samples from each distribution) choice of α(x) is
| (52) |
where si = Ni/(N1 + N2) (i = 1, 2), and N1,2 are the number of samples drawn from p(x|r) and pL(x|r), respectively. As this choice for α(x) itself depends on η, it suggests an iterative solution, namely
| (53) |
where xij (i = 1, 2) are samples drawn from p(x|r) and pL(x|r) respectively, and lij ≡ q(xij|r)/qL(xij|r). Since we expect Z ≈ ZL, we take η̂(0) = 1. In our calculations, we stopped the bridge sampling iterations when , where d is the stimulus dimension.
Figure 10 shows a plot of IL and δI per stimulus dimension, calculated as described above,15 as a function of the standard deviation of the filtered stimulus input (which, for the white noise stimulus, is the contrast, c, times the magnitude of ki(t)). It is seen that IL grows as c||ki|| grows, but δI does not change significantly, and remains small. Thus the Laplace approximation for the mutual information is very accurate for moderately large c||ki||. Furthermore, for vanishing c||ki||, the posterior is equal to the Gaussian prior in this case, and this approximation is exact. Therefore the error δI has a maximum at a finite value of the stimulus filter, away from which the Laplace approximation is accurate. For comparison with our real spike data example presented in Sec. 4 and Fig. 8, we note that in that case the standard deviation of the filtered stimulus to different cells was in the range c||k|| ~ 3 ± 1, depending on the cell, and the Laplace approximation did indeed provide an accurate approximation for the mutual information, with δI/IL = 0.09.
Figure 10.
Comparison of Laplace approximation to Mutual Information per stimulus dimension IL/d, and the correction, δI/d (see Eq. (47)), based on the MCMC estimate of the true value, for a pair of ON and OFF RGCs, as a function of the magnitude of the filtered stimulus input c||k||, where c is the contrast, and ||k|| is the norm of the stimulus filter. The computationally inexpensive Laplace approximation for the mutual information is accurate for moderately strong stimulus filters which give rise to sharp likelihoods. Furthermore, at c||k|| = 0, the likelihood has no dependence on x and the posterior is equal to the Gaussian prior, for which the Laplace approximation is exact. Thus for very small |k| also, IL becomes exact and the error, δI, has a maximum around c||k|| ≈ 2.5.
6 Effect of uncertainty in the model parameters
In the previous sections we assumed the values of the parameters involved in the GLM likelihood, Eqs. (3)–(4), were known exactly. Of course, in reality these parameters themselves are obtained by fitting the GLM to experimental data, and are thus only known with a finite accuracy. In this section we investigate the effect of uncertainty in the GLM parameters θ (see Sec. 2), on the posterior mean estimate for the stimulus. We represent this uncertainty by a probability distribution, p(θ). In the presence of parameter uncertainty, the posterior mean of the stimulus, x, is modified to
| (54) |
(in this section, unlike in sections 3–5, we write θ explicitly when a distribution is conditioned on it – when there is no θ in the argument of the distribution, it means it has been marginalized). We assume the uncertainty in the parameters is small enough that a Laplace approximation for p(θ) applies, i.e., it can be taken to be Gaussian with mean θML, and a small covariance I−1(θML). Here, θML is the maximum likelihood fit to data, and I−1(θML) is the Hessian of the negative log-likelihood (as a function of GLM parameters, given the experimental data) at θML. For simplicity we assume the GLM nonlinearity (see Eq. (3)) is exponential: f(u) = exp (u). We also assume that the prior stimulus ensemble is Gaussian, with probability distribution described by Eq. (10).
We would like to understand how the uncertainty in θ will affect the posterior estimate. This uncertainty broadens the likelihood (as a function of x) and therefore, we expect that as it increases the posterior estimate E(x|r) will move towards the prior mean (in our case zero). Intuitively, this is because as the Bayesian decoder’s knowledge of the encoding mechanism (represented by the parameters θ) decreases, it discounts the information that the observed spike train, r, carries about the stimulus and instead relies more strongly on its prior information. To verify this intuition analytically, we consider the case where E(x|r, θ) ≈ xMAP (r, θ) (e.g., as we saw in the last section, the Laplace approximation is often quite adequate in the case of Gaussian priors, and this approximation therefore holds in that case). Assuming this, we can replace E(x|r, θ) with xMAP(r, θ) in Eq. (6), and obtain
| (55) |
In the following we will drop r from the arguments of xMAP when it is understood. We will denote the average over p(θ) in Eq. (55) by 〈xMAP〉θ.
Using the Bayes rule in the form log p(x|r) = log p(x) + log p(r|x) + const., with Eq. (10), and Eqs. (3)-(4) with exponential nonlinearity, we obtain
| (56) |
up to an additive constant. Here, 𝒞 is the covariance of the Gaussian prior, Eq. (10), and θ = {bi, Ki, ℋij} are the GLM parameters introduced in Sec. 2. The MAP satisfies
| (57) |
which yields the equation
| (58) |
When the contrast or the stimulus filter are small (corresponding to the regime of low signal to noise ratio), the exponential can be expanded to first order in xMAP (θ), yielding the linear equation (a similar expansion also appeared in (Pillow et al., 2010))
| (59) |
where we defined
| (60) |
| (61) |
and
| (62) |
| (63) |
Notice that 𝒜(θ) is the Hessian of the negative log-posterior Eq. (56) at x = 0. Assuming the matrix 𝒜(θ) is invertible,16 we then obtain
| (64) |
We write θ = θML + δθ, where δθ has zero mean, and expand Eq. (64) in δθ up to second order, to obtain
| (65) |
where
| (66) |
and
| (67) |
| (68) |
such that is homogeneously of order n in δθ. Here, we defined 𝒜0 ≡ 𝒜(θML), ℬ0 ≡ ℬ (θML), and d𝒜 and dℬ (d2𝒜 and d2ℬ) are the random first (second) order variations of 𝒜(θ) and ℬ(θ) in δθ. The first order variations, d𝒜 and dℬ, are thus Gaussian with zero mean and a covariance determined by the covariance of θ. After averaging over θ, will vanish, and we have
| (69) |
To gain some intuition, we now set out to evaluate in the regime of small baseline firing rates, so that are small, and we also assume we can neglect the uncertainty of the baseline firing rates and the post-spike feedback filters (i.e., we set δbi = δℋij = 0). In this case, d2ℬ = 0, and ignoring terms beyond the leading order in , we take , and obtain
| (70) |
| (71) |
Here, we denoted the maximum likelihood fit for the stimulus filters by K̄i, and in deriving the second line, we used , and , and in the last line we assumed the stimulus is white, i.e., 𝒞 ∝ 1. Equation (6) is not very enlightening, so we look at the special case where δKi = αK̄i, and α is a noisy Gaussian scalar with zero mean (this arises for example in the case of delta function kernels, as in the example of the last section – or more generally when only the over all scale of Ki is uncertain). Replacing for δKi, and using Eq. (66) with to write , for this case we obtain
| (72) |
Therefore, to the first non-vanishing order, the change in the L2 norm of the estimate is
| (73) |
where the inequality followed from the fact that , and therefore are positive definite operators. Thus we see that, at least in the special regime that we considered, parameter uncertainty will shrink the norm of the posterior mean estimate sending it towards the prior mean at the origin. This result is in agreement with the intuition stated above, and was further corroborated by our numerical results in more general parameter regimes.
Figure 11 shows a numerical plot of the norm of the posterior estimate as a function of the size of the uncertainty in Ki. Here, δKi was not constrained to be proportional to K̄i. However, again, as uncertainty in model parameters increases, leading to broadening of the likelihood, the posterior mean moves towards the prior mean.
Figure 11.
Effect of parameter uncertainty on the posterior estimate for Gaussian white-noise stimuli. Panel (a) is a plot of (where d = 50 is the stimulus dimension) vs. relative uncertainty, α, in the stimulus filter ki(t). α is defined through , where ε(t) is a standard Gaussian white-noise. Unlike in Sec. 4, (the maximum likelihood fit for ki(t)) was taken to have a time width spreading over a few stimulus frames. Furthermore, its magnitude was taken to be large enough to give rise to a sharp posterior, satisfying Eq. (8) and thus E(x|r, θ) ≈ xMAP(r, θ). For each value of α, 100 samples of ε(t) were generated, and the MAP was decoded for each using the corresponding ki(t) and the fixed spike train. The sample average of those MAPs was taken as the estimate for 〈xMAP〉θ ≈ E(x|r). Panel (b) shows 〈xMAP〉θ (solid trace) for α = 0 (top plot) and α = 1 (bottom plot) and the true stimulus (dotted trace). It is seen that the main effect of the finite uncertainty is a shrinkage of the estimate towards zero, i.e., the mean of the prior Gaussian distribution.
7 Discussion
Markov chain Monte Carlo allows for the calculation of general, fully Bayesian posterior estimates. The main goal of this paper was to survey the performance of a number of efficient MCMC algorithms in the context of model-based neural decoding of spike trains. Using these methods, we also verified and extended the results of (Pillow et al., 2010) on MAP based decoding and information estimation via Laplace approximation, in GLM based neural decoding problems. Although MCMC integration is more general in this sense, it is at the same time significantly more computationally expensive than the optimization algorithms used to find the MAP. As we explained in Sec. 2, the MAP is in general a good estimator when the Laplace approximation is accurate. The MAP also comes with natural error bars estimated through the Hessian matrix of the log-posterior at MAP, Eq. (9). Furthermore, when it is valid, this approximation provides a very efficient way of estimating the mutual information through Eq. (39). Thus it is important to have a clear knowledge of when this approximation holds, since when it does, it can be exploited to dramatically reduce the computational cost of stimulus decoding or information estimation.
In Sec. 3, we introduced the RWM, HMC, Gibbs, and hit-and-run Markov chains, all special cases of the Metropolis-Hastings algorithm. Although these methods allow for sampling from general posterior distributions, regardless of the forward model, we also took advantage of the specific properties of the distributions involved in our GLM-based decoding to increase the efficiency of these chains. The ARS algorithm, which exploits the log-concavity property of the GLM likelihood and the prior distribution, was used to significantly reduce the computational cost of the one-dimensional sampling in each hit-and-run step. We took advantage of the Laplace approximation (or a regularized version of it in the at prior case), to shape the proposal distributions to roughly match the covariance structure of the underlying distribution. Furthermore, we were able to carry this out in 𝒪(T) computational time (i.e., scaling only linearly with the stimulus duration, T), by exploiting the bandedness of the log-posterior Hessian in these settings. To the best of our knowledge, the use of 𝒪(T), Laplace-enhanced HMC in neural applications is novel. Similarly, even though the hit-and-run algorithm is well-known in the statistics literature, we are unaware of any previous application of it in the context of high-dimensional neural decoding.
We mention that these chains with 𝒪(T), Laplace-based enhancement can also be implemented in decoding posterior distributions based on state-space models with Markovian structure; an example of such an application was presented in Fig. 9, based on the state-space model used in Smith et al. (2007). However, in cases where the posterior distribution turns out to be non-concave, obtaining the Laplace approximation may be unfeasible or it may not improve the chain’s mixing. Even though MCMC without this enhancement is still applicable in such cases, other methods such as sequential Monte Carlo (“particle-filtering”) (Doucet et al., 2001; Brockwell et al., 2004; Kelly and Lee, 2004; Godsill et al., 2004; Shoham et al., 2005; Ergun et al., 2007; Vogelstein et al., 2008; Huys and Paninski, 2009) which are solely applicable in models with Markovian structure may prove to be more efficient.
It is worth noting a connection between this 𝒪(T) non-isotropic MCMC sampling and the Bayesian adaptive regression splines (BARS) method (DiMatteo et al., 2001; Wallstrom et al., 2007), which has become a popular tool in neursocientific applications. The BARS algorithm is a powerful non-parametric regression method designed to infer the shape of a smooth underlying curve that has produced noisy observations. This method assumes the curve can be approximated by a spline, and outputs samples from the posterior distribution of the spline knots and coefficients. Specifically, in the case of neural spike trains, it is assumed that the observed spikes, r(t), are produced by an inhomogeneous Poisson process with a rate λ(t) = exp (B(t)Tβ) where Bi(t) is a cubic B-spline basis, and βi are the spline coefficients. Here, i runs from 1 to k + 2, where k is the number of spline knots with positions τi; the spline basis functions, Bi(t), implicitly depend on k and τi. Conditioned on fixed τi and k, the prior distribution of the spline coefficients β is taken to be Gaussian with zero mean and inverse covariance (a unit information prior). Thus conditioned on fixed spline knots, the BARS model involves Poisson observations from a Gaussian latent variable β; this is directly analogous to our GLM model with Gaussian stimuli, x, with β and B(t) replacing x and K in the analogy, respectively. In particular, sampling from the posterior distribution of the a priori Gaussian β (given τi and k) is very similar to sampling from the posterior over the Gaussian stimulus, x, in our examples in this paper. Furthermore, to obtain conditional samples of β, the BARS code uses an RWM chain (Wallstrom et al., 2007), which as in Eq. (19)–(20), employs the Hessian at the MAP point for the spline coefficients to produce non-isotropic proposals. Using the form of the prior covariance, mentioned above, and the standard likelihood expression for a Poisson process, the Hessian of the negative log-posterior for β (given r(t), τi and k) is given by
| (74) |
where the first term is the inverse prior covariance, and a is some positive constant. Since, by definition, Bi(t) is non-zero only when t ∈ [τi, τi+4], we see that Hij vanishes when |i − j| > 3, and hence, is banded. Again, the bandedness of the Hessian is exploited (Wallstrom et al., 2007) to obtain the RWM proposals in 𝒪(T) computational time, by the method described after Eq. (21). We note that the BARS package could potentially be improved by using a faster-mixing chain such as HMC, which can out-perform RWM by orders of magnitude (Fig. 4).
We compared the mixing rates of the mentioned MCMC chains, in sampling from the posterior stimulus distributions for GLM modeled neurons. In this setting, when the posterior is smooth throughout its support, the HMC algorithm outperforms the other chains by an order of magnitude. On the other hand, when sampling from posteriors based on at priors with sharp corners, the hit-and-run chain mixed consistently faster than the others.
In Sec. 4, we compared the performance of the MAP and the posterior mean, calculated using MCMC, in different settings. In one example, we decoded simulated spike trains (generated in response to Gaussian and flat white-noise stimuli), in a range of stimulus input strengths and for different numbers of identical cells. We also decoded the filtered stimulus input into six retinal ganglion cells, based on their experimentally recorded spike trains. The average squared error of the MAP and mean estimates were in general quite close in the case of Gaussian stimuli, justifying MAP decoding in this case. In the flat prior case, however, the posterior mean can often have a much smaller average squared error than the MAP.
In Sec. 5, we applied MCMC to the problem of estimating properties of the joint distribution p(x|r) which cannot be obtained form its low dimensional marginals. In particular, we investigated the reliability of the Laplace approximation for the mutual information between the stimulus and spike trains (model-based calculations of the mutual information with Gaussian priors have been previously presented in Barbieri et al. (2004)). We found that the Laplace approximation for the mutual information was adequate in the case of Gaussian priors, except in a small range of moderate stimulus input strengths.
In the last section we dealt with the effect of uncertainty in GLM parameters (e.g., based on fits to experimental data) on the decoding. Intuitively, it is expected that when the forward model parameters become uncertain, information coming from the spike train and hence the likelihood becomes less reliable, and therefore the estimate will rely more heavily on the prior information. Thus the posterior mean is expected to revert towards the prior mean as parameter uncertainty increases. We verified this intuition analytically in the special case of localized stimulus filters (with no band-pass filtering) and small baseline firing rates. Our numerics showed that indeed the main systematic effect of increasing parameter uncertainty on the mean estimate, E(x|r) is to shrink its magnitude (thus sending to the origin which was the prior mean in our case) in a wide range of parameter values.
The methods developed in this paper and in (Pillow et al., 2010) can be used for a variety of applications. In future work we plan to further apply these techniques to other experimental data, and to compare different “codebooks” (as mentioned in the introduction) based on different reductions of the full spike trains, according to their robustness and fidelity.
Figure 1.
Illustration of Bayesian decoding paradigm. (A) Bayesian decoding performs inference about the stimulus using the observed spike times and a specified encoding model. (B) Schematic of the encoding model (“Generalized Linear Model”) used for the decoding examples shown in this paper. The model parameters (ki and hij) can be easily fit using maximum likelihood. Once fit, the model provides a description of the data likelihood, p(r|x), which is combined with the prior p(x) to estimate x.
Acknowledgments
Thanks to M. Kennel, E. Simoncelli, E.J. Chichilnisky, T. Teravainen, and K. Rahnama Rad for helpful conversations. We are grateful to Y. Ding for her implementation of the bridge sampling method. YA is supported by the Robert Leet and Clara Guthrie Patterson Trust Postdoctoral Fellowship, Bank of America, Trustee. LP is supported by NEI R01 EY018003, an Alfred P. Sloan Research Fellowship, and the McKnight Scholar award. JP is supported by a Royal Society USA/Canada Research Fellowship.
Footnotes
The dimension of x is thus d = NT, where T is the number of time steps, and N is the total number of components at each time step.
That is, the kernels ki(t, n) and hij(t) vanish for t < 0.
We note that even though the nonlinearity, f(·), has to be an increasing function, with appropriately chosen negative post-spike feedback filters, hii, the mean firing rate of the GLM modeled neurons will still exhibit saturation as a function of the input strength, x.
That is, for fixed θ is a concave function of x, and vice versa, but in general not a concave function of (x, θ) jointly.
Let us mention, however, that no first principle dictates that the posterior distribution over a biologically or behaviorally relevant variable (e.g., an external variable that a part of the brain seeks to estimate) should be log-concave. In fact, distributions which are not log-concave, such as multi-modal or very fat-tailed distributions, can be highly relevant in biological settings.
White or correlated Gaussian priors are often used in neural applications (e.g., Gaussian stimuli are widely used in neurophysiological experiments). Flat priors with infinitely sharp boundaries, are less biologically motivated. However, at priors are the best log-concave approximation to binary priors, which are also quite common in sensory physiology – both as binary white noise and M-sequences (Pillow et al., 2008; Reid et al., 1997). In this paper, we consider the at prior mainly as a limiting case of concave log-priors with sharp derivatives when we check the accuracy of the Laplace approximation and compare the efficiency of various MCMC chains (see Sec. 3.5) in different regimes.
We are, of course, interested in calculating posterior expectations corresponding to the case π(x) = p(x|r), but as the present discussion is general, we will use π(x) in the rest of this section for ease of notation.
| (22) |
As with the Gibbs case, it can be shown again that this proposal leads to a MH acceptance probability of one. Hence hit-and-run is also a special case of MH.
Strictly speaking this independence is only true for Harris recurrent chains, but this is the case in most practical examples (see e.g., (Geyer, 1992) and (Tierney, 1991)).
Although this prior belongs to the class of product densities considered in (Roberts and Rosenthal, 1998), it does not satisfy the stringent smoothness conditions crucial for the part of their theorem regarding the (fast) mixing of MALA.
More precisely, “sharp” here means that the curvature of the log-posterior is large enough so that the Taylor expansion of the log-posterior involved in the Laplace approximation, Eq. (8), is accurate for deviations from xMAP on a scale determined by the inverse square root of the smallest eigenvalue of the Hessian matrix, Eq. (9).
This is obviously not the case, however, for parameter directions along which the data is non-informative, and the likelihood function does not vary much. In the RGC case, these correspond to stimulus features (directions in the stimulus space) that fall orthogonal to the cells’ spatiotemporal filters Ki.
For a similar reason a “brute force” method for computing Z, such as a simple Monte Carlo integration of the high-dimensional distribution q(x|r), gives rise to an estimate with slow convergence and a large error (Meng and Wong, 1996).
In principle, according to Eqs. (37)–(39), one has to average IL(r) and δI(r) over the marginal response distribution p(r). But due to the intensive nature of IL(r)/d and δI(r)/d, and the large d, they depended on the specific realization of r only weakly, and were to a good degree self-averaging, so that averaging over p(r) would not considerably alter the plot of Fig. 10. A similar argument appeared in Strong et al. (1998).
This is true, at least when when Si are not too large.
Contributor Information
Yashar Ahmadian, Email: yashar@stat.columbia.edu.
Jonathan W. Pillow, Email: pillow@mail.utexas.edu.
Liam Paninski, Email: liam@stat.columbia.edu.
References
- Abbott L, Dayan P. The effect of correlated variability on the accuracy of a population code. Neural Computation. 1999;11:91–101. doi: 10.1162/089976699300016827. [DOI] [PubMed] [Google Scholar]
- Barbieri R, Frank L, Nguyen D, Quirk M, Solo V, Wilson M, Brown E. Dynamic analyses of information encoding in neural ensembles. Neural Computation. 2004;16:277–307. doi: 10.1162/089976604322742038. [DOI] [PubMed] [Google Scholar]
- Bennett CH. Efficient estimation of free energy divergences from Monte Carlo data. Journal of Computational Physics. 1976;22:245–268. [Google Scholar]
- Berger J. Statistical Decision Theory and Bayesian Analysis. Springer; 1993. [Google Scholar]
- Bialek W, Rieke F, de Ruyter van Steveninck R, Warland D. Reading a neural code. Science. 1991;252:1854–1857. doi: 10.1126/science.2063199. [DOI] [PubMed] [Google Scholar]
- Boneh A, Golan A. Constrainsts’ redundancy and feasible region boundedness by random feasible point generator (rfpg). Presented at the Third European Congress on Operations Research (EURO III); Amsterdam. 1979. [Google Scholar]
- Brillinger D. Maximum likelihood analysis of spike trains of interacting nerve cells. Biological Cyberkinetics. 1988;59:189–200. doi: 10.1007/BF00318010. [DOI] [PubMed] [Google Scholar]
- Brillinger D. Nerve cell spike train data analysis: a progression of technique. Journal of the American Statistical Association. 1992;87:260–271. [Google Scholar]
- Brockwell A, Rojas A, Kass R. Recursive Bayesian decoding of motor cortical signals by particle filtering. Journal of Neurophysiology. 2004;91:1899–1907. doi: 10.1152/jn.00438.2003. [DOI] [PubMed] [Google Scholar]
- Brooks S, Gelman A. General methods for monitoring convergence of iterative simulations. J Comput Graph Statist. 1998;7:434–455. [Google Scholar]
- Brown E, Barbieri R, Eden U, Frank L. Likelihood methods for neural data analysis. In: Feng J, editor. Computational Neuroscience: A Comprehensive Approach. London: CRC; 2003. pp. 253–286. [Google Scholar]
- Brown E, Frank L, Tang D, Quirk M, Wilson M. A statistical paradigm for neural spike train decoding applied to position prediction from ensemble firing patterns of rat hippocampal place cells. Journal of Neuroscience. 1998;18:7411–7425. doi: 10.1523/JNEUROSCI.18-18-07411.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KS, Ledolter J. Monte Carlo EM estimation for time series models involving counts. Journal of the American Statistical Association. 1995;90(429):242–252. [Google Scholar]
- Chichilnisky E. A simple white noise analysis of neuronal light responses. Network: Computation in Neural Systems. 2001;12:199–213. [PubMed] [Google Scholar]
- Cronin B, Stevenson IH, Sur M, Kording KP. Hierarchical Bayesian modeling and Markov chain Monte Carlo sampling for tuning curve analysis. J Neurophysiol. 2009:00379.2009. doi: 10.1152/jn.00379.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R, Rodriguez-Yam G. Estimation for state-space models: an approximate likelihood approach. Statistica Sinica. 2005;15:381–406. [Google Scholar]
- Dayan P, Abbott L. Theoretical Neuroscience. MIT Press; Cambridge: 2001. [Google Scholar]
- DiMatteo I, Genovese C, Kass R. Bayesian curve fitting with free-knot splines. Biometrika. 2001;88:1055–1073. [Google Scholar]
- Donoghue J. Connecting cortex to machines: recent advances in brain interfaces. Nature Neuroscience. 2002;5:1085–1088. doi: 10.1038/nn947. [DOI] [PubMed] [Google Scholar]
- Doucet A, de Freitas N, Gordon N, editors. Sequential Monte Carlo in Practice. Springer; 2001. [Google Scholar]
- Duane S, Kennedy AD, Pendleton BJ, Roweth D. Hybrid monte carlo. Physics Letters B. 1987;195(2):216–222. [Google Scholar]
- Eden UT, Frank LM, Barbieri R, Solo V, Brown EN. Dynamic analysis of neural encoding by point process adaptive filtering. Neural Comput. 2004;16(5):971–998. doi: 10.1162/089976604773135069. [DOI] [PubMed] [Google Scholar]
- Ergun A, Barbieri R, Eden U, Wilson M, Brown E. Construction of point process adaptive filter algorithms for neural systems using sequential Monte Carlo methods. IEEE Transactions on Biomedical Engineering. 2007;54:419–428. doi: 10.1109/TBME.2006.888821. [DOI] [PubMed] [Google Scholar]
- Gamerman D. Sampling from the posterior distribution in generalized linear mixed models. Statistics and Computing. 1997;7(1):57–68. [Google Scholar]
- Gamerman D. Markov chain monte carlo for dynamic generalised linear models. Biometrika. 1998;85(1):215–227. [Google Scholar]
- Gelman A. Bayesian Data Analysis. Chapman and Hall/CRC; Boca Raton, FL: 2004. [Google Scholar]
- Geman S, Geman D. Stochastic relaxation, Gibbs distribution, and the Bayesian restoration of images. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1984;6:721–741. doi: 10.1109/tpami.1984.4767596. [DOI] [PubMed] [Google Scholar]
- Gerwinn S, Macke JH, Bethge M. Bayesian population decoding of spiking neurons. Front Comput Neurosci. 2009;3(21) doi: 10.3389/neuro.10.021.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer CJ. Practical Markov chain Monte Carlo. Statistical Science. 1992;7:473–483. [Google Scholar]
- Gilks W, Wild P. Adaptive rejection sampling for Gibbs sampling. Applied Statistics. 1992;41:337–348. [Google Scholar]
- Gilks WR. Derivative-free adaptive rejection sampling for Gibbs sampling. In: Bernardo J, Berger J, Dawid A, Smith A, editors. Bayesian Statistics. Vol. 4. Oxford University Press; Oxford: 1992. pp. 641–649. [Google Scholar]
- Godsill S, Doucet A, West M. Monte Carlo smoothing for non-linear time series. Journal of the American Statistical Association. 2004;99:156–168. [Google Scholar]
- Hastings WK. Monte Carlo sampling methods using Markov chains and their applications. Biometrika. 1970;57:97–109. [Google Scholar]
- Huys Q, Paninski L. Smoothing of, and parameter estimation from, noisy biophysical recordings. PLoS Comput Biol. 2009;5(5):e1000379. doi: 10.1371/journal.pcbi.1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishwaran H. Applications of hybrid monte carlo to bayesian generalized linear models: quasicomplete separation and neural networks. Journal of Computational and Graphical Statistics. 1999;8:779–799. [Google Scholar]
- Jacobs A, Grzywacz N, Nirenberg S. Decoding the parallel pathways of the retina. SFN Abstracts 2006 [Google Scholar]
- Jungbacker B, Koopman SJ. Monte Carlo Estimation for Nonlinear Non-Gaussian State Space Models. Biometrika. 2007;94(4):827–839. [Google Scholar]
- Karmeier K, Krapp H, Egelhaaf M. Population coding of self-motion: Applying Bayesian analysis to a population of visual interneurons in the fly. Journal of Neurophysiology. 2005;94:2182–2194. doi: 10.1152/jn.00278.2005. [DOI] [PubMed] [Google Scholar]
- Kass R, Tierney L, Raftery A. Laplace’s method in Bayesian analysis. In: Flournoy N, Tsutakawa R, editors. Statistical Multiple Integration. Providence: Springer; 1991. pp. 89–99. [Google Scholar]
- Kelly R, Lee T. Decoding V1 neuronal activity using particle filtering with Volterra kernels. Advances in Neural Information Processing Systems. 2004;15:1359–1366. [Google Scholar]
- Kennedy A, Edwards R, Mino H, Pendleton B. Tuning the generalized hybrid monte carlo algorithm. Nuclear Physics B - Proceedings Supplements. 1996;47(1–3):781–784. [Google Scholar]
- Kipnis C, Varadhan SRS. Central limit theorem for additive functionals of reversible Markov processes and applications to simple exclusions. Comm Math Phys. 1986;104:1–19. [Google Scholar]
- Litke A, Bezayiff N, Chichilnisky E, Cunningham W, Dabrowski W, Grillo A, Grivich M, Grybos P, Hottowy P, Kachiguine S, Kalmar R, Mathieson K, Petrusca D, Rahman M, Sher A. What does the eye tell the brain? development of a system for the large scale recording of retinal output activity. IEEE Trans Nucl Sci. 2004:1434–1440. [Google Scholar]
- Lovasz L, Vempala S. Hit-and-run from a corner. Proc. of the 36th ACM Symposium on the Theory of Computing (STOC ‘04); Chicago. 2004. [Google Scholar]
- Maynard E, Hatsopoulos N, Ojakangas C, Acuna B, Sanes J, Normann R, Donoghue J. Neuronal interactions improve cortical population coding of movement direction. Journal of Neuroscience. 1999;19:8083–8093. doi: 10.1523/JNEUROSCI.19-18-08083.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullagh P, Nelder J. Generalized linear models. Chapman and Hall; London: 1989. [Google Scholar]
- Meng XL, Wong WH. Simulating ratios of normalizing constants via a simple identity: A theoretical exploration. Statistica Sinica. 1996;6:831–860. [Google Scholar]
- Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E. Equation of state calculations by fast computing machines. J Chem Phys. 1953;21:1087–1092. [Google Scholar]
- Neal R. Number 118 in Lecture Notes in Statistics. Springer; 1996. Bayesian Learning for Neural Networks. [Google Scholar]
- Neal R. The harmonic mean of the likelihood: Worst Monte Carlo method ever. Radford Neal’s blog. 2008 http://radfordneal.wordpress.com/2008/08/17/the-harmonic-mean-of-the-likelihood-worst-monte-carlo-method-ever/
- Newton MA, Raftery AE. Approximate Bayesian inference with the weighted likelihood bootstrap. Journal of the Royal Statistical Society Series B (Methodological) 1994;56(1):3–48. [Google Scholar]
- Paninski L. Estimation of entropy and mutual information. Neural Computation. 2003;15:1191–1253. [Google Scholar]
- Paninski L. Maximum likelihood estimation of cascade point-process neural encoding models. Network: Computation in Neural Systems. 2004;15:243–262. [PubMed] [Google Scholar]
- Paninski L, Ahmadian Y, Ferreira D, Koyama S, Rahnama Rad K, Vidne M, Vogelstein J, Wu W. A new look at state-space models for neural data. Journal of Computational Neuroscience. 2010 doi: 10.1007/s10827-009-0179-x. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paninski L, Fellows M, Shoham S, Hatsopoulos N, Donoghue J. Superlinear population encoding of dynamic hand trajectory in primary motor cortex. J Neurosci. 2004;24:8551–8561. doi: 10.1523/JNEUROSCI.0919-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paninski L, Iyengar S, Kass R, Brown E. Stochastic Methods in Neuroscience. Oxford University Press; 2008. Statistical models of spike trains. [Google Scholar]
- Pillow J, Ahmadian Y, Paninski L. Model-based decoding, information estimation, and change-point detection in multi-neuron spike trains. Neural Computation. 2010 doi: 10.1162/NECO_a_00058. In press. [DOI] [PubMed] [Google Scholar]
- Pillow J, Shlens J, Paninski L, Sher A, Litke A, Chichilnisky E, Simoncelli E. Spatiotemporal correlations and visual signaling in a complete neuronal population. Nature. 2008 doi: 10.1038/nature07140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid RC, Victor JD, Shapley RM. The use of m-sequences in the analysis of visual neurons: Linear receptive field properties. Visual Neuroscience. 1997;14(6):1015– 1027. doi: 10.1017/s0952523800011743. [DOI] [PubMed] [Google Scholar]
- Rieke F, Warland D, de Ruyter van Steveninck R, Bialek W. Spikes: Exploring the neural code. MIT Press; Cambridge: 1997. [Google Scholar]
- Robert C, Casella G. Monte Carlo Statistical Methods. Springer; 2005. [Google Scholar]
- Roberts G, Rosenthal J. Optimal scaling of discrete approximations to Langevin diffusions. J R Statist Soc B. 1998;160:255–268. [Google Scholar]
- Roberts G, Rosenthal J. Optimal scaling for various Metropolis-Hastings algorithms. Statistical Science. 2001;16:351–367. [Google Scholar]
- Roberts GO, Tweedie RL. Exponential convergence of Langevin diffusions and their discrete approximations. Biometrika. 1996;2:341–363. [Google Scholar]
- Sanger T. Theoretical considerations for the analysis of population coding in motor cortex. Neural Computation. 1994;6:12–21. [Google Scholar]
- Shephard N, Pitt MK. Likelihood analysis of non-Gaussian measurement time series. Biometrika. 1997;84(3):653–667. [Google Scholar]
- Shlens J, Field GD, Gauthier JL, Grivich MI, Petrusca D, Sher A, Litke AM, Chichilnisky EJ. The Structure of Multi-Neuron Firing Patterns in Primate Retina. J Neurosci. 2006;26(32):8254–8266. doi: 10.1523/JNEUROSCI.1282-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoham S, Paninski L, Fellows M, Hatsopoulos N, Donoghue J, Normann R. Optimal decoding for a primary motor cortical brain-computer interface. IEEE Transactions on Biomedical Engineering. 2005;52:1312–1322. doi: 10.1109/TBME.2005.847542. [DOI] [PubMed] [Google Scholar]
- Simoncelli E, Paninski L, Pillow J, Schwartz O. The Cognitive Neurosciences. 3 MIT Press; 2004. Characterization of neural responses with stochastic stimuli. [Google Scholar]
- Smith AC, Frank LM, Wirth S, Yanike M, Hu D, Kubota Y, Graybiel AM, Suzuki WA, Brown EN. Dynamic Analysis of Learning in Behavioral Experiments. J Neurosci. 2004;24(2):447–461. doi: 10.1523/JNEUROSCI.2908-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AC, Wirth S, Suzuki WA, Brown EN. Bayesian Analysis of Interleaved Learning and Response Bias in Behavioral Experiments. J Neurophysiol. 2007;97(3):2516–2524. doi: 10.1152/jn.00946.2006. [DOI] [PubMed] [Google Scholar]
- Smith RL. Monte Carlo techniques for generating random feasible solutions to mathematical programs. Presented at the ORSA/TIMS conference; Washington D. C. 1980. [Google Scholar]
- Snyder D, Miller M. Random Point Processes in Time and Space. Springer-Verlag; 1991. [Google Scholar]
- Stanley G, Boloori A. Decoding in neural systems: stimulus reconstruction from nonlinear encoding. Proceedings of the Annual EMBS International Conference; 2001. p. 23. [Google Scholar]
- Strong S, Koberle R, de Ruyter van Steveninck R, Bialek W. Entropy and information in neural spike trains. Physical Review Letters. 1998;80:197–202. [Google Scholar]
- Theunissen F, David S, Singh N, Hsu A, Vinje W, Gallant J. Estimating spatio-temporal receptive fields of auditory and visual neurons from their responses to natural stimuli. Network: Computation in Neural Systems. 2001;12:289–316. [PubMed] [Google Scholar]
- Tierney L. Technical Report. Vol. 560. School of Statistics, University of Minnesota; 1991. Markov chains for exploring posterior distributions. [Google Scholar]
- Tierney L, Kadane JB. Accurate approximations for posterior moments and marginal densities. Journal of the American Statistical Association. 1986;81(393):82–86. [Google Scholar]
- Truccolo W, Eden U, Fellows M, Donoghue J, Brown E. A point process framework for relating neural spiking activity to spiking history, neural ensemble and extrinsic covariate effects. Journal of Neurophysiology. 2005;93:1074–1089. doi: 10.1152/jn.00697.2004. [DOI] [PubMed] [Google Scholar]
- Tyler DE. Statistical analysis for the angular central Gaussian distribution on the sphere. Biometrika. 1987;74:579–859. [Google Scholar]
- Vogelstein J, Babadi B, Watson B, Yuste R, Paninski L. Fast nonnegative deconvolution via tridiagonal interior-point methods, applied to calcium fluorescence data. Statistical analysis of neural data (SAND) conference.2008. [Google Scholar]
- Wallstrom G, Liebner J, Kass RE. An implementation of Bayesian adaptive regression splines (BARS) in C with S and R wrappers. Journal of Statistical Software. 2007;26(1):1–21. [PMC free article] [PubMed] [Google Scholar]
- Warland D, Reinagel P, Meister M. Decoding visual information from a population of retinal ganglion cells. Journal of Neurophysiology. 1997;78:2336–2350. doi: 10.1152/jn.1997.78.5.2336. [DOI] [PubMed] [Google Scholar]
- Wu W, Black MJ, Mumford D, Gao Y, Bienenstock E, Donoghue J. Modeling and decoding motor cortical activity using a switching Kalman filter. IEEE Transactions on Biomedical Engineering. 2004;51:933–942. doi: 10.1109/TBME.2004.826666. [DOI] [PubMed] [Google Scholar]
- Yu BM, Cunningham JP, Santhanam G, Ryu SI, Shenoy KV, Sahani M. Gaussian-Process Factor Analysis for Low-Dimensional Single-Trial Analysis of Neural Population Activity. J Neurophysiol. 2009;102(1):614–635. doi: 10.1152/jn.90941.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Ginzburg I, McNaughton B, Sejnowski T. Interpreting neuronal population activity by reconstruction: Unified framework with application to hippocampal place cells. Journal of Neurophysiology. 1998;79:1017–1044. doi: 10.1152/jn.1998.79.2.1017. [DOI] [PubMed] [Google Scholar]