Abstract
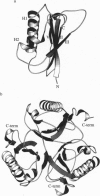
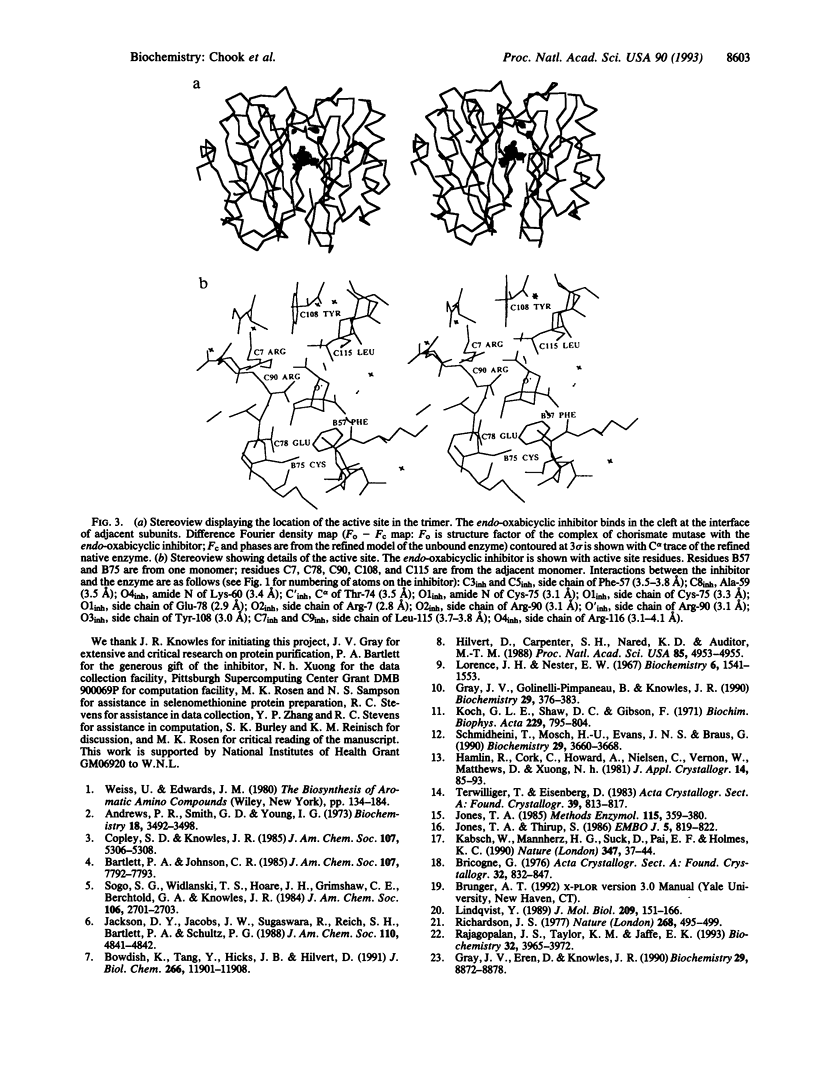
We have solved the structure of a chorismate mutase (chorismate pyruvatemutase, EC 5.4.99.5), the 1.9-A crystal structure of the monofunctional enzyme from Bacillus subtilis. The structure determination process was an unusual one, involving 12 monomers of the enzyme in the asymmetric unit. This structure was solved by the multiple isomorphous replacement method with partial structure phase combination and molecular averaging. The final model, which includes 1380 residues and 522 water molecules in an asymmetric unit, has been refined at 1.9 A and the current crystallographic R value is 0.201. The B. subtilis chorismate mutase is a homotrimer, with beta-sheets from each monomer packing to form the core of a pseudo-alpha beta-barrel with helices on the outside of the trimer. In addition, the active sites have been located by using data from a complex with an endo-oxabicyclic inhibitor that mimics the transition state of the reaction. The structure of this complex has been refined to 2.2 A with a current R value of 0.182 for a model that includes 1388 residues, 12 inhibitor molecules, and 530 water molecules in the asymmetric unit. In each trimer, three equivalent active sites are located at the interfaces of two adjacent subunits.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews P. R., Smith G. D., Young I. G. Transition-state stabilization and enzymic catalysis. Kinetic and molecular orbital studies of the rearrangement of chorismate to prephenate. Biochemistry. 1973 Aug 28;12(18):3492–3498. doi: 10.1021/bi00742a022. [DOI] [PubMed] [Google Scholar]
- Bowdish K., Tang Y., Hicks J. B., Hilvert D. Yeast expression of a catalytic antibody with chorismate mutase activity. J Biol Chem. 1991 Jun 25;266(18):11901–11908. [PubMed] [Google Scholar]
- Gray J. V., Eren D., Knowles J. R. Monofunctional chorismate mutase from Bacillus subtilis: kinetic and 13C NMR studies on the interactions of the enzyme with its ligands. Biochemistry. 1990 Sep 18;29(37):8872–8878. doi: 10.1021/bi00489a051. [DOI] [PubMed] [Google Scholar]
- Gray J. V., Golinelli-Pimpaneau B., Knowles J. R. Monofunctional chorismate mutase from Bacillus subtilis: purification of the protein, molecular cloning of the gene, and overexpression of the gene product in Escherichia coli. Biochemistry. 1990 Jan 16;29(2):376–383. doi: 10.1021/bi00454a011. [DOI] [PubMed] [Google Scholar]
- Hilvert D., Carpenter S. H., Nared K. D., Auditor M. T. Catalysis of concerted reactions by antibodies: the Claisen rearrangement. Proc Natl Acad Sci U S A. 1988 Jul;85(14):4953–4955. doi: 10.1073/pnas.85.14.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. A., Thirup S. Using known substructures in protein model building and crystallography. EMBO J. 1986 Apr;5(4):819–822. doi: 10.1002/j.1460-2075.1986.tb04287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W., Mannherz H. G., Suck D., Pai E. F., Holmes K. C. Atomic structure of the actin:DNase I complex. Nature. 1990 Sep 6;347(6288):37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- Koch G. L., Shaw D. C., Gibson F. The purification and characterisation of chorismate mutase-prephenate dehydrogenase from Escherichia coli K12. Biochim Biophys Acta. 1971 Mar 23;229(3):795–804. doi: 10.1016/0005-2795(71)90298-4. [DOI] [PubMed] [Google Scholar]
- Lindqvist Y. Refined structure of spinach glycolate oxidase at 2 A resolution. J Mol Biol. 1989 Sep 5;209(1):151–166. doi: 10.1016/0022-2836(89)90178-2. [DOI] [PubMed] [Google Scholar]
- Lorence J. H., Nester E. W. Multiple molecular forms of chorismate mutase in Bacillus subtillis. Biochemistry. 1967 May;6(5):1541–1553. doi: 10.1021/bi00857a041. [DOI] [PubMed] [Google Scholar]
- Rajagopalan J. S., Taylor K. M., Jaffe E. K. 13C NMR studies of the enzyme-product complex of Bacillus subtilis chorismate mutase. Biochemistry. 1993 Apr 20;32(15):3965–3972. doi: 10.1021/bi00066a017. [DOI] [PubMed] [Google Scholar]
- Richardson J. S. Schematic drawings of protein structures. Methods Enzymol. 1985;115:359–380. doi: 10.1016/0076-6879(85)15026-3. [DOI] [PubMed] [Google Scholar]
- Richardson J. S. beta-Sheet topology and the relatedness of proteins. Nature. 1977 Aug 11;268(5620):495–500. doi: 10.1038/268495a0. [DOI] [PubMed] [Google Scholar]
- Schmidheini T., Mösch H. U., Evans J. N., Braus G. Yeast allosteric chorismate mutase is locked in the activated state by a single amino acid substitution. Biochemistry. 1990 Apr 17;29(15):3660–3668. doi: 10.1021/bi00467a011. [DOI] [PubMed] [Google Scholar]