Abstract
There is accumulating evidence indicating that aldehyde dehydrogenase (ALDH) activity selects for cancer cells with increased aggressiveness, capacity for sustained proliferation, and plasticity in primary tumors. However, emerging data also suggests an important mechanistic role for the ALDH family of isoenzymes in the metastatic activity of tumor cells. Recent studies indicate that ALDH correlates with either increased or decreased metastatic capacity in a cellular context-dependent manner. Importantly, it appears that different ALDH isoforms support increased metastatic capacity in different tumor types. This review assesses the potential of ALDH as biological marker and mechanistic mediator of metastasis in solid tumors. In many malignancies, most notably in breast cancer, ALDH activity and expression appears to be a promising marker and potential therapeutic target for treating metastasis in the clinical setting.
Keywords: Aldehyde dehydrogenase, Metastasis, Solid tumors, Cancer stem cell, Biomarker
Introduction
Metastasis is a life-threatening systemic condition, with ninety percent of all cancer deaths resulting from cancer cell dissemination from the primary tumor to distant vital organs [1]. Navigation of the metastatic cascade is a complex, multistep process involving multiple tumor cell phenotypes, body compartments, and accelerated evolutionary cell trajectories [2]. Accordingly, in spite of enormous and earnest progress in elucidating the mechanisms that drive metastasis, the mortality of metastatic cancer has improved very little in the last several decades [3].
Despite the deadly nature of metastasis, it is a remarkably inefficient process. In fact, only a small fraction of cancer cells that survive in the systemic circulation are able to give rise to clinically relevant metastases [4]. Therefore, the identification, isolation, and characterization of potential metastasis-initiating cell (MIC) subpopulations has become a priority for many metastasis research groups including ours. One of the most attractive candidates for MICs are putative cancer stem cells (CSCs), which have been identified in a diverse array of hematopoietic and solid tumor types (reviewed in [5] and [6]). These CSC subpopulations can be defined by their capacity for sustained self-renewal and the ability to give rise to the heterogeneous population of cancer cells that make up a tumor. Importantly, it has also been shown that cells with a CSC phenotype characterized by high aldehyde dehydrogenase (ALDH) activity have an enhanced capacity for metastatic behavior in vitro (adhesion, colony formation, migration, and invasion) and/or metastasis in vivo [7–11], supporting the hypothesis that CSCs might act as the MIC subpopulation.
In the past several decades, increasing evidence has supported the role of ALDH as a biological marker for stem-like cancer cells and aggressive tumor cell behavior, as well as an indicator of poor clinical outcome with particular prominence in breast cancer experimental models and clinical studies (reviewed in [5, 12–15] ). In addition to its role as a detoxifying enzyme and key mediator of stem/progenitor cell expansion and differentiation, the functional and mechanistic involvement of ALDH in tumor initiation and progression has become a topic of considerable interest in the cancer field. While the involvement of ALDH in primary tumor formation, therapy resistance, and malignant behavior in vitro has been extensively described in the literature (reviewed in [5, 12–14, 16] ), the role of ALDH in metastasis has been less evident. The purpose of this review is to highlight the most recent evidence supporting a specific role for ALDH in metastasis, both in pre-clinical mechanistic studies and in vivo models, as well as in the clinical setting. Clarification of the tumor types affected, the isoforms implicated, and the underlying molecular mechanisms of ALDH in driving metastasis is necessary in order to achieve effective translational targeting of this important enzyme.
The human ALDH superfamily
Nineteen different ALDH functional genes and multiple splice variants have been characterized to date. Although they are widely expressed in multiple different tissues, these ALDH isoforms display tissue- and organ-specific expression patterns and have also been found in various cellular sub-compartments including the cytosol, nucleus, mitochondria, and endoplasmic reticulum (reviewed in [5] ). In these locations, ALDH catalyzes the oxidation of aldehydes to their corresponding carboxylic acids. For example, different ALDH families are capable of detoxifying highly reactive aldehydes that are products of lipid peroxidation (ALDH1, ALDH3, ALDH8) [17–19]. Others are critical regulators of the retinoic acid pathway through involvement in the catalysis of retinaldehyde to retinoic acid, and therefore play an important role in stem and progenitor cell expansion and differentiation (ALDH1A1, ALDH1A2, ALDH1A3) [20]. ALDH also has been found capable of inactivating xenobiotics, including the alkylating agent cyclophosphamide (CP) and analogous chemotherapeutic drugs (ALDH1A1, ALDH3A1) [21]. In addition, it has been observed that ALDH is mechanistically involved in other diverse cell activities including structural and osmoregulatory functions (ALDH1A1, ALDH3A1, ALDH7A1) [14, 22]. Importantly, the ability of ALDH to regulate cell proliferation and self-protection is believed to contribute its involvement in mediating CSC capabilities such tumor progression, phenotypical heterogeneity, and therapy resistance [5].
Functional role of ALDH in cancer
ALDH and retinoic acid signaling in cancer cells
The retinoic acid signaling pathway has been implicated in normal and cancer cell function including the regulation of gene expression and development [23–26]. In tumor biology, the retinoic acid pathway appears to play a dualistic role involving epigenetic context-dependent differential gene expression, mediation of apoptotic pathways, and immune system regulation [10, 20]. The human cytosolic ALDH1A subfamily (made up of ALDH1A1, ALDH1A2, and ALDH1A3) irreversibly oxidizes retinaldehyde to retinoic acid (RA). Subsequently, RA is translocated to the nucleus where it is able to regulate the transcriptional activity of more than 500 genes through activation of retinoic acid receptor (RAR), retinoic X receptor (RXR), and nuclear hormone receptors peroxisome proliferator activated receptor beta/delta (PPAR/β/δ). RA exerts differential effects on tumor growth in a cell context-dependent manner. It has been found that retinoic acid activation of RARs and RXRs is followed by cell cycle arrest and differentiation due to increased transcription of tumor suppressor factors, whereas RA activation of PPARs has been found to mediate the increased expression of oncogenes and subsequent cell cycle progression observed in other experimental model systems (reviewed in [20]). In addition, it has been observed that the influence of RA on tumor growth might be mediated by epigenetic silencing of RA-inducible tumor suppressors [10, 27].
Importantly, RA-producing ALDH isoforms, including ALDH1A1 and ALDH1A3 (but not ALDH1A2), are among the most common ALDH isoforms associated with a diverse arrange of hematopoietic and solid tumors. There is accumulating evidence that their increased expression selects for tumor cell subpopulations exhibiting stem-like or aggressive tumor cell phenotypes, indicating that the tumorigenic “branch” of the retinoic acid pathway might be co-opted in some tumors and/or that other non RA-dependent functional roles of ALDH1A1 and ALDH1A3 might be involved in tumorigenesis [7, 9, 10, 28]. For example, ALDH1A1 and ALDH1A3 have been shown to play functional roles in lung and breast cancer cell migration and invasion, although the mechanisms underlying this behavior have not been established [10, 29, 30]. It is of note that RA has been used to dramatically improve clinical outcome in patients with acute promyelocytic leukemia (APL); where ninety-eight percent of patients with this disease carry a fusion of the PML and RARα genes that impairs the ability of RARα to induce hematopoietic stem/progenitor cell differentiation at physiological levels of RA [31, 32]. However, attempts to use RA to induce tumor cell differentiation in other cancer types have shown mixed results at best in clinical trials (reviewed in [20] and [16]). The potential causes of the disparate results obtained after targeting the retinoic acid pathway in cancer are discussed later in this review.
ALDH plays a self-protective role in cancer cells
There is increasing evidence implicating ALDH in cancer cell self-protection against both endogenous and exogenous threats. Antioxidant and substrate-specific drug inactivation are among the key mechanisms underlying these capabilities. For example, ALDH has a NADPH recycling function that is believed to support antioxidant cell capabilities. In addition, it has been observed that ALDH is often co-expressed with antioxidant factors and drug efflux channels in cells with high ALDH activity [22, 33–39]. ALDH1A1 and ALDH1A3 are capable of enzymatic inactivation of alkylating agents such as CP and other oxazaphosphorines [40–43]. Moreover, ALDH appears to confer resistance to drugs other than CP and analogues including doxorubicin, cisplatin, arbinofuranosyl citidine (Ara-C), temozolemide and taxanes [28, 44–47], although the mechanisms underlying this are less clear. A comprehensive review of ALDH involvement in chemotherapy and radiotherapy resistance has recently been published by Januchowski et al. [21]. Thus, ALDH is not only involved in physiologic and cancer cell proliferation and differentiation, but also in promoting tumor cell survival through direct inactivation, indirect expulsion of xenobiotics, and enhancement of the oxidative stress resistance response.
ALDH as a marker for cancer stem cells
Assessment of ALDH in cancer research
Different approaches have been used to quantify ALDH in cancer cells and tumor tissue. Early methods for determining ALDH levels relied on measuring enzyme kinetics or immunoblotting of enzymes released after cell lysis, in addition to immunohistochemical detection. However, these methods are flawed by antibody cross-reactivity between enzyme isoforms and enzyme structural instability [48, 49]. An alternative and more recent approach consists of measuring ALDH activity in viable cells by quantifying the ALDH-mediated intracellular retention of the fluorescent compound BODIPY-aminoacetate (BAA−) using flow cytometry-based methods [7, 23, 50–52]. This functional assay is commercialized as the ALDEFLUOR™ assay and depends on the conversion of the uncharged ALDH substrate BODIPY amino acetaldehyde (BAAA), which freely diffuses in and out of the cell, into the negatively charged BAA-compound. As a result, after addition of BAAA, cancer cell subpopulations with elevated activity of ALDH (ALDHhi) become highly fluorescent and can be identified using flow cytometry gating criteria. The ALDHhi cell subset can be distinguished when compared to cells treated with the ALDH1 inhibitor 4-(diethylamino) benzaldehyde (DEAB), which is used as a reference control. Although the ALDEFLUOR™ assay works well in various human models, there is controversy about its adequacy for identifying cancer stem cells in murine models given disparate results among groups isolating murine HSCs using this technique [53–56]. However, enrichment of murine CSCs have been recently reported using the ALDEFLUOR™ assay in two different models of mouse breast cancer [57, 58]. In addition, attempts at in vivo stem cell labeling have employed ALDH radiolabeled substrates. However, the resultant charged compounds were not polar enough to be retained into the cells [59].
ALDH isoforms involved in the ALDEFLUOR™ assay
Early reports identified ALDH1 as the main ALDH enzyme family responsible for the enzymatic activity reported using the ALDEFLUOR™ assay [52, 60]. However, recent controversy has emerged regarding the family and subfamily isoforms acting on the BAAA substrate and thus responsible for the readout in this assay. This distinction is of importance given the ample and tissue-specific distribution of ALDH isoforms in normal tissue, the reliance on the ALDEFLUOR™ assay for isolation of viable CSCs for subsequent downstream assessment, and differential roles of different ALDH1 isoforms in tumorigenesis and metastasis [10]. In breast cancer, mixed results have been reported regarding the specific ALDH1A isoenzyme involved in tumorigenesis and the ALDEFLUOR™ assay, with some groups reporting ALDH1A1 as the main enzyme involved and others showing that ALDH1A3 is the accountable isoform [9, 10, 61, 62]. Recent research by our group using human breast cancer cell lines indicate that although both isoforms are involved in different phases of the metastatic cascade, 50 % of the ALDEFLUOR™ activity is driven by the ALDH1A3 isoform with no significant participation of ALDH1A1 in this assay (our unpublished data). In addition, other studies have found enzymatic participation of ALDH1A1, 1A2, 1A3, 2, 7A1 and 8 in the ALDEFLUOR™ assay in other tumor types [22, 63–65]. Thus, multiple ALDH isoforms may contribute to both the readout of the ALDEFLUOR™ assay and to cellular function in tumorigenesis in a tissue-specific manner.
It is important to clarify that, by current nomenclature, the term ALDH1 does not accurately describe any of the isoforms of the ALDH superfamily [12] and could refer to ALDH1A1, ALDH1A2, ALDH1A3, ALDH1B, ALDH1L1, or ALDH1L2. This is a major problem in the ALDH/cancer stem cell literature, where ALDH1A1 is often used interchangeably with ALDH1 or ALDH. Further confusion arises when ALDEFLUOR™ positivity is referred to as ALDH1 activity or positivity. To address this ambiguity, in the current review we will refer to ALDEFLUOR™ positive cells or ALDEFLUOR™ activity whenever the source refers to a phenotype identified using the ALDEFLOUOR™ assay. Notably, Pors et al. [12] recently reported that the specific isoform identified using BD Biosciences clone 44 or Abcam ab52492 antibodies against ALDH1 is ALDH1A1. In this review, we will therefore clarify this whenever ALDH1 expression is reported by authors using either of these antibodies. In addition, if the isoform specificity of the ALDH1 antibody could not be established, this will also be noted.
Recent evidence supporting ALDH as a CSC marker
Multiple markers have been used for enriching cells with stem-like properties from human tumor primary tissue and human cancer cell lines, and this is reviewed exhaustively elsewhere in the literature [6, 66–70]. We have previously reviewed the relevance of ALDH as a marker for normal and cancer stem cells [5]. Thus, for this review we have summarized the latest findings regarding the importance of ALDH as a cancer stem cell marker since 2010 (summarized in Table 1).
Table 1.
Recent evidence supporting ALDH as a marker of CSCs and cancer progression
| Tumor Type | ALDH isoform | Method of ALDH assessment | Functional/mechanistic observations associated with ALDH | Clinical/prognostic observations associated with ALDH |
|---|---|---|---|---|
| Breast cancer |
ALDH [9, 78, 87, 136, 148, 149] ALDH1 (ALDH1A1 as per [12] ) [78, 87, 119, 135, 149, 180, 181] ALDH1A1 [61, 62, 147] ALDH1A3 [9, 10, 182, 183] |
ALDEFLUOR™ [9, 78, 87, 136, 148] IHC [9, 10, 61, 62, 78, 87, 119, 135, 149, 180–184] Immunoblotting [147] qPCR [9, 10] |
Increased Notch and β-catenin levels, increased Ki67 [148] Increased HIF-1/2α expression [149] Increased HOXA1 and MUC4 expression [10] Increased in vitro invasion [10, 78], growth in 3D Matrigel™ [78], Increased primary tumor engraftment in patient derived xenograft (PDX) models [87] ALDH1A1 deacetylation regulated by Notch increases tumorigenicity in vivo [147] Dual role of ALDH1A3 in tumorigenicity and metastasis in vivo depending on epigenetic landscape [10] |
Poor clinical outcome alone [10, 135, 136, 149] or combined with CD44+CD24− phenotype [180] Increased ALDH expression in metastases vs. primary tumors [119] Tumor recurrence [181] and/or metastasis [9, 183] More common in younger woman [184] and those with TNBC [10, 62, 182, 184] Increased tumor stage [9, 62, 182] and/or grade [9] Poor response to chemotherapy [62] Familial risk for breast cancer [61] |
| Ovarian cancer | ALDH [79, 143, 185] ALDH1 [89] ALDH1 [80] (no isoform annotated) ALDH1A1 [90, 143] |
ALDEFLUOR™ [79, 80, 89, 90, 143] IHC [80, 185] Immunoblotting [143] qPCR [90, 143] |
Increased in vitro growth, migration, invasion [79, 80], colony formation, resistance to hypoxia [19] and therapy [79, 143], sphere-formation [80, 90], dormancy [143], DNA repair signalling [143], expression of β-catenin [90] Increased tumorigenicity in vivo [79, 80, 89, 90] Increased sphere formation, regulation of FoxM1 and Notch 1 [89] |
Poor clinical outcome [80, 143, 185] |
| Brain cancer | ALDH [91] ALDH1 [186] ALDH1 [92] (ALDH1A1 as per [12] ) ALDH1A3 [108, 187] |
ALDEFLUOR™ [91, 186] IHC (93) qPCR [108] DNA methylation profiling [187] |
Increased in vitro sphere formation [91, 186] asymmetric division [91, 92], growth [108], therapy resistance [108] Increased tumorigenicity in vivo [91, 92, 108] |
Poor clinical outcome [187] |
| Bone cancer | ALDH [85] | ALDEFLUOR™ [85] | Increased therapy resistance [85] | Increased metastases [85] |
| Prostate cancer | ALDH [11, 83, 86, 188] ALDH7A1 [11] ALDH3A1 [109] |
ALDEFLUOR™ [11, 83, 86, 188] IHC [11, 109] |
Increased in vitro colony-forming ability [11, 188], invasion [11, 83], sphere formation [11, 109], migration [11, 83] and therapy resistance [86] TGF-β [11] and Wnt/β-catenin [86] induce ALDH activity Increased in vivo tumorigenicity [11, 86, 188] |
Increased expression in metastases [109] |
| HNSCC | ALDH [93, 189] ALDH1 [189] (ALDH1A1 as per [12] ) |
ALDEFLUOR™ [93, 189] IHC [189] |
Increased in vitro sphere formation [93] Increased Nanog, Oct-3/4, and Stella expression [189] Increased Bmi-1 and Snail expression [93] Increased tumorigenicity in vivo [93, 189] |
Poor clinical outcome [93] |
| Colorectal cancer | ALDH [95] ALDH1 [94, 122] (ALDH1A1 as per [12]) ALDH1 [120, 190] (No isoform annotated) |
ALDEFLUOR™ [95] IHC [94, 120, 122, 190] |
Increased in vitro sphere formation [94, 95], in vitro colony formation [94] ALDH expression is correlated with β-catenin expression [122] |
Poor clinical outcome [94, 122, 190] Loss of ALDH expression in advanced stage disease [120] |
| Lung cancer | ALDH [96–98, 100] ALDH1A1 [29, 100, 101, 105, 191] ALDH7A1 [192] |
ALDEFLUOR™ [96–98, 100] IHC [29, 100, 101, 105] Immunoblotting [191, 192] |
Increased in vitro colony formation [29, 100], sphere formation [96–98] and migration [29] Increased in vivo tumorigenicity [29, 97, 100] Increased therapy resistance [105] Increased Notch [100, 101], Rac1 signaling [98] and RhoA/Rock signaling [191] Increased SOX2, CXCR4 and SDF-1 expression [96] Salinomycin targets ALDHhi cells and reduce xenograft growth [96] |
ALDH1A1 correlated with poor clinical prognosis [29, 100
Increased recurrence [192] |
| Cervical cancer | ALDH [99] | ALDEFLUOR™ [99] | Increased colony formation and sphere formation [99] Increased Twist 1, Twist 2, Snail 1, Snail 2 and Vimentin expression [99] |
Not assessed |
| Melanoma | ALDH [65, 106, 193] ALDH1A1 [65, 106] ALDH1A3 [65] |
ALDEFLUOR™ [65, 106, 193] Immunoblotting [106] qPCR [65] |
Increased in vivo tumorigenicity [65, 106, 193] Increased therapy resistance [65, 106] Increased RA response and stem cell gene expression [65] |
Not assessed |
| Endometrial cancer | ALDH1 [150] (ALDH1A1 as per [12] ) | ALDEFLUOR™ [150] IHC [150] |
Nodal inhibited ALDH expression trough ubiquitin–proteasome pathway [150] | Not assessed |
| Renal cancer | ALDH1 [142] (no isoform annotated) ALDH1A1 [104] |
IHC [104, 142] Immunoblotting [104] qPCR [104] |
Increased invasion [104] Increased therapy resistance [104] Increased in vitro tumorigenesis [104] |
Increased tumor grade [142] Poor clinical outcome [104] |
| Pancreatic cancer | ALDH1A1 [103] ALDH1A3 [107] |
qPCR [103] Chromatin immunoprecipitation [107] |
Increased colony formation [103] TGF-β/Smad4 inhibition of ALDH transcription [103] Correlation with cell adhesion, growth factor and receptor activity, transcription and differentiation [107] |
Not assessed |
| Hepatobiliary cancer | ALDH [151] ALDH1A3 [194] ALDH3A1 [110] |
ALDEFLUOR™ [151] IHC [110, 194] |
Increased in vitro cell proliferation [151] TGF-β induces ALDH expression [151] Increased in vivo tumorigenicity [151] Increased Wnt/β-catenin activity [110] |
Poor clinical outcome [151, 194] No correlation with clinical outcome [110] |
| Oesophageal cancer | ALDH [82, 102] ALDH1 [82] (no isoform annotated) ALDH1A1 [102] |
ALDEFLUOR™ [82, 102] IHC [82, 102] |
Increased sphere formation, therapy resistance and SOX9 and YAP1 gene expression [82] Increased colony formation and invasion [102] Increased expression of VIM, MMP2, MMP7 and MMP9 [102] Decreased expression of ECAD [102] Increased tumorigenicity in vivo [102] Increased therapy resistance [82] |
Poor clinical outcome [102] |
ALDH and metastasis
While there is growing evidence supporting the use of ALDH as a CSC marker and implicating it as having an important functional role in tumor cell self-protection, differentiation, expansion, and therapy resistance, less is known about its functional role in mediating metastasis. In this section we will review the mechanistic involvement of ALDH in metastasis based on experimental evidence derived from assessment of in vitro of cellular behaviors contributing to metastasis, in vivo animal models of metastasis, and patient-derived metastasis samples.
Functional association of ALDH with in vitro cell behaviors related to metastasis
Different in vitro assays have been used to model and estimate potential metastatic activity in vivo [71]. Clonogenic assays involving loss of substrate adherence or serum-free media, including colony formation in soft agar and tumorsphere growth in ultra-low attachment plates, are correlated with tumorigenicity and stemness. Therefore, they are often used to evaluate the capability of cancer cells to initiate metastatic growth in vivo [72–75]. Assessments of migration and invasion in vitro have also been used to estimate metastatic potential in vivo [71, 72]. In addition, given the intrinsic resistance of metastases to chemotherapy and radiotherapy, in vitro assessment of therapy response has also proven to be useful [72, 74, 76, 77]. ALDH and several of its isoforms have been recently evaluated for and positively correlated with multiple in vitro cell behaviors that are surrogates of metastatic behavior in vivo.
Tumor cells displaying high ALDEFLUOR™ activity have been demonstrated to have enhanced motility and ability to invade through a 3D basement membrane in breast cancer [7, 78], ovarian cancer [79, 80], osteosarcoma [36, 81], esophageal cancer [82], and prostate cancer [11, 83]; as well as increased therapy resistance in breast cancer [28, 84], ovarian cancer [79], osteosarcoma [85], and prostate cancer [86]. ALDEFLUOR™-positive cells have also been reported to exhibit increased capacity to form tumorspheres in breast cancer [8, 15, 52, 87], human malignant fibrous histiocytoma (HMFH) [88], ovarian cancer [80, 89, 90], brain tumors [91, 92], prostate cancer [11], head and neck squamous cell carcinoma (HNSCC) [93], colon cancer [94, 95], non-squamous cell lung cancer (NSCLC) [96–98], esophageal cancer [82], and cervical cancer [99]. Interestingly, ALDH-positive cells display additional stem-like behaviors such as resistance to hypoxia in ovarian cancer [79] and the capacity for asymmetric division in brain tumor cells [91].
Several specific ALDH isoforms have been correlated with in vitro metastatic behavior. For example, ALDH1A1 expression has been reported to correlate with increased in vitro clonogenic activity in NSCLC [29, 100, 101], esophageal cancer [102], ovarian cancer [90], pancreatic cancer [103], and renal cancer [104]. This ALDH isoenzyme has also been correlated with increased migratory capabilities in lung cancer [29], renal cancer [104], and esophageal cancer [29] and in vitro therapy resistance in lung cancer [105], melanoma [65, 106], and renal cancer [104]. Other ALDH isoforms have been mechanistically associated with metastatic behavior in vitro, including ALDH1A3 in breast cancer [9, 10], melanoma [65], pancreatic cancer [107] and brain cancer [108]; ALDH3A1 in prostate cancer [109] and liver cancer [110]; and ALDH7A1 in prostate cancer [11]. In summary, increasing evidence from in vitro studies suggests a mechanistic role for ALDH in metastasis and have laid the groundwork to further the study of the involvement of this enzyme in metastatic activity in vivo.
Functional association of ALDH with in vivo metastasis
Croker et al. [7] and Charafe-Jauffret et al. [111], using the ALDHhiCD44+CD24− and ALDHhi and phenotypes respectively, provided the first direct experimental evidence implicating ALDHhi cells in breast cancer metastasis in vivo. Moreover, it was shown that ALDHhiCD44+CD24− cells were the only cancer cell subpopulation capable of metastasizing beyond the lungs in a pattern that mirrors the clinical behavior of breast cancer [7]. Concurrently, it was also reported that ALDH1A1+CD44+CD24+ cells correlated with metastatic activity in matched primary and metastatic samples from pancreatic cancer [112]. Subsequent studies have shown that high ALDEFLUOR™ activity enriches for cells with increased metastatic capability in vivo in prostate cancer [83, 113, 114], breast cancer [15, 33], HNSCC [93], osteosarcoma [81], esophageal cancer [102], ovarian cancer [79], hepatic cancer [95], and adenoid cystic carcinoma [115]. More recently, other studies have further investigated the specific ALDH isoenzymes correlated with metastatic activity in vivo. For example, ALDH7A1 has been implicated in prostate cancer metastatic activity after left ventricular injection (LVI) of a prostate cancer cell line [11], while ALDH3A1 has been shown to be associated with metastasis using a mouse tail vein injection model [109]. ALDH1A3 has been observed to be involved in metastasis in vivo in breast cancer after cancer cell xenotransplantation into mouse models [10]. It has also been reported that inhibition of ALDH1A1 by RNA interference in melanoma cell results in reduced metastatic ability in vivo [106], providing functional validation and indicating that ALDH is not only a biological marker for enhanced metastatic ability, but also plays a functional role in metastasis in vivo. Interestingly, in one study, ALDH1A3 was found to promote or inhibit breast cancer metastasis depending on the specific epigenetic context framing retinoic acid signaling in vivo [10]. Another elegant study reported that immune targeting of breast CSCs cancer stem cells using ALDH1A1-specific CD8+ T cells resulted in reduced metastatic activity of breast cancer cells in vivo [116]. Interestingly, this study reported that ALDH1A1 was the main ALDH isoenzyme determining of ALDEFLUOR™ assay activity.
Association of ALDH and metastasis in clinical tissue samples
The association of ALDH and metastases in the clinical setting has been of interest since the late 1980s, when Marselos et al. showed increased enzymatic activity of ALDH (no isoform specified) in metastatic lesions from colon cancer compared with normal adjacent tissue [117]. This study would contrast with another early publication indicating that ALDH activity was not elevated in the serum of patients with metastatic hepatic cancer when compared with serum of patients with non-metastatic cancers [118]. ALDH expression has been assessed in matched primary tumor and metastases tissues in breast cancer [119], colorectal cancer [94, 120, 121], pancreatic cancer [112], and hepatic cancer [95]. The majority of these reports have shown a positive correlation between ALDH1 expression (ALDH1A1 as per [12] ) and metastasis. However, these results should be interpreted with caution due to technical and ethical limitations of working with metastatic tissue, including small numbers of samples analyzed and lack of consistency in staining and scoring methods. For example, diverse grading scales have been used in these studies to score ALDH staining in pathological specimens, including dichotomous scales and continuous scales with diverse cutoff points. This lack of consistency contrasts with the standardized in vivo functional evaluation of ALDH activity using the ALDEFLUOR™ assay. In a recent study using matched colon primary tumor and metastases, Fitzgerald et al. reported ALDH1 (ALDH1A1 as per [12] ) as a predictor of poor clinical outcome [94], in direct contrast with an earlier study that had found a negative correlation [120]. In a study that might shed light in this controversy, using non-matched primary tumor and metastases in colorectal cancer tissues, only homogeneous and intense expression of ALDH1A1 was correlated with metastasis [122].
In addition to ALDH activity/expression in tumor cells, ALDH expression has been recently also been observed in tumor-associated endothelial cells (TEC) from melanoma and oral cancers in vivo. TEC were shown to exhibit increased expression of angiogenic factors and angiogenic behavior in vitro, suggesting that there might be an ALDH-mediated CSC activity in the tumor vascular compartment which in turn could promote tumor cell escape to the intravascular space [123].
ALDH in circulating tumor cells (CTCs) and disseminated tumor cells (DTCs)
The detection of circulating tumor cells (CTCs) in blood and disseminated tumor cells (DTCs) in bone marrow has been associated with the presence of clinically relevant metastatic disease and poor clinical outcome for a diverse group of solid tumors [124–127]. However, given the high inefficiency of the metastatic cascade, it has been hypothesized that CTCs and/or DTCs may contain sub-populations of cells with enhanced capacity to initiate and maintain growth and give rise to clinically relevant metastases. The biological characteristics and markers of metastasis-initiating CTCs and DTCs are not completely understood. The preclinical and clinical data reviewed above indicate that ALDH is functionally involved in metastatic activity and is a determinant of cancer clinical outcome, and thus implicate ALDH as a potential biomarker to be assessed in correlation with CTC/DTC activity.
To our knowledge, the first attempt to assess ALDH activity in the systemic circulation in correlation with metastasis was published by Jelski et al. [118]. Evaluating serum alcohol dehydrogenase (ADH) and ALDH activity in a small sample of patients with metastatic hepatocarcinoma, this study found a correlation for ADH, but not for ALDH, with metastatic disease. However, early and subsequent attempts at identifying ALDH expression in CTCs/DTCs sub-populations have been successful in breast and endometrial cancer [128–133]. While the scope of these studies is complex given the multi-compartment distribution of CTCs/DTCs, in general they evaluate one or more of the following variables: (i) expression of ALDH in primary tumors in correlation with CTCs/DTCs, (ii) expression of ALDH in CTCs/DTCs as a stem cell marker and its correlation with clinical prognosis and metastatic activity; and/or (iii) co-expression of ALDH with other known markers of stem-like behavior and cancer progression in CTCs/DTCs. Interestingly, the results of these studies are mixed, but this is not surprising given the nascent nature these type of studies, the small cohorts evaluated, the diversity of assays and parameters used to identify and isolate CTCs/DTCs, the inherent technical difficulties of isolating rare cells form clinical samples, and uncertainty about the biological characteristics of clinically relevant CTCs/DTCs that would allow their identification.
In one study involving 502 non-metastatic breast cancer patients, although ALDH was expressed in CTCs and DTCs in 14 % of patients in the study cohort, it was not correlated with clinical prognosis or metastasis. In addition, this study showed no correlation of EMT markers in CTCs/DTCs with clinical outcome [134]. In another study, ALDH expression in primary breast tumors was not found to be correlated with the presence of CTCs, although it was correlated with clinical outcome in patients with non-metastatic disease [135]. In an additional cohort of non-metastatic breast cancer patients, although ALDH expression in DTCs was correlated with use of neoadjuvant chemotherapy and high tumor grade, it was not associated with metastatic recurrence [130]. Another study in a non-metastatic breast cancer cohort showed that the CD44+CD24− phenotype in DTCs was a better clinical predictor of cancer progression relative to ALDH expression [136]. In general, studies evaluating ALDH expression in CTCs/DTCs from patients with early non-metastatic breast cancer have shown less correlation of ALDH in CTCs/DTCs with clinical outcome and metastasis than studies performed in patients with metastatic disease [128, 129]. Therefore, although there is evidence supporting ALDH as a marker of CTC and DTC activity in advanced breast cancer, the functional and clinical implications of ALDH expression in CTCs/DTCs and eventual metastasis in breast cancer patients with early disease remains to be established. This suggests that the use of ALDH in CTCs/DTCs as a risk stratification marker might not be useful in patients with non-advanced disease. However, as described below, important technical considerations involving the methods to enrich CTCs/DTCs might change these results.
It is very important to note that in all the studies mentioned above, CTCs/DTCs were enriched using epithelial markers as primary criteria for their isolation. However, there is increasing evidence suggesting that a substantial proportion of CTCs/DTCs might display a more mesenchymal and aggressive phenotype that may not be picked up by standard CTC/DTC assays [124]. In fact, it has been shown that mesenchymal markers are over-expressed in CTCs and DTCs of metastatic and aggressive subtypes of breast cancer [137, 138]. Moreover, ALDH expression in CTCs has been correlated with poor clinical outcome, metastatic progression, and therapy response in patients with metastatic breast cancer [128, 129]. Interestingly, Liu et al. observed that breast CSCs transition between mesenchymal an epithelial states in order to fully develop metastasis, with ALDH participating during the epithelial and more proliferative phase of metastatic colonization in distant tissues. This suggests that ALDH may not be a key marker expressed during the intravascular and more mesenchymal phase of the metastatic cascade [139, 140]. Overall, the study of ALDH as a biomarker for metastasis-initiating cells and clinical outcome in CTCs/DTC is an emerging and promising field of research that is evolving at the pace of the refinement and validation of the technologies used to isolate this important group of cells navigating the metastatic cascade in the systemic circulation.
Molecular mechanisms associated with ALDH in metastasis promotion
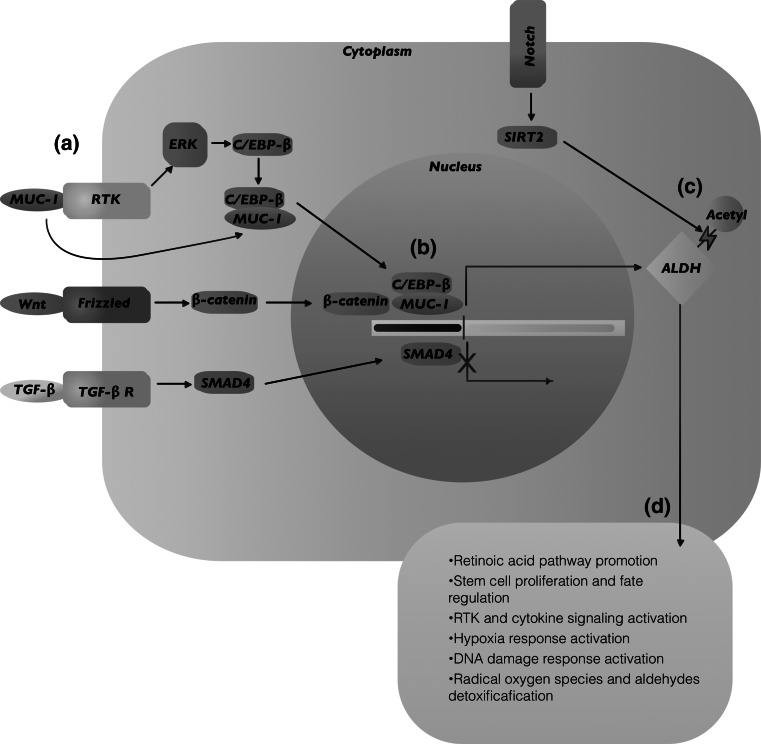
The study of the molecular mechanisms underlying the increased tumorigenicity of ALDHhi cells has revealed diverse co-expressed molecular factors and signaling pathways that might potentially also explain the observed increased metastatic behavior of ALDHhi cancer cells. Since 2007, when the Hedgehog pathway was shown upregulated in ALDHhi pancreatic cancer cells [141], there has been accumulating evidence of signaling pathways associated with ALDHhi cells and their malignant activity (Summarized in Fig. 1). In particular, pathways involved in stem cell proliferation and cell fate [93, 101, 110, 141–147]; EMT [11, 36, 78, 81, 86, 90, 100, 110, 112, 146–155]; retinoic acid pathway [10, 89]; hypoxia [33, 79, 149, 156] and DNA damage responses [143, 157, 158]; cytokine and receptor tyrosine kinase (RTK) [158–160] signaling activation; and cell migration [161], among others, have been implicated in promotion of aggressive behavior in ALDHhi cells. Different isoforms of the Notch receptor have been found to be upregulated in ALDHhi cells from breast cancer, ovarian cancer, lung cancer, and osteosarcoma [81, 89, 100, 144, 145, 147, 148]. In addition, ALDH1A1 has been found to be correlated with Notch expression in lung and breast CSCs [100, 147]. The Wnt-β-catenin pathway has been shown to be activated in cancer cells with high expression of ALDH1A1 and ALDH3A1 in prostate, ovarian, and liver cancer cells [86, 90, 110]; and the TGFβ pathway has been reported to facilitate therapy resistance in ALDHhi breast cancer cells, and to be involved in expression of ALDH1A1 in cholangiocarcinoma and pancreatic cancer [90, 103, 151, 152].
Fig. 1.
Molecular mechanisms associated with ALDH in metastasis promotion. The molecular mechanisms underlying the increased tumorigenicity and metastatic capacity of ALDHhi cancer cells involve diverse co-expressed molecular factors and signaling pathways. For example, in breast, ovarian, and pancreatic cancer, ALDH1A1 transcription has been shown to be regulated after binding of C/EBPβ, β-catenin, or Smad-4 to the ALDH1A1 promoter sequences (a, b). In breast cancer cells, Notch-induced deacetylation of ALDH1A1 can result in increased CSC capabilities (c). Taken together, these pathways influence downstream functional behaviors such as stem cell-related decisions regarding proliferation and cell fate, epithelial-to-mesenchymal transition, retinoic acid synthesis, hypoxia, DNA damage response, cytokine and RTK signaling activation, and cell migration (d), all of which may contribute to the role of ALDHhi cancer cells in metastasis promotion
Interestingly, for some of these signaling pathways, the evidence suggests dualistic roles in metastasis depending on the cell context. For example, Li et al. reported that ALDH1A1 was involved in lung CSC activity via suppression of the Notch/CDK2/CCNE pathway [101], and, in other study, it was found that Nodal had a tumor suppressor function in ALDHhi cells via the activation of TGFβ pathway [150]. It is of note that there is evidence supporting a more complex model of metastasis navigation in which not only the EMT process but also the mesenchymal-to epithelial-transition (MET) processes are both critical for forming clinically relevant metastases [139, 140]. In this regard, research from two independent groups has shown that ALDH may be a marker and potential mechanistic promoter of a more proliferative/epithelial CSC phenotype via MET, while CD44 may, in contrast, be a marker of a more invasive subpopulation of migratory/mesenchymal CSCs via EMT [78, 146]. In addition, Marcato et al. have reported that ALDH1A3 may have a dual role in breast cancer metastasis promotion depending in the epigenetic cell context through differential retinoic acid signaling [10].
Therapeutic potential of targeting ALDH in metastasis treatment and prevention
Although there are relatively few studies assessing the impact of pharmacological or immune targeting of ALDH on metastasis in vivo, the majority of them are consistent in reporting a decrease of metastatic burden after targeting of ALDH. It has been shown that after treatment with the ALDH inhibitors DEAB and the novel A37 compound, there is a decreased metastatic activity in murine models of breast and ovarian cancer [33, 90]. It has also been demonstrated that CSC targeting with ALDH1A1-specific CD8+ T cells is followed by decreased spontaneous metastatic burden of HNSCC, pancreatic, and breast cancer cells in vivo [116]. Treatment with RA results in downregulation of ALDH1A1/ALDH3A1 expression or decreased ALDEFLOUR™ activity [28, 42, 89], and, in one recent study it has been reported that there is a reduction of in vitro metastatic behavior and reduction of xenograft growth of ovarian cancer cells after RA treatment [89]. However, Marcato et al. have reported that RA treatment has dual effects in a mouse model of human breast cancer metastasis depending on the cell epigenetic context [10].
The small molecule compound disulfiram has been found to display tumor inhibitory activity attributed to different properties, including ALDH1A1 inhibition [35, 162, 163], inhibition of proteasome activity [164, 165], prevention of NF-κB translocation [165, 166], induction of reactive oxygen species generation [167, 168], blockade of the PI3K/PTEN/AKT signaling pathway [169], inactivation of tumor-associated enzymes [170–172], and suppression of metastasis-associated gene expression [170, 171, 173]. Interestingly, it has been reported that disulfiram may have a therapeutic role in the metastatic setting. In one study, cell growth of metastatic osteosarcoma patient-derived cells was significantly decreased after disulfiram treatment in vitro [85]. In another study, cell growth was significantly decreased after disulfiram treatment of a metastatic murine osteosarcoma cell line in vitro [36]. It has also been reported that treatment with this compound decreases spontaneous lung metastatic burden in a syngeneic pre-clinical model of metastatic murine breast cancer [174]. In addition, a recent Phase IIb clinical trial found that addition of disulfiram to chemotherapy was well tolerated and appears to improve survival in newly diagnosed patients with metastatic non-small cell lung cancer [175]. Currently two clinical trials are evaluating the effect of disulfiram in treating glioblastoma multiforme (NCT01907165 and NCT01777919; www.clinicaltrials.gov).
The multiplicity of ALDH isoforms and its widespread tissue/tumor distribution, combined with differential epigenetic landscapes and inconsistency among parameters evaluated in small cohort trials may explain discrepancies observed between preclinical and clinical outcomes. Moreover, selective druggability of different ALDH isoforms is a formidable pharmacological challenge that may also contribute to conflicting results. Physiological concentrations of ALDH are most highly expressed in kidney and liver, which in turn limits the use of nonspecific inhibitors of ALDH that may result in toxic side-effects in patients. In addition, the enzymatic oxidative reaction carried out by different ALDH isoforms is highly nonspecific. For example, the ALDH substrate BAA has been found to be metabolized by different ALDH isoenzymes, including ALDH1A1, 1A2, 1A3, 2, 3A1, and 7 [4, 39, 67, 85]. Thus, there are important biochemical barriers to device a specific ALDH family or isoform inhibitor.
Rationalized small molecule discovery has been proposed as a viable methodology to surmount these difficulties and has been successfully employed to target specific aldehyde oxidases of the cytochrome P450 family in cancer cells [176–179]. An array of new generation isoform-specific ALDH inhibitors are under development. Among them, duocarmycin analogues stand out due to their ultra-high alkylating potency and additional ability to specifically target ALDH1A1 [12]. Therefore, it is expected that novel, potent, and isoform-specific ALDH inhibitors could enter the pipeline of experimental and clinical assessment in cancer therapy in the coming years. Taken together, these results underscore the potential of ALDH as a therapeutic target against metastasis.
Conclusions and future perspectives
Tumors are heterogeneous at the genetic, epigenetic, and tumorigenic level. There is substantial evidence indicating that tumor cells with stem-like capabilities are responsible, at least in part, for heterogeneity at the tumorigenic and metastatic level. ALDH stands out among the expansive and diverse group of CSC markers because of its widespread association with different types of solid tumors and the multiplicity of its biological functions, including retinoic acid signaling, antioxidant protection, osmoregulation, drug metabolism, and structural support. However, validation of ALDH as a prognostic biomarker and/or therapeutic target in the clinical setting has not yet come fully to fruition. Moving forward, it is critical that future studies include better standardization of ALDH identification and scoring methods, patient characteristics, and cohort sizes. In addition, more attention must be drawn to the study of the therapeutic effects of ALDH isoenzyme inhibitors in CTCs, DTCs, and metastatic and migratory activity. We believe that only a consistent preclinical and clinical approach revolving around CSC-mediated metastasis and therapy resistance will reveal the therapeutic and biomarker potential of ALDH in solid tumors.
In conclusion, it is clear that ALDH is not only a marker for aggressive stem-like and metastatic cells, but that it is also mechanistically involved in these behaviors. Hence, the study of ALDH as a biomarker and functional mediator of metastasis in vivo is a promissory field for discovering targets that might interfere with solid tumor progression. However, with the heterogeneity of ALDH isoforms described as CSC markers in different tumor types, and the newly described cell context dependent tumorigenic function of ALDH, it is likely that different isoforms may contribute differently to metastasis in different types of solid tumors. Moreover, given that it has been shown that other isoenzymes besides ALDH1A1 and ALDH1A3 are responsible for the activity reported in the ALDEFLUOR™ assay, and the experimental evidence supporting multi-enzyme isoform participation in the same tumor, it is also likely that more than one ALDH isoform may be contributing to progression within the same tumor. Continued intensive investigation into the functional contribution of ALDH to cancer progression and metastasis will be important for tackling the enormous therapeutic challenge that such diverse landscape imposes.
Acknowledgments
We thank members of our laboratory and our collaborators for their research work and helpful discussions. The authors’ research on ALDH and CSCs is supported by research grants from the Canadian Breast Cancer Foundation (Ontario Region) and donor support from John and Donna Bristol and Richard and Susanne Shaftoe through the London Health Sciences Foundation (to ALA). MR-T is the recipient of a Vanier Doctoral Scholarship from the Canadian Institutes of Health Research (CIHR). ALA is supported by a CIHR New Investigator Award and an Early Researcher Award from the Ontario Ministry of Research and Innovation.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Redig A, McAllister S. Breast cancer as a systemic disease: a view of metastasis. J Internal Med. 2013;274(2):113–126. doi: 10.1111/joim.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chambers AF, Groom AC, MacDonald IC. Metastasis: dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2(8):563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 3.Lu J, Steeg PS, Price JE, Krishnamurthy S, Mani SA, Reuben J, Cristofanilli M, Dontu G, Bidaut L, Valero V. Breast cancer metastasis: challenges and opportunities. Cancer Res. 2009;69(12):4951–4953. doi: 10.1158/0008-5472.CAN-09-0099. [DOI] [PubMed] [Google Scholar]
- 4.Luzzi KJ, MacDonald IC, Schmidt EE, Kerkvliet N, Morris VL, Chambers AF, Groom AC. Multistep nature of metastatic inefficiency: dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am J Pathol. 1998;153(3):865–873. doi: 10.1016/S0002-9440(10)65628-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma I, Allan AL. The role of human aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell Rev. 2011;7(2):292–306. doi: 10.1007/s12015-010-9208-4. [DOI] [PubMed] [Google Scholar]
- 6.Beck B, Blanpain C. Unravelling cancer stem cell potential. Nat Rev Cancer. 2013;13(10):727–738. doi: 10.1038/nrc3597. [DOI] [PubMed] [Google Scholar]
- 7.Croker AK, Goodale D, Chu J, Postenka C, Hedley BD, Hess DA, Allan AL. High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J Cell Mol Med. 2009;13(8B):2236–2252. doi: 10.1111/j.1582-4934.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, Hur MH, Diebel ME, Monville F, Dutcher J, Brown M, Viens P, Xerri L, Bertucci F, Stassi G, Dontu G, Birnbaum D, Wicha MS. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69(4):1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcato P, Dean CA, Pan D, Araslanova R, Gillis M, Joshi M, Helyer L, Pan L, Leidal A, Gujar S, Giacomantonio CA, Lee PW. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem Cells. 2011;29(1):32–45. doi: 10.1002/stem.563. [DOI] [PubMed] [Google Scholar]
- 10.Marcato P, Dean CA, Liu RZ, Coyle KM, Bydoun M, Wallace M, Clements D, Turner C, Mathenge EG, Gujar SA, Giacomantonio CA, Mackey JR, Godbout R, Lee PW. Aldehyde dehydrogenase 1A3 influences breast cancer progression via differential retinoic acid signaling. Mol Oncol. 2015;9(1):17–31. doi: 10.1016/j.molonc.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van den Hoogen C, van der Horst G, Cheung H, Buijs JT, Pelger RC, van der Pluijm G. The aldehyde dehydrogenase enzyme 7A1 is functionally involved in prostate cancer bone metastasis. Clin Exp Metastasis. 2011;28(7):615–625. doi: 10.1007/s10585-011-9395-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pors K, Moreb JS. Aldehyde dehydrogenases in cancer: an opportunity for biomarker and drug development? Drug Discov Today. 2014;19(12):1953–1963. doi: 10.1016/j.drudis.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Alison MR, Guppy NJ, Lim SM, Nicholson LJ. Finding cancer stem cells: are aldehyde dehydrogenases fit for purpose? J Pathol. 2010;222(4):335–344. doi: 10.1002/path.2772. [DOI] [PubMed] [Google Scholar]
- 14.Vasiliou V, Thompson DC, Smith C, Fujita M, Chen Y. Aldehyde dehydrogenases: from eye crystallins to metabolic disease and cancer stem cells. Chem Biol Interact. 2013;202(1–3):2–10. doi: 10.1016/j.cbi.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charafe-Jauffret E, Ginestier C, Iovino F, Tarpin C, Diebel M, Esterni B, Houvenaeghel G, Extra JM, Bertucci F, Jacquemier J, Xerri L, Dontu G, Stassi G, Xiao Y, Barsky SH, Birnbaum D, Viens P, Wicha MS. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2010;16(1):45–55. doi: 10.1158/1078-0432.CCR-09-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koppaka V, Thompson DC, Chen Y, Ellermann M, Nicolaou KC, Juvonen RO, Petersen D, Deitrich RA, Hurley TD, Vasiliou V. Aldehyde dehydrogenase inhibitors: a comprehensive review of the pharmacology, mechanism of action, substrate specificity, and clinical application. Pharmacol Rev. 2012;64(3):520–539. doi: 10.1124/pr.111.005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell DY, Petersen DR. The oxidation of α-β unsaturated aldehydic products of lipid peroxidation by rat liver aldehyde dehydrogenases. Toxicol Appl Pharm. 1987;87(3):403–410. doi: 10.1016/0041-008x(87)90245-6. [DOI] [PubMed] [Google Scholar]
- 18.Lindahl R, Petersen DR. Lipid aldehyde oxidation as a physiological role for class 3 aldehyde dehydrogenases. Biochem Pharm. 1991;41(11):1583–1587. doi: 10.1016/0006-2952(91)90157-z. [DOI] [PubMed] [Google Scholar]
- 19.Singh S, Brocker C, Koppaka V, Chen Y, Jackson BC, Matsumoto A, Thompson DC, Vasiliou V. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic Biol Med. 2013;56:89–101. doi: 10.1016/j.freeradbiomed.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coyle K, Sultan M, Thomas M, Vaghar-Kashani A, Marcato P. Retinoid signaling in cancer and its promise for therapy. J Carcinog Mutagen S. 2013;7:16–18. [Google Scholar]
- 21.Januchowski R, Wojtowicz K, Zabel M. The role of aldehyde dehydrogenase (ALDH) in cancer drug resistance. Biomed Pharm. 2013;67(7):669–680. doi: 10.1016/j.biopha.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008;4(6):697–720. doi: 10.1517/17425250802102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chute JP, Muramoto GG, Whitesides J, Colvin M, Safi R, Chao NJ, McDonnell DP. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc Nat Acad Sci USA. 2006;103(31):11707–11712. doi: 10.1073/pnas.0603806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tonge PD, Andrews PW. Retinoic acid directs neuronal differentiation of human pluripotent stem cell lines in a non-cell-autonomous manner. Differentiation. 2010;80(1):20–30. doi: 10.1016/j.diff.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Z-y Su, Li Y, X-l Zhao, Zhang M. All-trans retinoic acid promotes smooth muscle cell differentiation of rabbit bone marrow-derived mesenchymal stem cells. J Zhejiang Univ Sci B. 2010;11(7):489–496. doi: 10.1631/jzus.B0900415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simandi Z, Balint BL, Poliska S, Ruhl R, Nagy L. Activation of retinoic acid receptor signaling coordinates lineage commitment of spontaneously differentiating mouse embryonic stem cells in embryoid bodies. FEBS Lett. 2010;584(14):3123–3130. doi: 10.1016/j.febslet.2010.05.052. [DOI] [PubMed] [Google Scholar]
- 27.Ren M, Pozzi S, Bistulfi G, Somenzi G, Rossetti S, Sacchi N. Impaired retinoic acid (RA) signal leads to RARβ2 epigenetic silencing and RA resistance. Mol Cell Biol. 2005;25(23):10591–10603. doi: 10.1128/MCB.25.23.10591-10603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Croker AK, Allan AL. Inhibition of aldehyde dehydrogenase (ALDH) activity reduces chemotherapy and radiation resistance of stem-like ALDHhiCD44(+) human breast cancer cells. Breast Cancer Res Treat. 2012;133(1):75–87. doi: 10.1007/s10549-011-1692-y. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Wan L, Geng J, Wu C-L, Bai X. Aldehyde dehydrogenase 1A1 possesses stem-like properties and predicts lung cancer patient outcome. J Thoracic Oncol. 2012;7(8):1235–1245. doi: 10.1097/JTO.0b013e318257cc6d. [DOI] [PubMed] [Google Scholar]
- 30.Moreb JS, Baker HV, Chang LJ, Amaya M, Lopez MC, Ostmark B, Chou W. ALDH isozymes downregulation affects cell growth, cell motility and gene expression in lung cancer cells. Mol Cancer. 2008;7:87. doi: 10.1186/1476-4598-7-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kakizuka A, Miller W, Umesono K, Warrell R, Frankel S, Murty V, Dmitrovsky E, Evans R. Chromosomal translocation t (15; 17) in human acute promyelocytic leukemia fuses RARα with a novel putative transcription factor, PML. Cell. 1991;66(4):663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 32.de Thé H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RARα fusion mRNA generated by the t (15; 17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66(4):675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 33.Kim R-J, Park J-R, Roh K-J, Choi A-R, Kim S-R, Kim P-H, Yu JH, Lee JW, Ahn S-H, Gong G. High aldehyde dehydrogenase activity enhances stem cell features in breast cancer cells by activating hypoxia-inducible factor-2α. Cancer Lett. 2013;333(1):18–31. doi: 10.1016/j.canlet.2012.11.026. [DOI] [PubMed] [Google Scholar]
- 34.Mizuno T, Suzuki N, Makino H, Furui T, Morii E, Aoki H, Kunisada T, Morishige K. Cancer stem-like cells of ovarian clear cell carcinoma are enriched in the ALDH-high population associated with an accelerated scavenging system in reactive oxygen species. Gynecol Oncol. 2015;137(2):299–305. doi: 10.1016/j.ygyno.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 35.Liu P, Brown S, Goktug T, Channathodiyil P, Kannappan V, Hugnot J, Guichet P, Bian X, Armesilla A, Darling J. Cytotoxic effect of disulfiram/copper on human glioblastoma cell lines and ALDH-positive cancer-stem-like cells. Br J Cancer. 2012;107(9):1488–1497. doi: 10.1038/bjc.2012.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mu X, Isaac C, Schott T, Huard J, Weiss K. Rapamycin inhibits ALDH activity, resistance to oxidative stress, and metastatic potential in murine osteosarcoma cells. Sarcoma. 2013;2013:480713. doi: 10.1155/2013/480713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vasiliou V, Nebert DW. Analysis and update of the human aldehyde dehydrogenase (ALDH) gene family. Hum Genom. 2005;2(2):138. doi: 10.1186/1479-7364-2-2-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jean E, Laoudj-Chenivesse D, Notarnicola C, Rouger K, Serratrice N, Bonnieu A, Gay S, Bacou F, Duret C, Carnac G. Aldehyde dehydrogenase activity promotes survival of human muscle precursor cells. J Cell Mol Med. 2011;15(1):119–133. doi: 10.1111/j.1582-4934.2009.00942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kitahara O, Katagiri T, Tsunoda T, Harima Y, Nakamura Y. Classification of sensitivity or resistance of cervical cancers to ionizing radiation according to expression profiles of 62 genes selected by cDNA microarray analysis. Neoplasia. 2002;4(4):295–303. doi: 10.1038/sj.neo.7900251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sladek NE, Kollander R, Sreerama L, Kiang DT. Cellular levels of aldehyde dehydrogenases (ALDH1A1 and ALDH3A1) as predictors of therapeutic responses to cyclophosphamide-based chemotherapy of breast cancer: a retrospective study. Rational individualization of oxazaphosphorine-based cancer chemotherapeutic regimens. Cancer Chemother Pharmacol. 2002;49(4):309–321. doi: 10.1007/s00280-001-0412-4. [DOI] [PubMed] [Google Scholar]
- 41.Russo JE, Hilton J. Characterization of cytosolic aldehyde dehydrogenase from cyclophosphamide resistant L1210 cells. Cancer Res. 1988;48(11):2963–2968. [PubMed] [Google Scholar]
- 42.Moreb JS, Gabr A, Vartikar GR, Gowda S, Zucali JR, Mohuczy D. Retinoic acid down-regulates aldehyde dehydrogenase and increases cytotoxicity of 4-hydroperoxycyclophosphamide and acetaldehyde. J Pharmacol Exper Ther. 2005;312(1):339–345. doi: 10.1124/jpet.104.072496. [DOI] [PubMed] [Google Scholar]
- 43.Moreb J, Zucali J, Zhang Y, Colvin M, Gross M. Role of aldehyde dehydrogenase in the protection of hematopoietic progenitor cells from 4-hydroperoxycyclophosphamide by interleukin 1β and tumor necrosis factor. Cancer Res. 1992;52(7):1770–1774. [PubMed] [Google Scholar]
- 44.Honoki K, Fujii H, Kubo A, Kido A, Mori T, Tanaka Y, Tsujiuchi T. Possible involvement of stem-like populations with elevated ALDH1 in sarcomas for chemotherapeutic drug resistance. Oncol Rep. 2010;24(2):501–505. doi: 10.3892/or_00000885. [DOI] [PubMed] [Google Scholar]
- 45.Landen CN, Goodman B, Katre AA, Steg AD, Nick AM, Stone RL, Miller LD, Mejia PV, Jennings NB, Gershenson DM. Targeting aldehyde dehydrogenase cancer stem cells in ovarian cancer. Mol Cancer Ther. 2010;9(12):3186–3199. doi: 10.1158/1535-7163.MCT-10-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawasoe M, Yamamoto Y, Okawa K, Funato T, Takeda M, Hara T, Tsurumi H, Moriwaki H, Arioka Y, Takemura M. Acquired resistance of leukemic cells to AraC is associated with the upregulation of aldehyde dehydrogenase 1 family member A2. Exp Hematol. 2013;41(7):597–603. doi: 10.1016/j.exphem.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee DH, Chung K, Song J-A, T-h Kim, Kang H, Huh JH, S-g Jung, Ko JJ, An HJ. Proteomic identification of paclitaxel-resistance associated hnRNP A2 and GDI 2 proteins in human ovarian cancer cells. J Proteome Res. 2010;9(11):5668–5676. doi: 10.1021/pr100478u. [DOI] [PubMed] [Google Scholar]
- 48.Stewart M, Malek K, Crabb DW. Distribution of messenger RNAs for aldehyde dehydrogenase 1, aldehyde dehydrogenase 2, and aldehyde dehydrogenase 5 in human tissues. J Investig Med. 1996;44(2):42–46. [PubMed] [Google Scholar]
- 49.Dipple K, Crabb D. The mitochondrial aldehyde dehydrogenase gene resides in an HTF island but is expressed in a tissue-specific manner. Biochem Biophys Res Commun. 1993;193(1):420–427. doi: 10.1006/bbrc.1993.1640. [DOI] [PubMed] [Google Scholar]
- 50.Storms RW, Trujillo AP, Springer JB, Shah L, Colvin OM, Ludeman SM, Smith C. Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc Nat Acad of Sci USA. 1999;96(16):9118–9123. doi: 10.1073/pnas.96.16.9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corti S, Locatelli F, Papadimitriou D, Donadoni C, Salani S, Del Bo R, Strazzer S, Bresolin N, Comi GP. Identification of a primitive brain–derived neural stem cell population based on aldehyde dehydrogenase activity. Stem Cells. 2006;24(4):975–985. doi: 10.1634/stemcells.2005-0217. [DOI] [PubMed] [Google Scholar]
- 52.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, Schott A, Hayes D, Birnbaum D, Wicha MS, Dontu G. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pearce DJ, Bonnet D. The combined use of Hoechst efflux ability and aldehyde dehydrogenase activity to identify murine and human hematopoietic stem cells. Exp Hematol. 2007;35(9):1437–1446. doi: 10.1016/j.exphem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 54.Levi BP, Yilmaz ÖH, Duester G, Morrison SJ. Aldehyde dehydrogenase 1a1 is dispensable for stem cell function in the mouse hematopoietic and nervous systems. Blood. 2009;113(8):1670–1680. doi: 10.1182/blood-2008-05-156752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muramoto GG, Russell JL, Safi R, Salter AB, Himburg HA, Daher P, Meadows SK, Doan P, Storms RW, Chao NJ. Inhibition of aldehyde dehydrogenase expands hematopoietic stem cells with radioprotective capacity. Stem Cells. 2010;28(3):523–534. doi: 10.1002/stem.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Armstrong L, Stojkovic M, Dimmick I, Ahmad S, Stojkovic P, Hole N, Lako M. Phenotypic characterization of murine primitive hematopoietic progenitor cells isolated on basis of aldehyde dehydrogenase activity. Stem Cells. 2004;22(7):1142–1151. doi: 10.1634/stemcells.2004-0170. [DOI] [PubMed] [Google Scholar]
- 57.Zhuang X, Zhang W, Chen Y, Han X, Li J, Zhang Y, Zhang Y, Zhang S, Liu B. Doxorubicin-enriched, ALDHbr mouse breast cancer stem cells are treatable to oncolytic herpes simplex virus type 1. BMC Cancer. 2012;12(1):549. doi: 10.1186/1471-2407-12-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang SH, Lu L, Fan Y, Wicha MS, Cao Z, Chang AE, J-c Xia, Baker JR, Jr, Li Q. Characterization of a novel transgenic mouse tumor model for targeting HER2+ cancer stem cells. Int J Biol Sci. 2014;10(1):25. doi: 10.7150/ijbs.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vaidyanathan G, Song H, Affleck D, McDougald DL, Storms RW, Zalutsky MR, Chin BB. Targeting aldehyde dehydrogenase: a potential approach for cell labeling. Nucl Med Biol. 2009;36(8):919–929. doi: 10.1016/j.nucmedbio.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, Fields JZ, Wicha MS, Boman BM. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009;69(8):3382–3389. doi: 10.1158/0008-5472.CAN-08-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Isfoss BL, Holmqvist B, Jernström H, Alm P, Olsson H. Women with familial risk for breast cancer have an increased frequency of aldehyde dehydrogenase expressing cells in breast ductules. BMC Clin Pathol. 2013;13(1):28. doi: 10.1186/1472-6890-13-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khoury T, Ademuyiwa FO, Chandraseekhar R, Jabbour M, DeLeo A, Ferrone S, Wang Y, Wang X. Aldehyde dehydrogenase 1A1 expression in breast cancer is associated with stage, triple negativity, and outcome to neoadjuvant chemotherapy. Mod Pathol. 2012;25(3):388–397. doi: 10.1038/modpathol.2011.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moreb JS, Ucar D, Han S, Amory JK, Goldstein AS, Ostmark B, Chang LJ. The enzymatic activity of human aldehyde dehydrogenases 1A2 and 2 (ALDH1A2 and ALDH2) is detected by Aldefluor, inhibited by diethylaminobenzaldehyde and has significant effects on cell proliferation and drug resistance. Chem Biol Interact. 2012;195(1):52–60. doi: 10.1016/j.cbi.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marcato P, Dean CA, Giacomantonio CA, Lee PW. Aldehyde dehydrogenase: its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle. 2011;10(9):1378–1384. doi: 10.4161/cc.10.9.15486. [DOI] [PubMed] [Google Scholar]
- 65.Luo Y, Dallaglio K, Chen Y, Robinson WA, Robinson SE, McCarter MD, Wang J, Gonzalez R, Thompson DC, Norris DA. ALDH1A isozymes are markers of human melanoma stem cells and potential therapeutic targets. Stem Cells. 2012;30(10):2100–2113. doi: 10.1002/stem.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xia P. Surface markers of cancer stem cells in solid tumors. Curr Stem Cell Res Ther. 2014;9(2):102–111. doi: 10.2174/1574888x09666131217003709. [DOI] [PubMed] [Google Scholar]
- 67.Tirino V, Desiderio V, Paino F, De Rosa A, Papaccio F, La Noce M, Laino L, De Francesco F, Papaccio G. Cancer stem cells in solid tumors: an overview and new approaches for their isolation and characterization. FASEB. 2013;27(1):13–24. doi: 10.1096/fj.12-218222. [DOI] [PubMed] [Google Scholar]
- 68.Fabian A, Vereb G, Szöllősi J. The hitchhikers guide to cancer stem cell theory: markers, pathways and therapy. Cytom A. 2013;83(1):62–71. doi: 10.1002/cyto.a.22206. [DOI] [PubMed] [Google Scholar]
- 69.Keysar SB, Jimeno A. More than markers: biological significance of cancer stem cell-defining molecules. Mol Cancer Ther. 2010;9(9):2450–2457. doi: 10.1158/1535-7163.MCT-10-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8(10):755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 71.Pouliot N, Pearson HB, Burrows A (2000) Investigating metastasis using in vitro platforms. In: Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000. http://www.ncbi.nlm.nih.gov/books/NBK100379/
- 72.Brooks SA, Schumacher U. Metastasis research protocols. Berlin: Springer; 2001. [Google Scholar]
- 73.Grimshaw MJ, Cooper L, Papazisis K, Coleman JA, Bohnenkamp HR, Chiapero-Stanke L, Taylor-Papadimitriou J, Burchell JM. Mammosphere culture of metastatic breast cancer cells enriches for tumorigenic breast cancer cells. Breast Cancer Res. 2008;10(3):R52. doi: 10.1186/bcr2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Soritău O, Tomuleasa C, Pall E, Virág P, Fischer-Fodor E, Foris V. Enhanced chemoresistance and tumor sphere formation as a laboratory model for peritoneal micrometastasis in epithelial ovarian cancer. Rom J Morphol Embryol. 2010;51(2):259–264. [PubMed] [Google Scholar]
- 75.Cioce M, Gherardi S, Viglietto G, Strano S, Blandino G, Muti P, Ciliberto G. Mammosphere-forming cells from breast cancer cell lines as a tool for the identification of CSC-like-and early progenitor-targeting drugs. Cell Cycle. 2010;9(14):2950–2959. [PubMed] [Google Scholar]
- 76.Kerbel R, Kobayashi H, Graham CH. Intrinsic or acquired drug resistance and metastasis: are they linked phenotypes? J Cell Biochem. 1994;56(1):37–47. doi: 10.1002/jcb.240560108. [DOI] [PubMed] [Google Scholar]
- 77.Wong ST, Goodin S. Overcoming drug resistance in patients with metastatic breast cancer. Pharmacotherapy. 2009;29(8):954–965. doi: 10.1592/phco.29.8.954. [DOI] [PubMed] [Google Scholar]
- 78.Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu Y, Martin-Trevino R, Shang L, McDermott SP, Landis MD, Hong S, Adams A, D’Angelo R, Ginestier C, Charafe-Jauffret E, Clouthier SG, Birnbaum D, Wong ST, Zhan M, Chang JC, Wicha MS. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Rep. 2014;2(1):78–91. doi: 10.1016/j.stemcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liao J, Qian F, Tchabo N, Mhawech-Fauceglia P, Beck A, Qian Z, Wang X, Huss WJ, Lele SB, Morrison CD. Ovarian cancer spheroid cells with stem cell-like properties contribute to tumor generation, metastasis and chemotherapy resistance through hypoxia-resistant metabolism. PLoS One. 2014;9(1):e84941. doi: 10.1371/journal.pone.0084941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuroda T, Hirohashi Y, Torigoe T, Yasuda K, Takahashi A, Asanuma H, Morita R, Mariya T, Asano T, Mizuuchi M, Saito T, Sato N. ALDH1-high ovarian cancer stem-like cells can be isolated from serous and clear cell adenocarcinoma cells, and ALDH1 high expression is associated with poor prognosis. PLoS One. 2013;8(6):e65158. doi: 10.1371/journal.pone.0065158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mu X, Isaac C, Greco N, Huard J, Weiss K. Notch signaling is associated with ALDH activity and an aggressive metastatic phenotype in murine osteosarcoma cells. Front Oncol. 2013;3:143. doi: 10.3389/fonc.2013.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ajani J, Wang X, Song S, Suzuki A, Taketa T, Sudo K, Wadhwa R, Hofstetter W, Komaki R, Maru D. ALDH-1 expression levels predict response or resistance to preoperative chemoradiation in resectable esophageal cancer patients. Mol Oncol. 2014;8(1):142–149. doi: 10.1016/j.molonc.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reeves K, van der Pluijm G, Cecchini M, Eaton C, Hamdy F, Brown N (2013) The influence of tumour aldehyde dehydrogenase activity on prostate cancer cell metastatic potential and interaction with the bone endothelium in vivo. In: Proceedings of the physiological society, 2013. The Physiological Society
- 84.Tanei T, Morimoto K, Shimazu K, Kim SJ, Tanji Y, Taguchi T, Tamaki Y, Noguchi S. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential Paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin Cancer Res. 2009;15(12):4234–4241. doi: 10.1158/1078-0432.CCR-08-1479. [DOI] [PubMed] [Google Scholar]
- 85.Greco N, Schott T, Mu X, Rothenberg A, Voigt C, McGough RL, 3rd, Goodman M, Huard J, Weiss KR. ALDH activity correlates with metastatic potential in primary sarcomas of bone. J Cancer Ther. 2014;5(4):331–338. doi: 10.4236/jct.2014.54040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cojoc M, Peitzsch C, Kurth I, Trautmann F, Kunz-Schughart LA, Telegeev GD, Stakhovsky EA, Walker JR, Simin K, Lyle S. Aldehyde dehydrogenase is regulated by β-Catenin/TCF and promotes radioresistance in prostate cancer progenitor cells. Cancer Res. 2015;75(7):1482–1494. doi: 10.1158/0008-5472.CAN-14-1924. [DOI] [PubMed] [Google Scholar]
- 87.Charafe-Jauffret E, Ginestier C, Bertucci F, Cabaud O, Wicinski J, Finetti P, Josselin E, Adelaide J, Nguyen T-T, Monville F. ALDH1-positive cancer stem cells predict engraftment of primary breast tumors and are governed by a common stem cell program. Cancer Res. 2013;73(24):7290–7300. doi: 10.1158/0008-5472.CAN-12-4704. [DOI] [PubMed] [Google Scholar]
- 88.Li D, Zhang T, Gu W, Li P, Cheng X, Tong T, Wang W. The ALDH1+ subpopulation of the human NMFH-1 cell line exhibits cancer stem-like characteristics. Oncol Rep. 2015;33(5):2291–2298. doi: 10.3892/or.2015.3842. [DOI] [PubMed] [Google Scholar]
- 89.Young M-J, Wu Y-H, Chiu W-T, Weng T-Y, Huang Y-F, Chou C-Y. All-trans retinoic acid downregulates ALDH1-mediated stemness and inhibits tumour formation in ovarian cancer cells. Carcinogenesis. 2015;36:498–507. doi: 10.1093/carcin/bgv018. [DOI] [PubMed] [Google Scholar]
- 90.Condello S, Morgan C, Nagdas S, Cao L, Turek J, Hurley T, Matei D. β-Catenin-regulated ALDH1A1 is a target in ovarian cancer spheroids. Oncogene. 2015;34(18):2297–2308. doi: 10.1038/onc.2014.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Choi SA, Lee JY, Phi JH, Wang K-C, Park C-K, Park S-H, Kim S-K. Identification of brain tumour initiating cells using the stem cell marker aldehyde dehydrogenase. Eur J Cancer. 2014;50(1):137–149. doi: 10.1016/j.ejca.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 92.Soehngen E, Schaefer A, Koeritzer J, Huelsmeyer V, Zimmer C, Ringel F, Gempt J, Schlegel J. Hypoxia upregulates aldehyde dehydrogenase isoform 1 (ALDH1) expression and induces functional stem cell characteristics in human glioblastoma cells. Brain Tumor Pathol. 2014;31(4):247–256. doi: 10.1007/s10014-013-0170-0. [DOI] [PubMed] [Google Scholar]
- 93.Yu C-C, Lo W-L, Chen Y-W, Huang P-I, Hsu H-S, Tseng L-M, Hung S-C, Kao S-Y, Chang C-J, Chiou SH (2010) Bmi-1 regulates snail expression and promotes metastasis ability in head and neck squamous cancer-derived ALDH1 positive cells. J Oncol 2011 [DOI] [PMC free article] [PubMed]
- 94.Fitzgerald TL, Rangan S, Dobbs L, Starr S, Sigounas G. The impact of Aldehyde dehydrogenase 1 expression on prognosis for metastatic colon cancer. J Surg Res. 2014;192(1):82–89. doi: 10.1016/j.jss.2014.05.054. [DOI] [PubMed] [Google Scholar]
- 95.James MI, Howells LM, Karmokar A, Higgins JA, Greaves P, Cai H, Dennison A, Metcalfe M, Garcea G, Lloyd DM. Characterization and propagation of tumor initiating cells derived from colorectal liver metastases: trials, tribulations and a cautionary note. PLoS One. 2015;10(2):e0117776. doi: 10.1371/journal.pone.0117776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Larzabal L, El-Nikhely N, Redrado M, Seeger W, Savai R, Calvo A. Differential effects of drugs targeting cancer stem cell (CSC) and non-CSC populations on lung primary tumors and metastasis. PLoS One. 2013;8(11):e79798. doi: 10.1371/journal.pone.0079798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liang D, Shi Y. Aldehyde dehydrogenase-1 is a specific marker for stem cells in human lung adenocarcinoma. Med Oncol. 2012;29(2):633–639. doi: 10.1007/s12032-011-9933-9. [DOI] [PubMed] [Google Scholar]
- 98.Akunuru S, Zhai QJ, Zheng Y. Non-small cell lung cancer stem/progenitor cells are enriched in multiple distinct phenotypic subpopulations and exhibit plasticity. Cell Death Dis. 2012;3(7):e352. doi: 10.1038/cddis.2012.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lin J, Liu X, Ding D. Evidence for epithelial-mesenchymal transition in cancer stem-like cells derived from carcinoma cell lines of the cervix uteri. Int J Clin Exper Pathol. 2015;8(1):847. [PMC free article] [PubMed] [Google Scholar]
- 100.Sullivan JP, Spinola M, Dodge M, Raso MG, Behrens C, Gao B, Schuster K, Shao C, Larsen JE, Sullivan LA. Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on notch signaling. Cancer Res. 2010;70(23):9937–9948. doi: 10.1158/0008-5472.CAN-10-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li Z, Xiang Y, Xiang L, Xiao Y, Li F, Hao P. ALDH maintains the stemness of lung adenoma stem cells by suppressing the Notch/CDK2/CCNE pathway. PLoS One. 2014;9(3):e92669. doi: 10.1371/journal.pone.0092669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang L, Ren Y, Yu X, Qian F, Bian BS, Xiao HL, Wang WG, Xu SL, Yang J, Cui W, Liu Q, Wang Z, Guo W, Xiong G, Yang K, Qian C, Zhang X, Zhang P, Cui YH, Bian XW. ALDH1A1 defines invasive cancer stem-like cells and predicts poor prognosis in patients with esophageal squamous cell carcinoma. Mod Pathol. 2014;27(5):775–783. doi: 10.1038/modpathol.2013.189. [DOI] [PubMed] [Google Scholar]
- 103.Hoshino Y, Nishida J, Katsuno Y, Koinuma D, Aoki T, Kokudo N, Miyazono K, Ehata S. Smad4 decreases the population of pancreatic cancer-initiating cells through transcriptional repression of ALDH1A1. Am J Pathol. 2015;185(5):1457–1470. doi: 10.1016/j.ajpath.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 104.Wang K, Chen X, Zhan Y, Jiang W, Liu X, Wang X, Wu B. Increased expression of ALDH1A1 protein is associated with poor prognosis in clear cell renal cell carcinoma. Med Oncol. 2013;30(2):1–9. doi: 10.1007/s12032-013-0574-z. [DOI] [PubMed] [Google Scholar]
- 105.Huang C-P, Tsai M-F, Chang T-H, Tang W-C, Chen S-Y, Lai H-H, Lin T-Y, Yang JC-H, Yang P-C, Shih J-Y. ALDH-positive lung cancer stem cells confer resistance to epidermal growth factor receptor tyrosine kinase inhibitors. Cancer Lett. 2013;328(1):144–151. doi: 10.1016/j.canlet.2012.08.021. [DOI] [PubMed] [Google Scholar]
- 106.Yue L, Huang Z-M, Fong S, Leong S, Jakowatz JG, Charruyer-Reinwald A, Wei M, Ghadially R. Targeting ALDH1 to decrease tumorigenicity, growth and metastasis of human melanoma. Melanoma Res. 2015;25(2):138–148. doi: 10.1097/CMR.0000000000000144. [DOI] [PubMed] [Google Scholar]
- 107.Jia J, Parikh H, Xiao W, Hoskins JW, Pflicke H, Liu X, Collins I, Zhou W, Wang Z, Powell J, Thorgeirsson SS, Rudloff U, Petersen GM, Amundadottir LT. An integrated transcriptome and epigenome analysis identifies a novel candidate gene for pancreatic cancer. BMC Med Genomics. 2013;6:33. doi: 10.1186/1755-8794-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mao P, Joshi K, Li J, Kim S-H, Li P, Santana-Santos L, Luthra S, Chandran UR, Benos PV, Smith L. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Nat Acad Sci. 2013;110(21):8644–8649. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yan J, De Melo J, Cutz J, Aziz T, Tang D. Aldehyde dehydrogenase 3A1 associates with prostate tumorigenesis. Br J Cancer. 2014;110(10):2593–2603. doi: 10.1038/bjc.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Calderaro J, Nault J-C, Bioulac-Sage P, Laurent A, Blanc J-F, Decaens T, Zucman-Rossi J. ALDH3A1 is overexpressed in a subset of hepatocellular carcinoma characterised by activation of the Wnt/ß-catenin pathway. Virchows Arch. 2014;464(1):53–60. doi: 10.1007/s00428-013-1515-0. [DOI] [PubMed] [Google Scholar]
- 111.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10(2):R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rasheed ZA, Yang J, Wang Q, Kowalski J, Freed I, Murter C, Hong S-M, Koorstra J-B, Rajeshkumar N, He X. Prognostic significance of tumorigenic cells with mesenchymal features in pancreatic adenocarcinoma. J Nat Cancer Inst. 2010;102(5):340–351. doi: 10.1093/jnci/djp535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.van den Hoogen C, van der Horst G, Cheung H, Buijs JT, Pelger RC, van der Pluijm G. Integrin αv expression is required for the acquisition of a metastatic stem/progenitor cell phenotype in human prostate cancer. Am J Pathol. 2011;179(5):2559–2568. doi: 10.1016/j.ajpath.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.van den Hoogen C, van der Horst G, Cheung H, Buijs JT, Lippitt JM, Guzman-Ramirez N, Hamdy FC, Eaton CL, Thalmann GN, Cecchini MG, Pelger RC, van der Pluijm G. High aldehyde dehydrogenase activity identifies tumor-initiating and metastasis-initiating cells in human prostate cancer. Cancer Res. 2010;70(12):5163–5173. doi: 10.1158/0008-5472.CAN-09-3806. [DOI] [PubMed] [Google Scholar]
- 115.Sun S, Wang Z. ALDH high adenoid cystic carcinoma cells display cancer stem cell properties and are responsible for mediating metastasis. Biochem Biophys Res Commun. 2010;396(4):843–848. doi: 10.1016/j.bbrc.2010.04.170. [DOI] [PubMed] [Google Scholar]
- 116.Visus C, Wang Y, Lozano-Leon A, Ferris RL, Silver S, Szczepanski MJ, Brand RE, Ferrone CR, Whiteside TL, Ferrone S. Targeting ALDHbright Human Carcinoma-Initiating Cells with ALDH1A1-Specific CD8+ T Cells. Clin Cancer Res. 2011;17(19):6174–6184. doi: 10.1158/1078-0432.CCR-11-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Marselos M, Michalopoulos G. Changes in the pattern of aldehyde dehydrogenase activity in primary and metastatic adenocarcinomas of the human colon. Cancer Lett. 1987;34(1):27–37. doi: 10.1016/0304-3835(87)90070-x. [DOI] [PubMed] [Google Scholar]
- 118.Jelski W, Zalewski B, Szmitkowski M. Alcohol dehydrogenase (ADH) isoenzymes and aldehyde dehydrogenase (ALDH) activity in the sera of patients with liver cancer. J Clin Lab Anal. 2008;22(3):204–209. doi: 10.1002/jcla.20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shao J, C-p Pan, Wang M-w Wu, X-h Ma B. Discordance of aldehyde dehydrogenase 1 and estrogen receptor expression between primary and metastatic focuses of breast cancer. Acta Anat Sin. 2013;2:224–228. [Google Scholar]
- 120.Hessman CJ, Bubbers EJ, Billingsley KG, Herzig DO, Wong MH. Loss of expression of the cancer stem cell marker aldehyde dehydrogenase 1 correlates with advanced-stage colorectal cancer. Am J Surg. 2012;203(5):649–653. doi: 10.1016/j.amjsurg.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sreerama L, Sladek NE. Cellular levels of class 1 and class 3 aldehyde dehydrogenases and certain other drug-metabolizing enzymes in human breast malignancies. Clin Cancer Res. 1997;3(11):1901–1914. [PubMed] [Google Scholar]
- 122.Vogler T, Kriegl L, Horst D, Engel J, Sagebiel S, Schäffauer AJ, Kirchner T, Jung A. The expression pattern of aldehyde dehydrogenase 1 (ALDH1) is an independent prognostic marker for low survival in colorectal tumors. Exp Mol Pathol. 2012;92(1):111–117. doi: 10.1016/j.yexmp.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 123.Ohmura-Kakutani H, Akiyama K, Maishi N, Ohga N, Hida Y, Kawamoto T, Iida J, Shindoh M, Tsuchiya K, Shinohara N. Identification of tumor endothelial cells with high aldehyde dehydrogenase activity and a highly angiogenic phenotype. PLoS One. 2014;9(12):e113910. doi: 10.1371/journal.pone.0113910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lowes LE, Allan AL. Recent advances in the molecular characterization of circulating tumor cells. Cancers. 2014;6(1):595–624. doi: 10.3390/cancers6010595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Giuliano M, Giordano A, Jackson S, De Giorgi U, Mego M, Cohen EN, Gao H, Anfossi S, Handy BC, Ueno NT. Circulating tumor cells as early predictors of metastatic spread in breast cancer patients with limited metastatic dissemination. Breast Cancer Res. 2014;16(5):440. doi: 10.1186/s13058-014-0440-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V, Klein C, Saini M, Bäuerle T, Wallwiener M. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol. 2013;31(6):539–544. doi: 10.1038/nbt.2576. [DOI] [PubMed] [Google Scholar]
- 127.Bednarz-Knoll N, Alix-Panabières C, Pantel K. Clinical relevance and biology of circulating tumor cells. Breast Cancer Res. 2011;13(6):228. doi: 10.1186/bcr2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Aktas B, Tewes M, Fehm T, Hauch S, Kimmig R, Kasimir-Bauer S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009;11(4):R46. doi: 10.1186/bcr2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gradilone A, Naso G, Raimondi C, Cortesi E, Gandini O, Vincenzi B, Saltarelli R, Chiapparino E, Spremberg F, Cristofanilli M. Circulating tumor cells (CTCs) in metastatic breast cancer (MBC): prognosis, drug resistance and phenotypic characterization. Ann Oncol. 2011;22(1):86–92. doi: 10.1093/annonc/mdq323. [DOI] [PubMed] [Google Scholar]
- 130.Reuben JM, Lee BN, Gao H, Cohen EN, Mego M, Giordano A, Wang X, Lodhi A, Krishnamurthy S, Hortobagyi GN, Cristofanilli M, Lucci A, Woodward WA. Primary breast cancer patients with high risk clinicopathologic features have high percentages of bone marrow epithelial cells with ALDH activity and CD44(+)CD24lo cancer stem cell phenotype. Eur J Cancer. 2011;47(10):1527–1536. doi: 10.1016/j.ejca.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Theodoropoulos PA, Polioudaki H, Agelaki S, Kallergi G, Saridaki Z, Mavroudis D, Georgoulias V. Circulating tumor cells with a putative stem cell phenotype in peripheral blood of patients with breast cancer. Cancer Lett. 2010;288(1):99–106. doi: 10.1016/j.canlet.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 132.Zirvi KA, Hill GJ, Hill HZ. Comparative studies of chemotherapy of human tumor cells in vitro by tritiated thymidine uptake inhibition and soft agar clonogenic assay. J Surg Oncol. 1986;31(2):123–126. doi: 10.1002/jso.2930310210. [DOI] [PubMed] [Google Scholar]
- 133.Alonso-Alconada L, Muinelo-Romay L, Madissoo K, Diaz-Lopez A, Krakstad C, Trovik J, Wik E, Hapangama D, Coenegrachts L, Cano A. Molecular profiling of circulating tumor cells links plasticity to the metastatic process in endometrial cancer. Mol Cancer. 2014;13(1):223. doi: 10.1186/1476-4598-13-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Krawczyk N, Meier-Stiegen F, Banys M, Neubauer H, Ruckhaeberle E, Fehm T. Expression of stem cell and epithelial-mesenchymal transition markers in circulating tumor cells of breast cancer patients. Biomed Res Int. 2014;2014:415721. doi: 10.1155/2014/415721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Woodward WA, Krishnamurthy S, Lodhi A, Xiao L, Gong Y, Cristofanilli M, Buchholz TA, Lucci A. Aldehyde dehydrogenase1 immunohistochemical staining in primary breast cancer cells independently predicted overall survival but did not correlate with the presence of circulating or disseminated tumors cells. J Cancer. 2014;5(5):360. doi: 10.7150/jca.7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Giordano A, Gao H, Cohen E, Anfossi S, Khoury J, Hess K, Krishnamurthy S, Tin S, Cristofanilli M, Hortobagyi G (2013) Clinical relevance of cancer stem cells in bone marrow of early breast cancer patients. Ann Oncol: mdt223 [DOI] [PMC free article] [PubMed]
- 137.Papadaki MA, Kallergi G, Zafeiriou Z, Manouras L, Theodoropoulos PA, Mavroudis D, Georgoulias V, Agelaki S. Co-expression of putative stemness and epithelial-to-mesenchymal transition markers on single circulating tumour cells from patients with early and metastatic breast cancer. BMC Cancer. 2014;14(1):651. doi: 10.1186/1471-2407-14-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Giordano A, Gao H, Anfossi S, Cohen E, Mego M, Lee B-N, Tin S, De Laurentiis M, Parker CA, Alvarez RH. Epithelial–mesenchymal transition and stem cell markers in patients with HER2-positive metastatic breast cancer. Mol Cancer Ther. 2012;11(11):2526–2534. doi: 10.1158/1535-7163.MCT-12-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 2012;22(6):725–736. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ocaña OH, Córcoles R, Fabra Á, Moreno-Bueno G, Acloque H, Vega S, Barrallo-Gimeno A, Cano A, Nieto MA. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell. 2012;22(6):709–724. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 141.Feldmann G, Dhara S, Fendrich V, Bedja D, Beaty R, Mullendore M, Karikari C, Alvarez H, Iacobuzio-Donahue C, Jimeno A. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67(5):2187–2196. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ozbek E, Calik G, Otunctemur A, Aliskan T, Cakir S, Dursun M, Somay A. Stem cell markers aldehyde dehydrogenase type 1 and nestin expressions in renal cell cancer. Archivio italiano di urologia, andrologia. 2012;84(1):7–11. [PubMed] [Google Scholar]
- 143.Meng E, Mitra A, Tripathi K, Finan MA, Scalici J, McClellan S, da Silva LM, Reed E, Shevde LA, Palle K. ALDH1A1 maintains ovarian cancer stem cell-like properties by altered regulation of cell cycle checkpoint and DNA repair network signaling. PLoS One. 2014;9(9):e107142. doi: 10.1371/journal.pone.0107142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Suman S, Das T, Damodaran C. Silencing NOTCH signaling causes growth arrest in both breast cancer stem cells and breast cancer cells. Br J Cancer. 2013;109(10):2587–2596. doi: 10.1038/bjc.2013.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.D’Angelo RC, Ouzounova M, Davis A, Choi D, Tchuenkam SM, Kim G, Luther T, Quraishi AA, Senbabaoglu Y, Conley SJ. Notch reporter activity in breast cancer cell lines identifies a subset of cells with stem cell activity. Mol Cancer Ther. 2015;14(3):779–787. doi: 10.1158/1535-7163.MCT-14-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chen X, Lingala S, Khoobyari S, Nolta J, Zern MA, Wu J. Epithelial mesenchymal transition and hedgehog signaling activation are associated with chemoresistance and invasion of hepatoma subpopulations. J Hepatol. 2011;55(4):838–845. doi: 10.1016/j.jhep.2010.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhao D, Mo Y, Li M-T, Zou S-W, Cheng Z-L, Sun Y-P, Xiong Y, Guan K-L, Lei Q-Y. NOTCH-induced aldehyde dehydrogenase 1A1 deacetylation promotes breast cancer stem cells. J Clin Investig. 2014;124(12):5453–5465. doi: 10.1172/JCI76611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cui J, Li P, Liu X, Hu H, Wei W. Abnormal expression of the Notch and Wnt/β-catenin signaling pathways in stem-like ALDHhiCD44+ cells correlates highly with Ki-67 expression in breast cancer. Oncol Lett. 2015;9(4):1600–1606. doi: 10.3892/ol.2015.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tiezzi DG, Clagnan WS, Mandarano LRM, de Sousa CB, Marana HRC, Tiezzi MG, de Andrade JM. Expression of aldehyde dehydrogenase after neoadjuvant chemotherapy is associated with expression of hypoxia-inducible factors 1 and 2 alpha and predicts prognosis in locally advanced breast cancer. Clinics. 2013;68(5):592–598. doi: 10.6061/clinics/2013(05)03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang Y, Jiang Y, Tian T, Hori Y, Wada N, J-i Ikeda, Morii E. Inhibitory effect of Nodal on the expression of aldehyde dehydrogenase 1 in endometrioid adenocarcinoma of uterus. Biochem Biophys Res Comm. 2013;440(4):731–736. doi: 10.1016/j.bbrc.2013.09.139. [DOI] [PubMed] [Google Scholar]
- 151.Shuang Z-Y, Wu W-C, Xu J, Lin G, Liu Y-C, Lao X-M, Zheng L, Li S. Transforming growth factor-β1-induced epithelial–mesenchymal transition generates ALDH-positive cells with stem cell properties in cholangiocarcinoma. Cancer Lett. 2014;354(2):320–328. doi: 10.1016/j.canlet.2014.08.030. [DOI] [PubMed] [Google Scholar]
- 152.Bhola NE, Balko JM, Dugger TC, Kuba MG, Sánchez V, Sanders M, Stanford J, Cook RS, Arteaga CL. TGF-β inhibition enhances chemotherapy action against triple-negative breast cancer. J Clin Invest. 2013;123(3):1348. doi: 10.1172/JCI65416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wei L, Liu T-T, Wang H-H, Hong H-M, Yu AL, Feng H-P, Chang W-W. Hsp27 participates in the maintenance of breast cancer stem cells through regulation of epithelial-mesenchymal transition and nuclear factor-kappaB. Breast Cancer Res. 2011;13(5):R101. doi: 10.1186/bcr3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lee D, Lee JW. Self-renewal and circulating capacities of metastatic hepatocarcinoma cells required for collaboration between TM4SF5 and CD44. BMB Rep. 2015;48(3):127. doi: 10.5483/BMBRep.2015.48.3.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Qian X, Anzovino A, Kim S, Suyama K, Yao J, Hulit J, Agiostratidou G, Chandiramani N, McDaid H, Nagi C. N-cadherin/FGFR promotes metastasis through epithelial-to-mesenchymal transition and stem/progenitor cell-like properties. Oncogene. 2014;33(26):3411–3421. doi: 10.1038/onc.2013.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Taiseer I, Samar AR, Abdelmonem H. Immunohistochemical Expression of Aldehyde Dehydrogenase-1 and Hypoxia- Inducible Factor-1 alpha in Breast Cancer. Int J Adv Res. 2014;2(7):822–830. [Google Scholar]
- 157.Serrano D, Bleau A-M, Fernandez-Garcia I, Fernandez-Marcelo T, Iniesta P, Ortiz-de-Solorzano C, Calvo A. Inhibition of telomerase activity preferentially targets aldehyde dehydrogenase-positive cancer stem-like cells in lung cancer. Mol Cancer. 2011;10(96):10.1186. doi: 10.1186/1476-4598-10-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lin L, Fuchs J, Li C, Olson V, Bekaii-Saab T, Lin J. STAT3 signaling pathway is necessary for cell survival and tumorsphere forming capacity in ALDH+/CD133+ stem cell-like human colon cancer cells. Biochem Biophys Res Commun. 2011;416(3):246–251. doi: 10.1016/j.bbrc.2011.10.112. [DOI] [PubMed] [Google Scholar]
- 159.Korkaya H, Paulson A, Iovino F, Wicha MS. HER2 regulates the mammary stem/progenitor cell population driving tumorigenesis and invasion. Oncogene. 2008;27(47):6120–6130. doi: 10.1038/onc.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Alam M, Ahmad R, Rajabi H, Kharbanda A, Kufe D. MUC1-C oncoprotein activates ERK → C/EBPβ signaling and induction of aldehyde dehydrogenase 1A1 in breast cancer cells. J Biol Chem. 2013;288(43):30892–30903. doi: 10.1074/jbc.M113.477158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rosenthal DT, Zhang J, Bao L, Zhu L, Wu Z, Toy K, Kleer CG, Merajver SD. RhoC impacts the metastatic potential and abundance of breast cancer stem cells. PLoS One. 2012;7(7):e40979. doi: 10.1371/journal.pone.0040979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Raha D, Wilson TR, Peng J, Peterson D, Yue P, Evangelista M, Wilson C, Merchant M, Settleman J. The cancer stem cell marker aldehyde dehydrogenase is required to maintain a drug-tolerant tumor cell subpopulation. Cancer Res. 2014;74(13):3579–3590. doi: 10.1158/0008-5472.CAN-13-3456. [DOI] [PubMed] [Google Scholar]
- 163.Kim SK, Kim H, D-h Lee, T-s Kim, Kim T, Chung C, Koh GY, Kim H, Lim D-S. Reversing the intractable nature of pancreatic cancer by selectively targeting ALDH-high, therapy-resistant cancer cells. PLoS One. 2013;8(10):e78130. doi: 10.1371/journal.pone.0078130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chen D, Cui QC, Yang H, Dou QP. Disulfiram, a clinically used anti-alcoholism drug and copper-binding agent, induces apoptotic cell death in breast cancer cultures and xenografts via inhibition of the proteasome activity. Cancer Res. 2006;66(21):10425–10433. doi: 10.1158/0008-5472.CAN-06-2126. [DOI] [PubMed] [Google Scholar]
- 165.Lövborg H, Öberg F, Rickardson L, Gullbo J, Nygren P, Larsson R. Inhibition of proteasome activity, nuclear factor-KB translocation and cell survival by the antialcoholism drug disulfiram. Int J Cancer. 2006;118(6):1577–1580. doi: 10.1002/ijc.21534. [DOI] [PubMed] [Google Scholar]
- 166.Cen D, Gonzalez RI, Buckmeier JA, Kahlon RS, Tohidian NB, Meyskens FL. Disulfiram induces apoptosis in human melanoma cells: a redox-related process1. Mol Cancer Ther. 2002;1(3):197–204. [PubMed] [Google Scholar]
- 167.Morrison BW, Doudican NA, Patel KR, Orlow SJ. Disulfiram induces copper-dependent stimulation of reactive oxygen species and activation of the extrinsic apoptotic pathway in melanoma. Melanoma Res. 2010;20(1):11–20. doi: 10.1097/CMR.0b013e328334131d. [DOI] [PubMed] [Google Scholar]
- 168.Yip N, Fombon I, Liu P, Brown S, Kannappan V, Armesilla A, Xu B, Cassidy J, Darling J, Wang W. Disulfiram modulated ROS–MAPK and NFκB pathways and targeted breast cancer cells with cancer stem cell-like properties. Br J Cancer. 2011;104(10):1564–1574. doi: 10.1038/bjc.2011.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang H, Chen D, Ringler J, Chen W, Cui QC, Ethier SP, Dou QP, Wu G. Disulfiram treatment facilitates phosphoinositide 3-kinase inhibition in human breast cancer cells in vitro and in vivo. Cancer Res. 2010;70(10):3996–4004. doi: 10.1158/0008-5472.CAN-09-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Shian S-G, Kao Y-R, Wu FY-H, Wu C-W. Inhibition of invasion and angiogenesis by zinc-chelating agent disulfiram. Mol Pharmacol. 2003;64(5):1076–1084. doi: 10.1124/mol.64.5.1076. [DOI] [PubMed] [Google Scholar]
- 171.Marikovsky M, Ziv V, Nevo N, Harris-Cerruti C, Mahler O. Cu/Zn superoxide dismutase plays important role in immune response. J Immunol. 2003;170(6):2993–3001. doi: 10.4049/jimmunol.170.6.2993. [DOI] [PubMed] [Google Scholar]
- 172.Marikovsky M, Nevo N, Vadai E, Harris-Cerruti C. Cu/Zn superoxide dismutase plays a role in angiogenesis. Int J Cancer. 2002;97(1):34–41. doi: 10.1002/ijc.1565. [DOI] [PubMed] [Google Scholar]
- 173.Cho H, Lee T, Park J, Park K, Sin D, Park Y, Moon Y, Lee K, Yeo J. Disulfiram suppresses invasive ability of osteosarcoma cells via the inhibition of MMP-2 and MMP-9 expression. J Biochem Mol Biol. 2007;40(6):1069. doi: 10.5483/bmbrep.2007.40.6.1069. [DOI] [PubMed] [Google Scholar]
- 174.Duan X, Xiao J, Yin Q, Zhang Z, Yu H, Mao S, Li Y. Multi-targeted inhibition of tumor growth and lung metastasis by redox-sensitive shell crosslinked micelles loading disulfiram. Nanotechnology. 2014;25(12):125102. doi: 10.1088/0957-4484/25/12/125102. [DOI] [PubMed] [Google Scholar]
- 175.Nechushtan H, Hamamreh Y, Nidal S, Gotfried M, Baron A, Shalev YI, Nisman B, Peretz T, Peylan-Ramu N. A phase IIb trial assessing the addition of disulfiram to chemotherapy for the treatment of metastatic non-small cell lung cancer. Oncologist. 2015;20(4):366–367. doi: 10.1634/theoncologist.2014-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Pors K, Loadman PM, Shnyder SD, Sutherland M, Sheldrake HM, Guino M, Kiakos K, Hartley JA, Searcey M, Patterson LH. Modification of the duocarmycin pharmacophore enables CYP1A1 targeting for biological activity. Chem Commun. 2011;47(44):12062–12064. doi: 10.1039/c1cc15638a. [DOI] [PubMed] [Google Scholar]
- 177.Sheldrake HM, Travica S, Johansson I, Loadman PM, Sutherland M, Elsalem L, Illingworth N, Cresswell AJ, Reuillon T, Shnyder SD. Re-engineering of the Duocarmycin structural architecture enables bioprecursor development targeting CYP1A1 and CYP2W1 for biological activity. J Med Chem. 2013;56(15):6273–6277. doi: 10.1021/jm4000209. [DOI] [PubMed] [Google Scholar]
- 178.Sutherland M, Gill JH, Loadman PM, Laye JP, Sheldrake HM, Illingworth NA, Alandas MN, Cooper PA, Searcey M, Pors K. Antitumor activity of a duocarmycin analogue rationalized to be metabolically activated by cytochrome P450 1A1 in human transitional cell carcinoma of the bladder. Mol Cancer Ther. 2013;12(1):27–37. doi: 10.1158/1535-7163.MCT-12-0405. [DOI] [PubMed] [Google Scholar]
- 179.Travica S, Pors K, Loadman PM, Shnyder SD, Johansson I, Alandas MN, Sheldrake HM, Mkrtchian S, Patterson LH, Ingelman-Sundberg M. Colon cancer-specific cytochrome P450 2W1 converts duocarmycin analogues into potent tumor cytotoxins. Clin Cancer Res. 2013;19(11):2952–2961. doi: 10.1158/1078-0432.CCR-13-0238. [DOI] [PubMed] [Google Scholar]
- 180.Neumeister V, Agarwal S, Bordeaux J, Camp RL, Rimm DL. In situ identification of putative cancer stem cells by multiplexing ALDH1, CD44, and cytokeratin identifies breast cancer patients with poor prognosis. Am J Pathol. 2010;176(5):2131–2138. doi: 10.2353/ajpath.2010.090712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhong Y, Shen S, Zhou Y, Mao F, Guan J, Lin Y, Xu Y, Sun Q. ALDH1 is a better clinical indicator for relapse of invasive ductal breast cancer than the CD44+/CD24− phenotype. Med Oncol. 2014;31(3):1–8. doi: 10.1007/s12032-014-0864-0. [DOI] [PubMed] [Google Scholar]
- 182.Opdenaker LM, Arnold KM, Pohlig RT, Padmanabhan JS, Flynn DC, Sims-Mourtada J. Immunohistochemical analysis of aldehyde dehydrogenase isoforms and their association with estrogen-receptor status and disease progression in breast cancer. Breast Cancer. 2014;6:205. doi: 10.2147/BCTT.S73674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Qiu Y, Pu T, Li L, Cheng F, Lu C, Sun L, Teng X, Ye F, Bu H. The expression of aldehyde dehydrogenase family in breast cancer. J Breast Cancer. 2014;17(1):54–60. doi: 10.4048/jbc.2014.17.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Tan EY, Thike AA, Breast Surgical Team at O. Tan PH. ALDH1 expression is enriched in breast cancers arising in young women but does not predict outcome. Br J Cancer. 2013;109(1):109–113. doi: 10.1038/bjc.2013.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Liu S, Liu C, Min X, Ji Y, Wang N, Liu D, Cai J, Li K. Prognostic value of cancer stem cell marker aldehyde dehydrogenase in ovarian cancer: a meta-analysis. PLoS One. 2013;8(11):e81050. doi: 10.1371/journal.pone.0081050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Rasper M, Schäfer A, Piontek G, Teufel J, Brockhoff G, Ringel F, Heindl S, Zimmer C, Schlegel J (2010) Aldehyde dehydrogenase 1 positive glioblastoma cells show brain tumor stem cell capacity. Neuro-oncology:noq070 [DOI] [PMC free article] [PubMed]
- 187.Zhang W, Yan W, You G, Bao Z, Wang Y, Liu Y, You Y, Jiang T. Genome-wide DNA methylation profiling identifies ALDH1A3 promoter methylation as a prognostic predictor in G-CIMP− primary glioblastoma. Cancer Lett. 2013;328(1):120–125. doi: 10.1016/j.canlet.2012.08.033. [DOI] [PubMed] [Google Scholar]
- 188.Gangavarapu KJ, Azabdaftari G, Morrison CD, Miller A, Foster BA, Huss WJ. Aldehyde dehydrogenase and ATP binding cassette transporter G2 (ABCG2) functional assays isolate different populations of prostate stem cells where ABCG2 function selects for cells with increased stem cell activity. Stem Cell Res Ther. 2013;4(5):132. doi: 10.1186/scrt343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wu J, Mu Q, Thiviyanathan V, Annapragada A, Vigneswaran N. Cancer stem cells are enriched in Fanconi anemia head and neck squamous cell carcinomas. Int J Oncol. 2014;45(6):2365–2372. doi: 10.3892/ijo.2014.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Deng Y, Zhou J, Fang L, Cai Y, Ke J, Xie X, Huang Y, Huang M, Wang J. ALDH1 is an independent prognostic factor for patients with stages II–III rectal cancer after receiving radiochemotherapy. Br J Cancer. 2014;110(2):430–434. doi: 10.1038/bjc.2013.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Patlolla JM, Qian L, Biddick L, Zhang Y, Desai D, Amin S, Lightfoot S, Rao CV. β-Escin Inhibits NNK-induced lung adenocarcinoma and ALDH1A1 and RhoA/Rock expression in A/J mice and growth of H460 human lung cancer cells. Cancer Prev Res. 2013;6(10):1140–1149. doi: 10.1158/1940-6207.CAPR-13-0216. [DOI] [PubMed] [Google Scholar]
- 192.Giacalone NJ, Den RB, Eisenberg R, Chen H, Olson SJ, Massion PP, Carbone DP, Lu B. ALDH7A1 expression is associated with recurrence in patients with surgically resected non-small-cell lung carcinoma. Future Oncol. 2013;9(5):737–745. doi: 10.2217/fon.13.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Boonyaratanakornkit JB, Yue L, Strachan LR, Scalapino KJ, LeBoit PE, Lu Y, Leong SP, Smith JE, Ghadially R. Selection of tumorigenic melanoma cells using ALDH. J Invest Dermatol. 2010;130(12):2799–2808. doi: 10.1038/jid.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Z-l Yang, Yang L, Zou Q, Yuan Y, Li J, Liang L, Zeng G, Chen S. Positive ALDH1A3 and negative GPX3 expressions are biomarkers for poor prognosis of gallbladder cancer. Dis Markers. 2013;35(3):10. doi: 10.1155/2013/187043. [DOI] [PMC free article] [PubMed] [Google Scholar]