Abstract
Single nucleotide polymorphisms (SNPs) in the interleukin-17 (IL-17) gene have been shown to be correlated with susceptibility to cancer. However, various studies report different results of this association. The aim of the present work was to clarify the effects of IL-17A G197A (rs2275913) and IL-17F T7488C (rs763780) polymorphisms on cancer risk. We performed systematic searches of the PubMed and CNKI databases to obtain relevant publications. Odds ratios (ORs) with 95% confidence intervals (CIs) were used to evaluate the association of rs2275913 and rs763780 polymorphisms with cancer risk. Data were extracted from the selected studies, and statistical analysis was conducted using the STATA software. Our results indicated that rs2275913 and rs763780 polymorphisms significantly increase cancer risk, especially in gastric cancers. Subgroup analysis suggested the existence of a significant correlation between rs763780 polymorphism and cancer susceptibility in Caucasian populations. This updated meta-analysis confirms that rs2275913 and rs763780 polymorphisms are highly associated with increased risk for multiple forms of cancer.
According to the latest global statistics on cancer, approximately 14.1 million new cancer cases and 8.2 million cancer-related deaths occurred worldwide in 20121. Cancer is currently the leading cause of death worldwide, and it represents a major global health concern. Although the pathogenic factors of cancer remain unknown, complex interactions between an individual’s genetic background and environmental factors have been suggested to be highly associated with cancer development2.
Inherited factors leading to the development of cancer are not clearly understood, but the roles of cytokines in tumour immunity and carcinogenesis have been well established3. Th17 cells, which were identified as a new subset of T helper cells4, play pivotal roles in both adaptive and innate immunity, by secreting the pro-inflammatory cytokine interleukin (IL)-175. IL-17 has six family members (IL17A-F) that bind to five receptors (IL-17RA-RD and SEF)6. Among all IL-17 family members, IL-17A is one of the most important cytokines, and it may play a role in autoimmune diseases, chronic inflammatory diseases, and malignancies7,8,9; IL-17A has been shown to induce the production of inflammatory chemokines and cytokines by macrophages and neutrophils. More recent studies have reported that IL-17F can also induce the expression of various chemokines, cytokines, and adhesion molecules involved in inflammation-related cancer10. The rs763780 variant in the IL-17F gene can lead to a His-to-Arg substitution at amino acid position 161, and thus, inhibit the function of wild-type IL-17F. This may contribute to increased risk of several malignant tumors including gastric cancer, colorectal cancer, and breast cancer11,12,13,14.
Meta-analysis is a statistical technique that combines results from different individual studies to produce a comprehensive assessment of the major findings with enhanced accuracy15. IL-17 polymorphism has been hypothesized to play a role in carcinogenesis, and numerous studies investigating the same have been published in the past few years11,12,13,14,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31. However, these published studies have reported mixed findings. Therefore, to clarify the role of IL-17A rs2275913 and IL-17F rs763780 polymorphisms in cancer risk, we conducted a comprehensive meta-analysis of all eligible case-control studies.
Results
Study characteristics
Through primary literature retrieval from Pubmed and CNKI databases, we identified 95 studies that investigated the effect of IL-17 polymorphisms on cancer risk. After screening the titles and abstracts according to the preferred reporting items for systematic reviews and meta-analyses (PRISMA, Fig. 1), 55 studies were excluded from our meta-analysis. Then remaining 40 articles were assessed for eligibility by reading the full-text; 14 articles were excluded owing to either lack of complete data or presence of irrelevant data that focused on other IL-17 polymorphisms. Finally, 26 studies with 7,872 cases and 9,646 cancer-free controls met the inclusion criteria for our meta-analysis for assessing the influence of rs2275913 and rs763780 polymorphisms on cancer risk. Among these, 20 studies were based on the Asian population, and 6 were based on Caucasian populations. The selected studies presented data on several different cancer types: gastric, colorectal, prostate, thyroid, cervical, breast, ovary, bladder, hepatocellular, lung, and oesophageal cancer, and acute myeloid leukaemia. The main characteristics of the included studies are presented in Table 1. The distributions of IL-17A rs2275913 and rs763780 polymorphisms among patients and controls are shown in Table 2.
Figure 1. Preferred reporting items for systematic reviews and meta-analyses flow diagram of the literature review process for IL-17 polymorphisms and cancer.
Table 1. Characteristics of the studies included in the meta-analysis.
| First author | Year | Ethnicity | Tumor type | case | control | Genotyping medthod | Source of control | No. of SNP | cancer risk |
|---|---|---|---|---|---|---|---|---|---|
| Hou | 2015 | Asian | GC | 326 | 326 | MassARRAY | Population | 1, 2 | No.1 yes |
| Nemati | 2015 | Caucasian | CRC | 202 | 203 | PCR-RFLP | Hospital | 1*, 2 | yes |
| Gao | 2015 | Asian | GC | 572 | 572 | PCR–RFLP | Hospital | 1, 2* | No.2 yes |
| Lv | 2015 | Asian | CC | 264 | 264 | PCR–RFLP | Population | 1, 2* | No.1 yes |
| Lee | 2015 | Asian | PTC | 94 | 260 | TaqMan | Population | 1 | no risk |
| Xi | 2015 | Asian | HCC | 155 | 171 | PCR–RFLP | Hospital | 1, 2 | no risk |
| Wang | 2014 | Asian | GC | 462 | 462 | PCR–RFLP | Population | 1, 2 | No.1 yes |
| Wróbel | 2014 | Caucasian | AML | 62 | 125 | PCR–RFLP | Population | 1, 2 | No.2 yes |
| Omrane | 2014 | Caucasian | CRC | 102 | 139 | TaqMan | Population | 1 | yes |
| Omrane | 2014 | Caucasian | CRC | 102 | 139 | TaqMan | Population | 2 | yes |
| Yin | 2014 | Asian | EC | 380 | 380 | SNPscan | Hospital | 1 | yes |
| Li | 2014 | Asian | HCC | 395 | 174 | PCR–RFLP | Hospital | 1 | yes |
| Kaabachi | 2014 | Caucasian | LC | 239 | 258 | PCR–RFLP | Population | 1, 2 | No.2 yes |
| Zhu | 2014 | Asian | GC | 311 | 611 | MassARRAY | Hospital | 1, 2* | No.1 yes |
| Zhang | 2014 | Asian | GC | 260 | 512 | MassARRAY | Hospital | 1*, 2* | yes |
| Bi | 2014 | Asian | GC | 99 | 150 | PCR-RFLP | Hospital | 1, 2 | no risk |
| Rafiei | 2013 | Caucasian | GC | 161 | 171 | PCR–RFLP | Hospital | 1 | yes |
| Zhou | 2013 | Asian | BLC | 301 | 446 | TaqMan | Hospital | 1, 2 | yes |
| Arisawa | 2012 | Asian | GC | 337 | 587 | PCR-SSCP | Hospital | 1 | yes |
| Quan | 2012 | Asian | CC | 311 | 463 | TaqMan | Hospital | 1, 2 | No.1 yes |
| Wang | 2012 | Asian | BC | 491 | 502 | SNaPshot | Population | 1, 2 | No.1 yes |
| Ruan | 2012 | Asian | OC | 92 | 38 | PCR-RFLP | Hospital | 1*, 2* | no risk |
| Chen | 2010 | Asian | GC | 1042 | 1090 | TaqMan | Population | 1 | no risk |
| Wu | 2010 | Asian | GC | 1010 | 800 | PCR–RFLP | Population | 1, 2 | No.2 yes |
| Luo | 2010 | Asian | GC | 24 | 50 | PCR-RFLP | Hospital | 1*, 2 | No.1 yes |
| Shibata | 2009 | Asian | GC | 287 | 524 | PCR–SSCP | Hospital | 1*, 2 | No.1 yes |
*The P-values of the Hardy-Weinberg equilibrium test of control group less than 0.05.
CRC: colorectal cancer; GC: gastric cancer; CC: cervical cancer; PTC: papillary thyroid cancer; HCC: hepatocellular carcinoma; LC: lung cancer; AML: acute myeloid leukemia; EC: esophageal cancer; BLC: bladder cancer; BC: breast cancer; OC: ovarian cancer; NA: not available; PCR-RFLP: polymerase chain reaction restriction fragment length polymorphism; SSCP: single strand conformation polymorphism; SNP: single-nucleotide polymorphisms; No. of SNP: No.1: rs2275913, No.2: rs763780.
Table 2. IL-17 polymorphisms Genotype Distribution and Allele Frequency in Cases and Controls.
| First author | Genotype (N,%) |
Allele frequency (N, %) |
MAF | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case |
Control |
Case |
Control |
||||||||||
| total | AA | AB | BB | total | AA | AB | BB | A | B | A | B | ||
| rs2275913 | |||||||||||||
| Hou 2015 | 326 | 121 | 149 | 56 | 326 | 161 | 136 | 29 | 391 | 261 | 458 | 194 | 0.40 |
| Nemati 2015 | 202 | 100 | 82 | 20 | 199 | 110 | 50 | 39 | 282 | 122 | 270 | 128 | 0.30 |
| Gao 2015 | 572 | 239 | 250 | 83 | 573 | 260 | 241 | 72 | 728 | 416 | 761 | 385 | 0.36 |
| Lv 2015 | 264 | 110 | 117 | 37 | 264 | 139 | 105 | 20 | 337 | 191 | 383 | 145 | 0.36 |
| Lee 2015 | 94 | 28 | 42 | 24 | 260 | 76 | 137 | 47 | 98 | 90 | 289 | 231 | 0.48 |
| Xi 2015 | 155 | 38 | 71 | 46 | 171 | 35 | 90 | 46 | 147 | 163 | 160 | 182 | 0.53 |
| Wang 2014 | 462 | 160 | 211 | 91 | 462 | 214 | 190 | 58 | 531 | 393 | 618 | 306 | 0.43 |
| Wróbel 2014 | 62 | 23 | 25 | 14 | 125 | 38 | 67 | 20 | 71 | 53 | 143 | 107 | 0.43 |
| Omrane 2014 | 102 | 48 | 51 | 3 | 139 | 95 | 38 | 6 | 147 | 57 | 228 | 50 | 0.28 |
| Yin 2014 | 364 | 104 | 180 | 80 | 370 | 117 | 174 | 79 | 388 | 340 | 408 | 332 | 0.47 |
| Li 2014 | 391 | 110 | 197 | 84 | 174 | 50 | 85 | 39 | 417 | 365 | 185 | 163 | 0.47 |
| Kaabachi 2014 | 239 | 147 | 80 | 12 | 258 | 166 | 79 | 13 | 374 | 104 | 411 | 105 | 0.22 |
| Zhu 2014 | 293 | 126 | 122 | 45 | 550 | 273 | 216 | 61 | 374 | 212 | 762 | 338 | 0.36 |
| Zhang 2014 | 260 | 110 | 102 | 48 | 512 | 258 | 187 | 67 | 322 | 198 | 703 | 321 | 0.38 |
| Bi 2014 | 99 | 32 | 39 | 28 | 150 | 41 | 69 | 40 | 103 | 95 | 151 | 149 | 0.48 |
| Rafiei 2013 | 161 | 56 | 61 | 44 | 171 | 78 | 72 | 21 | 173 | 149 | 228 | 114 | 0.46 |
| Zhou 2013 | 301 | 79 | 154 | 68 | 446 | 164 | 204 | 78 | 312 | 290 | 532 | 360 | 0.48 |
| Arisawa 2012 | 333 | 112 | 137 | 84 | 583 | 218 | 293 | 72 | 361 | 305 | 729 | 437 | 0.46 |
| Quan 2012 | 311 | 93 | 142 | 76 | 463 | 168 | 215 | 80 | 328 | 294 | 551 | 375 | 0.47 |
| Wang 2012 | 491 | 165 | 234 | 92 | 501 | 198 | 245 | 58 | 564 | 418 | 641 | 361 | 0.43 |
| Ruan 2012 | 92 | 20 | 60 | 12 | 38 | 12 | 24 | 2 | 100 | 84 | 48 | 28 | 0.46 |
| Chen 2010 | 1,042 | 300 | 522 | 220 | 1,090 | 325 | 541 | 224 | 1,122 | 962 | 1,191 | 989 | 0.46 |
| Wu 2010 | 945 | 210 | 485 | 250 | 768 | 193 | 371 | 204 | 905 | 985 | 757 | 779 | 0.52 |
| Luo 2010 | 24 | 11 | 12 | 1 | 530 | 58 | 426 | 46 | 34 | 14 | 542 | 518 | 0.29 |
| Shibata 2009 | 287 | 94 | 124 | 69 | 523 | 175 | 299 | 49 | 312 | 262 | 649 | 397 | 0.46 |
| rs763780 | |||||||||||||
| Hou 2015 | 326 | 266 | 38 | 22 | 326 | 278 | 33 | 15 | 570 | 82 | 589 | 63 | 0.13 |
| Nemati 2015 | 200 | 177 | 23 | 0 | 201 | 190 | 11 | 0 | 377 | 23 | 391 | 11 | 0.06 |
| Gao 2015 | 572 | 420 | 67 | 85 | 572 | 472 | 58 | 42 | 907 | 237 | 1002 | 142 | 0.21 |
| Lv 2015 | 264 | 209 | 35 | 20 | 264 | 223 | 30 | 11 | 453 | 75 | 476 | 52 | 0.14 |
| Xi 2015 | 155 | 100 | 46 | 9 | 171 | 105 | 63 | 3 | 246 | 64 | 273 | 69 | 0.21 |
| Wang 2014 | 462 | 349 | 98 | 15 | 462 | 362 | 90 | 10 | 796 | 128 | 814 | 110 | 0.14 |
| Wróbel 2014 | 62 | 42 | 15 | 5 | 125 | 114 | 11 | 0 | 99 | 25 | 239 | 11 | 0.20 |
| Omrane 2014 | 100 | 72 | 27 | 1 | 137 | 98 | 38 | 1 | 171 | 29 | 234 | 40 | 0.15 |
| Kaabachi 2014 | 239 | 204 | 34 | 1 | 258 | 236 | 22 | 0 | 442 | 36 | 494 | 22 | 0.08 |
| Zhu 2014 | 293 | 241 | 35 | 17 | 550 | 463 | 58 | 29 | 517 | 69 | 984 | 116 | 0.12 |
| Zhang 2014 | 260 | 209 | 30 | 21 | 512 | 429 | 53 | 30 | 448 | 72 | 911 | 113 | 0.14 |
| Bi 2014 | 100 | 69 | 22 | 9 | 150 | 108 | 35 | 7 | 160 | 40 | 251 | 49 | 0.20 |
| Zhou 2013 | 301 | 240 | 57 | 4 | 446 | 317 | 124 | 5 | 537 | 65 | 758 | 134 | 0.11 |
| Quan 2012 | 311 | 222 | 85 | 4 | 463 | 332 | 126 | 5 | 529 | 93 | 790 | 136 | 0.15 |
| Wang 2012 | 491 | 382 | 103 | 6 | 502 | 396 | 99 | 7 | 867 | 115 | 891 | 113 | 0.12 |
| Ruan 2012 | 92 | 13 | 69 | 10 | 38 | 2 | 34 | 2 | 95 | 89 | 38 | 38 | 0.48 |
| Wu 2010 | 927 | 540 | 332 | 55 | 777 | 527 | 214 | 36 | 1412 | 442 | 1268 | 286 | 0.24 |
| Luo 2010 | 24 | 14 | 10 | 0 | 230 | 176 | 51 | 3 | 38 | 10 | 403 | 57 | 0.21 |
| Shibata 2009 | 280 | 221 | 55 | 4 | 523 | 419 | 100 | 4 | 497 | 63 | 938 | 108 | 0.11 |
A represents the major allele, B represents the minor allele. MAF: minor allele frequencies.
Quantitative synthesis results
IL-17A G197A polymorphism (rs2275913).
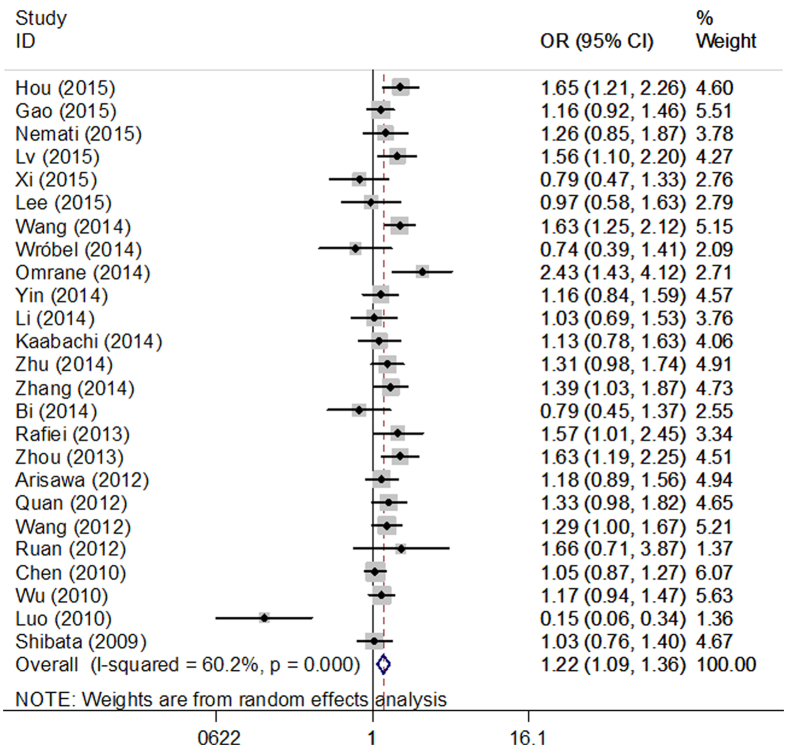
Overall, our meta-analysis found a borderline association between rs2275913 polymorphism and increased cancer risk in all genetic models (AA vs. GG: OR = 1.48, 95% CI = 1.25–1.74; AA vs. GG + GA: OR = 1.40, 95% CI = 1.19–1.65; AA + AG vs. GG: OR = 1. 22, 95% CI = 1.09–1.36, Fig. 2; GA vs. GG: OR = 1.13, 95% CI = 1.01–1.28; A vs. G: OR = 1.22, 95% CI = 1.13–1.32) for all cancer types. When only studies following the Hardy–Weinberg equilibrium (HWE) were included in the analysis, a significant association was also observed under all genetic models, and these results are shown in Table 3.
Figure 2. Forest plots of IL-17A rs2275913 polymorphism and cancer risk using a recessive genetic model (AA+AG vs. GG).
Table 3. Summary of ORs and 95% CI of IL-17A rs2275913 and IL-17F rs763780 polymorphisms with cancer risk.
| Comparisons | B vs A |
BB vs AA |
BB vs AB + AA |
BB + AB vs AA |
AB vs AA |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | P | OR (95%CI) | P | OR (95%CI) | P | OR (95%CI) | P | OR (95%CI) | P | |
| rs2275913 | ||||||||||
| Overall | 1.22 (1.13–1.32) | <0.001 | 1.48 (1.25–1.74) | <0.001 | 1.40 (1.19–1.65) | <0.001 | 1.22 (1.09–1.36) | 0.001 | 1.13 (1.01–1.28) | 0.04 |
| HWE | ||||||||||
| Yes | 1.23 (1.14–1.33) | <0.001 | 1.49 (1.27–1.75) | <0.001 | 1.39 (1.20–1.61) | <0.001 | 1.26 (1.14–1.38) | <0.001 | 1.17 (1.06–1.30) | 0.002 |
| Ethnicity | ||||||||||
| Asian | 1.22 (1.12–1.32) | <0.001 | 1.53 (1.29–1.81) | <0.001 | 1.45 (1.24–1.70) | <0.001 | 1.20 (1.06–1.35) | 0.003 | 1.10 (0.97–1.24) | 0.13 |
| Caucasian | 1.24 (0.94–1.63) | 0.13 | 1.16 (0.59–2.27) | 0.66 | 1.08 (0.51–2.28) | 0.84 | 1.34 (0.97–1.84) | 0.07 | 1.35 (0.90–2.03) | 0.14 |
| Cancer type | ||||||||||
| GC | 1.24 (1.10–1.40) | <0.001 | 1.62 (1.26–2.07) | <0.001 | 1.56 (1.23–1.99) | <0.001 | 1.17 (0.99–1.38) | 0.07 | 1.05 (0.89–1.25) | 0.56 |
| CRC | 1.25 (0.65–2.38) | 0.51 | 0.61 (0.35–1.07) | 0.09 | 0.48 (0.28–0.82) | 0.007 | 1.71 (0.90–3.24) | 0.10 | 2.10 (1.49–2.96) | <0.001 |
| CC | 1.38 (1.18–1.62) | <0.001 | 1.89 (1.35–2.64) | <0.001 | 1.66 (1.23–2.24) | 0.001 | 1.43 (1.14–1.80) | 0.002 | 1.29 (1.01–1.64) | 0.04 |
| HCC | 0.99 (0.818–1.20) | 0.89 | 0.96 (0.65–1.41) | 0.82 | 1.03 (0.75–1.42) | 0.85 | 0.94 (0.68–1.28) | 0.68 | 0.92 (0.66–1.29) | 0.63 |
| rs763780 | ||||||||||
| Overall | 1.28 (1.11–1.47) | 0.001 | 1.69 (1.40–2.04) | <0.001 | 1.64 (1.36–1.97) | <0.001 | 1.25 (1.07–1.47) | 0.001 | 1.17 (1.00–1.37) | 0.06 |
| HWE | ||||||||||
| Yes | 1.25 (1.02–1.52) | 0.03 | 1.70 (1.21–2.39) | 0.002 | 1.69 (1.20–2.38) | 0.002 | 1.21 (0.98–1.50) | 0.08 | 1.15 (0.93–1.41) | 0.20 |
| Ethnicity | ||||||||||
| Asian | 1.06 (0.95–1.19) | 0.28 | 1.54 (1.08–2.20) | 0.02 | 1.55 (1.09–2.20) | 0.02 | 1.04 (0.87–1.24) | 0.66 | 0.99 (0.83–1.19) | 0.95 |
| Caucasian | 2.08 (0.94–1.63) | 0.03 | 6.17 (1.50–30.0) | 0.01 | 6.19 (1.36–28.1) | 0.02 | 2.02 (1.08–3.76) | 0.03 | 1.83 (1.08–3.11) | 0.02 |
| Cancer type | ||||||||||
| GC | 1.37 (1.25–1.51) | <0.001 | 1.67 (1.35–2.06) | <0.001 | 1.59 (1.29–1.95) | <0.001 | 1.37 (1.22–1.53) | <0.001 | 1.28 (1.13–1.45) | <0.001 |
| CRC | 1.40 (0.65–3.00) | 0.38 | – | – | – | – | 1.43 (0.63–3.22) | 0.39 | 1.42 (0.63–3.24) | 0.40 |
A: the major allele; B: the minor allele; CI: confidence interval; OR: odds ratio; GC: Gastric cancer; CRC: colorectal cancer; CC: cervical cancer; HCC: hepatocellular carcinoma.
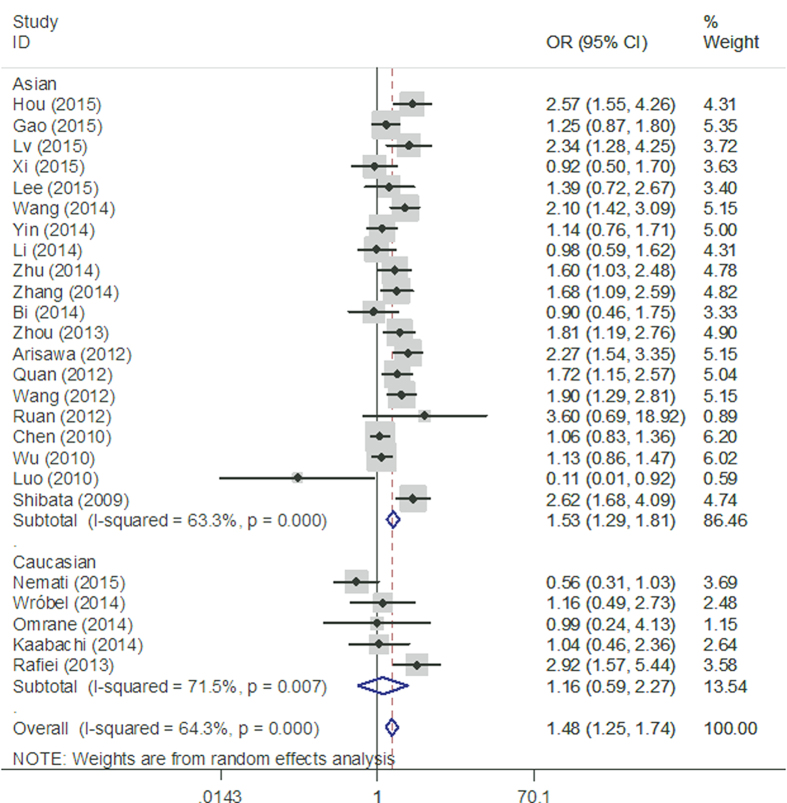
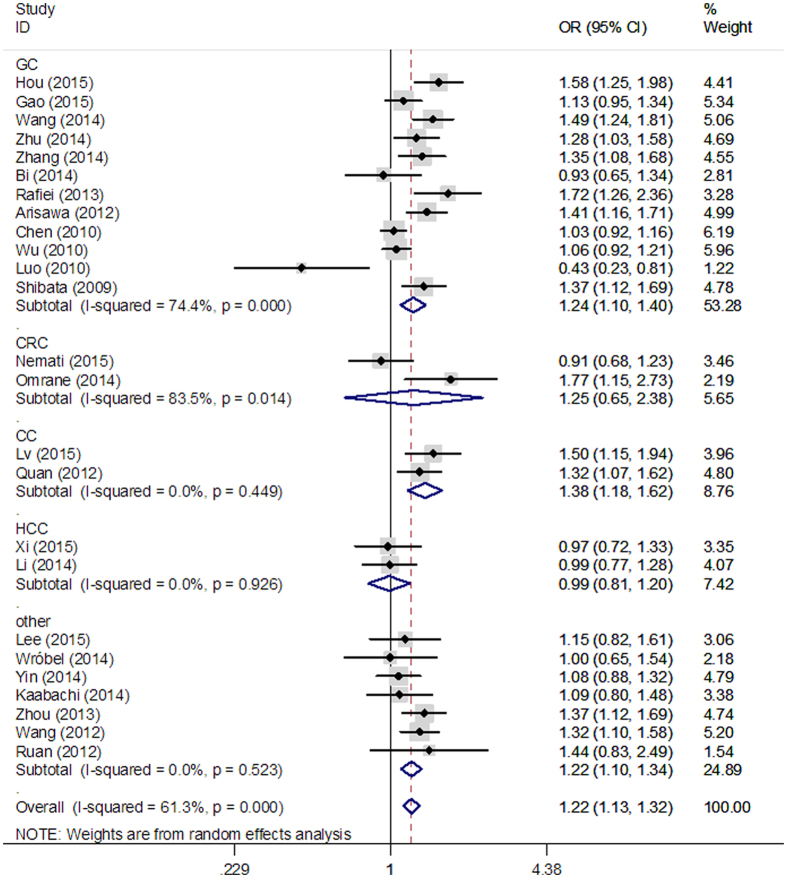
When subgroup analysis was performed based on ethnicity, no significant correlation was observed between rs2275913 polymorphism and cancer risk in Caucasians. However, statistically significant associations were found in the following genetic models in Asians: (AA vs. GG: OR = 1.53, 95% CI = 1.29–1.81 Fig. 3; AA vs. GG + GA: OR = 1.45, 95% CI = 1.24–1.70; AA + AG vs. GG: OR = 1. 20, 95% CI = 1.06–1.35; A vs. G: OR = 1.22, 95% CI = 1.12–1.32). When results were stratified by cancer type, we found a significant association between rs2275913 polymorphism and increased gastric cancer risk in three genetic models (AA vs. GG: OR = 1.62, 95% CI = 1.26–2.07; AA + AG vs. GG: OR = 1.56, 95% CI = 1.23–1.99; A vs. G: OR = 1.24, 95% CI = 1.10–1.40, Fig. 4). Moreover, significant associations were observed between rs2275913 polymorphism and cervical cancer in all 5 comparison models. All comparisons are listed in Table 3.
Figure 3. Stratified analysis based on ethnicity for the association between IL-17A rs2275913 polymorphism and cancer risk using a homozygote genetic model (AA vs. GG).
Figure 4. Stratified analysis based on the different cancer sites for the association between IL-17A rs2275913 polymorphism and cancer risk using an allele comparison model (A vs. G).
IL-17F T7488C polymorphism (rs763780).
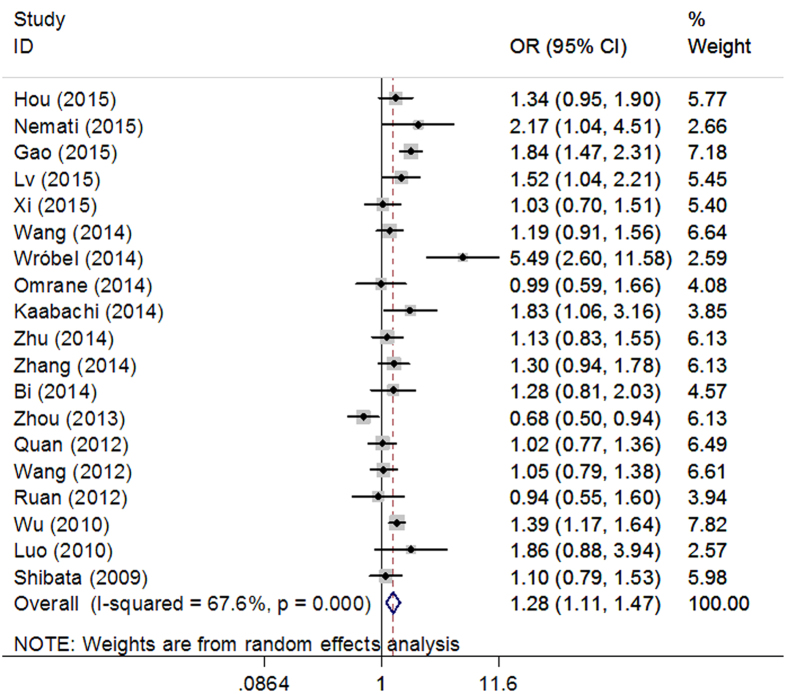
A significant association was found between rs763780 and cancer susceptibility in 3 different comparison models (CC vs. TT: OR = 1.69, 95% CI = 1.40–2.04; CC vs. TT + TC: OR = 1.64, 95% CI = 1.36–1.97; C vs. T: OR = 1.28, 95% CI = 1.11–1.47, Fig. 5). The same results were observed when studies with HW disequilibrium in controls were excluded.
Figure 5. Forest plots of IL-17F rs763780 polymorphism and cancer risk using an allele comparison model (C vs. T).
As shown in Table 3, when subgroup analyses was performed based on ethnicity, significant correlation was observed between rs763780 polymorphism and increased risk of cancer in both Caucasians and Asians. When results were stratified by cancer type, rs763780 polymorphism was found to be significantly associated with an increased risk for gastric cancer in all genetic models (CC vs. TT: OR = 1.67, 95% CI = 1.35–2.06; CC vs. TT + TC: OR = 1.59, 95% CI = 1.29–1.95; CC + TC vs. TT: OR = 1.37, 95% CI = 1.22–1.53; TC vs. TT: OR = 1.28, 95% CI = 1.13–1.45; C vs. T: OR = 1.37, 95% CI = 1.25–1.51).
Publication bias
Publication bias of the selected articles was assessed using Begg’s funnel plot and Egger’s test. As shown in Fig. 6, the funnel plot was symmetrical in shape, and the P-value of Egger’s test indicated a lack of publication bias for rs2275913 and rs763780 polymorphisms.
Figure 6. Funnel plot assessing evidence of publication bias from the eligible studies.
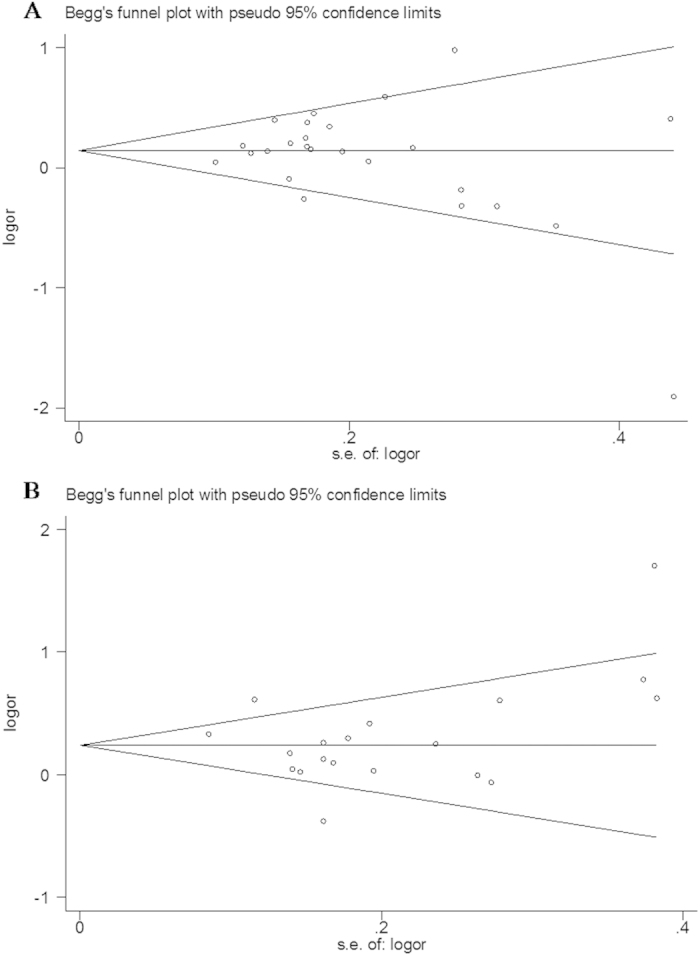
A. rs2275913; B. rs763780.
Heterogeneity and sensitivity analyses
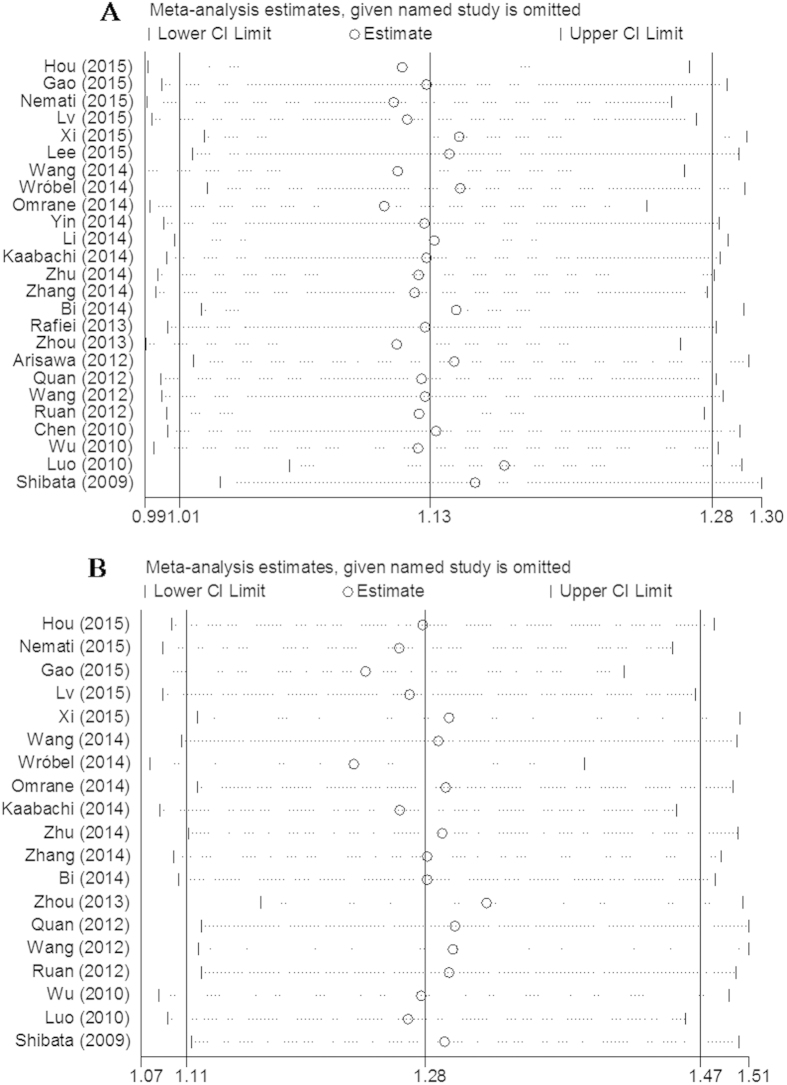
Significant heterogeneities in the data of IL-17A rs2275913 and IL-17F rs763780 polymorphisms were observed in the overall meta-analysis as well as subgroup analysis (Table 4). Due to significant heterogeneity across studies, individual studies used in the meta-analysis were sequentially omitted to to identify the source by sensitivity analysis. The results showed that no individual study skewed the pooled OR values for rs2275913 and rs763780 polymorphisms (Fig. 7).
Table 4. Heterogeneity-analysis results.
| Comparisons | B vs A |
BB vs AA |
BB vs AA+AB |
BB+AB vs AA |
AB vs AA |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I2 | P | EM | I2 | P | EM | I2 | P | EM | I2 | P | EM | I2 | P | EM | |
| rs2275913 | |||||||||||||||
| Overall | 61% | <0.001 | R | 64% | <0.001 | R | 70% | <0.001 | R | 60% | <0.001 | R | 62% | <0.001 | R |
| HWE | |||||||||||||||
| Yes | 58% | 0.001 | R | 57% | 0.001 | R | 60% | <0.001 | R | 43% | 0.02 | R | 39% | 0.04 | R |
| Ethnicity | |||||||||||||||
| Asian | 62% | <0.001 | R | 63% | <0.001 | R | 68% | <0.001 | R | 62% | <0.001 | R | 60% | <0.001 | R |
| Caucasian | 68% | 0.01 | R | 72% | 0.007 | R | 80% | 0.001 | R | 58% | 0.05 | R | 70% | 0.009 | R |
| Cancer type | |||||||||||||||
| GC | 74% | <0.001 | R | 75% | <0.001 | R | 79% | <0.001 | R | 73% | <0.001 | R | 71% | <0.001 | R |
| CRC | 84% | 0.01 | R | 0% | 0.48 | F | 0% | 0.61 | F | 74% | 0.05 | R | 14% | 0.28 | F |
| CC | 0% | 0.45 | F | 0% | 0.40 | F | 0% | 0.47 | F | 0% | 0.51 | F | 0% | 0.51 | F |
| HCC | 0% | 0.93 | F | 0% | 0.88 | F | 0% | 0.56 | F | 0% | 0.43 | F | 8% | 0.30 | F |
| rs763780 | |||||||||||||||
| Overall | 68% | <0.001 | R | 0% | 0.79 | F | 0% | 0.79 | F | 64% | <0.001 | R | 57% | 0.001 | R |
| HWE | |||||||||||||||
| Yes | 68% | <0.001 | R | 0% | 0.79 | F | 0% | 0.82 | F | 66% | <0.001 | R | 61% | 0.002 | R |
| Ethnicity | |||||||||||||||
| Asian | 37% | 0.12 | F | 0% | 0.94 | F | 0% | 0.93 | F | 43% | 0.08 | R | 44% | 0.08 | R |
| Caucasian | 78% | 0.003 | R | 17% | 0.30 | F | 5% | 0.35 | F | 72% | 0.02 | R | 59% | 0.06 | R |
| Cancer type | |||||||||||||||
| GC | 31% | 0.17 | F | 6% | 0.79 | F | 0% | 0.72 | F | 10% | 0.36 | F | 0% | 0.48 | F |
| CRC | 66% | 0.09 | R | – | – | – | – | – | – | 67% | 0.08 | R | 67% | 0.08 | R |
A: the major allele; B: the minor allele; EM: Effects model; F: fixed effects model; R: random effects model; GC: Gastric cancer; CRC: colorectal cancer; CC: cervical cancer; HCC: hepatocellular carcinoma.
Figure 7. Sensitivity analysis of association between the polymorphisms and cancer risk.
A. rs2275913; B. rs763780.
Re-sampling statistics
To obtain robust and replicable results, we performed the correlation analysis 10000 times using non-parametric bootstrap re-sampling method. As showed in Supplementary Table S1 & S2, the results indicated that rs2275913 and rs763780 polymorphism in IL-17 gene were consistently associated with cancer risk in different genetic models (P < 0.05).
Discussion
Recently, inflammatory factors have been shown to increase the risk of developing malignant tumors. IL-17 is a key pro-inflammatory cytokine originally produced by CD4+ memory T cells, and it is involved in both innate and acquired immune responses32,33. Studies indicate that IL-17 is activated by microbial products, and may promote tumor growth and progression via angiogenic functions34,35. Aberrant levels of IL-17 have been observed in gastric, colorectal, hepatocellular, ovarian, and breast cancers36,37,38,39,40.
The IL-17A rs22759133 polymorphism is located in close proximity to 2 nuclear factors activated T cell binding motifs, and it promotes production of high levels of IL-17, which in turn upregulates IL-17-mediated immune responses41. IL-17F, another important member of the IL-17 family, plays a key role in neutrophil recruitment and activation by inducing the secretion of cytokines and chemokines. IL-17F rs763780 polymorphism may inhibit the biological activity of IL-17F, and thus contribute to variations in host’s susceptibility to tumors. Data indicate that IL-17A and IL-17F gene polymorphism may play important roles in the pathogenesis of cancer11,13,16,18,24.
Our study indicated that the two variants of human IL-17 gene significantly increased the risk of cancer in the overall population. When we eliminated studies that deviated from the HWE, similar results were observed. Furthermore, subgroup analyses indicated that associations between these two polymorphisms and cancer risk were also ethnicity- and site-specific. According to the results, rs2275913 polymorphism was significantly associated with elevated cancer risk in Asians (mainly Chinese), but not Caucasians. When subgroup analysis was performed based on cancer types, a significant association was found between rs2275913 polymorphism and risk of gastric/cervical cancer. Interestingly, individuals with the rs2275913 AA genotype showed decreased risk of colorectal cancer as compared to individuals with the GG or GA genotypes. However, only two eligible studies examined IL-17 polymorphisms in colorectal cancer, and therefore, the results may need to be further confirmed. Interestingly, a significant association was found between the rs763780 variant and cancer risk in both the Asian and Caucasian populations. This meta-analysis is, to our knowledge, the first study showing that rs763780 polymorphism increases cancer risk in the Caucasian population.
A meta-analysis by Niu et al.42 suggested that IL-17 polymorphisms increase the risk of cancer, particularly gastric cancer, in Asian (especially Chinese) populations; our findings were partially in line with results from this meta-analysis. Another meta-analysis by Zhao et al.43 concluded that not rs763780, but rs2275913, polymorphism may contribute to cancer susceptibility in Asian populations. Long et al.44 found a positive association between the two polymorphisms and the occurrence of gastric cancer in a meta-analysis, which included 7 independent, case-control studies. Other meta-analyses focused on the association between IL-17F rs763780 polymorphism and cancer risk, and the results indicated that the CC allele might increase the risk of cancer, particularly gastric cancer, in Asian populations45,46. All of these previous meta-analyses included fewer than 10 eligible case-control studies, with few studies examining Caucasian populations. The present meta-analysis includes 25 independent case-control studies with 7,872 cancer cases and 9,646 cancer-free controls. In addition, 6 of the included studies were based on Caucasians, to more comprehensively evaluate the relationship between IL-17 polymorphisms and cancer risk in Caucasian populations14,16,24,27,31,47.
We conducted sensitivity analysis to confirm the validity of the results presented in our meta-analysis, and studies in which the genotype frequencies in the control group deviated from the HWE were excluded. Results showed that no individual study skewed the overall OR value.
Some limitations of the present meta-analysis should be addressed. First, although significant associations were found between the two polymorphisms and the risk of cancer in multiple genetic models, some potential sources of heterogeneity, such as source of controls, lifestyle, and environmental exposures, were not explored. In addition, some cancer types included in this meta-analysis were investigated only in 1 or 2 studies (Supplementary Table S3), which led to heterogeneity in quantitative analysis. Second, the study results included in this meta-analysis were based on unadjusted analyses, and therefore, we could not estimate the risk of cancer with respect to environmental factors, age, family history, lifestyle, and other risk factors that might have influenced the pooled results. Third, we did not include any studies on the African population, and therefore, the results should be interpreted with caution when extrapolating them to the overall population. Lastly, the study with relative smaller sample size is more likely to be lack of sufficient statistical power to influence the overall results.
Our study represents a comprehensive meta-analysis of the role of IL-17A rs2275913 and IL-17F rs763780 polymorphisms in cancer risk. The results demonstrated that these two polymorphisms significantly increase the risk of development of cancer, particularly gastric cancer. Further large-scale, multicentre studies are required to confirm the pre-diagnostic effect of IL-17 gene polymorphisms on the risk of cancer.
Material and Methods
Identification of eligible studies
Systematic article search and quantitative analysis were performed, and written reports were generated according to the Meta-analysis of Observational Studies in Epidemiology guidelines48. Eligible studies with publication dates up to March 2015 were obtained through the Pubmed and Chinese National Knowledge Infrastructure (CNKI) databases. No language or geographical restriction was placed for study selection. The keywords search was performed with or without the Medical Subject Headings (MeSH) terms for: ‘interleukin-17/IL-17’, ‘polymorphism’, and ‘cancer’. Additionally, the references in the retrieved articles were manually screened for potential eligible studies.
Inclusion and exclusion criteria
Studies included in our meta-analysis were required to meet the following criteria: (1) a case-control design; (2) the study goal was to evaluate the association of IL17A rs2275913 and IL-17F rs763780 polymorphisms with cancer risk; (3) the study offered available information on genotype frequency, (4) the controls used had no malignant disease. The following were used as our exclusion criteria: (1) the study was a repeat studies, reviews, or abstracts; (2) the study design was based on family cancers; (3) the study did not include a control group; (4) the study did not investigate the effect of polymorphism; (5) duplicate data.
Data extraction
Two authors independently selected the potentially relevant studies for data extraction. We screened the titles and abstracts of the studies that met our inclusion criteria. If the content of the abstract was relevant, full articles were read to extract related information. For each eligible study included in our meta-analysis, we obtained information pertaining to first author, years of publication, country of origin, racial ancestry, cancer types, source of control, genotyping method, total number of cases and controls, and P value of Hardy–Weinberg equilibrium (HWE). All cancers were confirmed by histology or pathology. All the case and control groups were well controlled.
Resampling
We applied re-sampling statistic to examine the robustness of the associations, and 10000 re-sampling analyses were conducted using the bootstrap re-sampling procedure49. All the re-sampling analyses were performed by R 3.2.2 software using non-parametric bootstrapping method. The 95% confidence intervals (95% CIs) were estimated by bias-corrected and accelerated (BCa) and overall odds ratios (ORs) were calculated containing all samples under the five different genetic models.
Statistical analysis
The strength of association between IL-17A rs2275913 and IL-17F rs763780 polymorphisms and cancer risk was assessed as ORs with corresponding 95% CIs based on the allele frequencies in cases and controls of each study selected. The summary OR was calculated according to Woolf’s method. Five different ORs were calculated: dominant model (BB+AB vs. AA), recessive model (BB vs. AA+AB), homozygote comparison (BB vs. AA), heterozygote comparison (AB vs. AA) and allele comparison (B vs. A), the A represents the major allele, and the B represents the minor allele. The Chi-square-based Q statistic was implemented to assess heterogeneity among the studies50. The controls that departures of the HWE were evaluated for each study using chi-square test.
The effect of heterogeneity using I2 test statistical and significance was considered at P < 0.10. In case of a significant heterogeneity, the pooled ORs were analyzed using a random-effects model, otherwise a fixed-effects model should be used. To evaluate the ethnicity-specific and control-specific effects, subgroup analyses were conducted by source of controls, cancer types, controls whether satisfied HWE or not, and features of the population such as ethnicity. Additionally, to estimate the possible sources of bias, we considered the Egger’s test and Begg’s funnel plot. All statistical analyses were calculated with the software STATA (Version 11.0; Stata Corp, College Station, TX). P-values less than 0.05 were considered statistically significant.
Additional Information
How to cite this article: Dai, Z.-M. et al. Role of IL-17A rs2275913 and IL-17F rs763780 polymorphisms in risk of cancer development: an updated meta-analysis. Sci. Rep. 6, 20439; doi: 10.1038/srep20439 (2016).
Supplementary Material
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 81471670); China Postdoctoral Science Foundation (No. 2014M560791); the Fundamental Research Funds for the Central Universities, China (No. 2014qngz-04) and the Science and Technology Foundation of Shaanxi Province, China (No. 2014K11020107).
Footnotes
Author Contributions Z.-M.D. and T.-S.Z. contributed equally to the work. Z.-J.D. designed the study. Z.-M.D., S.L. and W.-G.Z. wrote the main manuscript text, J.L., X.-M.C. and M.W. prepared figures and tables, H.-B.L., Z.-J.D., X.-H.L., K.L. and S.-L.L. reviewed the manuscript.
References
- Ferlay J. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136, E359–386, doi: 10.1002/ijc.29210 (2015). [DOI] [PubMed] [Google Scholar]
- Pharoah P. D., Dunning A. M., Ponder B. A. & Easton D. F. Association studies for finding cancer-susceptibility genetic variants. Nature Reviews Cancer 4, 850–860 (2004). [DOI] [PubMed] [Google Scholar]
- Smyth M. J., Cretney E., Kershaw M. H. & Hayakawa Y. Cytokines in cancer immunity and immunotherapy. Immunol Rev 202, 275–293, doi: 10.1111/j.0105-2896.2004.00199.x (2004). [DOI] [PubMed] [Google Scholar]
- Harrington L. E. et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol 6, 1123–1132, doi: 10.1038/ni1254 (2005). [DOI] [PubMed] [Google Scholar]
- Iwakura Y., Ishigame H., Saijo S. & Nakae S. Functional specialization of interleukin-17 family members. Immunity 34, 149–162, doi: 10.1016/j.immuni.2011.02.012 (2011). [DOI] [PubMed] [Google Scholar]
- Kawaguchi M., Adachi M., Oda N., Kokubu F. & Huang S. K. IL-17 cytokine family. J Allergy Clin Immunol 114, 1265-1273; quiz 1274, doi: 10.1016/j.jaci.2004.10.019 (2004). [DOI] [PubMed] [Google Scholar]
- Kirkham B. W. et al. Synovial membrane cytokine expression is predictive of joint damage progression in rheumatoid arthritis: a two-year prospective study (the DAMAGE study cohort). Arthritis Rheum 54, 1122–1131, doi: 10.1002/art.21749 (2006). [DOI] [PubMed] [Google Scholar]
- Fujino S. et al. Increased expression of interleukin 17 in inflammatory bowel disease. Gut 52, 65–70 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X. et al. Tumor microenvironments direct the recruitment and expansion of human Th17 cells. J Immunol 184, 1630–1641, doi: 10.4049/jimmunol.0902813 (2010). [DOI] [PubMed] [Google Scholar]
- Song X. & Qian Y. IL-17 family cytokines mediated signaling in the pathogenesis of inflammatory diseases. Cell Signal 25, 2335–2347, doi: 10.1016/j.cellsig.2013.07.021 (2013). [DOI] [PubMed] [Google Scholar]
- Wang L. et al. Association analysis of IL-17A and IL-17F polymorphisms in Chinese Han women with breast cancer. PLoS One 7, e34400, doi: 10.1371/journal.pone.0034400 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. et al. Association between polymorphisms in interleukin-17A and interleukin-17F genes and risks of gastric cancer. Int J Cancer 127, 86–92, doi: 10.1002/ijc.25027 (2010). [DOI] [PubMed] [Google Scholar]
- Shibata T., Tahara T., Hirata I. & Arisawa T. Genetic polymorphism of interleukin-17A and -17F genes in gastric carcinogenesis. Hum Immunol 70, 547–551, doi: 10.1016/j.humimm.2009.04.030 (2009). [DOI] [PubMed] [Google Scholar]
- Omrane I. et al. Significant association between interleukin-17A polymorphism and colorectal cancer. Tumour Biol 35, 6627–6632, doi: 10.1007/s13277-014-1890-4 (2014). [DOI] [PubMed] [Google Scholar]
- Zintzaras E. & Lau J. Synthesis of genetic association studies for pertinent gene-disease associations requires appropriate methodological and statistical approaches. J Clin Epidemiol 61, 634–645, doi: 10.1016/j.jclinepi.2007.12.011 (2008). [DOI] [PubMed] [Google Scholar]
- Nemati K., Golmoghaddam H., Hosseini S. V., Ghaderi A. & Doroudchi M. Interleukin-17FT7488 allele is associated with a decreased risk of colorectal cancer and tumor progression. Gene 561, 88–94, doi: 10.1016/j.gene.2015.02.014 (2015). [DOI] [PubMed] [Google Scholar]
- Gao Y. W., Xu M., Xu Y., Li D. & Zhou S. Effect of three common IL-17 single nucleotide polymorphisms on the risk of developing gastric cancer. Oncol Lett 9, 1398–1402, doi: 10.3892/ol.2014.2827 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Q. et al. Association between six genetic variants of IL-17A and IL-17F and cervical cancer risk: a case-control study. Tumour Biol, doi: 10.1007/s13277-015-3041-y (2015). [DOI] [PubMed] [Google Scholar]
- Lee Y. C. et al. Association between interleukin 17/interleukin 17 receptor gene polymorphisms and papillary thyroid cancer in Korean population. Cytokine 71, 283–288, doi: 10.1016/j.cyto.2014.11.011 (2015). [DOI] [PubMed] [Google Scholar]
- Xi X. E. et al. Interleukin-17A and interleukin-17F gene polymorphisms and hepatitis B virus-related hepatocellular carcinoma risk in a Chinese population. Med Oncol 32, 355, doi: 10.1007/s12032-014-0355-3 (2015). [DOI] [PubMed] [Google Scholar]
- Wang N. et al. IL-17 gene polymorphism is associated with susceptibility to gastric cancer. Tumour Biol 35, 10025–10030, doi: 10.1007/s13277-014-2255-8 (2014). [DOI] [PubMed] [Google Scholar]
- Yin J. et al. Interleukin 17A rs4711998 A>G polymorphism was associated with a decreased risk of esophageal cancer in a Chinese population. Dis Esophagus 27, 87–92, doi: 10.1111/dote.12045 (2014). [DOI] [PubMed] [Google Scholar]
- Li N. et al. IL17A gene polymorphisms, serum IL-17A and IgE levels, and hepatocellular carcinoma risk in patients with chronic hepatitis B virus infection. Mol Carcinog 53, 447–457, doi: 10.1002/mc.21992 (2014). [DOI] [PubMed] [Google Scholar]
- Kaabachi W. et al. Interleukin-17A and -17F genes polymorphisms in lung cancer. Cytokine 66, 23–29, doi: 10.1016/j.cyto.2013.12.012 (2014). [DOI] [PubMed] [Google Scholar]
- Qinghai Z., Yanying W., Yunfang C., Xukui Z. & Xiaoqiao Z. Effect of interleukin-17A and interleukin-17F gene polymorphisms on the risk of gastric cancer in a Chinese population. Gene 537, 328–332, doi: 10.1016/j.gene.2013.11.007 (2014). [DOI] [PubMed] [Google Scholar]
- Zhang X., Zheng L. & Sun Y. Analysis of the association of interleukin-17 gene polymorphisms with gastric cancer risk and interaction with Helicobacter pylori infection in a Chinese population. Tumour Biol 35, 1575–1580, doi: 10.1007/s13277-013-1217-x (2014). [DOI] [PubMed] [Google Scholar]
- Rafiei A. et al. Polymorphism in the interleukin-17A promoter contributes to gastric cancer. World J Gastroenterol 19, 5693–5699, doi: 10.3748/wjg.v19.i34.5693 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B. et al. Interleukin-17 gene polymorphisms are associated with bladder cancer in a Chinese Han population. Mol Carcinog 52, 871–878, doi: 10.1002/mc.21928 (2013). [DOI] [PubMed] [Google Scholar]
- Arisawa T. et al. Genetic polymorphisms of IL17A and pri-microRNA-938, targeting IL17A 3’-UTR, influence susceptibility to gastric cancer. Hum Immunol 73, 747–752, doi: 10.1016/j.humimm.2012.04.011 (2012). [DOI] [PubMed] [Google Scholar]
- Quan Y. et al. Association between IL17 polymorphisms and risk of cervical cancer in Chinese women. Clin Dev Immunol 2012, 258293, doi: 10.1155/2012/258293 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel T. et al. IL-17F gene polymorphism is associated with susceptibility to acute myeloid leukemia. J Cancer Res Clin Oncol 140, 1551–1555, doi: 10.1007/s00432-014-1674-7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T., Bettelli E., Oukka M. & Kuchroo V. K. IL-17 and Th17 Cells. Annu Rev Immunol 27, 485–517, doi: 10.1146/annurev.immunol.021908.132710 (2009). [DOI] [PubMed] [Google Scholar]
- Moseley T. A., Haudenschild D. R., Rose L. & Reddi A. H. Interleukin-17 family and IL-17 receptors. Cytokine Growth Factor Rev 14, 155–174 (2003). [DOI] [PubMed] [Google Scholar]
- Shime H. et al. Tumor-secreted lactic acid promotes IL-23/IL-17 proinflammatory pathway. J Immunol 180, 7175–7183 (2008). [DOI] [PubMed] [Google Scholar]
- Kato T. et al. Expression of IL-17 mRNA in ovarian cancer. Biochem Biophys Res Commun 282, 735–738, doi: 10.1006/bbrc.2001.4618 (2001). [DOI] [PubMed] [Google Scholar]
- Chen J. G. et al. Intratumoral expression of IL-17 and its prognostic role in gastric adenocarcinoma patients. Int J Biol Sci 7, 53–60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus D. S. TNFalpha and IL-17 cooperatively stimulate glucose metabolism and growth factor production in human colorectal cancer cells. Mol Cancer 12, 78, doi: 10.1186/1476-4598-12-78 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao R. et al. High expression of IL-17 and IL-17RE associate with poor prognosis of hepatocellular carcinoma. J Exp Clin Cancer Res 32, 3, doi: 10.1186/1756-9966-32-3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan C. et al. High density of IL-17-producing cells is associated with improved prognosis for advanced epithelial ovarian cancer. Cell Tissue Res 352, 351–359, doi: 10.1007/s00441-013-1567-0 (2013). [DOI] [PubMed] [Google Scholar]
- Zhu X. et al. IL-17 expression by breast-cancer-associated macrophages: IL-17 promotes invasiveness of breast cancer cell lines. Breast Cancer Res 10, R95, doi: 10.1186/bcr2195 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. K., Lin X. & Gaffen S. L. Crucial role for nuclear factor of activated T cells in T cell receptor-mediated regulation of human interleukin-17. J Biol Chem 279, 52762–52771, doi: 10.1074/jbc.M405764200 (2004). [DOI] [PubMed] [Google Scholar]
- Niu Y. M., Yuan H. & Zhou Y. Interleukin-17 gene polymorphisms contribute to cancer risk. Mediators Inflamm 2014, 128490, doi: 10.1155/2014/128490 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H. Y., Wang R. & Ma W. IL-17A G197A and IL-17F T7488C polymorphisms and cancer risk in Asian populations: a meta-analysis. J BUON 19, 562–566 (2014). [PubMed] [Google Scholar]
- Long Z. W. et al. Association of IL-17 polymorphisms with gastric cancer risk in Asian populations. World J Gastroenterol 21, 5707–5718, doi: 10.3748/wjg.v21.i18.5707 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao F. et al. Role of IL-17F T7488C polymorphism in carcinogenesis: a meta-analysis. Tumour Biol 35, 9061–9068, doi: 10.1007/s13277-014-2171-y (2014). [DOI] [PubMed] [Google Scholar]
- Chen X. J., Zhou T. Y., Chen M. & Pu D. Meta analysis of association of the IL-17F rs763780T>C gene polymorphism with cancer risk. Asian Pac J Cancer Prev 15, 8083–8087 (2014). [DOI] [PubMed] [Google Scholar]
- Omrane I. et al. Significant association between IL23R and IL17F polymorphisms and clinical features of colorectal cancer. Immunol Lett 158, 189–194, doi: 10.1016/j.imlet.2014.01.002 (2014). [DOI] [PubMed] [Google Scholar]
- Stroup D. F. et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA 283, 2008–2012 (2000). [DOI] [PubMed] [Google Scholar]
- Li J. et al. Identification of high-quality cancer prognostic markers and metastasis network modules. Nat Commun 1, 34, doi: 10.1038/ncomms1033 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J. P. T. & Thompson S. G. Quantifying heterogeneity in a meta-analysis. Statistics in Medicine 21, 1539–1558, doi: Doi 10.1002/Sim.1186 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.