Abstract Abstract
The monotypic genus Cryptomaster Briggs, 1969 was described based on individuals from a single locality in southwestern Oregon. The described species Cryptomaster leviathan Briggs, 1969 was named for its large body size compared to most travunioid Laniatores. However, as the generic name suggests, Cryptomaster are notoriously difficult to find, and few subsequent collections have been recorded for this genus. Here, we increase sampling of Cryptomaster to 15 localities, extending their known range from the Coast Range northeast to the western Cascade Mountains of southern Oregon. Phylogenetic analyses of mitochondrial and nuclear DNA sequence data reveal deep phylogenetic breaks consistent with independently evolving lineages. We use discovery and validation species delimitation approaches to generate and test species hypotheses, including a coalescent species delimitation method to test multi-species hypotheses. For delimited species, we use light microscopy and SEM to discover diagnostic morphological characters. Although Cryptomaster has a small geographic distribution, this taxon is consistent with other short-range endemics in having deep phylogenetic breaks indicative of species level divergences. Herein we describe Cryptomaster behemoth sp. n., and provide morphological diagnostic characters for identifying Cryptomaster leviathan and Cryptomaster behemoth.
Keywords: Short-Range Endemic, DNA barcoding, cryptic species, Bayes Factor Delimitation, genealogical congruence
Introduction
With more than 4100 described species (Kury 2013), the Opiliones suborder Laniatores is incredibly diverse, and many more species likely await discovery. Despite their diversity, Laniatores is understudied, and many aspects of their phylogeny and evolution remain unknown (Giribet and Sharma 2015). An example is the monotypic genus Cryptomaster Briggs, 1969 and its large-bodied (~4 mm) but cryptic species, Cryptomaster leviathan Briggs, 1969. Cryptomaster leviathan was described from a single locality near the coastal town of Gold Beach, Oregon in the (PNW) of the United States. Briggs noted that this new species is remarkably large in size relative to most other Nearctic Laniatores. However, Cryptomaster leviathan did not receive further study and remained known only from the type locality until recent published records from the Cascade Range (Derkarabetian et al. 2010) and north of the type locality in the Coast Range (Shear et al. 2014). Given that many Laniatores taxa show high species diversity in small geographic regions (e.g., Ubick and Briggs 1989, Briggs and Ubick 1989, Ubick and Briggs 2002, Derkarabetian and Hedin 2014), these extensions of the known distribution across different mountain ranges indicate the potential for additional species within Cryptomaster.
In this study we increase the number of Cryptomaster localities to 15, all from mountainous southwestern Oregon. We use multi-locus DNA sequence data to investigate population structure and divergence from samples throughout this range. We recover a deep and concordant phylogenetic split for five loci that is indicative of species level divergence. Discovery and validation species delimitation approaches are used to assess support for multiple species within Cryptomaster. Also, we examine morphological differentiation between the divergent genetic groups to provide diagnostic characters for species identification. We delimit two species within Cryptomaster, and here describe Cryptomaster behemoth sp. n. This research highlights the importance of short-range endemic arachnids for understanding biodiversity (Harvey 2002, Harvey et al. 2011, Keith and Hedin 2012), and further reveals mountainous southern Oregon as a hotspot for endemic animal species (e.g. Shelley 1995, Cokendolpher et al. 2005, Mead et al. 2005, Leonard et al. 2011, Griswold et al. 2012).
Methods
Specimen collection
We collected 77 Cryptomaster individuals from 14 localities in the Coast and Cascade Mountains of southern Oregon (Fig. 1, Suppl. material 1: Table S1), including from near the type locality of Cryptomaster leviathan (Gold Beach, OR). Cryptomaster are primarily found in mature coniferous forests under woody debris. We attempted collections specifically targeting Cryptomaster further north and south of our samples, but these were unsuccessful. Individuals were preserved in 100% EtOH (Koptec) or 80% EtOH for genetic or morphological analysis, respectively. Speleomaster lexi Briggs, 1974 was used as an outgroup, following both morphological (Briggs 1974) and molecular evidence (Derkarabetian et al. 2010) that indicate a Speleomaster + Cryptomaster sister group relationship. Locality data for all specimens are available on the Symbiota Collections of Arthropods Network (http://symbiota4.acis.ufl.edu/scan/portal/index.php). Specimens are housed in the (SDSU_TAC); type specimens are deposited at the California Academy of Sciences (CASENT9039221).
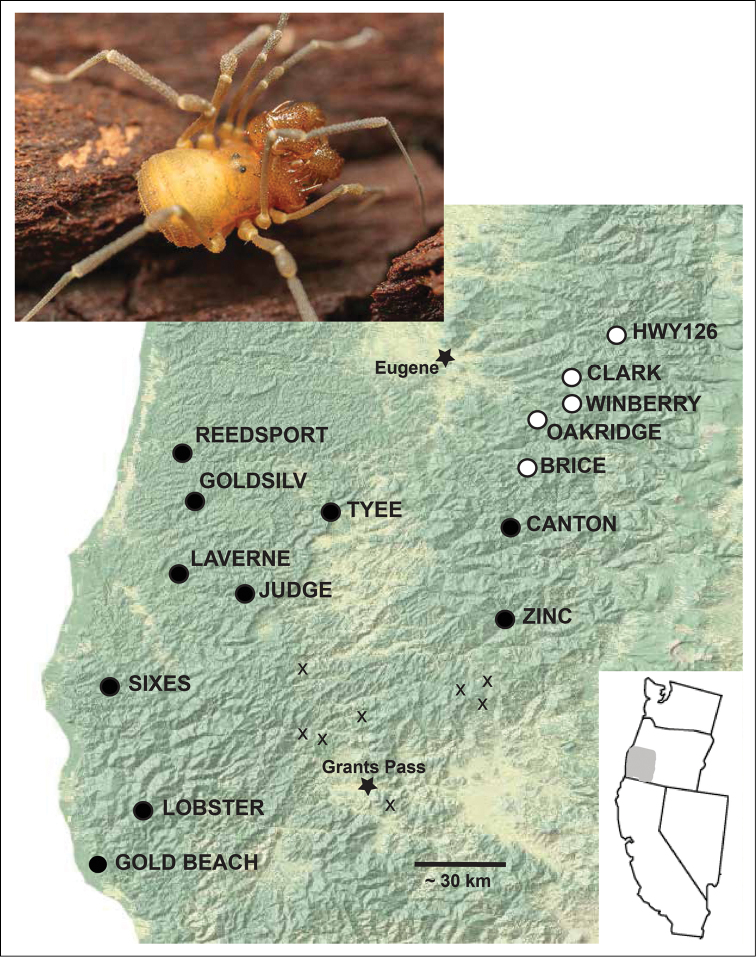
Figure 1.
Distribution of Cryptomaster leviathan (closed circles) and Cryptomaster behemoth (open circles) in southwestern Oregon. Small X’s represent locations in a potential Cascade to Coast corridor where we have collected other travunioids, but not Cryptomaster. Inset: male Cryptomaster leviathan from the Sixes River location.
Genetic sampling
Genomic DNA was extracted from multiple legs per specimen using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA) and the manufacturer’s protocol. Sequence data were obtained for the mitochondrial gene (COI), and four nuclear loci (Toll, putative; F-box/LRR-repeat protein, putative; Protein phosphatase 2A regulatory subunit A, putative; Neuromusculin, putative). DNA amplification was performed in a total volume of 25 µL with 1.6 units Platinum Taq (Invitrogen, Carlsbad, CA), 2.2 mM MgCl2, 1X PCR buffer, 0.2 mM each dNTP, and 0.4 µM of each primer (Suppl. material 1: Table S2). For COI, cycling conditions consisted of 94 °C for 2 minutes, then 30 cycles of 94 °C for 30 seconds, 50 °C (+ 0.2 °C/cycle) for 40 seconds, and 72 °C for 1.5 minutes, followed by 5 cycles of 94 °C for 30 seconds, 56 °C for 40 seconds, and 72 °C for 1.5 minutes, and then a final 72 °C for 5 minutes. For the nuclear loci, cycling conditions consisted of 94 °C for 3 minutes, then 10 cycles of 94 °C for 1 minute, 63 °C (-0.5 °C/cycle) for 1 minute, and 72 °C for 1 minute, followed by 30 cycles of 94 °C for 15 seconds, 58 °C for 1 minute, and 72 °C for 1 minute, and then a final 72 °C for 5 minutes. PCR products were purified using Montage filter plates and sequenced in both directions by Macrogen USA using the amplification primers. Sequences were edited in Geneious Pro 6 (Kearse et al. 2012) and aligned using MAFFT (Katoh 2013). For heterozygous nuclear sequences, Phase v.2.1.1 (Stephens et al. 2001, Stephens and Scheet 2005) was used to bioinformatically infer alleles.
Species discovery
Phylogenetic and genetic distance based discovery analyses were used to generate species hypotheses. (ML) analyses of individual loci were conducted with RAxML BlackBox v.8.1.11 (Stamatakis et al. 2008) with the GTR + Γ model on the CIPRES web server (Miller et al. 2010), with automatic bootstrap termination. For ML analysis of each locus and *BEAST analyses, the COI dataset was partitioned by codon position, while the less-variable nuclear loci datasets were not partitioned. Genetic diversity statistics were calculated using MEGA v6.06 (Tamura et al. 2013). (ABGD) was applied to the COI data using default settings and the transition/transversion ratio set at 2 (Puillandre et al. 2011). POFAD was run with all nuclear loci using standardized distances calculated with the genpofad option (Joly and Bruneau 2006). POFAD distance results were imported into SplitsTree4 (Huson and Bryant 2006) for reconstruction of a NeighborNet network.
Species validation
Based on the results of gene tree and species discovery approaches, we compared four alternative species hypotheses using Bayes Factor Delimitation (Grummer et al. 2014). *BEAST v1.8.1 (Drummond et al. 2012) was run with the four nuclear loci under four different species models (see Results, Table 1). Briefly, these species models consisted of: 1) a single species (Cryptomaster leviathan), 2) two species, following POFAD results, 3) three species, following the COI gene tree, and 4) four species, following ABGD results. Analyses were run with a strict molecular clock and sequence models determined by jModeltest2 (Guindon and Gascuel 2003, Darriba et al. 2012). Analyses were run for 100,000,000 generations with data stored every 10,000 generations. Log files for all *BEAST runs were visualized in Tracer v.1.6 (Rambaut et al. 2014). Analyses run with GTR sequence models failed to converge, and thus HKY models were applied with the other model parameters as determined by jModeltest2. The (MLE) was generated based on path sampling (Lartillot and Philippe 2006) and stepping stone (Xie et al. 2011) methods with a chain length of 100,000 generations and pathSteps set at 100. Analyses were run twice and the average MLE was taken for each species model. The Bayes factor was determined by 2*(-lnHypA - -lnHypB), with values greater than 10 indicating decisive support for a hypothesis (Kass and Raftery 1995).
Table 1.
Results of Bayes Factor Delimitation analysis. Species hypotheses are indicated in Fig. 2. (MLE) from (PS) and (SS) analyses are shown, with corresponding (BF) values.
| MLE (PS) | BF | MLE (SS) | BF | |
|---|---|---|---|---|
| 4 Species | -4939.87 | 12.14 | -4942.67 | 13.03 |
| 3 Species | -4940.62 | 13.63 | -4943.17 | 14.03 |
| 2 Species | -4933.80 | - | -4936.16 | - |
| 1 Species | -5010.68 | 153.75 | -5014.01 | 155.70 |
Morphological methods
Linear measurements were taken as in Derkarabetian and Hedin (2014) using an Olympus SZX12 dissecting microscope with an ocular micrometer. For SEM imaging, genitalia were extended from the body by opening the genital and anal opercula, and inserting a small blunt insect pin through the anal operculum and pushing the genitalia out. Specimens were dried using a critical point dryer, mounted on Ted Pella aluminum SEM stubs using copper conductive tape and coated with a 0.6 nm platinum coat. Multiple coats were applied in order to ensure proper coverage and to prevent charging. Coated specimens were imaged on a Quanta 450 SEM at the San Diego State University Electron Microscope Facility. Additionally, genitalia were examined using an Olympus BX40 compound microscope with a drawing tube. Genitalia were dissected from the body as above, then cleared in 10% KOH before viewing.
Results
Gene trees and species discovery
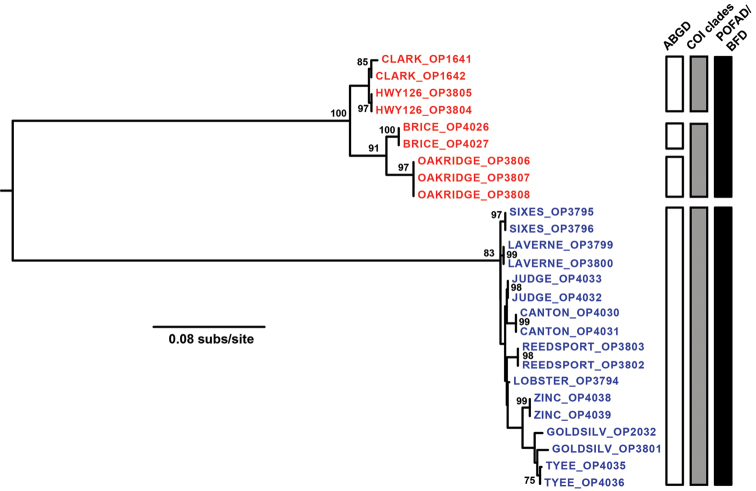
Genetic sampling results, GenBank accession numbers, and genetic diversity statistics are summarized in Suppl. material 1: Table S3. Sequence alignments of phased data have been submitted to DRYAD doi: 10.5061/dryad.76rb1. Maximum likelihood analysis of each locus revealed a deep and concordant phylogenetic split within Cryptomaster (Figs 2, 3, Suppl. material 2: Figs S1–5). All loci show strong support (bootstrap >83%) for a clade distributed in the Coast Range and southwestern Cascade Range. A second clade with a relatively restricted distribution occurs further northeast in the western Cascade Range (Figs 1–3). Within these clades there is little supported phylogenetic structure with the exception of COI, which further divides the northern Cascade group into a well-supported clade (BS = 91%) consisting of Oakridge+Brice, and a clade with low support (BS = 57%) consisting of Clark+HWY126. The mean p-distance between the two primary clades for COI was 15.5%, and ranged from 3.6-8.2% for the nuclear loci (Suppl. material 1: Table S3).
Figure 2.
Maximum likelihood COI gene tree with vertical bars showing results of discovery and validation analyses. Blue text = Cryptomaster leviathan, red text = Cryptomaster behemoth. Numbers adjacent to nodes indicate bootstrap support greater than 70%. The tree was rooted with Speleomaster lexi (not shown, see Suppl. material 2: Fig. S1).
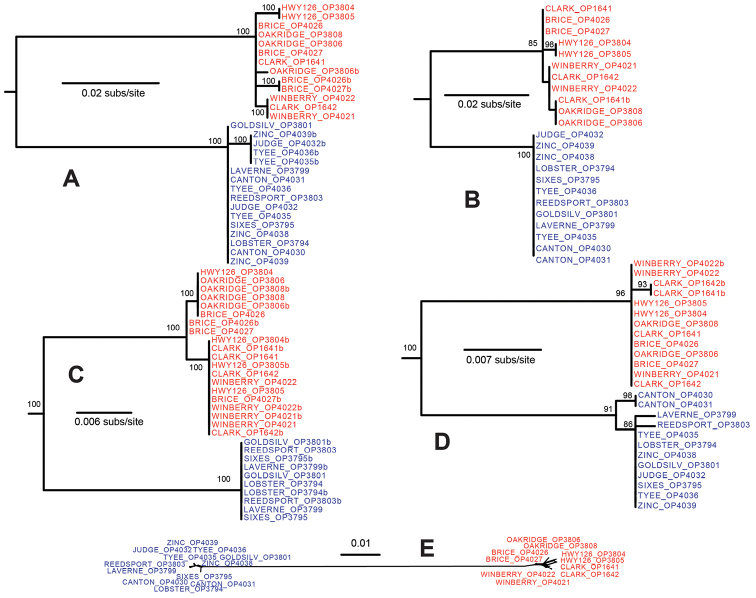
Figure 3.
A–D Maximum likelihood nuclear gene trees; numbers adjacent to nodes indicate bootstrap support greater than 70%. Trees were rooted with Speleomaster lexi (not shown, see Suppl. material 2: Fig. S2–5). Blue text = Cryptomaster leviathan, red text = Cryptomaster behemoth. A Toll B F-box/LRR-repeat protein C phosphatase 2A protein D Neuromusculin E NeighborNet network generated from summary distances (ingroup nuclear data only) calculated in POFAD.
ABGD analysis of the COI data supported a four species hypothesis (Fig. 2). One species consists of the well-supported COI clade that is distributed in the Coast Range and southwestern Cascades (= Cryptomaster leviathan). The remaining three ABGD species occur further north in the Cascade Range and consist of Clark+HWY126, Oakridge, and Brice. POFAD analysis of the nuclear data resulted in two clusters with minimal internal divergence, consistent with the deep phylogenetic split found in all nuclear loci (Fig. 3).
Species validation
Multiple species hypothesis models were tested using Bayes Factor Delimitation (Fig. 2, Table 1). These hypotheses consisted of 1) a single species of Cryptomaster, following Briggs 1969, 2) two species, following POFAD results, 3) three species following the major splits observed in the COI gene tree (Cryptomaster leviathan, Clark+HWY126, Oakridge+Brice), and 4) the four species indicated by the ABGD analysis. BF results based on MLE from both path sampling and stepping stone methods (Table 1) indicate decisive support for the two species hypothesis over alternative hypotheses, with a single-species hypothesis most strongly rejected.
Taxonomy
We note that within each species of Cryptomaster two forms are present, a larger and a smaller form, that show a bimodal size distribution (Fig. 4). The basis for these two forms is unknown – the different forms can be found in both sexes, in both species, and from the same localities. Additionally, the two forms are not genetically divergent as COI sequences from different individuals from the same locality are typically most closely related (Fig. 2), and little intraspecific variation exists for the nuclear genes (Fig. 3). Since the original diagnostic characters for the genus Cryptomaster (hind claws meeting in 180° opposition, distal swelling on tibia of second leg of males) apply to all examined specimens, we do not redescribe the genus. Here we redescribe Cryptomaster leviathan (Fig. 5A, C) from type specimens held at the (CAS) and newly collected material, increasing our understanding of Cryptomaster leviathan with new localities (Fig. 1, Suppl. material 1: Table S1) and morphological data (Suppl. material 1: Table S4) from across its range. Additionally, we describe the new species Cryptomaster behemoth sp. n. (Fig. 5B, D), providing diagnostic characters for both species (Fig. 6).
Figure 4.
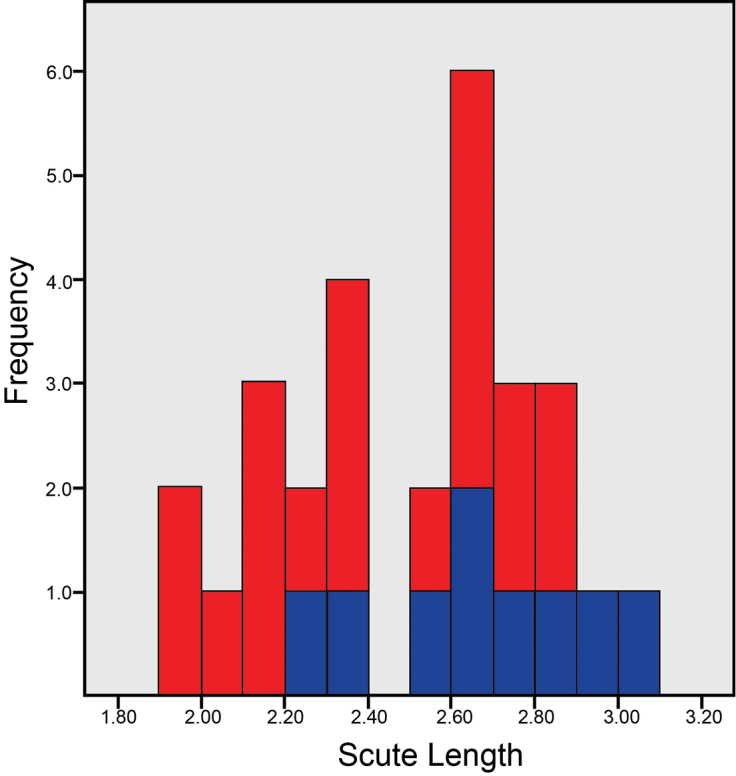
Scute length (mm) distribution of Cryptomaster. Figure made using SPSS 22 (IBM Corp. 2013). Blue = Cryptomaster leviathan, red = Cryptomaster behemoth.
Figure 5.
Cryptomaster dorsal coloration. A Male Cryptomaster leviathan (SDSU_OP4039) B Holotype male Cryptomaster behemoth (CASENT9039221, SDSU_OP4026) C Female Cryptomaster leviathan (SDSU_OP4037) D Allotype female Cryptomaster behemoth (CASENT9039221, SDSU_OP4029). Scale bars: 1 mm.
Figure 6.
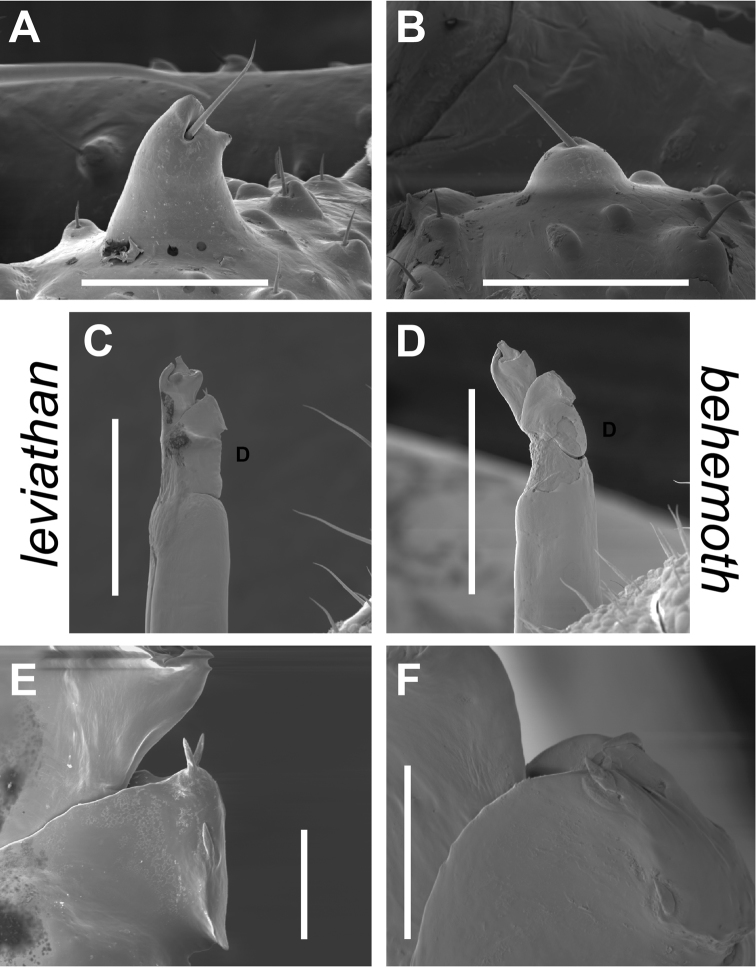
Cryptomaster diagnostic morphological characters. A–B Lateral view of seta-bearing tubercle found on ventral side of palpal trochanter A Cryptomaster leviathan, Judge Hamilton B Cryptomaster behemoth, Brice Creek C–D Lateral view of male penis (“D” indicates dorsal side) C Cryptomaster leviathan, Laverne County Park. D Cryptomaster behemoth, Oakridge E–F Close-up lateral view of penis tip showing spines on dorsal plate E Cryptomaster leviathan, Laverne County Park, detail showing erect apical spines F Cryptomaster behemoth, Oakridge, detail showing appressed apical spines. Scale bar: 200 µm (A, B); 300 µm (C–D); 40 µm (E–F).
Morphological abbreviations: DCS, GO, LII, OC, PCS, PF, PT, SBT. All measurements are in millimeters. Morphological images have been submitted to Morphbank.
Cryptomaster leviathan
Briggs, 1969
Figures: map 1 ; habitus 5A, C , 7A, C, E ; somatic 6A , 8A, C, E ; penis 6C, E , 9A, C, E ; ovipositor 10A–E
Figure 7.
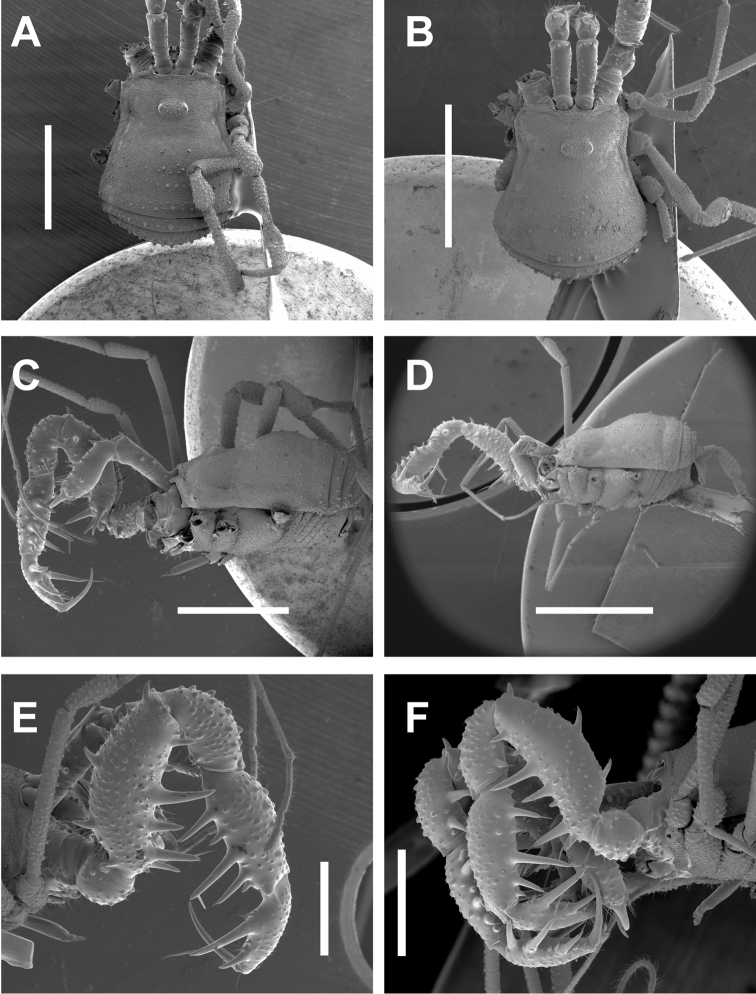
Cryptomaster habitus. A–B Habitus, dorsal A Cryptomaster leviathan, Judge Hamilton B Cryptomaster behemoth, Brice Creek C–D Habitus, lateral C Cryptomaster leviathan, Judge Hamilton D Cryptomaster behemoth, Oakridge (female) E–F Pedipalp, retrolateral E Cryptomaster leviathan, Judge Hamilton F Cryptomaster behemoth, Oakridge. Scale bar: 2 mm (A, B, C, D); 1 mm (E–F).
Figure 8.
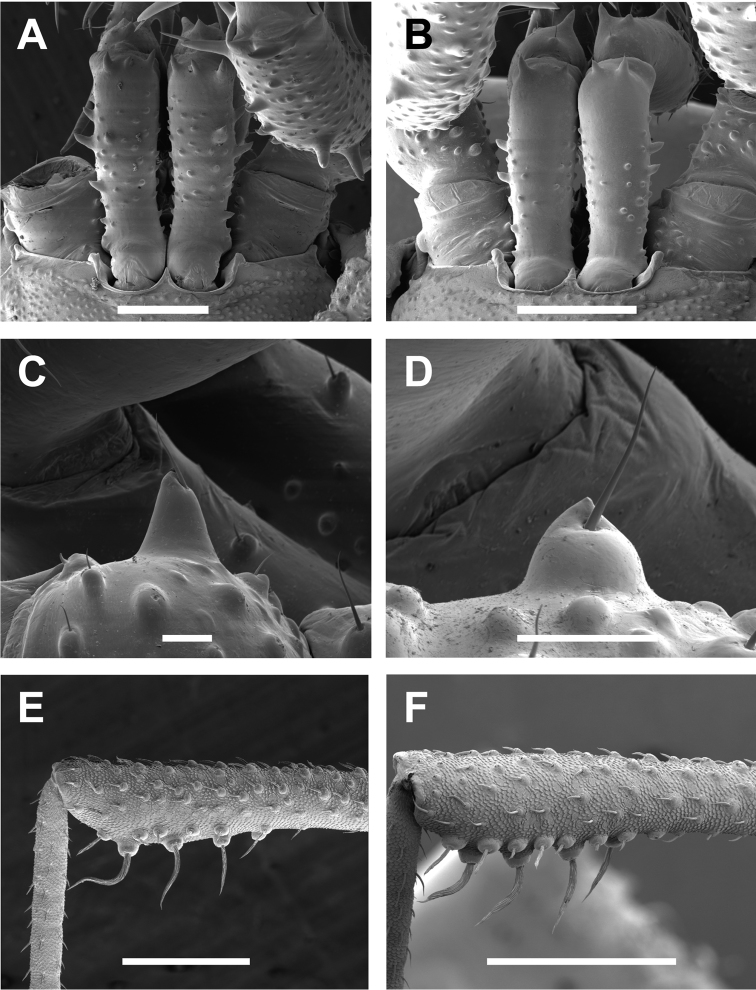
Cryptomaster appendages. A–B Proximal cheliceral segment, dorsal A Cryptomaster leviathan, Reedsport B Cryptomaster behemoth, Oakridge C–D SBT of palpal trochanter, lateral C Cryptomaster leviathan, Laverne County Park D Cryptomaster behemoth, Oakridge E–F Leg II tibia, distal swelling, retrolateral E Cryptomaster leviathan, Judge Hamilton F Cryptomaster behemoth, Oakridge. Scale bar: 500 µm (A, B); 100 µm (C, D); 400 µm (E, F).
Figure 9.
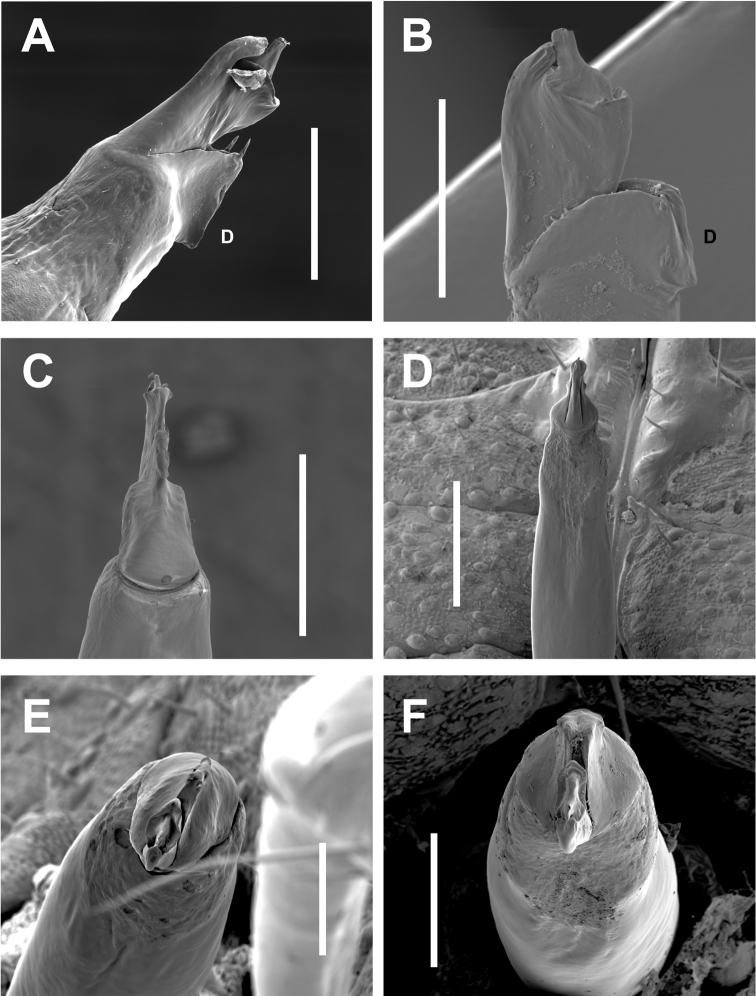
Cryptomaster penises. A–B Apicolateral (“D” indicates dorsal side) A Cryptomaster leviathan, Laverne County Park B Cryptomaster behemoth, Brice Creek C–D Apicodorsal C Cryptomaster leviathan, Judge Hamilton D Cryptomaster behemoth, Oakridge E–F Apical E Cryptomaster leviathan, Judge Hamilton F Cryptomaster behemoth, Brice Creek. Scale bar: 100 µm (A, B, E, F); 200 µm (C, D).
Figure 10.
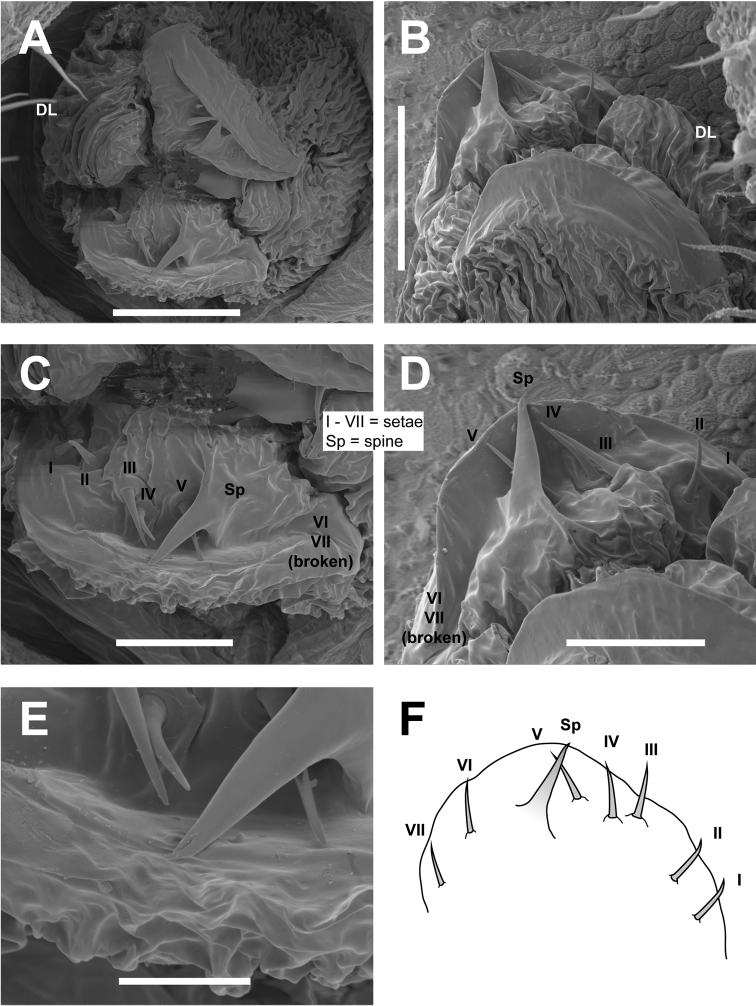
Cryptomaster ovipositors. A–E Cryptomaster leviathan, Reedsport A Apical (“DL” indicates dorsal lobe) B Lateral C Apical, emphasizing setae and spine arrangement D Lateral, emphasizing setae and spine arrangement E Apical, spine F Cryptomaster behemoth, Oakridge, sagittal view, emphasizing setae and spine arrangement, 40×. Scale bar: 100 µm (A, B); 50 µm (C, D); 25 µm (E).
Cryptomaster leviathan Briggs, 1969: 41–43, figures 15–25.
Type material examined.
Holotype male and five female paratypes from 4.5 miles south of Gold Beach, Curry County, Oregon, 29 January 1967, under spruce bark in virgin spruce forest, coll. T. Briggs, V. Lee, and K. Hom.
Diagnosis.
This species differs from Cryptomaster behemoth in having the enlarged SBT of PT acute (Fig. 6A), and keel-shaped protrusion of dorsal plate of penis with apical pair of spines fully erect and directed along the longitudinal axis of the penis (Fig. 6C, E).
Genetic data.
GenBank Accession numbers: KU059639-KU059655, KU059667-KU059678, KU059690-KU059701, KU059713-KU059717, KU059729-KU059740.
Morphbank images.
SDSU_TAC000021, Morphbank Specimen ID: 855927
<http://www.morphbank.net/?id=855927>, 14 SEM images
SDSU_TAC000022, Morphbank Specimen ID: 855928
<http://www.morphbank.net/?id=855928>, 7 SEM images
SDSU_TAC000027, Morphbank Specimen ID: 855931
<http://www.morphbank.net/?id=855931>, 17 SEM images
SDSU_TAC000204, Morphbank Specimen ID: 855933
<http://www.morphbank.net/?id=855933>, 19 SEM images
SDSU_TAC000248, Morphbank Specimen ID: 856245
<http://www.morphbank.net/?id=856245>, 4 SEM images
Redescription.
MALE: Measurements of holotype male, with the average and range of all three specimens measured in parentheses (Suppl. material 1: Table S4).
Body length 3.44, scute length 2.75 (2.71; 2.5–2.89), scute width 3.06 (2.75; 2.5–3.06), prosoma width 2.05 (1.95; 1.81–2.05). Shoulder tubercles present but small. Scute microgranulate. Holotype discolored due to preservation; for other specimens, integument color contrasts dorsally at midline between prosoma and opisthosoma, although not as strongly as in females. OC width 0.59 (0.55; 0.49–0.59). Ventral surface microgranulate. GO missing in holotype, length 0.3–0.34, width 0.28–0.31.
PT with acute mesal SBT. PF length 2.01 (1.98; 1.8–2.13), PF depth 0.72 (0.69; 0.64–0.72), with 7 (sometimes 5–6) spines, with the basal pair prominent, 3 (sometimes 4) enlarged dorsal spines, and 2 enlarged prolateral spines distally. PCS with 3 anterior spines dorsally and with 2 or 3 small retrolateral spines; DCS with 2 rows of small, dorsal, forward-facing acute SBTs. PCS width 0.42, DCS length 1.6, DCS width 0.46.
Trochanter 0.57, femur 0.92, patella 0.7, tibia 1.48, metatarsus 1.72, tarsus 1.04. LII length 11.19 (11.31; 10.69–12.06): trochanter 0.62 (0.6; 0.58–0.62), femur 2.73 (2.73; 2.55–2.91), patella 0.94 (0.89; 0.77–0.95), tibia 2.3 (2.37; 2.28–2.55), metatarsus 2.49 (2.61; 2.49–2.8); tarsus 2.1 (2.11; 1.99–2.25); tibia distally and ventrally swollen, with 5 rounded SBTs, 1–3 with setae twisted. Tarsal count 5-[11–15]-5–6.
Penis elongate; glans laterally compressed, dorsal plate extending outward into a more angled and acute keel shaped protrusion, with two pairs of spines, apical pair erect (pointing along the longitudinal axis of the penis), subapical pair appressed to dorsal plate; ventral plate cultriform with dorsally curved apical process.
FEMALE: Nineteen total individuals examined, including five paratypes; average measurements taken for subset (Suppl. material 1: Table S4), with range of all seven specimens measured in parentheses. Descriptive characters as for males unless otherwise noted.
Scute length 2.82 (2.69–3.03), scute width 3.01 (2.84–3.13), prosoma width 1.96 (1.8–2.14). Relative to males very dark, with strong contrast at midline between light-brown prosoma and dark-brown opisthosoma. OC width 0.56 (0.51–0.6). GO length 0.37 (0.33–0.39), width 0.38 (0.34–0.4).
PT mesal SBT acute. PF length 1.84 (1.68–1.94), PF depth 0.63 (0.58–0.67), usually with 5 (up to 7) ventral spines, 4 dorsal spines (2 to 6), and 2 distal prolateral spines. PCS with 2–3 anterior spines dorsally, with 1–4 small retrolateral spines.
LII length 10.23 (9.61–10.81): trochanter 0.58 (0.54–0.67), femur 2.53 (2.35–2.63), patella 0.83 (0.76–0.89), tibia 2.16 (1.99–2.29), metatarsus 2.39 (2.16–2.55), tarsus 1.92 (1.81–2.0); tibia without distal ventral swelling.
Ovipositor with four lobes, lateral lobes largest with seven apical setae, and a single large spine with a bifurcate tip, ventral lobe smallest.
Other material examined.
See Suppl. material 1: Table S1 and S4 for locality information of all specimens examined.
Distribution and habitat.
For specific localities, habitats, and microhabitats see Suppl. material 1: Table S1. This species is distributed in southwestern Oregon including throughout the southern Oregon Coast Range from the Umpqua River to the Coquille River, and south into the Klamath Mountains to the Rogue River. The range extends east to the southern Oregon Cascade Mountains in the South Umpqua and North Umpqua River Basins. Cryptomaster leviathan is typically associated with mature coniferous or mixed coniferous and hardwood forests, but has also been found in disturbed forests and forests with few conifers. Specimens are most often found under large woody debris associated with decaying logs and stumps, and in Acer and Polystichum leaf litter.
Cryptomaster behemoth
Starrett & Derkarabetian sp. n.
http://zoobank.org/6F8DCC84-A59D-4FEC-AB7C-0A05678D8223
Figures: map 1 ; habitus 5B, D , 7B, D, F ; somatic 6B , 8B, D, F ; penis 6D, F , 9B, D, F ; ovipositor 10F
Cryptomaster leviathan [partim] Derkarabetian et al. (2010)
Cryptomaster leviathan [partim] Shear et al. (2014)
Etymology.
The specific epithet is a noun in apposition, which refers to the large size of this species. Like leviathan, the specific epithet behemoth is derived from Hebrew; these are the names of two large and powerful beasts mentioned in the Book of Job.
Type material.
Holotype male and allotype female (deposited in CAS, CASENT9039221; SDSU_OP4026, SDSU_OP4029) from near Brice Creek, Brice Creek Road, 3.3 miles southeast of Forest Service Road 17, Umpqua National Forest, Lane County, Oregon; N43.6749°, W122.7290°; elevation 418 m; 29 March 2015; habitat: Acer macrophyllum, Thuja plicata, Pseudotsuga menziesii, Polystichum munitum forest; in and under large woody debris and other forest litter; collectors: J. Starrett, S. Derkarabetian, A. Cabrero, C. Richart. Paratypes: One female (deposited in CAS) from identical locality and information as holotype and allotype. Three females (two deposited in CAS, 1 deposited in SDSU_TAC; SDSU_TAC0000023) from Goodman Creek Road, off OR 58, northwest of Oakridge, Lane County, Oregon; N43.8429°, W122.6854°; elevation 340 m; 19 August 2014; habitat: old growth Douglas fir forest/woody debris; collectors: M Hedin, E Ciaccio, A Cabrero, J Starrett, S Derkarabetian. Two females (one each deposited in CAS and SDSU_TAC; SDSU_TAC0000234) from Brice Creek, Brice Creek Road, 3.3 miles southeast of FS 17, Umpqua National Forest, Lane County, Oregon; N43.6760°, W122.7290°; elevation 418 m; 29 March 2015; habitat: Acer macrophyllum, Thuja plicata, Pseudotsuga menziesii, Polystichum munitum forest; woody debris and litter; collectors: J Starrett, S Derkarabetian, A Cabrero, C Richart. One female (deposited in SDSU_TAC; SDSU_TAC0000028) from Highway 126, near Quartz Creek Road, Lane County, Oregon; N44.1248°, W122.3846°; elevation 300 m; 19 August 2014; habitat: decent Pseudotsuga menziesii forest; woody debris; collectors: M Hedin, E Ciaccio, A Cabrero, J Starrett, S Derkarabetian.
Diagnosis.
This species differs from Cryptomaster leviathan by having the enlarged SBT of PT rounded (Fig. 6B), and keel-shaped protrusion of dorsal plate of penis with apical pair of spines appressed and perpendicular to the longitudinal axis of the penis (Fig. 6D, F).
Genetic data.
GenBank Accession numbers: HM056724, KU059631-KU59638, KU059657-KU059666, KU059680-KU059689, KU059703-KU059712, KU059719-KU59728.
Morphbank images.
CASENT9039221, Holotype, Morphbank Specimen ID: 855951
<http://www.morphbank.net/?id=855951>, 1 image
CASENT9039221, Paratype, Morphbank Specimen ID 855929,
<http://www.morphbank.net/?id=855929>, 1 image
SDSU_TAC000023.5, Morphbank Specimen ID: 855929
<http://www.morphbank.net/?id=855929>, 1 SEM image
SDSU_TAC000023.6, Morphbank Specimen ID: 855930
<http://www.morphbank.net/?id=855930>, 12 SEM images
SDSU_TAC000203, Morphbank Specimen ID: 855932
<http://www.morphbank.net/?id=855932>, 16 SEM images
SDSU_OP1641, GUID: 38c9a86e-088d-4040-8988-af37fa74ad84
<http://symbiota4.acis.ufl.edu/scan/portal/collections/individual/index.php?occid=14702249>, 1 image
SDSU_OP1641, Morphbank Specimen ID: 835725
<http://www.morphbank.net/?id=835725>, 2 images
SDSU_OP1642, GUID: 8558ef80-a8c7-439d-bd93-dba8ec8d11d4
<http://symbiota4.acis.ufl.edu/scan/portal/collections/individual/index.php?occid=14702250>, 1 image
Description.
MALE: Measurements of holotype male, with the average and range of all nine specimens measured in parentheses (Suppl. material 1: Table S4).
Body length 3.40, scute length 2.69 (2.46; 1.97–2.75), scute width 2.58 (2.41; 1.97–2.75), prosoma width 1.88 (1.77; 1.48–1.94). Shoulder tubercles present but small. Scute microgranulate. Integument color without contrast dorsally at midline between prosoma and opisthosoma in all individuals. OC a low broad mound; height 0.14; width 0.59 (0.52; 0.39–0.59). Eye color dark brown; surrounding integument with black pigment. Ventral surface microgranulate. GO length 0.32 (0.31; 0.27–0.32), width 0.28 (0.27; 0.26–0.29).
Mesal SBT of PT relatively low, rounded, with seta near apex of tubercle. PF length 2.0 (1.78; 1.38–2.03), PF depth 0.76 (0.65; 0.44–0.8), with 4–6 ventral spines, with the basal pair prominent, usually 3 enlarged dorsal spines (sometimes 4), and 2 enlarged distal prolateral spines. Pedipalp patella with 2 (one specimen with 3) enlarged prolateral spines and 1 ventroretrolateral spine; tibia with rows of 5 enlarged pro- and retrolateral spines; tarsus with 3 prolateral and 2 retrolateral enlarged spines. PCS with 2 dorsal anterior spines (sometimes 1–3); and with 2 small retrolateral spines (sometimes 1); DCS with 2 rows of small, dorsal, forward-facing acute SBTs. PCS width 0.37, DCS length 1.63, DCS width 0.47.
Leg II length 10.59 (9.88; 8.15–11.0); trochanter 0.58 (0.53; 0.41–0.59), femur 2.66 (2.45; 1.99–2.72), patella 0.84 (0.8; 0.64–0.89), tibia 2.26 (2.14; 1.73–2.4), metatarsus 2.33 (2.22; 1.82–2.48), tarsus 1.93 (1.81; 1.54–1.97); tibia distally and ventrally swollen, with 3–5 rounded SBTs, 2–4 with setae twisted. Tarsal claw as for genus. Tarsal count 5–13–5–6; variation exists in the number of LII tarsal segments.
Penis elongate; glans laterally compressed, dorsal plate extending outward into a more rounded keel shaped protrusion, with two pairs of spines, both pairs appressed to plate (perpendicular to the longitudinal axis of the penis); ventral plate cultriform with dorsally curved apical process.
FEMALE: Measurements of allotype female, with the average and range of all 10 specimens measured in parentheses (Suppl. material 1: Table S4). Descriptive characters as for males unless otherwise noted.
Body length 2.94, scute length 2.35 (2.29; 2.05–2.69), scute width 2.5 (2.52; 2.23–2.75), prosoma width 1.62 (1.61; 1.49–1.76). Integument color darker than males, usually with light contrast dorsally at midline between prosoma and opisthosoma (in 7 of 8 individuals). OC height 0.12; width 0.47 (0.47; 0.4–0.52).
Mesal SBT of PT relatively low, rounded, with seta near apex of tubercle. PF length 1.53 (1.48; 1.39–1.64), PF depth 0.54 (0.53; 0.47–0.61), with 6 or 7 ventral spines (sometimes 5), with the basal pair prominent, with 3 enlarged dorsal spines (sometimes 4), and 2 enlarged distal prolateral spines (sometimes 1). Pedipalp patella with 2 enlarged prolateral spines and 1 ventroretrolateral spine. PCS with 2 anterior spines dorsally, with 2 small retrolateral spines; PCS length 0.30; DCS length 1.19, width 0.28.
LII length 8.66 (8.51; 8.15–9.38); trochanter 0.5 (0.49; 0.44–0.59), femur 2.15 (2.06; 1.95–2.29), patella 0.68 (0.69; 0.66–0.76), tibia 1.78 (1.79; 1.68–1.96), metatarsus 1.95 (1.91; 1.82–2.1), tarsus 1.6 (1.57; 1.55–1.69); Tarsal count 5–11–5-6; variation exists in the number of LII tarsal segments.
GO length 0.3 (0.3; 0.27–0.34), width 0.29 (0.3; 0.27–0.36).
Ovipositor with four lobes, lateral lobes largest with seven apical setae, and a single large spine with a bifurcate tip, ventral lobe smallest.
Other material examined.
See Suppl. material 1: Tables S1 and S4 for locality information of all specimens examined.
Distribution and Habitat.
For specific localities, habitats, and microhabitats see Suppl. material 1: Table S1. This species is distributed in the central Cascade Mountains of Oregon east and southeast of Eugene from Brice Creek in the Row River Drainage north to the north side of the McKenzie, with all known localities in Lane County. It is possible that populations occur further north in the western Cascades (Fig. 1). Habitats and microhabitats do not obviously differ from Cryptomaster leviathan, found in mature coniferous or mixed coniferous and hardwood forests, most often associated with large woody debris and bark.
Discussion
Species delimitation and short range endemic taxa
Cryptomaster exhibits a deep molecular phylogenetic break consistent with species level divergence, similar to that observed in many other harvestmen taxa (Boyer et al. 2007a, Thomas and Hedin 2008, Hedin and Thomas 2010, Schönhofer and Martens 2010, Derkarabetian et al. 2011, Richart and Hedin 2013, Fernández and Giribet 2014). Based on analyses of genetic data using discovery and validation approaches, we conclude that Cryptomaster consists of two species. We found complete genealogical concordance in the mitochondrial and nuclear loci sampled, indicating strong evidence for long-term reproductive isolation between Cryptomaster leviathan and Cryptomaster behemoth (Avise and Ball 1990). While gene tree and ABGD analyses of COI data indicate additional potential species, mitochondrial datasets are known to over-split short range endemic arachnid taxa (e.g., Keith and Hedin 2012, Satler et al. 2013, Derkarabetian and Hedin 2014, Fernández and Giribet 2014, Hamilton et al. 2014, Leavitt et al. 2015, Hedin et al. 2015, Wachter et al. 2015). Thus, we favor the more conservative two-species hypothesis, which is supported by the multi-species coalescent validation approach using four independent nuclear loci.
Our sampling efforts greatly increased the known range of Cryptomaster, yet both species still appear to have limited distributions. Interestingly, the range for Cryptomaster leviathan extends across multiple mountain ranges, yet little to no genetic structure exists between these ranges. Conversely, Cryptomaster behemoth has a particularly small range, yet harbors higher population genetic structure. This could be due to the greater topographic complexity of the central Cascade Range, or these populations may have persisted in their current locations for a longer time compared to Cryptomaster leviathan populations. A pattern of deep population structure in topographically complex regions is consistent with numerous other arachnid taxa (Hendrixson and Bond 2005, Boyer et al. 2007a, Thomas and Hedin 2008, Bryson et al. 2013, Hedin et al. 2013, Esposito et al. 2015).
Biogeographic Uncertainty
Short-range endemic taxa have been shown to help elucidate ancient biogeographic processes (Boyer et al. 2007b, Bryson et al. 2013, Hedin et al. 2013). However, given the deep genetic break between Cryptomaster leviathan and Cryptomaster behemoth, contrasting with minimal population structure within these taxa, it is difficult to decipher the processes that led to the division of these species. Cryptomaster leviathan is comparatively much more widespread with populations in the Klamath Mountains, southern Oregon Coast Ranges, and the Cascade Mountains, although these ranges appear to be connected by corridors of possibly suitable habitat (Fig. 1). Speciation may have occurred while Cryptomaster leviathan and Cryptomaster behemoth inhabited separate mountain ranges and Cryptomaster leviathan later dispersed from the coast northeast to the Cascade Range. Alternatively, the two species may have diverged within the Cascade Range. Under this scenario, Cryptomaster leviathan could have already been present in the Coast Range or dispersed west from the Cascades subsequent to speciation. These species currently occupy different river drainage systems, separated by the relatively high-elevation Calapooya Divide, but it remains unclear whether this represents a primary or secondary barrier to dispersal. Sampling of faster evolving loci and additional fine-scale geographic sampling is needed to test these alternative hypotheses.
Supplementary Material
Acknowledgements
We thank Adrienne Richart and Erik Ciaccio for assistance with specimen collection, and Erik Ciaccio helped with the SCAN database. Darrell Ubick, Charles Griswold, and Vic Smith provided assistance with morphological analyses and imaging. Stephanie Castillo assisted with molecular data collection. Dean Leavitt and Dave Carlson helped with molecular analyses. Steve Barlow provided SEM support at SDSU. Sara Griffith from the OR Parks & Recreation Department assisted with collecting permits. Comments from Darrell Ubick, Gonzalo Giribet, and one anonymous reviewer helped to improve the manuscript. This research was supported by an NSF grant awarded to M Hedin (DEB 1354558).
Citation
Starrett J, Derkarabetian S, Richart CH, Cabrero A, Hedin M (2016) A new monster from southwest Oregon forests: Cryptomaster behemoth sp. n. (Opiliones, Laniatores, Travunioidea). ZooKeys 555: 11–35. doi: 10.3897/zookeys.555.6274
Supplementary materials
Supplementary Table 1–4
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
James Starrett, Shahan Derkarabetian, Casey H. Richart, Allan Cabrero, Marshal Hedin
Data type: Excel Table
Explanation note:
Table S1. Locality data
Table S2. PCR primer information
Table S3. GenBank accession information and genetic diversity statistics
Table S4. Morphological measurements
Supplementary Figures 1–5
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
James Starrett, Shahan Derkarabetian, Casey H. Richart, Allan Cabrero, Marshal Hedin
Data type: figures
Explanation note:
Figure S1. Maximum likelihood gene tree for (COI) locus. Numbers adjacent to nodes indicate bootstrap support greater than 70%.
Figure S2. Maximum likelihood gene tree for Toll locus. Numbers adjacent to nodes indicate bootstrap support greater than 70%.
Figure S3. Maximum likelihood gene tree for F-box/LRR-repeat locus. Numbers adjacent to nodes indicate bootstrap support greater than 70%.
Figure S4. Maximum likelihood gene tree for phosphatase 2A locus. Numbers adjacent to nodes indicate bootstrap support greater than 70%.
Figure S5. Maximum likelihood gene tree for Neuromusculin locus. Numbers adjacent to nodes indicate bootstrap support greater than 70%.
References
- Avise JC, Ball RM. (1990) Principles of genealogical concordance in species concepts and biological taxonomy. Oxford Surveys in Evolutionary Biology 7: 45–67. [Google Scholar]
- Boyer SL, Baker JM, Giribet G. (2007a) Deep genetic divergences in Aoraki denticulata (Arachnida, Opiliones, Cyphophthalmi): a widespread ‘mite harvestman’ defies DNA taxonomy. Molecular Ecology 16: 4999–5016. doi: 10.1111/j.1365-294X.2007.03555.x [DOI] [PubMed] [Google Scholar]
- Boyer SL, Clouse RM, Benavides LR, Sharma P, Schwendinger PJ, Karunarathna I, Giribet G. (2007b) Biogeography of the world: a case study from cyphophthalmid Opiliones, a globally distributed group of arachnids. Journal of Biogeography 34: 2070–2085. doi: 10.1111/j.1365-2699.2007.01755.x [Google Scholar]
- Briggs TS. (1969) A new holarctic family of laniatorid phalangids. Pan-Pacific Entomologist 45(1): 35–50. [Google Scholar]
- Briggs TS. (1974) Troglobitic harvestmen recently discovered in North American lava tubes (Travuniidae, Erebomastridae, Triaenonychidae: Opiliones). Journal of Arachnology 1: 205–214. [Google Scholar]
- Briggs TS, Ubick D. (1989) The harvestman family Phalangodidae. 2. The new genus Microcina (Opiliones, Laniatores). Journal of Arachnology 17: 207–220. [Google Scholar]
- Bryson Jr RW, Riddle BR, Graham MR, Smith BT, Prendini L. (2013) As old as the hills: Montane scorpions in Southwestern North America reveal ancient associations between biotic diversification and landscape history. PLoS ONE 8(1): . doi: 10.1371/journal.pone.0052822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokendolpher JC, Peck RW, Niwa CG. (2005) Mygalomorph spiders from southwestern Oregon, USA, with descriptions of four new species. Zootaxa 1058: 1–34. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9(8): . doi: 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkarabetian S, Steinmann DB, Hedin M. (2010) Repeated and time-correlated morphological convergence in cave-dwelling harvestmen (Opiliones, Laniatores) from montane western North America. PLoS ONE 5(5): . doi: 10.1371/journal.pone.0010388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkarabetian S, Ledford J, Hedin M. (2011) Genetic diversification without obvious genitalic morphological divergence in harvestman (Opiliones, Laniatores, Sclerobunus robustus) from montane sky islands of western North America. Molecular Phylogenetics and Evolution 61: 844–853. [DOI] [PubMed] [Google Scholar]
- Derkarabetian S, Hedin M. (2014) Integrative taxonomy and species delimitation in harvestmen: a revision of the Western North American genus Sclerobunus (Opiliones: Laniatores: Travunioidea). PLoS ONE 9(8): . doi: 10.1371/journal.pone.0104982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution 29: 1969–1973. doi: 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito LA, Bloom T, Caicedo-Quiroga L, Alicea-Serrano AM, Sánchez-Ruíz JA, May-Collado LJ, Binford GJ, Agnarsson I. (2015) Islands within islands: Diversification of tailless whip spiders (Amblypygi, Phrynus) in Caribbean caves. Molecular Phylogenetics and Evolution 93: 107–117. doi: 10.1016/j.ympev.2015.07.005 [DOI] [PubMed] [Google Scholar]
- Fernández R, Giribet G. (2014) Phylogeography and species delimitation in the New Zealand endemic, genetically hypervariable harvestman species, Aoraki denticulata (Arachnida, Opiliones, Cyphophthalmi). Invertebrate Systematics 28: 401–414. [Google Scholar]
- Giribet G, Sharma PP. (2015) Evolutionary biology of harvestmen (Arachnida, Opiliones). Annual Review of Entomology 60: 157–175. doi: 10.1146/annurev-ento-010814-021028 [DOI] [PubMed] [Google Scholar]
- Griswold CE, Audisio T, Ledford JM. (2012) An extraordinary new family of spiders from caves in the Pacific Northwest (Araneae, Trogloraptoridae, new family). ZooKeys 215: 77–102. doi: 10.3897/zookeys.215.3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummer JA, Bryson Jr RW, Reeder TW. (2014) Species delimitation using Bayes factors: simulations and application to the Sceloporus scalaris species group (Squamata: Phrynosomatidae). Systematic Biology 63(2): 119–133. doi: 10.1093/sysbio/syt069 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. (2003) A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Systematic Biology 52: 696–704. doi: 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Hamilton CA, Hendrixson BE, Brewer MS, Bond JE. (2014) An evaluation of sampling effects on multiple DNA barcoding methods leads to an integrative approach for delimiting species: A case study of the North American tarantula genus Aphonopelma (Araneae, Mygalomorphae, Theraphosidae). Molecular Phylogenetics and Evolution 71: 79–93. doi: 10.1016/j.ympev.2013.11.007 [DOI] [PubMed] [Google Scholar]
- Harvey MS. (2002) Short-range endemism amongst the Australian fauna: some examples from non-marine environments. Invertebrate Systematics 16: 555–570. doi: 10.1071/IS02009 [Google Scholar]
- Harvey MS, Rix MG, Framenau VW, Hamilton ZR, Johnson MS, Teale RJ, Humphreys G, Humphreys WF. (2011) Protecting the innocent: studying short-range endemic taxa enhances conservation outcomes. Invertebrate Systematics 25: 1–10. doi: 10.1071/IS11011 [Google Scholar]
- Hedin M, Thomas SM. (2010) Molecular systematics of eastern North American Phalangodidae (Arachnida: Opiliones: Laniatores), demonstrating convergent morphological evolution in caves. Molecular Phylogenetics and Evolution 54: 107–121. doi: 10.1016/j.ympev.2009.08.020 [DOI] [PubMed] [Google Scholar]
- Hedin M, Starrett J, Hayashi C. (2013) Crossing the uncrossable: novel trans-valley biogeographic patterns revealed in the genetic history of low-dispersal mygalomorph spiders (Antrodiaetidae, Antrodiaetus) from California. Molecular Ecology 22: 508–526. doi: 10.1111/mec.12130 [DOI] [PubMed] [Google Scholar]
- Hedin M, Carlson D, Coyle F. (2015) Sky island diversification meets the multispecies coalescent – divergence in the spruce-fir moss spider (Microhexura montivaga, Araneae, Mygalomorphae) on the highest peaks in southern Appalachia. Molecular Ecology 24: 3467–3484. doi: 10.1111/mec.13248 [DOI] [PubMed] [Google Scholar]
- Hendrixson BE, Bond JE. (2005) Testing species boundaries in the Antrodiaetus unicolor complex (Araneae: Mygalomorphae: Antrodiaetidae): “Paraphyly” and cryptic diversity. Molecular Phylogenetics and Evolution 36: 405–416. doi: 10.1016/j.ympev.2005.01.021 [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. (2006) Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution 23(2): 254–267. doi: 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- Joly S, Bruneau A. (2006) Incorporating allelic variation for reconstructing the evolutionary history of organisms from multiple genes: an example from Rosa in North America. Systematic Biology 55(4): 623–636. doi: 10.1080/10635150600863109 [DOI] [PubMed] [Google Scholar]
- Kass RE, Raftery AE. (1995) Bayes factors. Journal of the American Statistical Association 90: 773–795. doi: 10.1080/01621459.1995.10476572 [Google Scholar]
- Katoh D, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. doi: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Mentjies P, Drummond A. (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12): 1647–1649. doi: 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith R, Hedin M. (2012) Extreme mitochondrial population subdivision in southern Appalachian paleoendemic spiders (Araneae: Hypochilidae: Hypochilus), with implications for species delimitation. Journal of Arachnology 40: 167–181. doi: 10.1636/A11-49.1 [Google Scholar]
- Kury AB. (2013) Order Opiliones Sundevall, 1833. In: Zhang Z-Q. (Ed.) Animal Biodiversity: An outline of higher–level classification and survey of taxonomic richness. Zootaxa 3703: 27–33. doi: 10.11646/zootaxa.3703.1.7 [DOI] [PubMed]
- Lartillot N, Philippe H. (2006) Computing Bayes factors using thermodynamic integration. Systematic Biology 55(2): 195–207. doi: 10.1080/10635150500433722 [DOI] [PubMed] [Google Scholar]
- Leavitt DH, Starrett J, Westphal MF, Hedin M. (2015) Multilocus sequence data reveal dozens of putative cryptic species in a radiation of endemic Californian mygalomorph spiders (Araneae, Mygalomorphae, Nemesiidae). Molecular Phylogenetics and Evolution 91: 56–67. doi: 10.1016/j.ympev.2015.05.016 [DOI] [PubMed] [Google Scholar]
- Leonard WP, Chichester L, Richart CH, Young TA. (2011) Securicauda hermani and Carinacauda stormi, two new genera and species of slug from the Pacific Northwest of the United States (Gastropoda: Stylommatophora: Arionidae), with notes on Gliabates oregonius Webb 1959. Zootaxa 2746: 43–56. [Google Scholar]
- Mead LS, Clayton DR, Nauman RS, Olson DH, Pfrender ME. (2005) Newly discovered populations of salamanders from Siskiyou County California represent a species distinct from Plethodon stormi. Herpetologica 61(2): 158–177. doi: 10.1655/03-86 [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), 14 N. 2010, New Orleans, LA, 1–8. doi: 10.1109/GCE.2010.5676129 [Google Scholar]
- Puillandre N, Lambert A, Brouillet S, Achaz G. (2012) ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Molecular Ecology 21: 1864–1877. doi: 10.1111/j.1365-294X.2011.05239.x [DOI] [PubMed] [Google Scholar]
- Rambaut A, Suchard MA, Xie D, Drummond AJ. (2014) Tracer v1.6. http://beast.bio.ed.ac.uk/Tracer
- Richart CH, Hedin M. (2013) Three new species in the harvestmen genus Acuclavella (Opiliones, Dyspnoi, Ischyropsalidoidea), including description of male Acuclavella quattuor Shear, 1986. ZooKeys 311: 19–68. doi: 10.3897/zookeys.311.2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satler J, Carstens BC, Hedin M. (2013) Multilocus species delimitation in a complex of morphologically conserved trapdoor spiders (Mygalomorphae, Antrodiaetidae, Aliatypus). Systematic Biology 62(6): 805–823. doi: 10.1093/sysbio/syt041 [DOI] [PubMed] [Google Scholar]
- Schönhofer AL, Martens J. (2010) Hidden Mediterranean diversity: Assessing species taxa by molecular phylogeny within the opilionid family Trogulidae (Arachnida, Opiliones). Molecular Phylogenetics and Evolution 54: 59–75. doi: 10.1016/j.ympev.2009.10.013 [DOI] [PubMed] [Google Scholar]
- Shear WA, Jones TH, Guidry HM, Derkarabetian S, Richart CH, Lewis JJ. (2014) Chemical defenses in the opilionid infraorder Insidiatores: divergence in chemical defenses between Triaenonychidae and Travunioidea and within travunioid harvestmen (Opiliones) from eastern and western North America. Journal of Arachnology 42: 248–256. doi: 10.1636/B14-41.1 [Google Scholar]
- Shelley RM. (1995) The milliped family Hirudisomatidae in the new world (Polyzoniida). Brimleyana 23: 103–143. [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. (2008) A rapid bootstrap algorithm for the RAxML web-servers. Systematic Biology 75: 758–771. doi: 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- Stephens M, Scheet P. (2005) Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. The American Journal of Human Genetics 76: 449–462. doi: 10.1086/428594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith N, Donnelly P. (2001) A new statistical method for haplotype reconstruction from population data. The American Journal of Human Genetics 68: 978–989. doi: 10.1086/319501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution 30: 2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SM, Hedin M. (2008) Multigenic phylogeographic divergence in the paleoendemic southern Appalachian opilionid Fumontana deprehendor Shear (Opiliones, Laniatores, Triaenonychidae). Molecular Phylogenetics and Evolution 46: 645–658. doi: 10.1016/j.ympev.2007.10.013 [DOI] [PubMed] [Google Scholar]
- Ubick D, Briggs TS. (1989) The harvestmen family Phalangodidae. 1. The new genus Calicina, with notes on Sitalcina (Opiliones: Laniatores). Proceedings of the California Academy of Sciences 46(4): 95–136. [Google Scholar]
- Ubick D, Briggs TS. (2002) The harvestman family Phalangodidae. 4. A review of the genus Banksula (Opiliones, Laniatores). Journal of Arachnology 30: 435–451. doi: 10.1636/0161-8202(2002)030[0435:THFPAR]2.0.CO;2 [Google Scholar]
- Wachter GA, Muster C, Arthofer W, Günther R, Föttinger P, Komposh C, Steiner FM, Schlick-Steiner BC. (2015) Taking the discovery approach in integrative taxonomy: decrypting a complex of narrow-endemic Alpine harvestmen (Opiliones: Phalangiidae: Megabunus). Molecular Ecology 24: 863–889. doi: 10.1111/mec.13077 [DOI] [PubMed] [Google Scholar]
- Xie WG, Lewis PO, Fan Y, Kuo L, Chen MH. (2011) Improving marginal likelihood estimation for Bayesian phylogenetic model selection. Systematic Biology 60: 150–160. doi: 10.1093/sysbio/syq085 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1–4
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
James Starrett, Shahan Derkarabetian, Casey H. Richart, Allan Cabrero, Marshal Hedin
Data type: Excel Table
Explanation note:
Table S1. Locality data
Table S2. PCR primer information
Table S3. GenBank accession information and genetic diversity statistics
Table S4. Morphological measurements
Supplementary Figures 1–5
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
James Starrett, Shahan Derkarabetian, Casey H. Richart, Allan Cabrero, Marshal Hedin
Data type: figures
Explanation note:
Figure S1. Maximum likelihood gene tree for (COI) locus. Numbers adjacent to nodes indicate bootstrap support greater than 70%.
Figure S2. Maximum likelihood gene tree for Toll locus. Numbers adjacent to nodes indicate bootstrap support greater than 70%.
Figure S3. Maximum likelihood gene tree for F-box/LRR-repeat locus. Numbers adjacent to nodes indicate bootstrap support greater than 70%.
Figure S4. Maximum likelihood gene tree for phosphatase 2A locus. Numbers adjacent to nodes indicate bootstrap support greater than 70%.
Figure S5. Maximum likelihood gene tree for Neuromusculin locus. Numbers adjacent to nodes indicate bootstrap support greater than 70%.