Abstract
The thale cress Arabidopsis thaliana is a powerful model organism for studying a wide variety of biological processes. Recent advances in sequencing technology have resulted in a wealth of information describing numerous aspects of A. thaliana genome function. However, there is a relative paucity of computational systems for efficiently and effectively using these data to create testable hypotheses. We present CressInt, a user-friendly web resource for exploring gene regulatory mechanisms in A. thaliana on a genomic scale. The CressInt system incorporates a variety of genome-wide data types relevant to gene regulation, including transcription factor (TF) binding site models, ChIP-seq, DNase-seq, eQTLs, and GWAS. We demonstrate the utility of CressInt by showing how the system can be used to (1) Identify TFs binding to the promoter of a gene of interest; (2) identify genetic variants that are likely to impact TF binding based on a ChIP-seq dataset; and (3) identify specific TFs whose binding might be impacted by phenotype-associated variants. CressInt is freely available at http://cressint.cchmc.org.
Keywords: Arabidopsis, functional genomics, transcription factors, gene regulation, systems biology, web server, computational tools
Introduction
The sequencing of the Arabidopsis thaliana genome over 15 years ago [1] enabled a new era of scientific exploration of this versatile model organism. As “next generation” sequencing technologies continue to mature, datasets capable of measuring function on a genome-wide scale continue to become more prevalent. Despite an exponential increase in our ability to generate data probing function on a genome-scale, there remains a lag in our analytical capability to effectively analyze these data to attain new biological insights.
Several useful bioinformatics tools are currently in widespread use in the Arabidopsis community (see de Lucas et al. [2], Bassel et al. [3], and Brady et al. [4] for reviews). However, as more complex and higher resolution data types become available, there is an increasing need for the development of user-friendly computational tools for their analysis. In the past five years alone, Arabidopsis data have been released describing genetic variants associated with particular traits [5] or altered gene expression levels [6], open chromatin regions in multiple tissue types and conditions [7], and DNA binding specificities for hundreds of transcription factors (TFs) [8]. Collectively, these data offer new opportunities to probe gene regulation and genome function. However, access to the wide range of analytical capabilities afforded by these data remains largely limited to bioinformaticians.
We present CressInt (thale cress data intersector), a user-friendly, freely accessible web server for integrating and analyzing genome-scale A. thaliana gene datasets. Conceptually, CressInt is similar to visually analyzing data in a genome browser such as those provided by UC Santa Cruz [9] or Ensembl [10], with the key differences that (1) up to thousands of loci of interest can be queried at once; (2) quality-controlled data specific to A. thaliana are pre-loaded into the CressInt system; and (3) results are downloadable in formats easily amenable to further downstream analysis. CressInt combines data from a wide variety of sources, including TF genomic binding regions (from ChIP-seq), TF DNA binding specificities (from Protein Binding Microarrays (PBMs) [11]), chromatin accessibility (DNAse-seq), and genetic variants associated with specific phenotypes (from GWAS) or genotype-dependent gene expression levels (i.e., expression quantitative trait loci or eQTLs). The CressInt system enables a wide range of queries, from simple (e.g., identifying all datasets intersecting genomic regions of interest), to complex (e.g., identifying genetic variants of interest likely to affect the binding of specific TFs). To our knowledge, there is currently no web server capable of performing these operations on A. thaliana datasets that are already integrated into the system. This not only enables easy access to these data for non-computational experts, but also saves hours of time that would otherwise be spent identifying, obtaining, quality checking, and re-formatting the various data sets.
CressInt’s intuitive graphical user interface is designed to be easy to use for non-bioinformaticians, while maintaining sufficient power and capabilities to enable downstream computational analysis. Using three case studies, we demonstrate the ability of CressInt to effectively use functional genomics data to generate testable hypotheses involving genes or phenotypes of interest. The CressInt web server is freely available at https://cressint.cchmc.org (Login for review: reviewer; Password: wrlcressint).
Materials and methods
Data and code availability
All source code developed for the web server is available on Bitbucket (https://bitbucket.org/weirauchlab/tf-tools-cressint). All datasets are available from the authors upon request.
Data collection and quality control
We obtained data from a variety of sources (Table 1). All genome-based datasets are organized by plant tissue type (e.g., seedling, leaf, inflorescence, etc.), and stored as UC Santa Cruz BED6 files [9]. DNase-seq data indicating open chromatin regions in A. thaliana seedlings exposed to heatshock, darkness, and light were taken from Sullivan et al. [7]. 4,355,790 naturally occurring genetic variants and eQTLs derived from seedlings were obtained from Gan et al. [6]. The eQTL set was filtered to only include SNPs with P-values < 0.001. GWAS data were obtained from Atwell et al. [5], and genetic variants were included in our set of phenotype-associated variants if they either (1) have associations exceeding genome-wide significance (P < 2.75×10−7, which corresponds to the Bonferroni-corrected P<0.05 cutoff used in the original study; 178 SNPs in total) or (2) are among the top 10 most strongly associated variants for each phenotype, regardless of P-value (943 SNPs in total). TF binding specificity models were taken from build 1.01 of the CisBP database [8] (http://cisbp.ccbr.utoronto.ca/).
Table 1.
Datasets incorporated into the CressInt system
| Data type | Source | Description |
|---|---|---|
| TF DNA binding specificity models | Weirauch et al., 2014 [8] | CisBP database, which contains thousands of TF binding models across eukaryotes |
| DNase-seq | Sullivan et al., 2014 [7] | Genome-wide mapping of DNase hypersensitive sites in A. thaliana seedlings |
| Genetic variants and eQTLs | Gan et al., 2011 [6] | Multiple reference genomes and transcriptomes for 19 A. thaliana strains |
| GWAS | Atwell et al., 2010 [5] | Genome-wide association study of 107 phenotypes in A. thaliana inbred lines |
| ChIP-seq | Chica et al., 2013 [24] | H3K4me3 and H3K27me3 marks in leaves |
| ChIP-seq | Willing et al., 2015 [25] | H3K27me1 marks in leaves |
| ChIP-seq | Ómaoiléidigh et al., 2013 [26] | AG binding in inflorescences |
| ChIP-seq | Pajoro et al., 2014 [27] | AP1 and SEP3 binding in inflorescences |
| ChIP-seq | Wuest et al., 2012 [28] | AP3 and PI binding in flowers |
| ChIP-seq | Oh et al., 2014 [29] | ARF6 binding in seedlings |
| ChIP-seq | Heyman et al., 2013 [30] | ERF115 binding in dark growing cells |
| ChIP-seq | Fan et al., 2014 [31] | HBI1 binding in seedlings |
| ChIP-seq | Zhiponova et al., 2014 [32] | IBL1 binding in seedlings |
| ChIP-seq | Moyroud et al., 2011 [33] | LFY binding in seedlings |
| ChIP-seq | Pfeiffer et al., 2014 [34] | PIF1 binding in seedlings |
| ChIP-seq | Zhang et al., 2013 [35] | PIF3 binding in seedlings |
| ChIP-seq | Brandt et al., 2012 [36] | REV binding in seedlings |
| ChIP-seq | Huang et al., 2012 [37] | TOC1 binding in seedlings |
We obtained ChIP-seq data from the Gene Expression Omnibus (GEO) [12]. Beginning with all 26 A. thaliana ChIP-seq datasets available in GEO in March of 2015, we used a three-step quality control procedure to ensure that only high-quality datasets are included in the CressInt system. First, we removed any datasets whose peak regions cover > 5% of the A. thaliana genome, deeming them too non-specific (with the exception of ChIP-seq for histone marks, which mark general regulatory regions and tend to have wider peaks). Next, we removed any datasets where the number of peaks obtained from the GEO dataset did not match the number of peaks reported in the publication associated with the data – this step is necessary because both GEO datasets and methods sections of manuscripts are often insufficiently documented to reproduce the reported peak calls. Finally, we ran all peak sets through the TF DNA binding motif enrichment algorithm used by HOMER [13], and only included datasets where the ChIP’ed TF’s motif ranks in the top three of enriched motifs. A total of 16 ChIP-seq datasets, taken from 13 different studies, passed our QC process (Table 1).
Differential binding of transcription factors to genetic variants
We used PBM data describing the DNA binding specificities of 575 A. thaliana TFs taken from Weirauch et al. [8], and a similar procedure used in that study and another recent study [14] to identify TFs whose binding might be affected by the alleles of 4,355,790 naturally occurring A. thaliana genetic variants [6]. One type of data produced by a PBM experiment is the E-score, which ranges from −0.50 to +0.50, and quantifies the relative preference of the binding of the tested TF to each of the 32,896 possible 8 base sequences [11]. We constructed a matrix containing the PBM 8-mer E-scores for 534 PBM experiments (267 constructs, each assayed on two independent array designs). 466 of these experiments directly assay the DNA binding specificity an A. thaliana TF. 68 of them measure a related TF in another organism that has a similar DNA binding domain (DBD) to at least one A. thaliana TF (68 experiments). Each PBM experiment was mapped to its “closest” A. thaliana TF by either (1) assigning it to the A. thaliana TF that was directly measured (trivial); or (2) (for PBMs measuring non-A. thaliana TFs) assigning it to the A. thaliana TF with the most similar DBD (based on percent amino acid identity in DBD alignments - see Weirauch et al. [8] for details of how thresholds for these inferred binding specificities are established).
We then scored the alleles of each genetic variant using the resulting 8-mer E-score matrix. For a given variant, we first determined all 8-mers in the reference genome sequence overlapping each allele - for example, a SNP will overlap eight 8-mers, plus their reverse complements, for each allele. For each PBM experiment, we then identified the highest scoring 8-mer E-score attained by any of the reference allele sequences (Eref), and the highest attained by any non-reference allele (Enon-ref). We then identified all PBM experiments where only one of Eref and Enon-ref has an E-score value exceeding 0.45 (values above this threshold will likely be strongly bound by the given TF [15]). All experiments meeting this criterion were then assigned a final score Efinal, which is the maximum value of (Eref and Enon-ref). Finally, we also calculated the predicted difference in binding strength between the two alleles as Edelta = |Eref - Enon-ref|. We then created a final ranked list of TFs (sorted by Efinal) whose binding is likely to be affected by the alleles of a given SNP (e.g., strongly binding to one allele, but not binding to the other).
Web server implementation
The user interface to the CressInt analysis pipeline is served by a GNU/Linux virtual machine running CentOS 6 and the Apache 2.2 web server. The web front-end is implemented primarily as HTML “templates” rendered through the use of a PHP library (http://twig.sensiolabs.org/), which maintains a separation of concerns between interface and application logic. Client-side JavaScript manages interaction among input form elements in the web front-end, and the form submission is done asynchronously (via Ajax), allowing certain types of validation errors such as missing inputs or malformed BED files to be detected and reported without a page reload. Input data for analysis is received and processed by a Perl CGI (Common Gateway Interface) script, which in turn interfaces with an in-house high-performance computing (HPC) cluster (currently containing over 700 processing cores) through a set of locally-developed Perl modules, generating shell scripts for batch processing. These Perl modules abstract away the implementation details of the batch facility (IBM’s Load Sharing Facility [LSF]) and allow interfaces to be written for other local or remote HPC load-sharing systems without impacting the front-end web service. Intersection analyses are performed using the BedTools suite [16], along with custom-written code written in C++. A user may optionally provide an email address to receive notification of a completed CressInt analysis, or may simply leave the web browser open and wait for the job to complete.
Results
Overview of the CressInt system
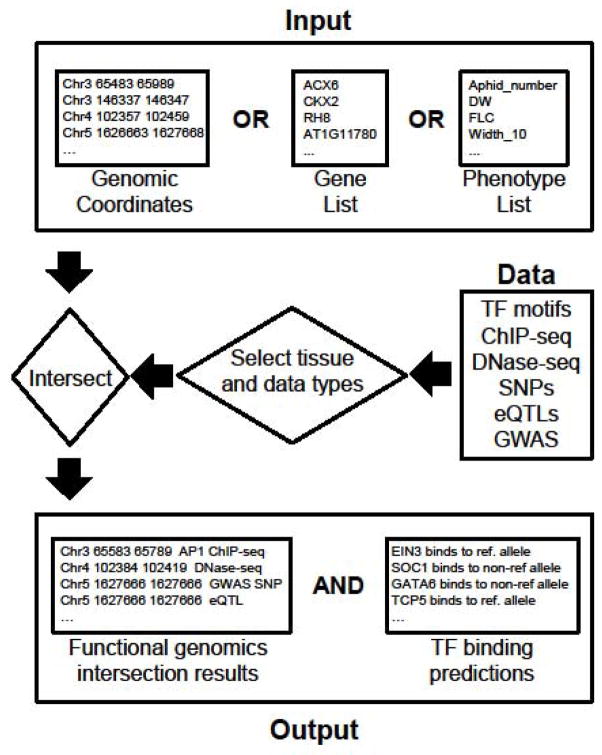
CressInt is designed to be easily useable for non-computational experts, while also maintaining sufficient power to be suitable for advanced downstream computational analyses. The system accepts one of three different types of inputs (Figure 1, top): (1) Genomic coordinates (in UC Santa Cruz BED3, BED4, BED5, or BED6 formats); (2) Gene lists (either common gene names or TAIR IDs; or (3) A set of phenotypes of interest, taken from a recent GWAS study [5]. The user can also choose to include or exclude functional genomics datasets based on data or tissue type (Figure 1, middle). After error and format checking, CressInt converts the input into a set of labeled genomic coordinates (in BED file format) and intersects these coordinates with the selected datasets. Two sets of results are presented to users (Figure 1, bottom): (1) The intersection results, which indicate all data sets in the system whose coordinates overlap with the input set; and (2) TF differential binding predictions, which identify genetic variants that might impact the binding of specific TFs.
Figure 1. Overview of the CressInt workflow.
As input, users can supply genomic coordinates, gene names, gene IDs, or phenotypes of interest (top). CressInt contains a wide range of genome-indexed data sources, which can be selected based on the data type or source tissue (middle – see also Table 1). Upon submitting a job, the user input is “intersected” with the selected data types, and results are displayed providing overlapping genomic coordinates (bottom left) and TF binding sites that overlap genetic variants (bottom right).
The CressInt system currently includes several different types of datasets relevant to gene regulation (Table 1). TF DNA binding specificity models taken from the CisBP database [8] are used to identify the specific TFs that might differentially bind a given genetic variant. The models are based on a large collection of universal PBM experiments covering 575 A. thaliana TFs [8]. Briefly, universal PBMs are double-stranded microarrays whose probes are designed such that all possible 10 base sequences occur exactly once, and hence all non-palindromic 32,896 8-base sequences occur 32 times in diverse flanking sequence contexts [11]. The resulting data, which track well with binding affinity [15], therefore offer a robust estimate of the binding of the assayed TF to every possible 8-base sequence. Although in vitro-derived TF binding specificities are in general reflective of in vivo specificities [17], we note that there can be exceptions (e.g., in cases where a TF’s binding is modified by a co-factor). Using this collection of TF binding models, CressInt has the capability to systematically scan the alleles of a given genetic variant to identify the particular TFs whose binding it might affect (see Methods).
In addition to TF binding models, CressInt incorporates ChIP-seq datasets taken from a variety of studies assaying either the binding of specific TFs (14 datasets), or histone marks that are indicative of chromatin state (three datasets) (Table 1). All ChIP-seq datasets were subjected to a rigorous three step quality control procedure before being considered for inclusion in the system (see Methods). CressInt also includes DNase-seq datasets taken from a recent large-scale study [7] and the Plant Regulome database (http://plantregulome.org/public/). DNase-seq is a next-generation sequencing assay that identifies DNase hypersensitive regions on a genome scale, and hence is capable of identifying regions of open chromatin in a certain tissue type [18]. Thus, DNase-seq data are useful for identifying areas of the genome that are likely to function as regulatory regions bound by TFs.
CressInt also includes the full set of 4,355,790 genetic variants identified in a recent study comparing the genomic sequences of 19 A. thaliana strains [6]. Among these variants, 317,570 are cis expression quantitative trait loci (eQTLs) taken from the same study (at a cutoff of P<0.001). Cis eQTLs are variants that affect the expression of a nearby gene as a function of genotype. Thus, eQTLs are useful for identifying functional variants that are likely to affect the binding of TFs. CressInt also includes a set of 1,004 genetic variants that are associated with one of 107 traits and phenotypes analyzed in a recent genome-wide association study (GWAS) [5] (see Methods). Such variants provide important clues for understanding genome function and biological diversity, due to their ability to modulate a phenotype in a genotype-dependent manner.
In the following sections, we demonstrate the power of CressInt by presenting case studies of how a user might use the system. First, we show how it can be used to identify TFs that bind to the promoter of a gene of interest. Next, we demonstrate how a user can input their own ChIP-seq data in order to identify genetic variants within the ChIP peaks that might impact the binding of the ChIP’ed TF. Finally, we show how CressInt can be used to find TF binding sites that might be impacted by genetic variants associated with a particular phenotype.
Case study 1: Identifying TFs binding to a promoter of interest
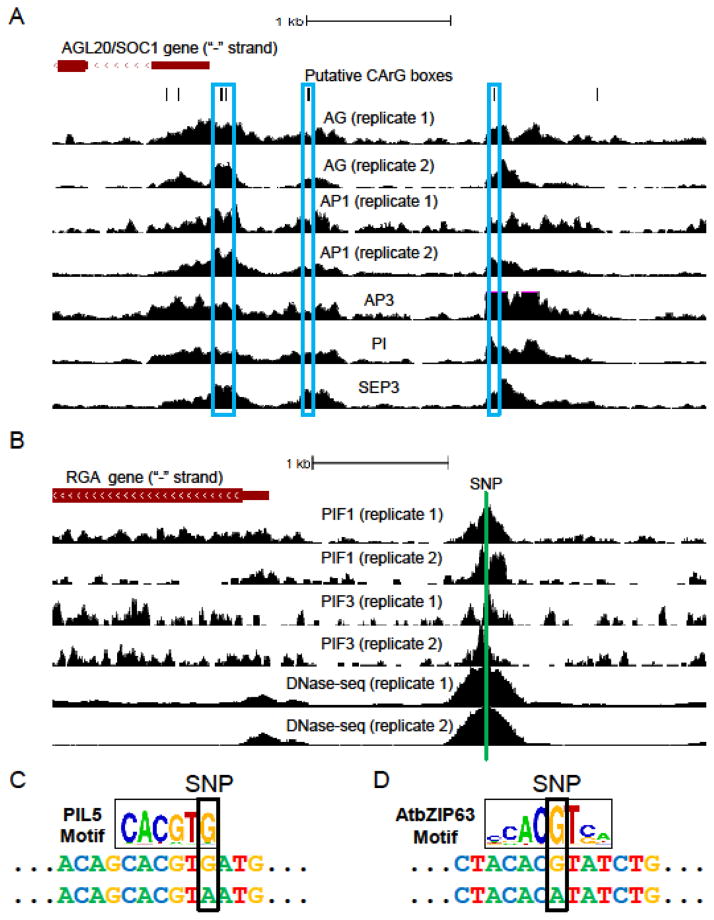
A fundamental, powerful feature of CressInt is its ability to query genomic regions of interest to generate hypotheses. For example, consider the case where a user is interested in identifying potential regulators of the AGL20/SOC1 gene, which encodes a MADS box family TF that controls the flowering process. To identify possible candidates, the user simply enters “AGL20” and defines the desired promoter search space (for this example, we use the region starting at the AGL20 TSS and extending 1000 bases upstream). Upon completion of the job, the user is provided with data indicating that five different MADS box family TFs all bind the AGL20 promoter, based on ChIP-seq experiments performed in flower and inflorescence tissues taken from three different studies (Figure 2A). Strikingly, all five TFs also play established roles in flower development. Further, MADS box TFs form homo- and hetero-dimers upon binding DNA [19], suggesting that these TFs might cooperatively regulate AGL20. Further supporting the CressInt-generated hypothesis, MADS box TFs recognize the CArG-box upon binding DNA [20], and there are five putative CArG boxes in the AGL20 promoter region (Figure 2A). A pair is located directly within the peak summits of all five TFs, with a third and fourth located just upstream, also near peak summits for all five TFs. In summary, through this simple query, we have identified specific binding sites for TFs likely to regulate a gene of interest.
Figure 2. Sample applications of CressInt.
A. Discovery of TFs binding a promoter of interest, as discussed in “case study 1”. UC Santa Cruz genome browser [9] screenshot depicting locations of (top to bottom): AGL20/SOC1 gene, putative CArG boxes (recognized by MADS box family TFs), and ChIP-seq data for seven experiments describing the genome-wide binding of five MADS-family TFs in flower and inflorescence tissues. Blue boxes, which contain CArG boxes and ChIP-seq peaks for multiple TFs, indicate likely binding sites. B. Identification of genetic variants likely to impact the binding of a ChIP’ed TF, as discussed in “case study 2”. See (A) for explanation. Green vertical line indicates location of the SNP discussed in “case study 2”. C. Data supporting the differential binding of the PIL5 TF to the “case study 2” SNP. Sequence logo [23] at top indicates the preferred base at each position of the PIL5 DNA recognition sequence – taller nucleotides indicate preference for the corresponding base at the corresponding position. DNA sequences below indicate the two alleles of the SNP, along with flanking genomic bases. Note that the reference allele (top) is a strong match to the PIL5 DNA binding motif, but the non-reference sequence (bottom) is not. D. Data supporting the differential binding of the AtbZIP63 TF to the “case study 3” SNP. See (C) for explanation.
Case study 2: Identifying genetic variants likely to affect the binding of a ChIP’ed TF
To demonstrate how CressInt can generate specific hypotheses from a user-provided genome-wide dataset, we submitted PIF1/PIL5 ChIP-seq peak regions in seedlings to CressInt as input, and asked the system to identify all likely PIF1 binding sites within these peaks that overlap naturally occurring genetic variants. In total, CressInt identified 53 variants that have a strong predicted binding site (E-score > 0.45) in the Col-0 (reference) strain, and a weak site (E-score < 0.30) in at least one other strain. For example, one variant, located in the promoter region of the RGA gene (Figure 2B), has a reference allele that is predicted to be very strongly bound (E-score = 0.499), with weak binding predicted for the alternative allele (E-score = 0.172). Figure 2C depicts the reference and non-reference allele sequences of this variant, along with flanking genomic bases. The reference allele perfectly matches the ideal PIF1 binding site (top sequence), while the non-reference allele “breaks” this site (bottom sequence). Both PIF1 and RGA are TFs involved in negative regulation of seed germination through participation in the gibberellic acid-mediated signaling pathway. Further, PIF1 directly increases the expression of RGA by binding to the same site identified by CressInt, with binding being abolished upon mutation of this site [21]. This example demonstrates that CressInt can be used to identify naturally occurring genetic variants that might impact the functional binding of a particular TF. Specifically, it shows how a genome-wide ChIP-seq dataset can be used to formulate the specific hypothesis that in the Mt-0 strain, which harbors the alternative allele of this variant, the direct regulation of RGA by PIF1 is likely attenuated, due to decreased PIF1 binding at this site. Intriguingly, this locus also overlaps ChIP-seq peaks for PIF3/POC1, a bHLH family TF that also recognizes G-box motifs (Figure 2B). Like PIF1, PIF3 is a member of the phytochrome interacting factor family of TFs, and the two proteins form heterodimers upon binding the G-box [22]. Thus, by intersecting the PIF1 ChIP-seq dataset with other datasets available in CressInt, we have arrived at the testable hypothesis that PIF1 and PIF3 might cooperatively bind the RGA promoter, and that this interaction might be impacted by a naturally occurring genetic variant.
Case study 3: Identifying TF binding sites likely to be affected by genetic variants associated with a phenotype of interest
As a final illustration, consider a user that is interested in the molecular mechanisms underlying the genetic determinants of flowering time. This user would start by selecting relevant phenotypes (e.g. “FT_field”, which counts the number of days between germination date and appearance of the first flower). CressInt then finds all genetic variants associated with the selected phenotypes, and identifies potential TFs whose binding they might affect. One such example is illustrated in Figure 2D, for a variant located 130 bases upstream of the ATCAF1B gene, which is expressed in flowers and plays a putative role in mRNA deadenylation. Although no clues for the function of this variant can be gleaned from the available functional genomics data (since it does not overlap any datasets), the expression of ATCAF1B is known to be affected by the genotype of the variant (i.e., it was identified as an eQTL in Gan et al. [6]), suggesting that its functional impact on flowering time is likely due to TF binding events specific to one of its alleles. Based on CressInt’s output, the top TF candidate is AtbZIP63, which is predicted to strongly bind the “G” allele (E-score = 0.463), and not bind the “A” allele (E-score = 0.138) (Figure 2D). Importantly, AtbZIP63 is also expressed in flowers. Further, a different genetic variant also associated with flowering time is located proximal to the AtbZIP63 gene locus. Collectively, these results implicate a potential role for AtbZIP63 in flowering time determination, and specifically suggest that a flowering time-associated variant located in the promoter of ATCAF1B acts by causing differential binding of this TF. Further, they demonstrate how CressInt can be used to generate testable hypotheses for mechanisms underlying a particular phenotype, even without a priori knowledge of specific genes or genomic regions of interest.
The CressInt web interface
CressInt is available at http://cressint.cchmc.org (Login for review: reviewer; Password: wrlcressint). The CressInt web server has been tested on several web browsers, including Google Chrome, Firefox, and Internet Explorer 10 and 11. In addition to the main page for creating a new job, the web site has several additional useful features, including Help and FAQ pages, details on the incorporated datasets, an update log, links to a variety of other Arabidopsis web resources, and a contact page.
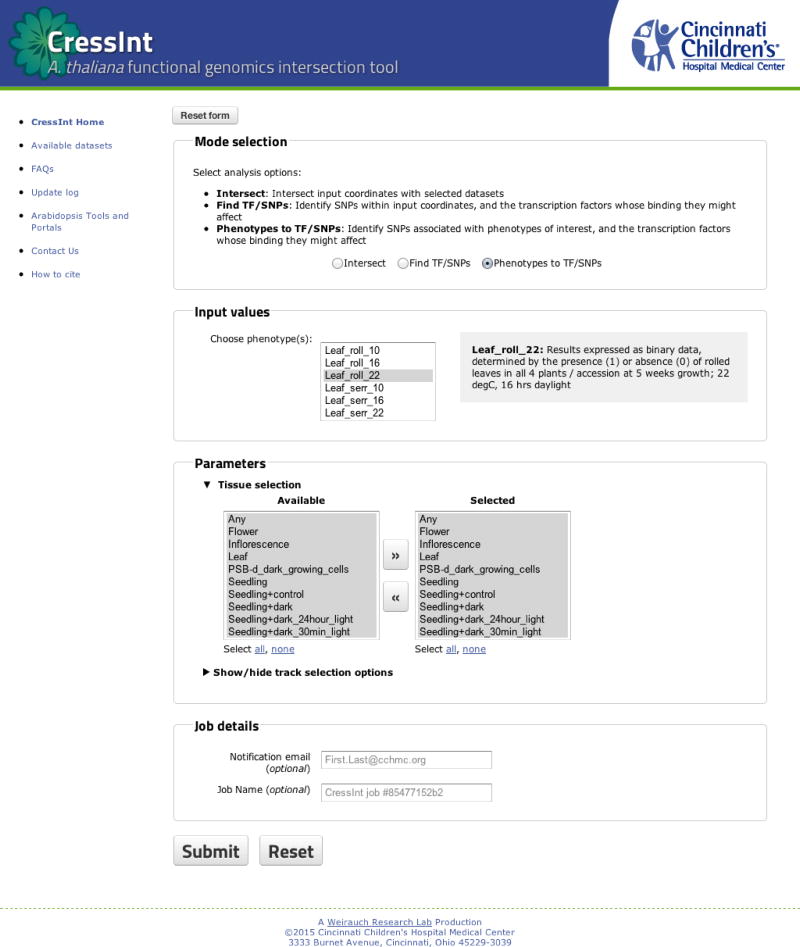
The CressInt web interface was designed to be easy to use, yet flexible (Figure 3). At the top of the page, a user can select between three modes of operation (corresponding to the three case studies above): Intersect, Find TF/SNPs, and Phenotypes to TF/SNPs. Intersect identifies all datasets that overlap the user query. Find TF/SNPs identifies genetic variants located within the user query regions, and predicts the TFs whose binding they might affect. Phenotypes to TF/SNPs starts with a phenotype of interest, and identifies TFs whose binding might be affected by variants associated with that phenotype. As described above, users can select from multiple input format options, including genomic regions, gene names, and phenotypes. Users can paste or type entries into the online form, or upload text files. In the ‘Parameters’ section, users can select the data and tissue types to be included in the query (by default, all datasets are included) (Figure 3, middle). Before submitting a job, a user has the option to provide their email address for automatic notification upon completion of their job. There is also an option to name the job for future reference. Upon submitting a job, a new page appears that automatically refreshes while the jobs run, and posts the final results when they are ready.
Figure 3. The CressInt web server front page.
Screenshot of the CressInt user interface. Here, users can select the “mode” they would like to run (top), enter input data (middle top), select from available datasets (middle bottom), and provide information on the job being submitted (bottom).
There are two basic output pages of CressInt (Figure 1): intersection results indicate all data in the system whose coordinates overlap with the input set, while TF differential binding predictions identify genetic variants that might impact the binding of specific TFs. Both outputs are formatted to be easily human-readable, with multiple visualization options. All input files and parameter choices are saved and documented for reference. Data can be easily sorted or filtered, and are downloadable in tab-delimited text format for processing in another application such as Microsoft Excel, or for additional downstream computational analysis.
Discussion and Conclusions
We present the CressInt web server, a user-friendly system for leveraging A. thaliana functional genomics datasets to formulate testable hypotheses about gene regulation and genome function. Through three case studies, we offer examples of how the CressInt system can be used to explore Arabidopsis biology. The power of CressInt lies in its combination of intuitive design and its inclusion of a wide range of diverse genome-scale datasets. The flexibility of the CressInt system design enables easy inclusion of additional datasets as they become available, and we encourage members of the plant community to use the provided links to alert us of additional useful datasets. In the future, we plan to extend the CressInt system to other plant and non-plant model organisms. We expect that CressInt will be a useful addition to the Arabidopsis genomic toolkit, and anticipate that it will enable numerous insights into the function of plant genomes.
Acknowledgments
We thank Siddharth Dixit and Tuhin Mukherjee for computational support, Phillip Drewe for providing us with eQTL data, and Jeremy Riddell and other members of the Weirauch lab for helpful suggestions. We thank Leah Kottyan, Nathan Salomonis, and Stephen Waggoner for critical feedback on the manuscript. This work was supported in part by NIH grant R21HG008186 to MTW.
Abbreviations
- TF
transcription factor
- ChIP-seq
Chromatin immunoprecipitation followed by sequencing
- DNase-seq
sequencing of DNase I hypersensitive sites
- eQTL
expression quantitative trait locus
- GWAS
genome-wide association study
- PBM
protein binding microarray
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Arabidopsis Genome, I. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408(6814):796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 2.de Lucas M, Provart NJ, Brady SM. Bioinformatic tools in Arabidopsis research. Methods Mol Biol. 2014;1062:97–136. doi: 10.1007/978-1-62703-580-4_5. [DOI] [PubMed] [Google Scholar]
- 3.Bassel GW, et al. Systems analysis of plant functional, transcriptional, physical interaction, and metabolic networks. Plant Cell. 2012;24(10):3859–75. doi: 10.1105/tpc.112.100776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brady SM, Provart NJ. Web-queryable large-scale data sets for hypothesis generation in plant biology. Plant Cell. 2009;21(4):1034–51. doi: 10.1105/tpc.109.066050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atwell S, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465(7298):627–31. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gan X, et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011;477(7365):419–23. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan AM, et al. Mapping and Dynamics of Regulatory DNA and Transcription Factor Networks in A. thaliana. Cell Rep. 2014 doi: 10.1016/j.celrep.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 8.Weirauch MT, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158(6):1431–43. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kersey PJ, et al. Ensembl Genomes 2013: scaling up access to genome-wide data. Nucleic Acids Res. 2014;42(Database issue):D546–52. doi: 10.1093/nar/gkt979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berger MF, et al. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol. 2006;24(11):1429–35. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrett T, et al. NCBI GEO: archive for functional genomics data sets--10 years on. Nucleic Acids Res. 2011;39(Database issue):D1005–10. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kottyan LC, et al. The IRF5-TNPO3 association with systemic lupus erythematosus has two components that other autoimmune disorders variably share. Hum Mol Genet. 2015;24(2):582–96. doi: 10.1093/hmg/ddu455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berger MF, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133(7):1266–76. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quinlan AR, I, Hall M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franco-Zorrilla JM, et al. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci U S A. 2014;111(6):2367–72. doi: 10.1073/pnas.1316278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyle AP, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132(2):311–22. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaufmann K, Melzer R, Theissen G. MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene. 2005;347(2):183–98. doi: 10.1016/j.gene.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 20.Huang K, et al. Solution structure of the MEF2A-DNA complex: structural basis for the modulation of DNA bending and specificity by MADS-box transcription factors. Embo J. 2000;19(11):2615–28. doi: 10.1093/emboj/19.11.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh E, et al. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell. 2007;19(4):1192–208. doi: 10.1105/tpc.107.050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bu Q, et al. Dimerization and blue light regulation of PIF1 interacting bHLH proteins in Arabidopsis. Plant Mol Biol. 2011;77(4–5):501–11. doi: 10.1007/s11103-011-9827-4. [DOI] [PubMed] [Google Scholar]
- 23.Crooks GE, et al. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–90. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chica C, et al. Profiling spatial enrichment of chromatin marks suggests an additional epigenomic dimension in gene regulation. Frontiers in Life Science. 2013;7(1–2):80–87. [Google Scholar]
- 25.Willing E-V, et al. Genome expansion of Arabis alpina linked with retrotransposition and reduced symmetric DNA methylation. Nature Plants. 2015:14023. doi: 10.1038/nplants.2014.23. [DOI] [PubMed] [Google Scholar]
- 26.DSOM, et al. Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant Cell. 2013;25(7):2482–503. doi: 10.1105/tpc.113.113209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pajoro A, et al. Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. 2014;15(3):R41. doi: 10.1186/gb-2014-15-3-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wuest SE, et al. Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc Natl Acad Sci U S A. 2012;109(33):13452–7. doi: 10.1073/pnas.1207075109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oh E, et al. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. Elife. 2014:3. doi: 10.7554/eLife.03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heyman J, et al. ERF115 controls root quiescent center cell division and stem cell replenishment. Science. 2013;342(6160):860–3. doi: 10.1126/science.1240667. [DOI] [PubMed] [Google Scholar]
- 31.Fan M, et al. The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis. Plant Cell. 2014;26(2):828–41. doi: 10.1105/tpc.113.121111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhiponova MK, et al. Helix-loop-helix/basic helix-loop-helix transcription factor network represses cell elongation in Arabidopsis through an apparent incoherent feed-forward loop. Proc Natl Acad Sci U S A. 2014;111(7):2824–9. doi: 10.1073/pnas.1400203111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moyroud E, et al. Prediction of regulatory interactions from genome sequences using a biophysical model for the Arabidopsis LEAFY transcription factor. Plant Cell. 2011;23(4):1293–306. doi: 10.1105/tpc.111.083329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfeiffer A, et al. Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol Plant. 2014;7(11):1598–618. doi: 10.1093/mp/ssu087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, et al. A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 2013;9(1):e1003244. doi: 10.1371/journal.pgen.1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brandt R, et al. Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J. 2012;72(1):31–42. doi: 10.1111/j.1365-313X.2012.05049.x. [DOI] [PubMed] [Google Scholar]
- 37.Huang W, et al. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science. 2012;336(6077):75–9. doi: 10.1126/science.1219075. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All source code developed for the web server is available on Bitbucket (https://bitbucket.org/weirauchlab/tf-tools-cressint). All datasets are available from the authors upon request.