RNA helicases function in all organisms to manage the conformational states of RNAs of all stripes. The largest family of RNA helicases is the DEAD-box family, with 37 members in humans [1]. DEAD-box proteins interact with and manipulate RNAs that range from rRNAs, as they fold and assemble into ribosomes, to mRNAs as they are exported from the nucleus, prepared for translation, and eventually destroyed. One of the best-studied and versatile DEAD-box proteins is DDX3X (Ded1 in yeast), which is implicated in processes including transcription, mRNA export, translation, storage, and decay [2–4].
Perhaps not surprisingly given the many links between these functions and gene expression, defects in several DEAD-box proteins have been linked to human diseases including cancer [4, 5]. DDX3X in particular is frequently mutated in a variety of cancer types, and the positions and frequencies of these mutations have recently been probed by next-generation sequencing of the exomes of various tumors [4, 5]. These approaches were used recently to detect DDX3X mutations in medulloblastoma [6–9], a type of malignant brain tumor that is more common in children than adults. In addition, a subgroup of medulloblastoma that is strongly linked to DDX3X displays activated Wnt signaling, and DDX3X has also been shown to participate in Wnt signaling by activating a kinase [10]. Intriguingly, many of the DDX3X mutations are single nucleotide changes in conserved regions of the protein, suggesting that these mutations may alter the functional properties of DDX3X rather than changing its expression level.
Sequencing methods are immensely powerful for establishing correlations between mutations and disease, but they do not reveal whether the mutations are a cause or consequence of the disease. For DDX3X, we do not even know what roles the protein may play in the maintenance and progression of tumor development. In this issue of JMB, Epling et al use an array of structural and functional approaches to probe several medulloblastoma-linked mutants of DDX3X [11]. The results from an in vitro analysis show how key mutations affect the biochemical activities of DDX3X, providing insights into possible contributors to disease, and the work establishes a complementation assay in S. pombe and shows that the in vitro defects from these mutations correlate with a loss of function in S. pombe. Along the way, the results also raise interesting questions about the mechanisms of DDX3X and even DEAD-box proteins more broadly.
DEAD-box proteins all share a conserved helicase core composed of two RecA-like domains, called D1 and D2, connected by a flexible linker. The ATPase domains interact weakly in the free protein [12–17] but engage each other to form a closed complex upon binding a double-stranded RNA helix (dsRNA) and ATP. As the domains engage each other, one turn of the RNA unwinds so that one strand can bind tightly to the closed complex while the other strand is expelled [18–21]. Once the ATP is hydrolyzed and inorganic phosphate is released, the remaining strand of RNA is released so that the DEAD-box protein is recycled for additional rounds of RNA unwinding [19, 22]. These properties result in an unwinding mechanism that is different from that of other helicases [23], as DEAD-box proteins unwind short RNA helices without translocating along the helix and without requiring a stretch of single-stranded RNA (ssRNA) adjacent to the helix to serve as a landing pad. This mechanism allows DEAD-box proteins to promote local rearrangements in diverse RNAs without disrupting their global structures [24–26].
A large fraction of the medulloblastoma-linked mutations in DDX3X are within D1, and Epling et al. focus on these (Fig. 1). In DDX3X and other DEAD-box proteins, D1 harbors the ATP-binding site [27] and part of the RNA-binding site [13, 28–30]. Most medulloblastoma-linked mutations are in the vicinity of the RNA-binding site and well separated from the ATP binding site. This location contributed to an earlier suggestion that the mutated proteins might suffer from altered RNA binding properties that promote tumor cell proliferation [8, 9]. The authors were able to express in E. coli and purify core domain constructs for four of the mutants, which cluster spatially in two groups; G302V and G325E have mutations in amino acids that contact ssRNA in crystal structures of presumed ‘product complexes’ that follow RNA unwinding [13, 28–30], and L353F and D354V impact amino acids that are close to ssRNA and are in a region that contacts dsRNA in related proteins that function in cellular defense [31].
Fig. 1.
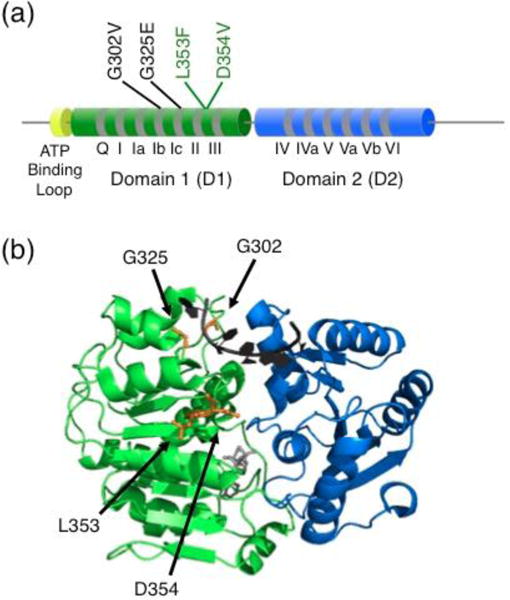
Medulloblastoma-linked DDX3X mutations studied in vitro and in S. pombe. (a) Schematic with domain architecture of DDX3X. Mutations that resulted in compromised RNA-dependent ATPase activity and failed to complement a lack of functional Ded1 in S. pombe are labeled in black. Mutations that did not reveal detectable defects are labeled in green. The study included five additional mutations, not shown, that were probed in vivo but could not be purified in soluble form to study in vitro. The newly-revealed ATP-binding loop is shown in yellow. The cylinders and line connecters are not to scale. (b) Approximate locations of mutations in the closed core complex. The structure depicts D1 and D2 of Vasa [28], and the amino acids that are homologous to those mutated by Epling et al are colored orange and labeled with DDX3X numbering. Bound ssRNA is shown in black and bound AMP-PNP is shown in gray.
These four mutant proteins were subjected to in vitro tests to determine whether specific steps of ATP-dependent function were compromised relative to the wild-type protein. Two of the mutants, G302V and G325E, were significantly defective in RNA-dependent ATPase activity (~5-fold). Interestingly, despite the location of the mutated amino acids near the known RNA binding site, dsRNA binding was minimally compromised in these mutants. Although RNA unwinding was not measured, the defect in RNA-dependent ATPase activity almost certainly results in a defect in RNA unwinding, which is strongly dependent on ATPase activity for Ded1 and other DEAD-box proteins [20, 32]. Interestingly the other two mutants have little or no defect in dsRNA binding or in RNA-stimulated ATPase activity (L353F and D354V), providing no evidence for a role for these amino acids in RNA unwinding by DDX3X.
To determine how the defects revealed in the in vitro experiments correlate with a phenotype in vivo, the authors developed a complementation assay in S. pombe. They first showed that expression of the wild-type DDX3X complements the defects of mutants of the S. pombe DDX3X homolog Ded1, as is the case in S. cerevisiae [33]. The two mutants that showed defects in the in vitro assays (G302V and G325E) fail to complement Ded1 at the non-permissive temperature. Cells harboring these mutants have reduced levels of a cyclin protein, perhaps because the mutations lead to a defect in its translation. Ded1 exerts both positive and negative effects on translation, and the positive effects require ATP while the negative effects do not [34]. Therefore, a mutant that is deficient in RNA-stimulated ATP hydrolysis might be a potent translational inhibitor, consistent with the S. pombe results. It is also possible that by binding RNA but failing to elicit RNA-dependent ATPase activity, these mutants may have increased lifetimes bound to RNA, inhibiting processes that require the RNA to be available for binding or translocation of machinery.
The mutants that are functional in vitro (L353F and D354V) are able to complement, as is another medulloblastoma-linked mutant with a substitution in a neighboring residue that could not be purified from E. coli (R351W). Thus it is not at all clear why these mutations are linked to medulloblastoma. Although it is possible that the linkage is coincidental, the clustering of these two mutations in the three-dimensional structure of DDX3X and the presence of a third mutation in proximity would seem to make this possibility unlikely. It seems more likely that the mutations affect a function of DDX3X that is not shared by Ded1 and not probed in the in vitro experiments, such as protein-protein interactions necessary for its roles in signaling or transcriptional regulation [5].
In the course of studying the defects of these specific mutant proteins, the authors obtained two intriguing results that raise questions about mechanism of DDX3X and DEAD-box proteins more broadly. First, Epling et al solved a crystal structure of one of the mutants, which revealed that an N-terminal extension from D1 is dynamic and can lie atop the ATP binding site. A similar loop was observed previously in a related DEAD-box protein, Vasa, but its function was not determined [28]. They used NMR chemical shift perturbation and isothermal titration calorimetry to show that this N-terminal loop contributes to ATP binding by DDX3X, albeit modestly, and has a very large effect on RNA-stimulated ATPase activity. This finding explains previous observations that core constructs of DDX3X that lack this ATP-binding loop (ABL) do not display detectable RNA-dependent ATPase activity [27]. It may be involved in communication between the ATP and RNA binding sites, sensing bound RNA or stabilizing the closed complex when ATP and RNA are bound. It should also be noted that the ATPase rate measurements revealed only an identical upper limit for the wild-type and ABL-deleted proteins in the absence of RNA, so it remains possible that the ABL contributes to ATP hydrolysis in the absence of bound RNA as well.
Second, the NMR analysis also revealed that dsRNA can bind to a construct containing only D1. Homology modeling against several other DEAD-box protein domains shows that the region is in the vicinity of the known ssRNA binding site and a potential dsRNA binding surface, raising the possibility that this binding represents a functional interaction. This finding could add a wrinkle to a model for the DEAD box mechanism binding that was put forth based on results from the yeast mitochondrial protein Mss116 [35, 36]. In this model, the process begins with ATP binding to D1 and dsRNA binding to D2. Domain closure then modifies the RNA binding site by engaging structural elements of D1, rendering the site incompatible with dsRNA binding and leading to unwinding of the RNA helix and release of one of the strands. The results of Epling et al suggest that in DDX3X the initial binding of dsRNA might be mediated by D1. Additional evidence exists for dsRNA binding to D1 in the bacterial DEAD-box protein Hera [37], and it is possible that different DEAD-box proteins have evolved modifications of the basic pathway for RNA unwinding with a different initial binding site.
However, more information will be necessary to understand the meaning of the observed dsRNA interaction with D1. First, the exact binding site is not yet clear, as the NMR signals may arise from indirect effects propagated by structural changes of the protein. In addition, it remains to be demonstrated that the observed dsRNA binding to D1 at the relatively high concentrations needed for NMR experiments indeed represents functional binding. It may be useful to identify mutations that modulate the affinity of dsRNA and to determine whether the efficiency of RNA unwinding for the intact protein tracks with these affinity changes. Last, it is important to note that due to technical limitations, D2 of DDX3X could not be probed for dsRNA binding, so it remains possible that D2 binds dsRNA as or more strongly than D1.
While these mechanistic questions are intriguing, the central contribution is the progress made toward defining specific biochemical defects caused by cancer-linked mutations of DDX3X. The work has shown clearly that cancer-linked point mutations of DDX3X result in specific biochemical defects and abrogate the ability of DDX3X to function properly in cells. These results and experimental systems set up by this work make DDX3X a leading candidate for further studies probing how specific changes in the functional activities of a DEAD-box protein can lead to cancer. Key challenges are now to define the relevant partner RNAs and proteins for DDX3X and to link specific changes in the mutants to cellular defects. It will be exciting to learn more in the coming years about the roles of this versatile protein, both in healthy physiology and in cancer.
Acknowledgments
I thank Luke Ward for assistance with Fig. 1 and Alan Lambowitz, Andreas Matouschek and members of the Russell lab for helpful comments on the manuscript. Research in the Russell lab is supported by grants from the National Institutes of Health (R01 GM070456 and P01 GM066275) and the Welch Foundation (F-1563).
References
- 1.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–24. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tarn WY, Chang TH. The current understanding of Ded1p/DDX3 homologs from yeast to human. RNA Biol. 2009;6:17–20. doi: 10.4161/rna.6.1.7440. [DOI] [PubMed] [Google Scholar]
- 3.Soto-Rifo R, Ohlmann T. The role of the DEAD-box RNA helicase DDX3 in mRNA metabolism. Wiley Interdiscip Rev RNA. 2013;4:369–85. doi: 10.1002/wrna.1165. [DOI] [PubMed] [Google Scholar]
- 4.Sharma D, Jankowsky E. The Ded1/DDX3 subfamily of DEAD-box RNA helicases. Crit Rev Biochem Mol Biol. 2014;49:343–60. doi: 10.3109/10409238.2014.931339. [DOI] [PubMed] [Google Scholar]
- 5.Fuller-Pace FV. DEAD box RNA helicase functions in cancer. RNA Biol. 2013;10:121–32. doi: 10.4161/rna.23312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–9. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–5. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–8. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–10. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruciat CM, Dolde C, de Groot RE, Ohkawara B, Reinhard C, Korswagen HC, et al. RNA helicase DDX3 is a regulatory subunit of casein kinase 1 in Wnt-beta-catenin signaling. Science. 2013;339:1436–41. doi: 10.1126/science.1231499. [DOI] [PubMed] [Google Scholar]
- 11.Epling LB, Grace CR, Lowe BR, Partridge JF, Enemark EJ. Cancer-associated mutants of RNA helicase DDX3X are defective in RNA-stimulated ATP hydrolysis. J Mol Biol. 2015 doi: 10.1016/j.jmb.2015.02.015. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caruthers JM, Johnson ER, McKay DB. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc Natl Acad Sci U S A. 2000;97:13080–5. doi: 10.1073/pnas.97.24.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–72. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 14.Wang S, Overgaard MT, Hu Y, McKay DB. The Bacillus subtilis RNA helicase YxiN is distended in solution. Biophys J. 2008;94:L01–3. doi: 10.1529/biophysj.107.120709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Theissen B, Karow AR, Kohler J, Gubaev A, Klostermeier D. Cooperative binding of ATP and RNA induces a closed conformation in a DEAD box RNA helicase. Proc Natl Acad Sci U S A. 2008;105:548–53. doi: 10.1073/pnas.0705488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hilbert M, Karow AR, Klostermeier D. The mechanism of ATP-dependent RNA unwinding by DEAD box proteins. Biol Chem. 2009;390:1237–50. doi: 10.1515/BC.2009.135. [DOI] [PubMed] [Google Scholar]
- 17.Mallam AL, Jarmoskaite I, Tijerina P, Del Campo M, Seifert S, Guo L, et al. Solution structures of DEAD-box RNA chaperones reveal conformational changes and nucleic acid tethering by a basic tail. Proc Natl Acad Sci U S A. 2011;108:12254–9. doi: 10.1073/pnas.1109566108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell. 2007;28:253–63. doi: 10.1016/j.molcel.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 19.Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci U S A. 2008;105:20209–14. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Potratz JP, Tijerina P, Del Campo M, Lambowitz AM, Russell R. DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc Natl Acad Sci U S A. 2008;105:20203–8. doi: 10.1073/pnas.0811075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jarmoskaite I, Russell R. DEAD-box proteins as RNA helicases and chaperones. WIREs: RNA. 2011;2:135–52. doi: 10.1002/wrna.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henn A, Cao W, Licciardello N, Heitkamp SE, Hackney DD, De La Cruz EM. Pathway of ATP utilization and duplex rRNA unwinding by the DEAD-box helicase, DbpA. Proc Natl Acad Sci U S A. 2010;107:4046–50. doi: 10.1073/pnas.0913081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jarmoskaite I, Russell R. RNA helicase proteins as chaperones and remodelers. Annu Rev Biochem. 2014;83:697–725. doi: 10.1146/annurev-biochem-060713-035546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan C, Russell R. Roles of DEAD-box proteins in RNA and RNP folding. RNA Biol. 2010;7:667–76. doi: 10.4161/rna.7.6.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jarmoskaite I, Bhaskaran H, Seifert S, Russell R. DEAD-box protein CYT-19 is activated by exposed helices in a group I intron RNA. Proc Natl Acad Sci U S A. 2014;111:E2928–36. doi: 10.1073/pnas.1404307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan C, Potratz JP, Cannon B, Simpson ZB, Ziehr JL, Tijerina P, et al. DEAD-box helicase proteins disrupt RNA tertiary structure through helix capture. PLoS Biol. 2014;12:e1001981. doi: 10.1371/journal.pbio.1001981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Högbom M, Collins R, van den Berg S, Jenvert RM, Karlberg T, Kotenyova T, et al. Crystal structure of conserved domains 1 and 2 of the human DEAD-box helicase DDX3X in complex with the mononucleotide AMP. J Mol Biol. 2007;372:150–9. doi: 10.1016/j.jmb.2007.06.050. [DOI] [PubMed] [Google Scholar]
- 28.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 29.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–25. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 30.Del Campo M, Lambowitz AM. Structure of the yeast DEAD box protein Mss116p reveals two wedges that crimp RNA. Mol Cell. 2009;35:598–609. doi: 10.1016/j.molcel.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo D, Kohlway A, Pyle AM. Duplex RNA activated ATPases (DRAs): platforms for RNA sensing, signaling and processing. RNA Biol. 2013;10:111–20. doi: 10.4161/rna.22706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- 33.Senissar M, Le Saux A, Belgareh-Touze N, Adam C, Banroques J, Tanner NK. The DEAD-box helicase Ded1 from yeast is an mRNP cap-associated protein that shuttles between the cytoplasm and nucleus. Nucleic Acids Res. 2014;42:10005–22. doi: 10.1093/nar/gku584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hilliker A, Gao Z, Jankowsky E, Parker R. The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol Cell. 2011;43:962–72. doi: 10.1016/j.molcel.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mallam AL, Del Campo M, Gilman B, Sidote DJ, Lambowitz AM. Structural basis for RNA-duplex recognition and unwinding by the DEAD-box helicase Mss116p. Nature. 2012;490:121–5. doi: 10.1038/nature11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mallam AL, Sidote DJ, Lambowitz AM. Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase. Elife. 2014;3:e04630. doi: 10.7554/eLife.04630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samatanga B, Klostermeier D. DEAD-box RNA helicase domains exhibit a continuum between complete functional independence and high thermodynamic coupling in nucleotide and RNA duplex recognition. Nucleic Acids Res. 2014;42:10644–54. doi: 10.1093/nar/gku747. [DOI] [PMC free article] [PubMed] [Google Scholar]


