Abstract
MLK4 is a member of the mixed-lineage family of kinases that regulate the JNK, p38, and ERK kinase signaling pathways. MLK4 mutations have been identified in various human cancers including frequently in colorectal cancer, where their function and pathobiological importance has been uncertain. In this study, we assessed the functional consequences of MLK4 mutations in colon tumorigenesis. Biochemical data indicated that a majority of MLK4 mutations are loss-of-function (LOF) mutations that can exert dominant negative effects. In seeking to understand the abrogated activity of these mutants, we elucidated a new MLK4 catalytic domain structure. To determine whether MLK4 is required to maintain the tumorigenic phenotype, we reconstituted its signaling axis in colon cancer cells harboring MLK4 inactivating mutations. We found that restoring MLK4 activity reduced cell viability, proliferation, and colony formation in vitro and delayed tumor growth in vivo. Mechanistic investigations established that restoring the function of MLK4 selectively induced the JNK pathway and its downstream targets, cJUN, ATF3 and the cyclin-dependent kinase inhibitors CDKN1A and CDKN2B. Our work indicates that MLK4 is a novel tumor suppressing kinase harboring frequent LOF mutations that lead to diminished signaling in the JNK pathway and enhanced proliferation in colon cancer.
Introduction
MLK4 is a member of the mixed-lineage kinase (MLK) family of kinases that serve as MAP3Ks and regulate MKK4/7 and MKK3/6 leading to JNK and p38 pathway activation, respectively (1). Recently we have identified a novel function of the MLKs as MEK kinases that are able to directly activate the ERK pathway in a kinase dependent manner (2). MLK4, the least characterized member of the MLK family, has two isoforms; α and β that differ in size by the presence of a unique C-terminal sequence in MLK4β that lies within a region showing a high degree of variability between all the MLKs (1).
The role of MLK4 downstream targets, MKK4/7 and JNK, in cancer development appears complex and contradictory, as previous findings indicate that they may be tumor suppressors or enhancers depending on the specific genetic environment (3, 4). The presence of multiple loss-of-function mutations in MKK4 as well as the documented tumor suppressive roles of MKK4 and JNK indicate that these kinases are tumor suppressors in several tumor types, including lung, pancreatic and ovarian cancers (5-11). The JNK signaling pathway has also been described to have pro-apoptotic effects in Ras-transformed cells (12), where signaling in the JNK pathway suppresses tumor development in mouse models of breast and prostate cancer (13-15). By contrast, JNK is required for cellular transformation by oncogenic RAS, and promotes lung cancer, lymphoma or intestinal cancer (16-20). These examples demonstrate that the role of JNK signaling in tumor development is tissue- and cell-type specific, and therefore understanding the status of upstream regulators and downstream targets is necessary for design of tissue appropriate therapeutic intervention strategies.
The role of MLK4 in cancer is poorly understood and likely to be complex, since the kinase regulates various MAPK pathways. Large consortium cancer genomic studies have revealed multiple somatic mutations in MLK4 in primary tumors and cancer cell lines, including colorectal cancer (CRC) at a frequency of 7% (21-23). It is important to assess the functional impact of mutations in MLK4 and gain an insight into the role this kinase plays in colorectal tumorigenesis. The functional consequences of MLK4 mutations in CRC have not been determined so far, except for one report, where the authors state that enzymatic activity of mutated MLK4 is increased in KRAS-driven colon cancer (24). Here, we present contradictory findings where we demonstrate that MLK4 mutations in colon cancer are loss-of-function (LOF). Reconstitution of CRC cell lines that harbor these LOF mutations in MLK4 with functional MLK4-WT (wild-type) slows the growth of the cancer cells and reduces tumor size in vivo. Furthermore, we shed light on a novel signaling-axis, whereby MLK4 suppresses the proliferation of colon cancer cells.
Materials and methods
Cell lines and reagents
HEK293T cells were obtained from ATCC and cultured in DMEM supplemented with 10% FCS, 1% penicillin/streptomycin and 2 mM L-glutamine. HCT15 and HT115 cells were obtained from European Collection of Cell Culture (ECACC) in October of 2012 and the HT115 was again obtained in November of 2014 due to loss of viability of original cell line. LoVo and HCT116 cells were obtained from ATCC in January of 2013 and July of 2013 respectively. The ECACC and ATCC authenticate cell lines by STR profiling and cell lines were validated throughout the study by sequencing for specific mutations within six months of the latest experiments. Cells were cultured in RPMI supplemented with 10% FCS, 1% penicillin/streptomycin and 2 mM L-glutamine. AZD6244 was purchased from Selleck Chemicals. Inhibitor was dissolved in DMSO (20 mM) and stored at −20°C.
Protein lysate preparation and immunoblots
Cells were lysed with Triton X-100 lysis buffer (Cell Signaling Technology) supplemented with protease inhibitor tablet (Roche). Proteins were resolved by SDS-PAGE and analyzed by western blotting. Primary antibodies: p-ERK1/2 (T202/Y204), p-MKK4 (S257/T261), MKK4, MKK7, p-JNK (T183/Y185), JNK, p-cJUN (S63), p-cJUN (T91), cJUN, p21, PARP, HA (Cell Signaling Technology), ATF3, p15 (Santa Cruz), MLK4 (Bethyl), tubulin, flag M2 (Sigma). All antibodies were used at the dilution of 1:1,000 except tubulin (1:10,000) and flag M2 (1:5,000).
Anchorage-dependent colony formation assay
Cells were seeded at approximately 100 cells/well in a 6-well plate format. Cells were left to grow for 2 weeks with or without 1 μg/ml tetracycline. Growth media was replaced every 2-3 days. Colonies formed were fixed with ice-cold methanol, stained with 0.5 % crystal violet (Sigma) solution in 25 % methanol. Wells were washed and air-dried. For quantification, 10 % acetic acid was added to each well, incubated for 20 minutes with shaking and absorbance values read at 590 nm.
Anchorage-independent colony formation assay
Cells were seeded at approximately 10.000 cells/well in a 6-well plate format in 0.35 % soft agar with or without 1 μg/ml tetracycline. Plates were previously coated with 0.6 % soft agar with or without 1 μg/ml tetracycline containing no cells. Growth media with or without 1 μg/ml tetracycline was added to the wells and it was replaced every few days. After 2 weeks cells were stained using 0.05 % crystal violet (Sigma) solution made up in 25 % methanol.
Mouse xenografts and in vivo studies
All procedures were approved by Animal Welfare and Ethical Review (University of Glasgow) and carried out in accordance with the Animals Scientific Procedures Act 1986 and guidelines of the Committee of the National Cancer Research Institute. 11-week old BALB/c-Nude female mice were injected subcutaneously with 5 × 106 HCT15 or HT115 cells (8 mice/group). Doxycycline was administered in drinking water from the first day after injections. Tumor formation was monitored and tumor volume based on caliper measurements was calculated by the formula: tumor volume = (length x width x width)/2. Mice were culled when tumors reached max permitted volume 1200 mm3 or after 12 weeks post-injection.
Statistical analysis
For xenograft studies, structural analysis, MTT, BrdU incorporation, apoptosis assay and colony formation assays P values were calculated by using two-tailed paired Student’s t-test.
For additional Methods, please see Supplementary Information.
Results
MLK4 mutations identified in colon cancer are loss-of-function
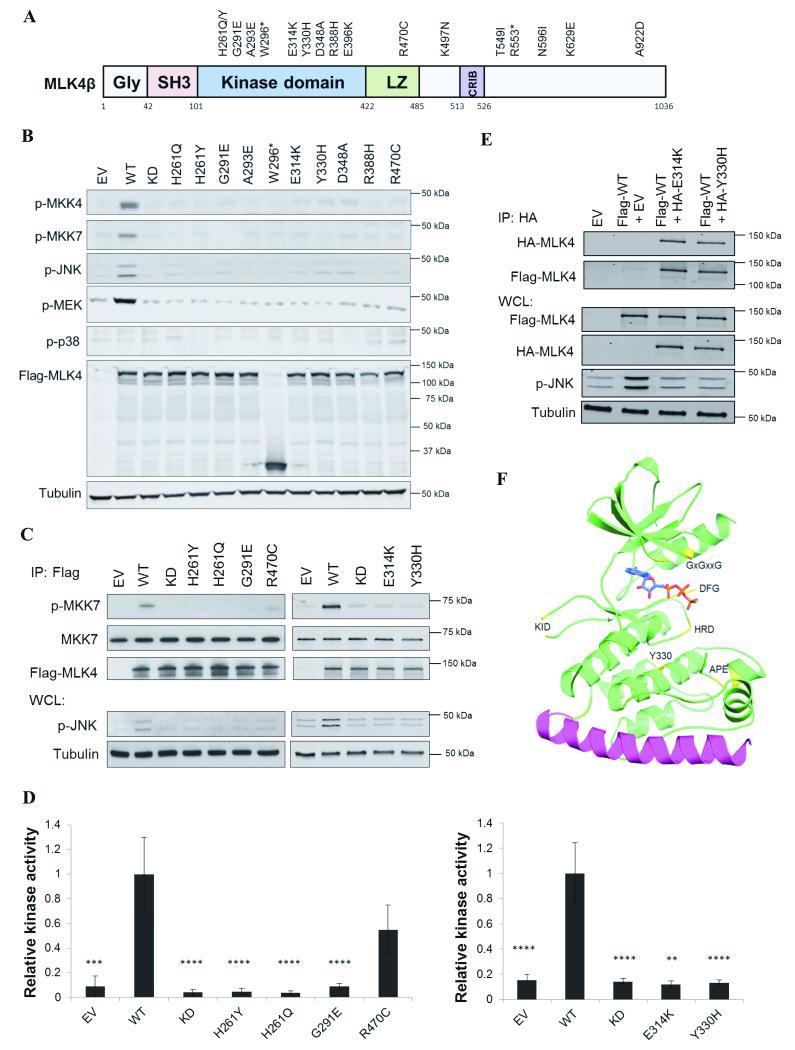
We examined the functional consequences of 17 point mutations in MLK4 identified in CRC cell lines or primary tumors (22, 23) (Table 1). Many of the mutations are located within the kinase catalytic domain including several mutations present in residues critical for kinase activity, e.g. in the DFG and HRD motifs (Fig. 1A). Firstly, we used three bioinformatic tools to analyze the impact of the mutations on the protein function (25-27). The majority of mutations located in the kinase domain were predicted to have a significant impact on protein function providing the impetus to biochemically characterize these cancer mutants (Table 1). We generated a panel of human MLK4 mutants by site-directed mutagenesis and compared their activity against WT and kinase dead (KD) MLK4 (Fig. 1B and Supplementary Fig. S1A). We observed that 10 mutations (59% of the mutations evaluated) ablated the kinase activity of MLK4 towards the JNK and ERK pathways, as assessed by measuring MKK4, MKK7, JNK and MEK phosphorylation in transiently transfected HEK293T cells (Fig. 1B). We did not detect any increase in phosphorylation of p38 by MLK4-WT indicating that the JNK and ERK pathways are the primary signaling cascades activated by MLK4 (Fig. 1B). Some cancer mutants displayed a neutral phenotype and had comparable activity to MLK4-WT (Supplementary Fig. S1A). We further evaluated several LOF cancer mutants in an in vitro kinase assay using inactive MKK7 as a substrate. Mutations located in the kinase domain (H261Y, H261Q, G291E, E314K, Y330H) showed complete loss-of-function towards phosphorylation of MKK7, while a mutation located in the leucine zipper (R470C) displayed weak phosphorylation of MKK7 in the in vitro kinase assay (Fig. 1C and D). Based on a previous study that characterized MLK4 as an oncogene with gain-of-function mutations (H261Y, G291E, R470C and R555*) (24), we performed additional blinded verification of WT and mutant MLK4 activity in an independent laboratory (L.P and T.H), which confirmed the LOF phenotype of MLK4 mutations (Supplementary Fig. S1B). Overall, our data imply that the mutations located in the MLK4 kinase domain abrogate the enzymatic activity of the kinase.
Table 1.
Bioinformatics analysis of MLK4 mutations identified in colon cancer.
| Mutation | Reference | PolyPhen-2 Prediction | SIFT | Mutation assessor |
|---|---|---|---|---|
| H261Q | Bardelli et al. | probably damaging | affect protein function | high |
| H261Y | Bardelli et al. | probably damaging | affect protein function | medium |
| G291E | Bardelli et al. | probably damaging | affect protein function | high |
| A293E | Bardelli et al. | probably damaging | affect protein function | high |
| W296* | Bardelli et al. | N/A | N/A | N/A |
| E314K | CCLE | probably damaging | affect protein function | high |
| Y330H | CCLE | probably damaging | affect protein function | medium |
| D348A | CCLE | probably damaging | affect protein function | neutral |
| R388H | CCLE | probably damaging | affect protein function | high |
| E396K | CCLE | benign | tolerant | neutral |
| R470C | Bardelli et al./CCLE | probably damaging | affect protein function | medium |
| K497N | CCLE | probably damaging | affect protein function | medium |
| T549I | CCLE | benign | tolerant | low |
| R553* | Bardelli et al. | N/A | N/A | N/A |
| N596I | Bardelli et al. | benign | affect protein function | low |
| K629E | Bardelli et al. | possibly damaging | tolerant | medium |
| A922D | CCLE | benign | tolerant | neutral |
Figure 1.
MLK4 mutations are loss-of-function. A, Schematic representation of MLK4 and location of mutations. B, HEK293T were transiently transfected with empty vector (EV), MLK4-WT, kinase dead (KD) or mutants. Whole cell lysates (WCL) were analyzed by western blot. C, MLK4-WT, KD, mutants or EV were transiently transfected into HEK293T. Flag-MLK4 were immunoprecipitated and subjected to a kinase assay with MKK7. Immunoprecipitated samples (IP) and WCL were analyzed by western blot. D, Densitometry analysis of kinase assays. Results are representative of three independent experiments. Error bars indicate ±SEM, ** p<0.01, *** p<0.001, **** p<0.0001. Densitometry was performed using ImageJ. The graphs show the ratio of p-MKK7 to MKK7. E, Flag-MLK4-WT was co-expressed with EV or HA-MLK4 mutants in HEK293T. HA-MLK4 was immunoprecipitated and analyzed by western blot together with WCL. F, Cartoon depiction of MLK4 structure (4UYA) - the kinase domain (green) with highlighted features (yellow), bonds of ATPγS (light blue carbon atoms), putative LZ1 (magenta) and the protein section linking the C-terminus of the kinase domain with the N-terminus of LZ1 (green line).
We next focused on two colorectal cancer cell lines, HCT15 and HT115, harboring MLK4 LOF mutations, Y330H and E314K, respectively (Supplementary Table S1). We have confirmed the presence of these heterozygous mutations by sequencing (Supplementary Fig. S1C). The mutational profiles of both of these cell lines evaluated in CCLE (Cancer Cell Line Encyclopedia) indicate that there are additional mutations in MLK4 and MLK1 that may impact JNK signaling. The HT115 cell line expresses double mutations in MLK4 (E314K and E396K) (Supplementary Table S1), including the LOF mutation (E314K) and a neutral mutation (E396K) (Fig. 1B and Supplementary Fig. S1A). Therefore, we generated a construct that had a double mutation (MLK4-E314K/E396K) and determined the activity of this mutant compared to MLK4-WT, kinase dead, or MLK4-E314K in HEK293T cells. Expression of the double mutant did not result in activation of the JNK pathway, indicating the LOF mutation is dominant over the neutral mutation (Supplementary Fig. S1D). We then characterized the mutations observed in MLK1 (MLK1-D353G mutation in HT115 and MLK1-Q423* in HCT15) (Supplementary Table S1) which revealed that the D353G mutation is LOF and the Q423* is a neutral mutation towards the JNK pathway (Supplementary Fig. S1E). These data suggest that colon cancer cells may acquire multiple LOF mutations in JNK pathway activating kinases to promote inactivation of the pathway.
MLK3 homodimerization is a key step in auto-phosphorylation required for activation of the kinase (28), therefore we investigated if MLK4 cancer mutants that lacked activity could suppress the function of the wild-type kinase, since a majority of MLK4 mutations are heterozygous. We co-expressed MLK4-WT together with MLK4-E314K or MLK4-Y330H, and observed that these mutants could act in a dominant negative manner to suppress the activity of the WT kinase and subsequently the phosphorylation of JNK (Fig. 1E). In order to check if MLK4-E314K and MLK4-Y330H form a complex with MLK4-WT, we immunoprecipitated LOF mutants and found that MLK4-WT and the LOF mutants interacted (Fig. 1E). These data indicate that MLK4 LOF mutants act in dominant fashion to suppress the activity of the WT kinase.
Structure and modeling of MLK4
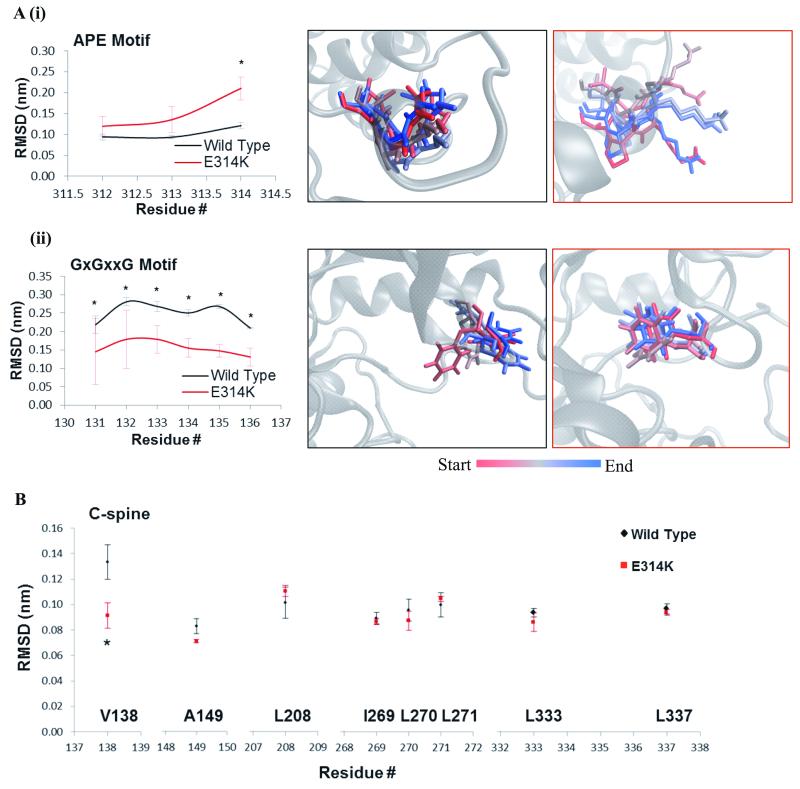
To analyze the impact of these mutations on the structure of the kinase, molecular dynamics (MD) simulations were performed using a newly elucidated MLK4 catalytic domain structure. We have obtained the crystal structure of MLK4 kinase domain as a monomer in an inactive kinase confirmation (PDB ID: 4UY9; Figure 1F). The crystallised MLK4 constructs lacked the kinase insert domain (KID – residues 215-229), which is only present in MLK4, and included the first leucine zipper (LZ1) on the C-terminal end of the construct (Supplementary Results and Supplementary Figure S1F). Next, we used MD simulations to compare the average movement (root-mean-squared deviation; RMSD) of each residue within key regions of the kinase domain of the WT structure and derivative structures where individual residues had been changed to a residue found in a LOF mutant (Fig. 2-3 and Supplementary Fig. S2-4). The MLK4-E314K construct exhibited increased movement within the APE motif, where the mutation is located (Fig. 2A(i)). This region is known to associate with residues from the F-helix to provide a rigid platform for substrate binding (29). Changing the glutamate within this motif to a lysine significantly increases the movement of this motif as observed in time-lapse images (Fig. 2A(i)). This increased bulk and movement could prevent the MLK4 kinase binding to its substrate resulting in a LOF phenotype. An increase in movement within the APE motif was also observed for both the H261Y and G291E mutants (Supplementary Fig. S2B(i) and S2C(i)), likely resulting in disrupted substrate binding. Furthermore, a significantly decreased movement was observed across the whole glycine rich (or GxGxxG) motif (Fig. 2A(ii)). This region functions to orientate the ATP molecule into the binding pocket. From the time-lapse images of the simulation we observe the phenylalanine within the glycine rich motif protrudes into the nucleotide binding pocket (Fig. 2A(ii)), blocking the ATP binding site (Supplementary Fig. S3A), which is not observed for MLK4-WT. Similar changes within the glycine rich motif were observed within MLK4-H261Y (Supplementary Fig. S2B(ii) and S3C). An increased movement was observed for MLK4-H261Q (Supplementary Fig. S2A); however, this mutation still appears to cause occlusion of the nucleotide binding site (Supplementary Fig. S3B). These changes in the APE and glycine rich motif suggest these mutations could prevent ATP binding and disrupt substrate binding to suppress the activity of the kinase.
Figure 2.
Structural analysis of MLK4-E314K mutant. A, Time lapse images show movements within the APE motif (i) and GxGxxG motif (ii) of MLK4-E314K (red) and WT (black) over the course of the simulation. Movement determined by the average RMSD for each residue within these structures. Values are representative of three independent simulations. Error bars indicate ±SEM, * p<0.05. B, Graph shows the average movement observed for each residue of the C-spine during the course of the simulations of MLK4-E314K (red) compared to MLK4-WT (black). Data is the average of three independent repeats, error bars show ±SEM, * p<0.05.
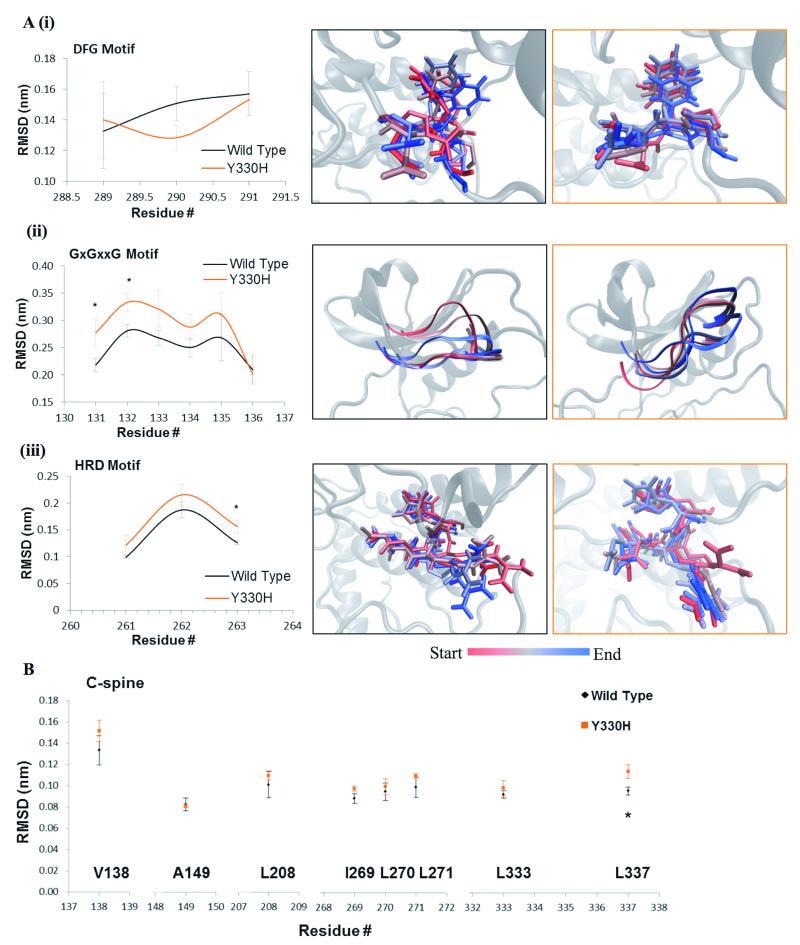
Figure 3.
Structural analysis of MLK4-Y330H mutant. A, Time lapse images highlight the changes occurring within the phenylalanine of the DFG motif (i), residues of the GxGxxG motif (ii) and HRD motif (iii) of MLK4-Y330H (orange) and WT (black) over the course of the simulation. Movement determined by the average RMSD for each residue within these structures. Values are representative of three independent simulations. Error bars indicate ±SEM, * p<0.05. B, Graph shows the average movement observed for each residue of the C-spine during the course of the simulations of MLK4-Y330H (orange) compared to MLK4-WT (black). Data is the average of three independent repeats, error bars show ±SEM, * p<0.05.
MD simulations of MLK4-Y330H showed a decrease in the movement of the phenylalanine within the DFG motif (Fig. 3A(i)). In the time-lapse images of MLK4-WT the phenylalanine demonstrates flexibility, which will allow for the flip from the inactive (D-in/F-out) to the active (D-out/F-in) conformation (Fig. 3A(i)). MLK4-Y330H shows a reduced movement that could indicate that this flip from the inactive to the active conformation would be harder to achieve. In addition, time-lapse images show the GxGxxG motif of the Y330H construct moving up and away from the nucleotide binding pocket of the kinase domain (Fig. 3A(ii) and Supplementary Fig. S3D) compared with the WT construct, which despite its movement, remains close to the site of ATP binding. This could result in the ATP binding being less stable, compared to WT. Finally, a significant increase in movement was also observed within the catalytic HRD motif of the MLK4-Y330H construct (Fig. 3A(iii)). The HRD motif is required for catalysis, functioning to orientate the substrate, support the activation loop configuration, and bind to the DFG motif. Overall, this provides a central scaffold for the active kinase conformation. The increase in movement seen across the HRD motif within the Y330H construct (Fig. 3A(iii)), could prevent substrate binding and provide less stability to the active conformation. Similar results were observed for MLK4-G291E (Supplementary Fig S2C(ii)). Combined, these results agree with biochemical data suggesting the MLK4-Y330H is a LOF mutation.
Interestingly, all of these mutations cause significant disturbances in one residue within either the C-spine (Fig. 2B, Fig. 3B and Supplementary Fig. S4A-B) or the R-spine (Supplementary Fig. S4C), structures that span the kinase domain providing stability to the core of the activated kinase, suggesting these mutants will be less stable in the active state.
Reintroduction of MLK4-WT decreases cell viability and tumor growth in vivo
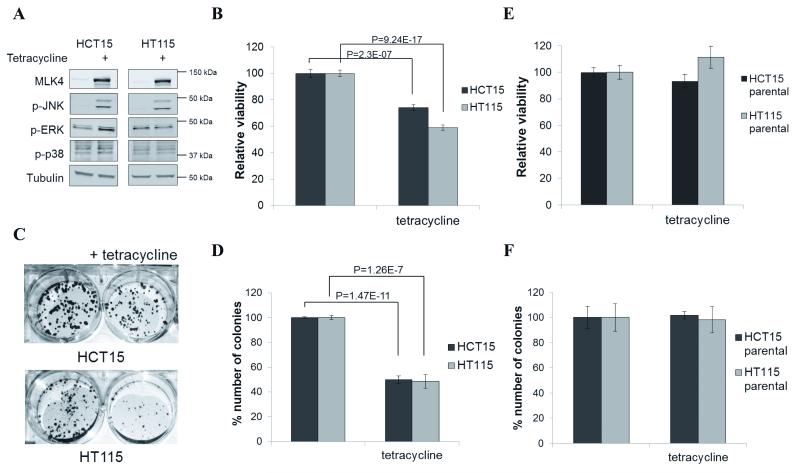
To evaluate the biological consequences of MLK4 LOF mutations in colon cancer, we generated HCT15 and HT115 cell lines where MLK4 expression could be induced in response to tetracycline (HCT15 and HT115 cells harbor the Y330H and E314K LOF mutations respectively). Induced expression of MLK4-WT led to strong activation of the JNK pathway (Fig. 4A). The MEK/ERK pathway was constitutively active in both cell lines and there was only a slight increase in ERK phosphorylation in the HCT15 cell line after MLK4-WT expression (Fig. 4A). This constitutive activation of the ERK pathway is likely due to additional mutations within the MEK/ERK pathway in these cells (e.g. KRAS, NRAS or BRAF, Supplementary Table S1), thus the expression of MLK4-WT does not lead to significant changes in activation of the ERK pathway. We found no activation of the p38 pathway in HCT15 and HT115 cells (Fig. 4A), which is consistent with our results from transfected HEK293T cells (Fig. 1B).
Figure 4.
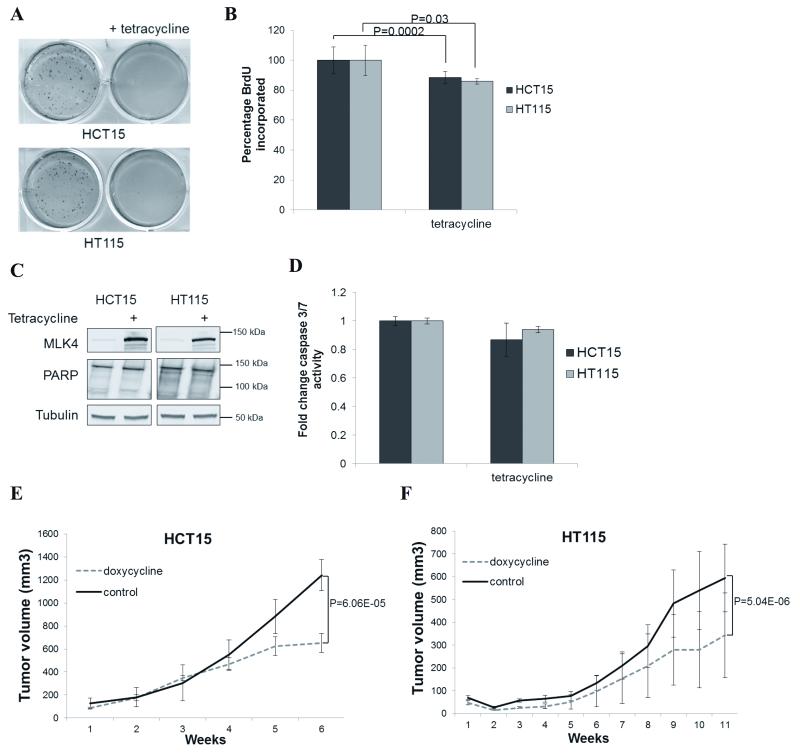
Reintroduction of MLK4-WT decreases cell viability. A, Expression of MLK4-WT in HCT15 and HT115 was induced by tetracycline for 1 day. WCL were analyzed by western blot. B, Expression of MLK4-WT in HCT15 and HT115 was induced by tetracycline for 4 days. Viability was determined by MTT assay. Error bars indicate ±SEM from three independent experiments performed in triplicate (n=9). C and D, HCT15 and HT115 were seeded at low density and expression of MLK4 was induced by tetracycline. After 2 weeks cells were stained with crystal violet (C) and results were quantified by absorbance (D). Error bars indicate ±SEM from three independent experiments (n=3). E, Parental HCT15 and HT115 were treated with tetracycline for 4 days. Viability was determined by MTT assay. Error bars indicate ±SEM from three independent experiments performed in triplicate (n=9). F, Parental HCT15 and HT115 were seeded at low density and expression of MLK4 was induced by tetracycline. After 2 weeks cells were stained with crystal violet and results were quantified by absorbance. Error bars indicate ±SEM from three independent experiments (n=3).
To determine if reconstitution of HCT15 and HT115 cells with MLK4-WT affects the viability of these cancer cells, we induced expression of MLK4 and this resulted in decreased viability in both cell lines (Fig. 4B). Similarly, the ability to form colonies in a long-term anchorage-dependent assay was also significantly impaired (Fig. 4C-D). Additionally, we used the LoVo cell line, which is a colorectal cancer cell line harboring a MLK4-R470C LOF mutation (Fig. 1B) to generate an additional MLK4-WT inducible cell line. Consistent with the results obtained in HCT15 and HT115 cells, the induction of MLK4-WT in LoVo cells activated the JNK pathway (Supplementary Fig. S5A), and led to a reduction in cell viability and colony number in an anchorage-dependent assay (Supplementary Fig. S5B-C).
In addition, we performed control experiments where we tested if tetracycline alone has any effect on viability of these cell lines. We treated parental HT115 and HCT15 cells with tetracycline, but no significant reduction in cell viability or colony number was observed (Fig. 4E-F). Moreover, when we induced expression of LOF MLK4 mutants identified in the respective cancer cell lines, HCT15 – Y330H and HT115 – E314K (Supplementary Fig. S6A-F), but we did not observe any decrease in cell viability (Supplementary Fig. S6A-D). Furthermore, when we knocked down endogenous MLK4 in parental HCT15 and HT115 cells, there were no pronounced effects on viability in these cells (Supplementary Fig. S6G). Finally, in order to provide an additional control, we used a colorectal cancer cell line lacking MLK4 mutations, HCT116, to generate MLK4-WT inducible cells. Induced expression of MLK4-WT in this cell line did not cause any changes in cell viability or colony number in a long-term anchorage-dependent assay (Supplementary Fig. S7A-C). This provides further evidence that the effects observed in tetracycline-inducible HCT15, HT115 and LoVo cell lines are specifically due to reactivation of the MLK4 signaling axis by a functional MLK4 kinase in cells with impaired MLK4 activity.
We next tested whether re-introduction of MLK-WT in colon cancer cells could alter their oncogenic potential. We induced expression of MLK4-WT by tetracycline in HCT15 and HT115 and assessed their ability to form colonies in soft agar. We observed a significant decrease in colony number suggesting that restoring the function of MLK4 leads to reduction in anchorage-independent growth, one of the hallmarks of cellular transformation (Fig. 5A). To investigate if the observed effects in CRC cells reconstituted with MLK4-WT were due to decreased proliferation or increased cell death, we performed BrdU incorporation assays or monitored for changes in cell death markers. Upon MLK4-WT expression in HCT15 and HT115 cells, we observed a reduction in the rate of proliferation that was modest but significant (Fig. 5B). MLK4-WT reconstitution did not induce apoptosis under these specific conditions as tested by caspase3/7 assay and confirmed by the lack of cleaved PARP in cell lysates (Fig. 5C-D). This indicates that the presence of the functional MLK4 decreases the rate of proliferation rather than induces apoptosis in CRC cells.
Figure 5.
Reintroduction of MLK4-WT reduces anchorage-independent colony growth, proliferation and tumor growth in vivo. A, HCT15 and HT115 were seeded in soft agar. Expression of MLK4-WT was induced by tetracycline and after 2 weeks cells were stained with crystal violet. B, HCT15 and HT115 were seeded in 96-well plates. Expression of MLK4-WT was induced by tetracycline for 2 days. Plates were subjected to BrdU assay. Error bars indicate ±SEM from three independent experiments performed in triplicate (n=9). C, Expression of MLK4-WT in HCT15 and HT115 was induced by tetracycline for 4 days. After that time cells were lysed and WCL were analyzed by western blot. D, HCT15 and HT115 were seeded in 96-well plates. Expression of MLK4-WT was induced by tetracycline for 4 days. Plates were subjected to caspase3/7 activity apoptosis assay. Error bars indicate ±SEM from three independent experiments performed in triplicate (n=9). E and F, HCT15 and HT115 were engrafted in nude mice. MLK4-WT expression by doxycycline induction started on the day of injection. Error bars indicate ±SEM (n = 8 mice/group).
Lastly, we aimed to determine if MLK4-WT could suppress tumor growth in a xenograft mouse model. We injected mice with the HT115 and HCT15 cells that had stably incorporated doxycycline-inducible MLK4. In both cases, doxycycline-induced expression of MLK4-WT slowed tumor formation in xenograft mouse models, which supports our in vitro results (Fig. 5E-F). Altogether, these data imply that the presence of LOF mutations in MLK4 suppresses activation of the JNK pathway to promote CRC proliferation.
MLK4 regulates signaling in the JNK-cJUN pathway to suppress growth
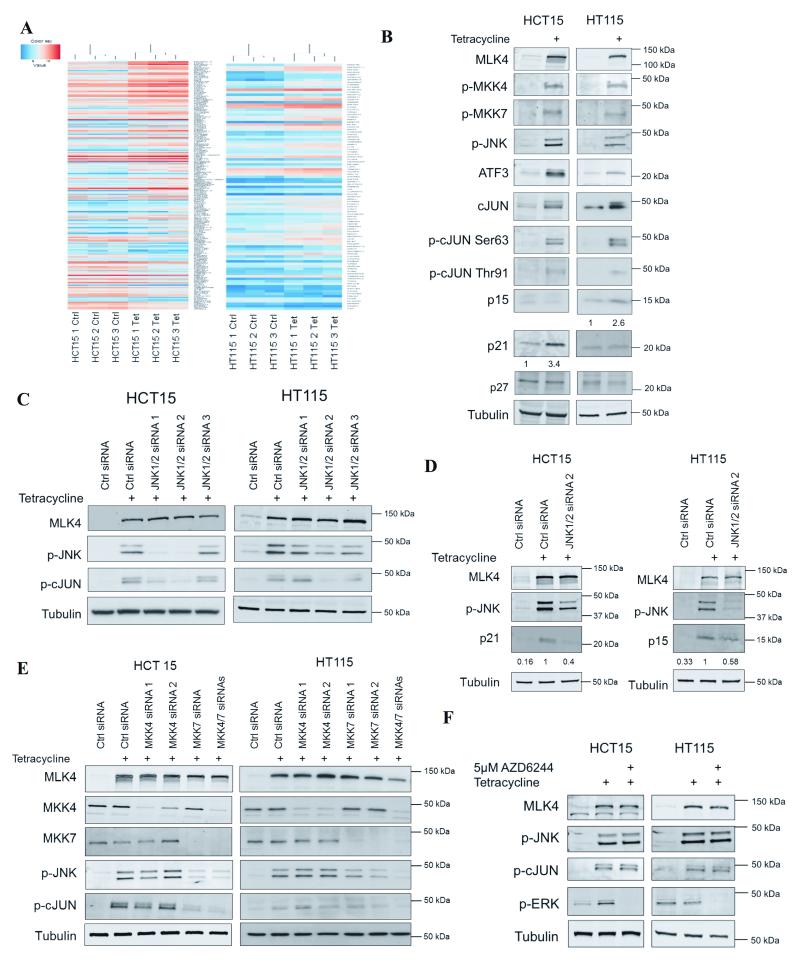
To investigate the downstream targets of MLK4 that mediate the above effects, we transcriptionally profiled HCT15 and HT115 cell lines after MLK4-WT reconstitution. Triplicate RNA samples were subjected to microarray analysis and we found that 143 and 93 genes were upregulated in HCT15 and HT115 cells, respectively (p<0.05) (Fig. 6A and Supplementary Tables S2 and S3). As a control, we also checked gene expression upon induction of Y330H in HCT15 and E314K in HT115, and found that only a few genes (less than 10) were upregulated (Supplementary Fig. S6E-F and Supplementary Table S4 and S5). We validated microarray results by assessing the expression and phosphorylation of the known JNK targets identified by microarray, cJUN and ATF3 (Fig. 6B). Because we observed the effects on cell proliferation by MLK4-WT re-introduction in CRC cells in previous experiments (Fig. 4B and 5B) we focused on the upregulation of the cyclin-dependent kinase inhibitor, p15 (CDKN2B) that was detected by microarray in HT115 cells (Supplementary Table S3). We confirmed increased expression of p15 in HT115 cells, but we did not observe any changes in HCT15 cells (Fig. 6B). Therefore, we tested expression of other Cdk inhibitors and we found that p21Cip1/Waf1 (CDKN1A) was highly upregulated in HCT15 cells (Fig. 6B). Enhanced expression of p21 in HCT15 was also detected by our microarray analysis in all triplicate samples for this cell line; however, the p-value was not significant (p=0.063). In order to test if upregulation of p21 or p15 results in cell-cycle arrest, we performed cell-cycle analysis. Reconstitution of CRC cells harboring LOF mutations in MLK4 with MLK4-WT resulted in a delay in the G1 phase of the cell cycle, although the result was modest in the HCT15 cell line (Supplementary Fig. S7D). This is in agreement with our BrdU experiments where we observed a moderate but significant reduction of proliferation (Fig. 5B); however, long-term MLK4-WT reconstitution had striking effects on colony formation assays and in in vivo mouse models (Fig. 4C-D, 5A and 5E-F). Taken together this indicates that expression of functional MLK4 leads to reduction in cell proliferation, likely due to upregulation of Cdk inhibitors (p15 or p21) (Fig. 6B).
Figure 6.
MLK4 downstream targets and signaling pathways. A, Differential gene expression. MLK4-WT expression was induced by treatment with tetracycline for 1 day. Cells were harvested and mRNA was isolated. Triplicate samples were hybridized to Human Plus 2.0 array. Heat maps were generated using expression values of the selected DE genes. B, Expression of MLK4-WT in HCT15 and HT115 was induced by tetracycline for 1 day. WCL were analyzed by western blot. Densitometry of p21 and p15 was performed using ImageJ. P21 or p15 were normalized to tubulin. Results were averaged from 3 experiments. C -E, HCT15 and HT115 were transfected using siRNA against JNK1/2 (C and D) or MKK4, MKK7 (E). MLK4-WT induction was induced by tetracycline the next day and cells were lysed the following day. WCL were subjected to western blot analysis (C-E). Densitometry was performed using ImageJ; p21 or p15 were normalized to tubulin and compared directly with lane 2 (JNK1/2 siRNA 2 with tetracycline); results were averaged from 3 experiments (D). F, Phosphorylation of cJUN is not MEK-dependent. Expression of MLK4-WT was induced by treatment with tetracycline. After 24 hours HCT15 and HT115 were treated with 5 μM AZD6244 for 2 hours. Cells were lysed and WCL were subjected to western blot analysis. All results are representative of three independent experiments.
To strengthen our model, we queried data compiled by TCGA to determine associations between colon cancer patients with MLK4 mutations and expression levels of Cdk inhibitors. This revealed that MLK4 mutations correlate with decreased mRNA levels of the Cdk inhibitor p27Kip1 (CDKN1B) in patient samples (21, 30) (Supplementary Fig. S8A). Interestingly, there was no significant decrease in levels of mRNAs for other Cdk inhibitors, including p21 or p15 in patient samples with MLK4 mutations (Supplementary Fig. S8A). We checked if the level of p27 was increased after MLK4-WT induction in HCT15 and HT115 but we did not find any changes of p27 in these cell lines (Fig. 6B). Furthermore, we assessed if MLK4 expression affected the levels of Cdk inhibitors in LoVo cells and we found that p21, but not p15 or p27, was increased upon induction of MLK4-WT (Supplementary Fig. S5A). The molecular mechanisms that dictate the differences in expression levels of CDK inhibitors are unclear at this stage; however, it might be explained by differences in the genetic background of patient samples and cell lines.
Lastly, we investigated whether the mechanism by which MLK4-WT promotes inhibition of cell proliferation depends on the JNK pathway, rather than the MEK/ERK pathway. We explored the effects of JNK1/2 and MKK4/7 depletion in cells, and monitored phosphorylation of cJUN-Ser63 and the level of p21 and p15 in HCT15 and HT115 cells as a downstream output. Knock-down of JNK1/2 validated that MLK4-induced phosphorylation of cJUN at Ser63 occurred via a JNK-dependent mechanism (Fig. 6C). Furthermore, the most effective siRNA against JNK1/2 was used to quantify the effects on p21 in HCT15 and p15 in HT115 cells in order to show that MLK4-induced upregulation of p21 and p15 is JNK-dependent (Fig. 6D). Similarly MKK7 depletion led to a decrease of p-JNK but surprisingly, knock-down of MKK4 did not reduce the phosphorylation of JNK (Fig. 6E). The results indicate that MLK4 promotes activation of JNK leading to cJUN-Ser63 phosphorylation through direct regulation of MKK7 (Fig. 6E). However, the correlation between JNK phosphorylation and activation of cJUN in HT115 cell line was less conclusive (Fig. 6E). Importantly, the use of AZD6244 (MEK inhibitor) did not have any effect on phosphorylation of cJUN-Ser63 (Fig. 6F). This suggests that phosphorylation and upregulation of MLK4 downstream targets does not depend on the MEK/ERK pathway. Collectively, these data highlight a novel signaling node where activation of the MLK4-JNK-cJUN pathway promotes inhibition of proliferation, therefore loss of signaling through this pathway will promote CRC cell proliferation and tumorigenesis.
Discussion
MLK4 is an understudied protein kinase that is frequently mutated in cancer, specifically colon cancer, and it is essential to understand the role of this kinase in tumorigenesis. To date, there has only been one report evaluating the role of MLK4 in cancer (24). In this study mutations in MLK4 were described as activating, and the authors suggested that MLK4 was oncogenic and promoted tumorigenesis in a mutant KRAS background in CRC (24). In contrast, we have characterized the same mutations in MLK4 as loss-of-function and our biochemical, structural, and independent blinded verification unequivocally indicated that these mutations abrogate the activity of the kinase. We also showed that MLK4 LOF mutants suppress the function of the WT allele leading to inactivation of signaling downstream of MLK4. The presence of additional mutations in the members of the MLK family (including loss-of-function MLK1-D353G) in colon cancer cell lines reinforces the idea that tumor progression favors inactivation of JNK signaling in colorectal cancer. Consistent with our data, a previous report analyzing mutations in MKK4, which lies upstream of JNK, showed that 5 out of 8 mutations identified in human colorectal cancer are loss-of-function (with two being neutral and one gain-of-function) (5), suggesting that inactivation of the JNK pathway may occur at different levels. Confirming the functional importance of MLK4 LOF mutations, we demonstrate that re-introduction of MLK4-WT into CRC cells carrying MLK4 LOF mutations decreases anchorage-independent growth, a hallmark of cellular transformation, and slows tumor growth in in vivo mouse xenograft models. We determined the molecular mechanism by which MLK4 promotes these phenotypes by demonstrating that restoration of MLK4-WT function led to activation of the JNK pathway, increased expression and phosphorylation of cJUN, and elevated expression of Cdk inhibitors, p21 and p15, leading to a reduction in proliferation and a cell-cycle arrest. Interestingly, loss of the p21 Cdk inhibitor was previously linked with colon cancer formation and an inability of colon cancer cells to arrest in G1 phase of the cell cycle (31, 32).
Colorectal cancer remains one of the most commonly diagnosed cancers and leading causes of cancer mortality worldwide. The nature of colorectal cancer has been described as extremely heterogeneous where activation of different molecular pathways leads to different phenotypes (33). Multiple gene mutations have been associated with colorectal carcinogenesis, including APC, KRAS, BRAF and p53 (33, 34). The presence of these mutations is sufficient to initiate primary events in transformation; however, additional mutations are required for the progression to a malignant or metastatic phenotype (35-37). It is well established that cancer cells need to accumulate other mutations and/or epigenetic alterations in order to overcome senescence and achieve full transformation (35-37). It is possible that inactivation of the JNK pathway in colorectal cancer is required to escape oncogene-induced senescence (OIS) that could be caused by KRAS GOF mutants or loss of APC, and one mechanism to overcome OIS will be loss of signaling downstream of MLK4 leading to inactivation of the JNK pathway (Supplementary Fig. S8B). Utilizing cancer genomics to identify novel signaling pathways where loss- or gain-of-function is required to promote tumorigenesis is a critical next step in translating better therapies into the clinic. In summary, we demonstrate that MLK4 is a novel colon cancer tumor suppressor harboring frequent LOF mutations that promote cancer cell proliferation by suppressing signaling in the JNK-cJUN-p21/p15 pathway.
Supplementary Material
Acknowledgments
We would like to thank Bohdan Waszkowycz for advice on ATP docking.
Financial Support:
This research was supported by Cancer Research UK (A.A. Marusiak, N.L. Stephenson, H. Baik, E.W. Trotter, Y. Li, K. Blyth, S. Mason, P. Chapman, E. Testoni, C.J. Miller, O.J. Sansom, J. Brognard). This work was also supported by USPHS grants from NCI to L. Puto and T. Hunter (CA14195 and CA82683). T. Hunter is a Frank and Else Schilling American Cancer Society Professor, and holds the Renato Dulbecco Chair in Cancer Research.
Footnotes
Disclosure of Potential Conflicts of Interest:
No potential conflicts of interest were disclosed by the authors.
References
- 1.Gallo KA, Johnson GL. Mixed-lineage kinase control of JNK and p38 MAPK pathways. Nature reviews Molecular cell biology. 2002;3:663–72. doi: 10.1038/nrm906. [DOI] [PubMed] [Google Scholar]
- 2.Marusiak AA, Edwards ZC, Hugo W, Trotter EW, Girotti MR, Stephenson NL, et al. Mixed lineage kinases activate MEK independently of RAF to mediate resistance to RAF inhibitors. Nature communications. 2014;5:3901. doi: 10.1038/ncomms4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kennedy NJ, Davis RJ. Role of JNK in tumor development. Cell cycle. 2003;2:199–201. [PubMed] [Google Scholar]
- 4.Whitmarsh AJ, Davis RJ. Role of mitogen-activated protein kinase kinase 4 in cancer. Oncogene. 2007;26:3172–84. doi: 10.1038/sj.onc.1210410. [DOI] [PubMed] [Google Scholar]
- 5.Ahn YH, Yang Y, Gibbons DL, Creighton CJ, Yang F, Wistuba II, et al. Map2k4 functions as a tumor suppressor in lung adenocarcinoma and inhibits tumor cell invasion by decreasing peroxisome proliferator-activated receptor gamma2 expression. Molecular and cellular biology. 2011;31:4270–85. doi: 10.1128/MCB.05562-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su GH, Song JJ, Repasky EA, Schutte M, Kern SE. Mutation rate of MAP2K4/MKK4 in breast carcinoma. Human mutation. 2002;19:81. doi: 10.1002/humu.9002. [DOI] [PubMed] [Google Scholar]
- 7.Su GH, Hilgers W, Shekher MC, Tang DJ, Yeo CJ, Hruban RH, et al. Alterations in pancreatic, biliary, and breast carcinomas support MKK4 as a genetically targeted tumor suppressor gene. Cancer research. 1998;58:2339–42. [PubMed] [Google Scholar]
- 8.Teng DH, Perry WL, 3rd, Hogan JK, Baumgard M, Bell R, Berry S, et al. Human mitogen-activated protein kinase kinase 4 as a candidate tumor suppressor. Cancer research. 1997;57:4177–82. [PubMed] [Google Scholar]
- 9.Yeasmin S, Nakayama K, Rahman MT, Rahman M, Ishikawa M, Katagiri A, et al. MKK4 acts as a potential tumor suppressor in ovarian cancer. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2011;32:661–70. doi: 10.1007/s13277-011-0166-5. [DOI] [PubMed] [Google Scholar]
- 10.Yeasmin S, Nakayama K, Rahman MT, Rahman M, Ishikawa M, Katagiri A, et al. Loss of MKK4 expression in ovarian cancer: a potential role for the epithelial to mesenchymal transition. International journal of cancer Journal international du cancer. 2011;128:94–104. doi: 10.1002/ijc.25332. [DOI] [PubMed] [Google Scholar]
- 11.Schramek D, Kotsinas A, Meixner A, Wada T, Elling U, Pospisilik JA, et al. The stress kinase MKK7 couples oncogenic stress to p53 stability and tumor suppression. Nature genetics. 2011;43:212–9. doi: 10.1038/ng.767. [DOI] [PubMed] [Google Scholar]
- 12.Kennedy NJ, Sluss HK, Jones SN, Bar-Sagi D, Flavell RA, Davis RJ. Suppression of Ras-stimulated transformation by the JNK signal transduction pathway. Genes & development. 2003;17:629–37. doi: 10.1101/gad.1062903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cellurale C, Weston CR, Reilly J, Garlick DS, Jerry DJ, Sluss HK, et al. Role of JNK in a Trp53-dependent mouse model of breast cancer. PloS one. 2010;5:e12469. doi: 10.1371/journal.pone.0012469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cellurale C, Girnius N, Jiang F, Cavanagh-Kyros J, Lu S, Garlick DS, et al. Role of JNK in mammary gland development and breast cancer. Cancer research. 2012;72:472–81. doi: 10.1158/0008-5472.CAN-11-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hubner A, Mulholland DJ, Standen CL, Karasarides M, Cavanagh-Kyros J, Barrett T, et al. JNK and PTEN cooperatively control the development of invasive adenocarcinoma of the prostate. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:12046–51. doi: 10.1073/pnas.1209660109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cellurale C, Sabio G, Kennedy NJ, Das M, Barlow M, Sandy P, et al. Requirement of c-Jun NH(2)-terminal kinase for Ras-initiated tumor formation. Molecular and cellular biology. 2011;31:1565–76. doi: 10.1128/MCB.01122-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hess P, Pihan G, Sawyers CL, Flavell RA, Davis RJ. Survival signaling mediated by c-Jun NH(2)-terminal kinase in transformed B lymphoblasts. Nature genetics. 2002;32:201–5. doi: 10.1038/ng946. [DOI] [PubMed] [Google Scholar]
- 18.Sancho R, Nateri AS, de Vinuesa AG, Aguilera C, Nye E, Spencer-Dene B, et al. JNK signalling modulates intestinal homeostasis and tumourigenesis in mice. The EMBO journal. 2009;28:1843–54. doi: 10.1038/emboj.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nateri AS, Spencer-Dene B, Behrens A. Interaction of phosphorylated c-Jun with TCF4 regulates intestinal cancer development. Nature. 2005;437:281–5. doi: 10.1038/nature03914. [DOI] [PubMed] [Google Scholar]
- 20.Behrens A, Jochum W, Sibilia M, Wagner EF. Oncogenic transformation by ras and fos is mediated by c-Jun N-terminal phosphorylation. Oncogene. 2000;19:2657–63. doi: 10.1038/sj.onc.1203603. [DOI] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas N Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bardelli A, Parsons DW, Silliman N, Ptak J, Szabo S, Saha S, et al. Mutational analysis of the tyrosine kinome in colorectal cancers. Science. 2003;300:949. doi: 10.1126/science.1082596. [DOI] [PubMed] [Google Scholar]
- 23.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martini M, Russo M, Lamba S, Vitiello E, Crowley EH, Sassi F, et al. Mixed lineage kinase MLK4 is activated in colorectal cancers where it synergistically cooperates with activated RAS signaling in driving tumorigenesis. Cancer research. 2013;73:1912–21. doi: 10.1158/0008-5472.CAN-12-3074. [DOI] [PubMed] [Google Scholar]
- 25.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nature methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic acids research. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic acids research. 2003;31:3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leung IW, Lassam N. Dimerization via tandem leucine zippers is essential for the activation of the mitogen-activated protein kinase kinase kinase, MLK-3. The Journal of biological chemistry. 1998;273:32408–15. doi: 10.1074/jbc.273.49.32408. [DOI] [PubMed] [Google Scholar]
- 29.Kornev AP, Taylor SS, Ten Eyck LF. A helix scaffold for the assembly of active protein kinases. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14377–82. doi: 10.1073/pnas.0807988105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.el-Deiry WS, Tokino T, Waldman T, Oliner JD, Velculescu VE, Burrell M, et al. Topological control of p21WAF1/CIP1 expression in normal and neoplastic tissues. Cancer research. 1995;55:2910–9. [PubMed] [Google Scholar]
- 32.Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–84. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 33.Al-Sohaily S, Biankin A, Leong R, Kohonen-Corish M, Warusavitarne J. Molecular pathways in colorectal cancer. Journal of gastroenterology and hepatology. 2012;27:1423–31. doi: 10.1111/j.1440-1746.2012.07200.x. [DOI] [PubMed] [Google Scholar]
- 34.Zenonos K, Kyprianou K. RAS signaling pathways, mutations and their role in colorectal cancer. World journal of gastrointestinal oncology. 2013;5:97–101. doi: 10.4251/wjgo.v5.i5.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer cell. 2004;5:375–87. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 36.Janssen KP, el-Marjou F, Pinto D, Sastre X, Rouillard D, Fouquet C, et al. Targeted expression of oncogenic K-ras in intestinal epithelium causes spontaneous tumorigenesis in mice. Gastroenterology. 2002;123:492–504. doi: 10.1053/gast.2002.34786. [DOI] [PubMed] [Google Scholar]
- 37.Velho S, Moutinho C, Cirnes L, Albuquerque C, Hamelin R, Schmitt F, et al. BRAF, KRAS and PIK3CA mutations in colorectal serrated polyps and cancer: primary or secondary genetic events in colorectal carcinogenesis? BMC cancer. 2008;8:255. doi: 10.1186/1471-2407-8-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.