Abstract
WRKY proteins are plant specific transcription factors involved in various developmental and physiological processes, especially in biotic and abiotic stress resistance. Although previous studies suggested that WRKY proteins in soybean (Glycine max var. Williams 82) involved in both abiotic and biotic stress responses, the global information of WRKY proteins in the latest version of soybean genome (Wm82.a2v1) and their response to dehydration and salt stress have not been reported. In this study, we identified 176 GmWRKY proteins from soybean Wm82.a2v1 genome. These proteins could be classified into three groups, namely group I (32 proteins), group II (120 proteins), and group III (24 proteins). Our results showed that most GmWRKY genes were located on Chromosome 6, while chromosome 11, 12, and 20 contained the least number of this gene family. More GmWRKY genes were distributed on the ends of chromosomes to compare with other regions. The cis-acting elements analysis suggested that GmWRKY genes were transcriptionally regulated upon dehydration and salt stress. RNA-seq data analysis indicated that three GmWRKY genes responded negatively to dehydration, and 12 genes positively responded to salt stress at 1, 6, and 12 h, respectively. We confirmed by qRT-PCR that the expression of GmWRKY47 and GmWRKY 58 genes was decreased upon dehydration, and the expression of GmWRKY92, 144 and 165 genes was increased under salt treatment.
Keywords: codon usage bias, dehydration stress, Glycine max, salt stress, WRKY protein
Introduction
WRKY transcription factors were first identified from plant species and were thought to be plant specific (Eulgem et al., 2000; Rushton et al., 2010). However, increasing studies identified WRKY proteins from non-plant species, including Caenorhabditis elegans, Dictyostelium discoideum, Drosophila melanogaster, Giardia lamblia, Klebsormidium flaccidum, and Saccharomyces cerevisiae (Riechmann et al., 2000; Zhang and Wang, 2005; Rinerson et al., 2015). WRKY proteins were named after a conserved WRKY domain, containing the WRKYGQK heptapeptide, followed by a zinc-finger motif (CX4−5CX22−23HXH or CX7CX23HXC; Eulgem et al., 2000; Rushton et al., 2010). Based on the number of WRKY domains and the type of zinc-finger motif, WRKY proteins were classified into three groups (group I–III; Eulgem et al., 2000; Rushton et al., 2010). Group I WRKY proteins contained two WRKY domains and a zinc-finger motif (CX4−5CX22−23HXH or CX7CX23HXC). Group II WRKY proteins which could be divided into five subgroups (IIa–IIe), contained a single WRKY domain and a CX4−5CX22−23HXH zinc-finger motif (Eulgem et al., 2000; Rushton et al., 2010). Group III WRKY proteins had a single WRKY domain and a CX7CX23HXC zinc-finger motif (Eulgem et al., 2000; Rushton et al., 2010).
To date, genome-wide WRKY analysis has been performed in many plant species including Arabidopsis thaliana (Eulgem et al., 2000), Brachypodium distachyon (Tripathi et al., 2012; Wen et al., 2014), Hordeum vulgare (Liu et al., 2014), Lotus japonicus (Song et al., 2014), Medicago truncatula (Song and Nan, 2014), Oryza sativa (Wu et al., 2005), Vitis vinifera (Wang et al., 2014), Zea mays (Wei et al., 2012), Gossypium (Ding et al., 2015), and Populus (He et al., 2012; Jiang et al., 2014). Over the past 15 years, studies demonstrated that WRKY proteins played crucial roles in pathogen defense and insect resistance (Eulgem and Somssich, 2007; Grunewald et al., 2008; Skibbe et al., 2008; Rushton et al., 2010). WRKY proteins were implicated to modulate plant development such as seed development (Luo et al., 2005), trichome morphogenesis (Johnson et al., 2002), senescence (Robatzek and Somssich, 2002), dormancy and germination (Zhang et al., 2004; Zentella et al., 2007; Zou et al., 2008). Recently, studies demonstrated that WRKY proteins were involved in response to abiotic stresses, such as salt, drought, and cold (Wu et al., 2009; Ren et al., 2010; Zou et al., 2010; Jiang et al., 2012; Rushton et al., 2012). WRKY proteins were involved in signal transductions mediated by plant hormones, for example, abscisic acid (ABA) (Rushton et al., 2012). In Arabidopsis, WRKY proteins are involved in regulation of ABA-responsive genes, such as MYB2, DREB1a, DREB2a, and RAB18 (Rushton et al., 2012). WRKY proteins could increase drought and salt tolerance in plants. Overexpression of ZmWRKY33 in Arabidopsis could improve salt stress tolerance of the transgenic plants (Li et al., 2013). Overexpression of OsWRKY11 under the control of HSP101 promoter led to enhanced drought tolerance (Wu et al., 2009). Moreover, overexpression of TaWRKY10 in tobacco resulted in enhanced drought and salt tolerance (Wang et al., 2013).
Soybean (Glycine max), as one of the important protein and oil crop, is planted worldwide. Zhou et al. (2008) identified 64 GmWRKY genes before the soybean genome was sequenced, and confirmed that GmWRKY13, 21, and 54 genes were involved in abiotic stresses. Bencke-Malato et al. (2014) identified 182 GmWRKY genes including 33 pseudogenes using the whole genome sequence information (Wm82.a1.v1; Schmutz et al., 2010). Among the 149 non-pseudogenized GmWRKY genes, 72 genes were differentially expressed during fungal infection based on SAGE, RNA-seq and microarray experiments (Bencke-Malato et al., 2014). However, the role of GmWRKY gene in dehydration and salt stress are largely unknown. We carried out a comprehensive analysis of GmWRKY genes base on the newly released genome Wm82.a2v1, and investigated their response to dehydration and salt stress. Totally, we identified 176 putative GmWRKY genes from soybean Wm82.a2v1 genome using bioinformatics approach. The chromosomal location, codon usage bias, cis-elements, and gene expression in different tissues and under dehydration and salt stress were analyzed. These results provided new insight into the roles of soybean WRKY genes in abiotic stress responses.
Materials and methods
Identification and analysis of WRKY genes in soybean
Sequences of the soybean genome (Wm82.a2.v1) were downloaded from Phytozome 10.0 database (http://www.phytozome.org). The Hidden Markov Model (HMM) profile of WRKY domain (PF03106) was downloaded from Pfam protein family database (http://pfam.janelia.org) and was used to survey all soybean proteins by HMMER program (Finn et al., 2011). To verify the reliability of searched results, each protein sequence was checked in Pfam database.
A. thaliana WRKY proteins (http://www.arabidopsis.org) were obtained and used for phylogenetic analysis. To categorize GmWRKY proteins, we used AtWRKY domains as query sequences to constructed phylogenetic tree. MAFFT 7.0 program was applied to multiple sequences alignment (Katoh and Standley, 2013). The phylogenetic trees were inferred using MEGA 6.0 with the neighbor-joining method (Tamura et al., 2013). Bootstrap values were calculated for 1000 iterations.
All GmWRKY genes were mapped to soybean chromosomes based on information available from SoyBase (http://soybase.org/). The map was drafted using MapInspect software (http://mapinspect.software.informer.com/).
Soybean EST sequences were downloaded from GenBank (http://www.ncbi.nlm.nih.gov/est). GmWRKY genes were used as query to blast against soybean ESTs using BLASTN with sequence similarity >96%, length >= 200 bp, and E-value <10−10.
Analysis of codon usage bias
Codon usage bias was derived from cDNA sequences encoding full-length proteins. To avoid sampling bias, CDS sequences were filtered based on the following criteria: (1) full-length CDS sequences shorter than 300 bp were excluded from this analysis; (2) the presence of a start codon (ATG) beginning and a stop codon (TAA, TAG, and TGA) ending in each CDS was required. Codon usage bias was calculated from sequences using the Codon W 1.4 program (http://codonw.sourceforge.net) and Perl scripts.
Expression profiles of GmWRKY genes
The normalized data (Reads/kb/Million, RPKM) for six different tissues from different growth periods was reported by Severin et al. (2010), and available from SoyBase website. A gene was considered expressed if the RPKM value was greater than or equal to two in an expression atlas (Belamkar et al., 2014). The RPKM normalized read count data of expressed genes was log2-transformed and displayed in the form of heatmaps in R script. To survey the involvement of GmWRKY genes in dehydration and salt stress responses, transcriptome sequencing data of soybean under dehydration, and salt stress at three time points (1, 6, and 12 h) was downloaded from described by Belamkar et al. (2014). The satisfied criteria of differentially expressed genes was as follows: (1) P-value adjusted for multiple testing correction using Benjamini and Hochberg method (Benjamini and Hochberg, 1995) to be <0.05, (2) two fold or greater fold change, (3) residual variance quotients of both the control and treatment samples of <20.
cis-Acting elements analysis in GmWRKY promoters
PlantCARE online program (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to predict cis-acting elements in GmWRKY promoters. Sequence of 2000 bp upstream of the start codon was used for cis-acting elements analysis. The sequences were obtained from Phytozome 10.0 database.
Plant material and stress treatments
The seeds of soybean (Williams 82) were germinated on wet filter paper in growth chamber at 28°C, and grown for 2 weeks at room temperature (about 32°C). For salt treatment, roots of the seedlings were transferred into 100 mM NaCl solution at room temperature. For dehydration treatment, seedlings were removed from the wet filter paper and kept in air at room temperature. Roots were harvested after 0, 6, 12, 24, and 48 h exposure to NaCl and dehydration treatment.
Quantitative real-time RT-PCR
Total RNA was extracted using the CTAB method (Chang et al., 1993). The first-strand cDNAs were synthesized using 2 μg RNA using the Reverse Transcriptase M-MLV System (Takara, Dalian, China). Primers for quantitative real-time RT-PCR (qRT-PCR) analysis were listed in Table S1. Metalloprotease gene (forward primer: 5′-ATGAATGACGGTTCCCATGTA-3′; reverse peimer: 5′-GGCATTAAGGCAGCTCACTCT-3′) was used as a reference gene (Libault et al., 2008). qRT-PCR was carried out using Fast Start Universal SYBR Green Master (ROX) with a 7500 real-time PCR machine (ABI). The reactions were carried out using the following program: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, 60°C for 30 s. Melting curve was generated from the following program: 95°C for 15 s, 60°C for 60 s, 95°C for 30 s, and 60°C for 15 s. Three biological replicates were used for qRT-PCR analysis. The ΔΔCt method was used for quantification (Livak and Schmittgen, 2001).
Results
WRKY proteins in soybean
We predicted 192 WRKY sequences from soybean genome using HMMER program. Seven of these protein sequences were not WRKY sequences, and other nine proteins were excluded in this study because each of them contained an incomplete WRKY domain (Table S2). The remaining 176 proteins were identified as putative WRKY proteins in soybean genome Wm82.a2v1. GmWRKY proteins could be divided into three groups based on the number of WRKY domains and the type of zinc-finger structure. Group I, II, and III contained 32, 120, and 24 proteins, respectively (Table 1 and Table S2). The conserved WRKY domain from Arabidopsis and soybean were used to reconstruct a neighbor-joining phylogenetic tree (Figure S1). Soybean Group II WRKY proteins could be further divided into five subgroups, namely IIa, IIb, IIc, IId, and IIe, and each containing 12, 30, 41, 15, and 22 sequences, respectively (Figure S1 and Table 1). Notably, we identified one novel WRKY (Glyma.14G085500), which was named GmWRKY183 (Table S2).
Table 1.
Number of WRKY genes identified in plants.
| Species | Group I | Subgroup IIa | Subgroup IIb | Subgroup IIc | Subgroup IId | Subgroup IIe | Group III | Total | Genome size(Mb) |
|---|---|---|---|---|---|---|---|---|---|
| Glycine max | 32 | 12 | 30 | 41 | 15 | 22 | 24 | 176 | 978 |
| Arabidopsis thalianaa | 13 | 4 | 7 | 18 | 7 | 9 | 14 | 72 | 125 |
| Brachypodium distachyonb | 17 | 3 | 6 | 21 | 6 | 10 | 23 | 86 | 272 |
| Cucumis sativusa | 10 | 4 | 4 | 16 | 8 | 7 | 6 | 55 | 367 |
| Lotus japonicusc | 12 | 5 | 8 | 13 | 5 | 9 | 7 | 59 | 472 |
| Medicago truncatulac | 19 | 6 | 8 | 19 | 8 | 8 | 13 | 81 | 375 |
| Oryza sativaa | 34 | 4 | 8 | 7 | 11 | 0 | 36 | 100 | 480 |
| Populus trichocarpad | 50 | 5 | 9 | 13 | 13 | 4 | 10 | 104 | 485 |
| Vitis viniferaa | 12 | 4 | 8 | 16 | 7 | 6 | 6 | 59 | 487 |
| Gossypium raimondiie | 20 | 7 | 16 | 26 | 16 | 13 | 14 | 112 | 737 |
| Gossypium arboreume | 19 | 7 | 16 | 26 | 14 | 13 | 14 | 109 | 1746 |
The WRKYGQK sequence is considered to be important for recognizing and binding to W-box elements (C/TTGACT/C) in the promoter of target genes (Eulgem et al., 2000). Previous studies showed that variation of WRKYGQK sequences was observed in many species (Wu et al., 2005; He et al., 2012; Liu et al., 2014; Song et al., 2014). Besides the most common WRKYGQK sequence, we found seven other heptapeptide variants in GmWRKY, namely, WRKYGKK, WRKYGEK, WRKYGKR, WRKYEDK, WKKYGQK, WRKYGKK, and WHQYGLK (Table S2). WRKYGKK sequence appeared with the highest frequency among them, which belong to subgroup IIc. WRKYGKK sequence is the most common variant not only in soybean, also in Solanum lycopersicum (Huang et al., 2012), L. japonicus (Song et al., 2014), and Brassica oleracea var. capitata (Yao et al., 2015). WRKYGKK sequence in tobacco WRKY could bind specifically to WK-box (TTTTCCAC), which was significantly different from the consensus sequence of W-box (van Verk et al., 2008). Three WRKYGEK sequences were found in GmWRKY5 (group I), GmWRKY67 (group I), and GmWRKY25 (subgroup IIc), respectively. Two WKKYGQK sequences were identified in GmWRKY80 (subgroup IIa) and GmWRKY102 (subgroup IIa), respectively. WRKYGKP, WRKYEDK, and WHQYGLK were identified in GmWRKY91 (group III), GmWRKY148 (group I), and GmWRKY130 (subgroup IIc), respectively (Table S2). WHQYGLK sequence, with the most divergent variation among these seven variants, might execute new biological functions.
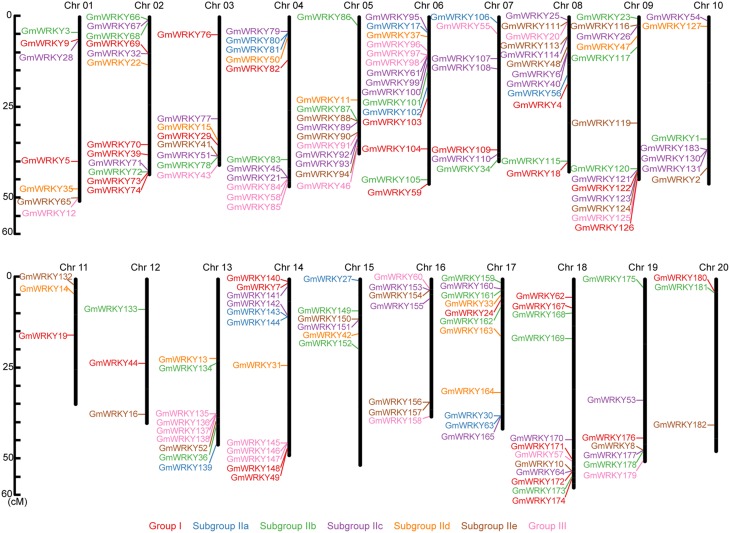
These 176 GmWRKY genes were randomly distributed throughout the 20 soybean chromosomes (Figure 1). There were more GmWRKY genes (15 genes) on chromosome 6, and chromosome 11, 12, and 20 each contained only three GmWRKY genes. We found that more GmWRKY genes were located at both ends of chromosomes (Figure 1). No group I GmWRKY gene was found in chromosome 5, 10, 13, 15, and 16. Group III GmWRKY gene was not detected in chromosome 2, 10, 11, 12, 15, 17, and 20. Group II genes were distributed in 20 chromosomes. Chromosome 10 and 15 contained only group II genes.
Figure 1.
Chromosomal location of GmWRKY genes. The chromosome numbers were shown at the top of each chromosome (black bars). The names on the left side of each chromosome correspond to the approximate location of each WRKY gene.
Publicly available ESTs were considered as a useful source for gene expression study (Ohlrogge and Benning, 2000). A total of 1,468,526 soybean ESTs were downloaded from GenBank. A total of 127 GmWRKY genes were obtained from these ESTs which were generated from leaf, seed, and other tissues (Table S3). These ESTs were sequenced from soybean plants under different stresses. We found that the expression of GmWRKY4, 5, 6, 9, 31, 46, 50, 56, 96, 106, 155, and 160 genes was responsive to water deficit stress, and GmWRKY10, 49, 121, and 155 genes showed altered expression in response to salt stress.
Codon usage bias analysis
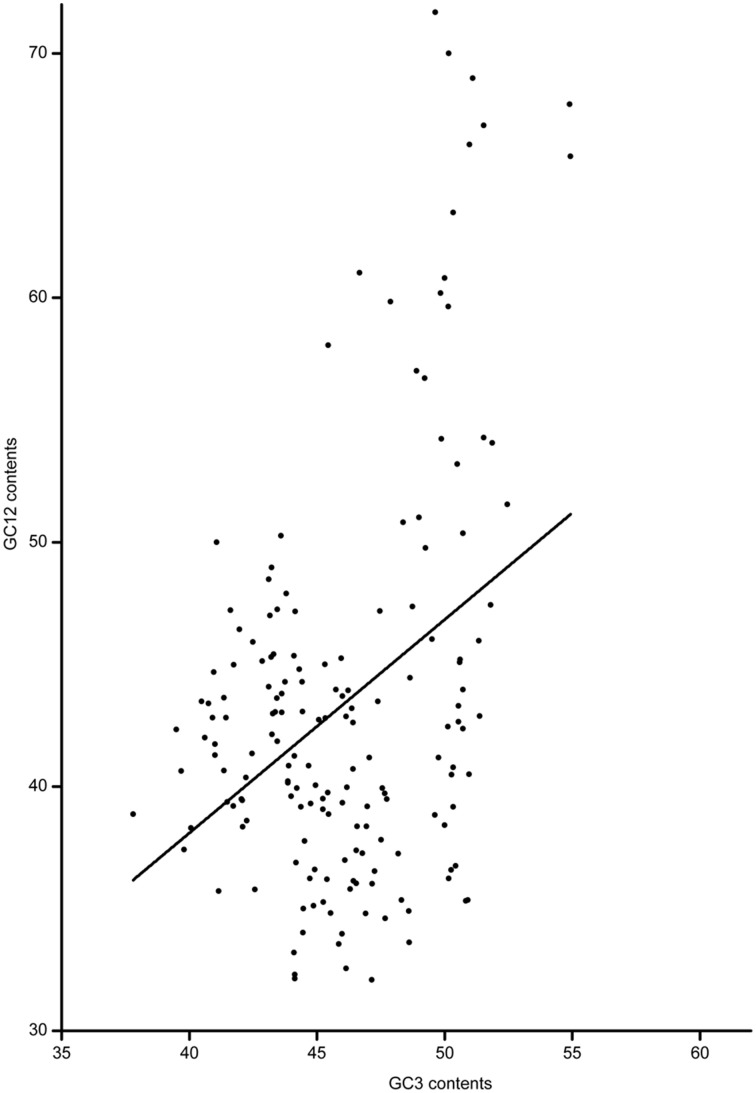
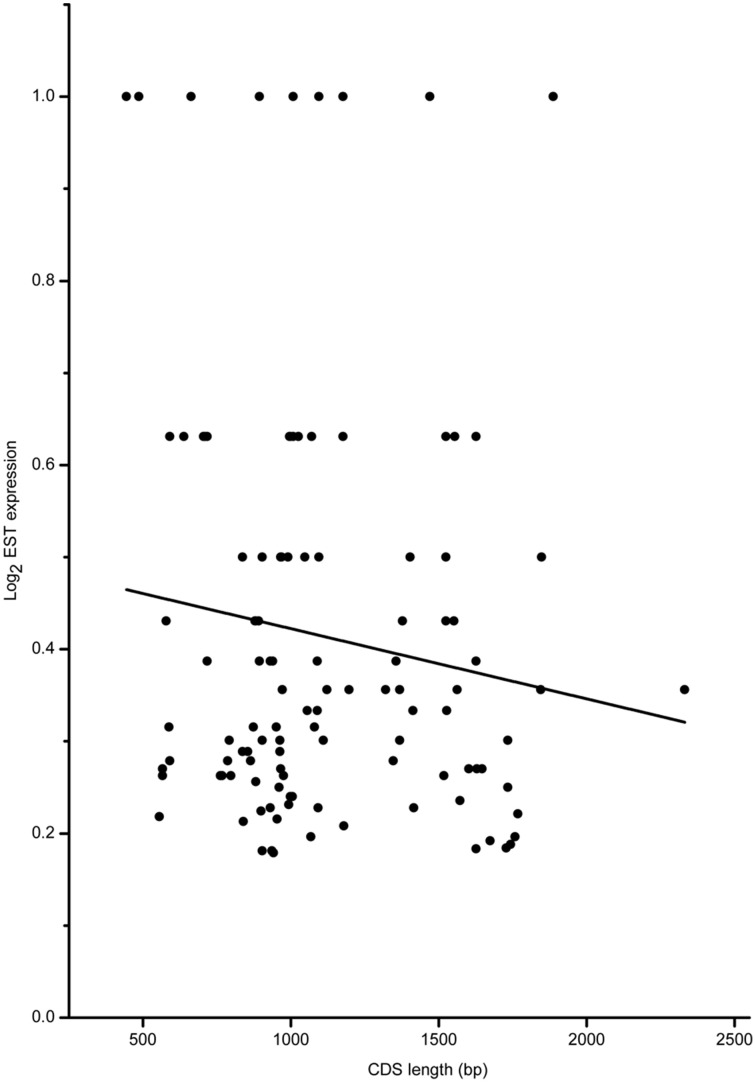
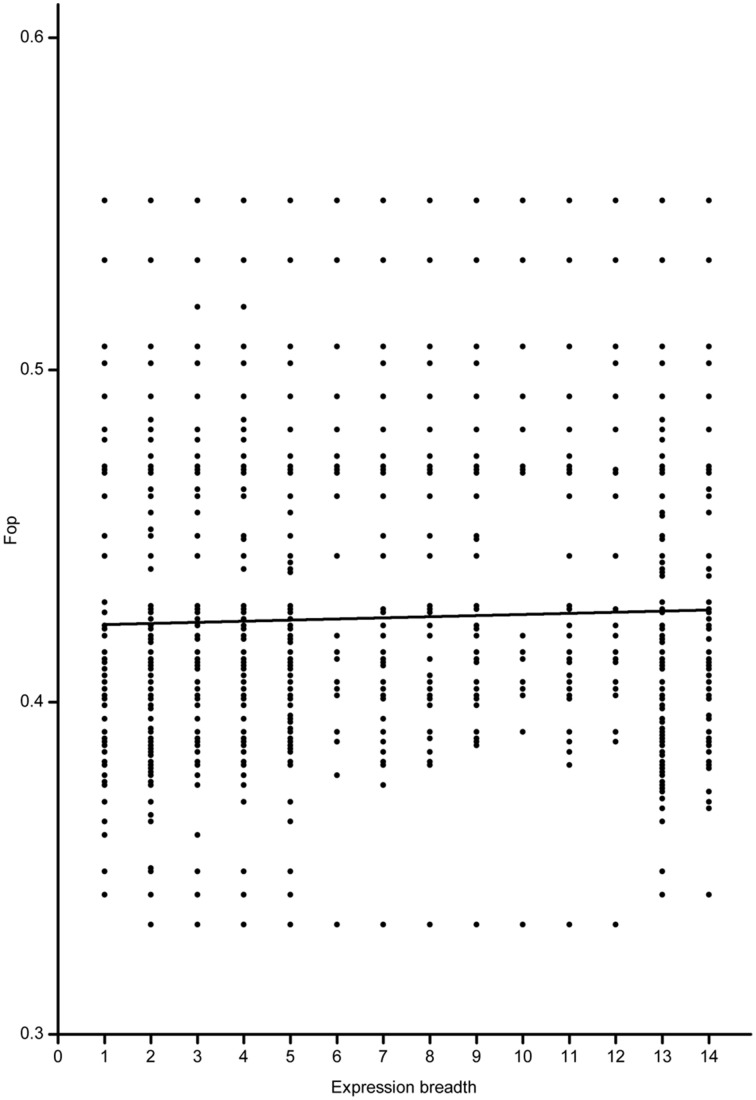
Based on the full-length CDS sequences of 171 GmWRKY genes, GC content in three codon positions was analyzed using the Perl scripts. The GC1 value (48.30) was higher than that of GC2 (43.80) and GC3 (43.37). The average GC content of all codons was 45.15. The AT content (54.85) was higher than GC content in GmWRKY genes. Neutrality plots (GC12 vs. GC3) were used to analyze the relationship among three codon positions. We detected a positive correlation (P < 0.05) between GC12 and GC3 (Figure 2), indicating GC mutational bias leading to similar GC content in all codon positions. Optimal codons of GmWRKY genes showed a greater preference for a C or G in the third base position (Table S6), while accounting for its lower GC in third base when compared with the AT content. Song et al. (2015) reported that AT content were higher than GC content in MtWRKY genes, but the 3rd position exclusively used G or C in optimal codons. We found a significant negative correlation (P < 0.05) between EST expression data and length of CDS sequences (Figure 3), indicating a tendency of higher level expression for genes with shorter CDS, and lower level expression for longer CDS genes. The correlation between codon bias and expression breadth was significant positive (P < 0.05; Figure 4), indicating that GmWRKY with larger expression breadth showed a high degree of codon usage bias.
Figure 2.
Neutrality plots (GC12 vs. GC3s). The regression line: y = 0.87345X + 3.15405.
Figure 3.
Relationship between gene expression and CDS length. The regression line: y = −0.000000762534X + 0.49866.
Figure 4.
Relationship between fop and expression breadth. The regression line: y = 0.000337875X + 0.42302. The number of 1–14 indicated yongg_leaf, flower, one.cm.pod, pod. shell. 10 DAF, pod. shell. 14 DAF, seed. 10 DAF, seed. 14 DAF, seed. 21 DAF, seed. 25 DAF, seed. 28 DAF, seed. 35 DAF, seed. 42 DAF, root, and nodule, respectively.
RNA-seq analysis of GmWRKY expression
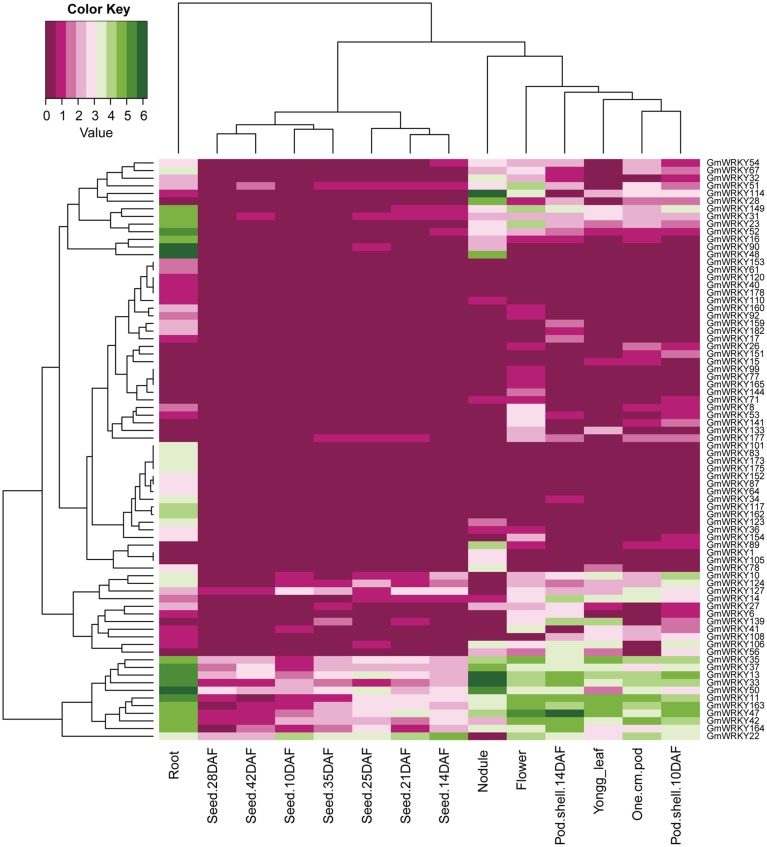
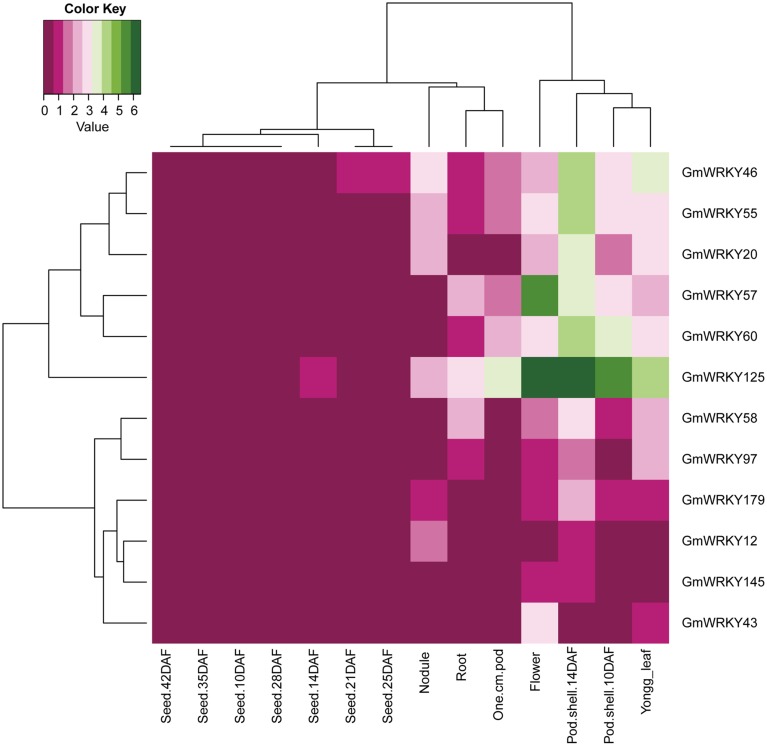
One hundred and three GmWRKY genes were expressed at various developmental stages in leaf, flower, pod, seed, root, and nodule of soybean. Fifteen, 76, and 12 of GmWRKY genes belonged to group I, II, and III, respectively. Group I genes were highly expressed in root, leaf, flower, nodule, and pod. Five group I genes (GmWRKY44, 59, 70, 82, and 103) expressed in all these tissues at various development stages (Figure 5). Group II GmWRKY23, 31, 52, and 149 genes were mainly expressed in leaf, flower, nodule, and pod. GmWRKY11, 13, 33, 35, 37, 42, 47, 50, and 127 genes in group II were expressed in all six tissues at various development stages (Figure 6). Group III GmWRKY genes were mainly expressed in root, leaf, flower, nodule, and pod, for example, GmWRKY46, 55, and 125 genes. However, for an individual group III gene, none of them was detected in all six tissues (Figure 7 and Table S2). Experimental evidence is lacking for the involvement of WRKY gene in floral development or organogenesis. However, most GmWRKY genes were highly expressed in flower, suggesting their roles in floral development.
Figure 5.
Expression pattern of GmWRKY group I genes in different tissues. The Reads/Kb/Million (RPKM) normalized values of expressed genes was log2-transformed. The abbreviation “DAF” in the tissue label indicates “Days after flowering.”
Figure 6.
Expression pattern of GmWRKY group II genes in different tissues. The Reads/Kb/Million (RPKM) normalized values of expressed genes was log2-transformed. The abbreviation “DAF” in the tissue label indicates “Days after flowering.”
Figure 7.
Expression pattern of GmWRKY group III genes in different tissues. The Reads/Kb/Million (RPKM) normalized values of expressed genes was log2-transformed. The abbreviation “DAF” in the tissue label indicates “Days after flowering.”
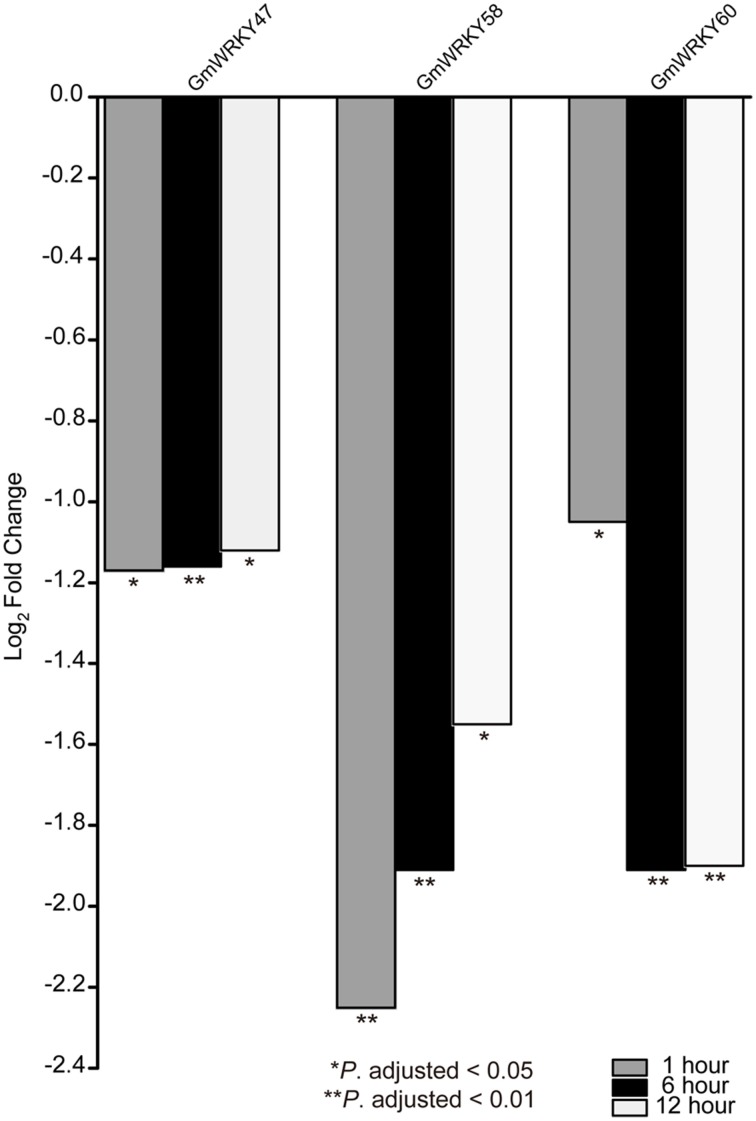
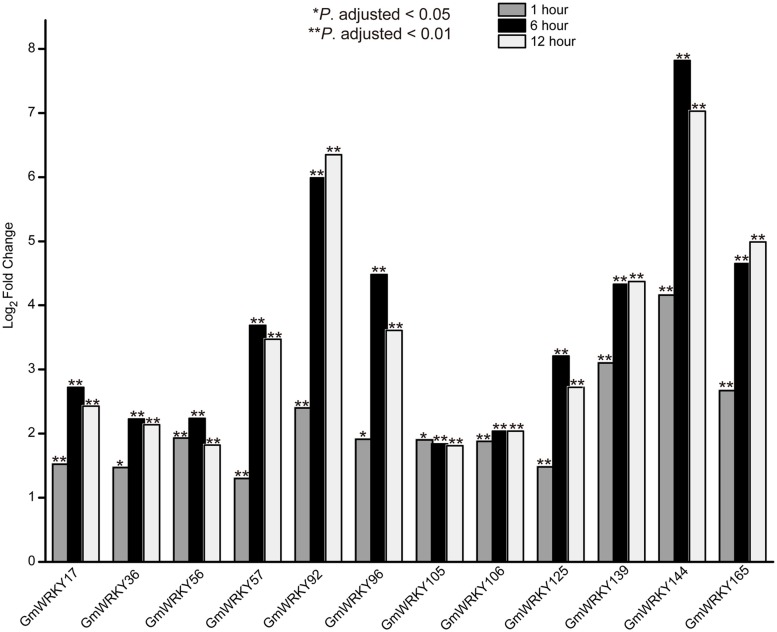
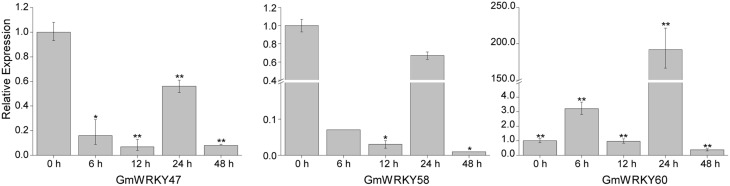
RNA-seq data was used to screen GmWRKY genes that are responsive to dehydration and salt stress. A total number of 31 and 65 GmWRKY genes are considered differentially expressed at least at one of the three time points under dehydration or NaCl treatment, respectively (Tables S4, S5). Dehydration induced down-regulation of most GmWRKY genes except GmWRKY56, 106, 120, and 139 genes at 1 h (Table S4). GmWRKY47, GmWRKY58, and GmWRKY60 genes were differentially expressed at least at one of three time points under dehydration. These genes were significantly down-regulated (Figure 8). NaCl treatment resulted in up-regulation of most GmWRKY genes (Figure 9 and Table S5). Twelve GmWRKY genes were differentially expressed at all three time points under salt (Figure 9). Most of the differentially expressed genes belong to group II (dehydration: 17/31; salt: 45/65), and followed by group III (dehydration: 9/31; salt: 13/65). Less group I genes (dehydration: 5/31; salt: 7/65) were found responsive to salt and dehydration stresses. These results were consistent with results from cotton WRKY that most group II and III GhWRKY genes are highly expressed under stress condition (Dou et al., 2014).
Figure 8.
Different expression of GmWRKY genes in three time point under dehydration stress based on RNA-seq data.
Figure 9.
Different expression of GmWRKY genes in three time point under salt stress based on RNA-seq data.
cis-Acting element analysis
Ninety-five GmWRKY genes are differentially expressed under dehydration and/or salt stresses (Tables S4, S5). Removal of three genes (GmWRKY175, GmWRKY53, and 155) with incomplete sequences or low quality sequences, we extracted 2000 bp promoter regions of 92 GmWRKY genes. Various types of cis-acting elements were detected in the promoter region of 92 WRKY genes, suggesting that the same type of GmWRKY could perform different functions. Most of W-box elements could be distributed in promoters of GmWRKY (Tables S7, S8). Some WRKY genes, contained majority of W-box elements, are auto-regulated by itself and cross-regulated, indicating a self-feedback or mutual manipulation channel might exist among WRKY genes (Chi et al., 2013). AtWRKY18, 40, and 60 genes were reported to be self- and cross-regulated based on W-box elements (Yan et al., 2013). Similarly, OsWRKY24, 53, and 70 genes were predicted self- and cross-regulated according to the presence of W-box clusters in their promoters (Zhang et al., 2015). Based on these reports, we speculated that the presence of W-box elements in GmWRKY gene might have the similar regulatory mode. ABRE and MBS elements that respond to dehydration or salt stress were distributed in promoter region of most GmWRKY genes (Tables S7,S8). These results suggested that GmWRKY genes were transcriptionally regulated upon dehydration and salt stress.
GmWRKY genes expression in response to dehydration and salt stress using qRT-PCR
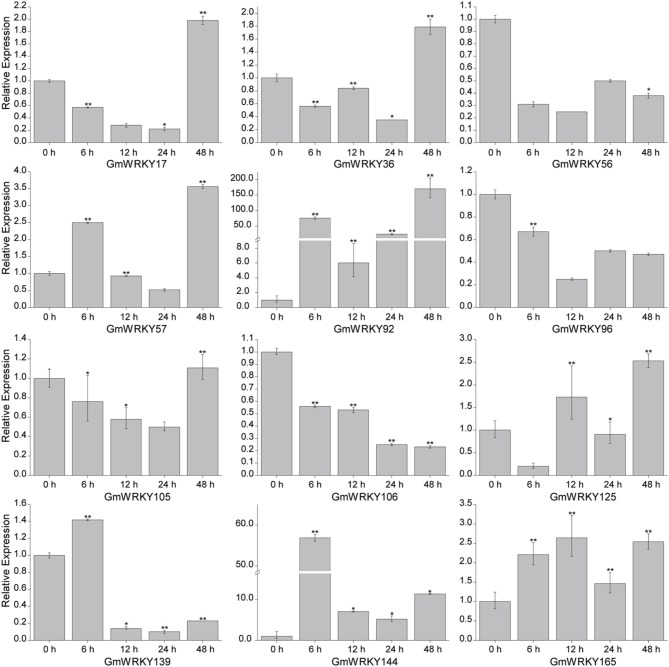
To validate the expression patterns of GmWRKY genes revealed by RNA-seq, 15 GmWRKY genes were selected for expression analysis by qRT-PCR. The RNA-seq results showed that 3 and 12 GmWRKY genes were responsive to dehydration and salt stress at all three time points, respectively. The qRT-PCR results showed the expression of GmWRKY47 and 58 genes was down-regulated at 6, 12, 24, and 48 h under dehydration stress (Figure 10). Expression of GmWRKY60 gene was significantly increased at 6 and 24 h, but significantly suppressed at 48 h under dehydration stress. The results showed that GmWRKY47 and 58 genes were consistent with RNA-seq data while GmWRKY60 gene was not. The expression of GmWRKY92, 144, and 165 genes was enhanced at all time points using qRT-PCR (Figure 11). The results were consistent with the RNA-seq data. Unexpectedly, the expression of remaining nine GmWRKY genes was not consistent with RNA-seq data. For example, the expression of GmWRKY56, 96, and 106 genes was reduced at all four time points (Figure 11), but they showed up-regulation in RNA-seq analysis.
Figure 10.
Expression pattern of selected GmWRKY genes under dehydration stress. The Y-axis indicates the relative expression; X-axis (0, 6, 12, 24, and 48 h) indicates hours of dehydration treatment. The standard errors are plotted using vertical lines. *significant difference at P < 0.05, **significant difference (P < 0.01).
Figure 11.
Expression pattern of selected GmWRKY genes under salt stress. The Y-axis indicates the relative expression; X-axis (0, 6, 12, 24, and 48 h) indicates hours of salt treatment. The standard errors are plotted using vertical lines. *significant difference at P < 0.05, **significant difference (P < 0.01).
Discussion
Identification and characterization of WRKY
In this study, we identified 176 WRKY proteins in soybean Wm82.a2v1 genome. Compared to present study, Zhou et al. (2008) found 64 GmWRKY sequences and Bencke-Malato et al. (2014) identified 149 GmWRKY in Wm82.a1v1 genome. The new assembled Wm82.a2v1 genome constructed using the latest ARACHNE assemble is more accurate. Annotation of eight GmWRKY has been changed in the new assembled genome (Table 2). For example, GmWRKY148, 171, and 172 belonged to subgroup IIc in Wm82.a1v1, but belong to group I in Wm82.a2v1. Moreover, GmWRKY57 and 67 were classified into group I in Wm82.a1v1, but they were in group III and subgroup IIc in Wm82.a2v1, respectively. These results showed that there were differences between Wm82.a1v1 and Wm82.a2v1 genomes. Therefore, it is important to update the global information of GmWRKY in the latest version of soybean genome.
Table 2.
Annotation revised in GmWRKY genes.
| Name | Previous annotation | Present annotation |
|---|---|---|
| GmWRKY10 | III | IIe |
| GmWRKY57 | I | III |
| GmWRKY64 | IIe | IIc |
| GmWRKY67 | I | IIc |
| GmWRKY148 | IIc | I |
| GmWRKY170 | IIb | IIc |
| GmWRKY171 | IIc | I |
| GmWRKY172 | IIc | I |
Compared to other plants, soybean genome contained the highest number of WRKY members. The expansion of WRKY proteins in soybean might be due to the following reasons. (1) soybean genome experienced at least three rounds of whole genome duplication (WGD) events that could produce a large number of paralogs (Conant and Wolfe, 2008). The dicotyledons, such as Arabidopsis, grape, soybean, and M. truncatula, share a general “gamma” genome triplication event about 117 million years ago (Mya; Schmutz et al., 2010). Subsequently, soybean and Medicago experience a common legume-specific WGD event around 59 Mya, and soybean has undergone an additional glycine-specific genome duplication event approximately 13 Mya (Schmutz et al., 2010). (2) The number of duplicated genes was mainly determined by segmental duplication events because genes generated by segmental duplicated events have more chance to be retained (Wang et al., 2005). Tandem duplication events play an important role in generating new duplicated genes (Cannon et al., 2004), whereas segmental duplication events may widely distribute duplicated genes across the genome (Baumgarten et al., 2003). Segmental duplication events could result in lost of many functional redundant genes to avoid fitness cost (Song et al., 2014). Yin et al. (2013) found that GmWRKY genes were generated mainly through segmental duplication events, which may lead to neofunctionalization or subfunctionalization (Moore and Purugganan, 2005). Gene duplication events could improve plant resistance to pathogens by allowing the functional diversification of genes (Moore and Purugganan, 2005). It was reported that 75 GmWRKY genes were involved in response to fungal infection (Bencke-Malato et al., 2014). (3) Positive selection play a key role in preserving duplicated genes, and can act at very early stage of gene duplication process (Moore and Purugganan, 2005). Site model and branch-site model analysis showed that group I, IIc, IIe, and III GmWRKY underwent positive selection (Yin et al., 2013). Positive selection promotes constant expansion of GmWRKY. Similarly, group IIc and III WRKY from eggplant and turkey berry were speculated to undergo positive selection (Yang et al., 2015). In contrast, Group III WRKY from L. japonicus (Song et al., 2014), M. truncatula (Song and Nan, 2014), and Cucumis sativus (Ling et al., 2011) appear to be under purifying selection. Purifying selection may generate genes with conserved functions or pseudogenization (Zhang, 2003). The genome size and the number of WRKY family members are not necessarily correlated. For example, the genome size of soybean is 978 Mb containing 176 WRKY, while the genome size of Gossypium arboretum is 1746 Mb containing 109 WRKY. The genome size of G. arboreum (1746 Mb) was about three times greater than the Populus trichocarpa genome size (485 Mb). These two plants have approximately same number of WRKY (109 vs. 104; Table 1).
GmWRKY expression in different tissues and stress conditions
Dou et al. (2014) reported that most Gossypium hirsutum WRKY genes expressed at low levels in all developmental stages, while a few GhWRKY expressed highly in specific organs. Huang et al. (2012) found that 10 S. lycopersicum WRKY genes were constitutively expressed in nearly all tissues. Our results showed that GmWRKY genes expressed with distinct temporal and spatial patterns. Sixteen GmWRKY genes from group II were expressed in root, flower, or nodule with tissue-specific manner (Figure 6). The expression of a particular GmWRKY gene in a given tissue may differ at different developmental stages. For example, the expression of GmWRKY71 and 154 genes was observed in 10 DAF pod shell, but hardly detected in other developmental stages. GmWRKY54, 62, 125, and 180 genes were expressed in 14 DAF seeds, but not in other seed developmental stages (Figure 6).
Although a little evidence demonstrated the involvement of GmWRKY genes in flower development, many GmWRKY genes were highly expressed in flowers (Figures 5–7). Recently, Luo et al. (2013b) reported that heterologous expression of WRKY20 from Glycine soja in Arabidopsis resulted in earlier flower. GsWRKY20 is orthologous of AtWRKY53, GmWRKY20, 46, and 55. We found that GmWRKY20, 46, and 55 genes were highly expressed in flower (Figure 7). This was consistent with the results from Brassica rapa, where most of BrWRKY highly expressed in flower buds (Kayum et al., 2015). Here, we speculated that these three GmWRKY genes might play roles in flower development.
Stomata and roots were involved in plant responses to dehydration and salt stress (Song et al., 2009; Chen et al., 2012; Belamkar et al., 2014). The RNA-seq data showed that 15 GmWRKY genes were differentially expressed under dehydration and salt stress in root. Zhou et al. (2008) found that GmWRKY54 genes confer tolerance to salt and drought stresses in Arabidopsis, possibly through the regulation of DREB2A and STZ/Zat10 genes. Heterologous expression of GmWRKY13 genes could increase the sensitivity of Arabidopsis to salt tolerance (Zhou et al., 2008). However, we failed to detect the response of these two genes to dehydration and salt stress. The possible explanation might be due to tissue-specific regulation; GmWRKY13 and 54 genes were cloned from leaf, while the RNA-seq data were from root.
Under dehydration stress, GmWRKY47, 58, and 60 genes were considered as differentially expressed genes. Their orthologous genes in Arabidopsis are AtWRKY11, 41, and 70, respectively (Table S2). WRKY11 from Vitis vinifera is orthologous gene of AtWRKY11. Transgenic Arabidopsis expressing VvWRKY11 showed higher tolerance to drought stress, indicating its involvement in response to dehydration stress (Liu et al., 2011). Overexpression of GsWRKY20 (orthologous of AtWRKY 70) in Arabidopsis and Medicago sativa could increase drought tolerance of the transgenic Arabidopsis, and enhance salt and drought tolerance of transgenic Medicago (Luo et al., 2013a; Tang et al., 2014). VvWRKY11 and GsWRKY20 promoted dehydration tolerance. However, our results from qRT-PCR and RNA-seq data showed that GmWRKY47 (AtWRKY11) and 60 (AtWRKY70) genes were negatively regulated by dehydration. AtWRKY41 (orthologous of GmWRKY58) could promote disease resistance (Higashi et al., 2008). It could promote seed dormancy through regulation of ABI3 gene (Ding et al., 2014). We first reported the observation that GmWRKY58 gene was involved in dehydration response.
Twelve GmWRKY genes were differentially expressed under salt stress. Their orthologous genes are AtWRKY6, 30, 40, 50, 51, and 70 in Arabidopsis, respectively (Table S2). The orthologous genes of AtWRKY40 are GmWRKY17, 56, 106, 139, and 144; Both of GmWRKY36 and 105, which are orthologous genes of AtWRKY6; and GmWRKY57 and 125 shared a common orthologous gene, AtWRKY70. AtWRKY6 was identified as target gene of AtWRKY53 (Miao et al., 2004). The expression of AtWRKY53 was up-regulated in Arabidopsis sos2 mutant under salt stress (Kamei et al., 2005), indicating its involvement in salt tolerance. Scarpeci et al. (2013) showed that overexpression of AtWRKY30 enhanced salt tolerance in Arabidopsis during early growth stages. ChIP experiments showed that AtWRKY40 directly targeted a number of known ABA-responsive genes, including ABI4, ABI5, ABF4, MYB2, DREB1A, and RAB18 genes (Shang et al., 2010), indicating that AtWRKY40 could promote salt tolerance. Previous study showed that AtWRKY50 and 51 played crucial roles in jasmonic acid (JA) pathway (Chen et al., 2012), which was a key component in pathogen tolerance. We found the expression of GmWRKY92 (orthologous of AtWRKY51) and 165 (orthologous of AtWRKY50) was up-regulated under 100 mM NaCl treatment (Figure 11), indicating their involvement in multiple stress responses.
There are many opposite results between qRT-PCR and RNA-seq analysis. We speculated that growing condition and seed dormancy time probably caused different expression pattern between qRT-PCR and RNA-seq data. WRKY genes are involved in varies of plant development, including seed dormancy and germination (Zhang et al., 2004; Zentella et al., 2007; Zou et al., 2008). Moreover, some WRKY genes have different functions (Rushton et al., 2010; Tang et al., 2013), indicating the selected GmWRKY genes probably involved in multiple biological pathway.
Conclusions
In this study, we identified 176 GmWRKY proteins in soybean genome (Wm82.a2v1) using bioinformatics approach. There are more WRKY proteins in soybean genome than other plant species. We found that no positive correlation exists between the genome size and the number of WRKY. Expression analysis showed that some GmWRKY genes were involved in response to dehydration and salt stress. Our results will be helpful for understanding the roles of WRKY gene family in soybean.
Author contributions
HS wrote the manuscript and performed the field and laboratory assays. PW performed the phylogentic analysis and reviewed the manuscript. LH, SZ, CZ, HX, and PL performed the RNA-seq analysis. YZ and XB provied help in analysis of qRT-PCR. XW served as the principal investigator, facilitated the project, and assisted in manuscript preparation and revision.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to the providers who submitted the RNA-seq data to the public expression databases. The authors thank Prof. Lei Zhang and Dr. Yongxiang Lin (Anhui Academy of Agricultural Sciences, China) for providing seeds of soybean (Williams 82). The authors also thank Dr. Jer-Young Lin (UCLA) and Dr. Dilin Liu (Guangdong Academy of Agricultural Sciences, China) for providing valuable suggestions and comments. This study was supported by National High Tech Project, NSFC Project (2011BAD35B04, 2013AA102602, 31500217), Taishan Scholar grant (tshw20100416), Young Talents Training Program of Shandong Academy of Agricultural Sciences and Shandong Province Germplasm Innovation and Utilization Project.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00009
qRT-PCR primers in this study.
Annotation of GmWRKY genes. The names GmWRKY1-64 and GmWRKY65-182 genes are given according to Zhou et al. (2008) and Bencke-Malato et al. (2014); GmWRKY183-186 genes are given according to the chromosome order in this study.
Digital expression analysis of GmWRKY genes.
GmWRKY genes differentially expressed under dehydration stress at 1, 6, and 12 h.
GmWRKY genes differentially expressed under salt stress at 1, 6, and 12 h.
Optimal codons analysis in GmWRKY genes. Number of codons in high bias dataset 2265; Number of codons in low bias dataset 3255; High bias was assigned to the dataset with the lower average NEC (effective number of codons); RSCU: relative synonymous codon usage; Codon UUC (Phe) chi value was 37.913; Codon UCC (Ser) chi value was 112.912; Codon UAC (Tyr) chi value was 19.766; Codon UGC (Cys) chi value was 5.112; Codon UCG (Ser) chi value was 22.781; Codon CUC (Leu) chi value was 83.801; Codon CCC (Pro) chi value was 7.139; Codon CAC (His) chi value was 60.096; Codon CGC (Arg) chi value was 17.807; Codon CCG (Pro) chi value was 61.074; Codon CGG (Arg) chi value was 13.621; Codon AUC (Ile) chi value was 9.526; Codon ACC (Thr) chi value was 28.927; Codon AAC (Asn) chi value was 43.255; Codon ACG (Thr) chi value was 23.339; Codon GUC (Val) chi value was 10.427; Codon GCC (Ala) chi value was 41.279; Codon GAC (Asp) chi value was 38.179; Codon GGC (Gly) chi value was 27.092; Codon GUG (Val) chi value was 19.760; Codon GCG (Ala) chi value was 39.804; Codon GAG (Glu) chi value was 13.304; Codon GGG (Gly) chi value was 10.177.
cis-element in differentially expressed genes of dehydration.
cis-element in differentially expressed genes of salt.
Phylogenetic tree of AtWRKY and GmWRKY domain. The phylogenetic tree was constructed using MAGE 6.0 by the Neighbor-Joining (NJ) method with 1000 bootstrap replicates.
References
- Baumgarten A., Cannon S., Spangler R., May G. (2003). Genome-level evolution of resistance genes in Arabidopsis thaliana. Genetics 165, 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belamkar V., Weeks N. T., Bharti A. K., Farmer A. D., Graham M. A., Cannon S. B. (2014). Comprehensive characterization and RNA-Seq profiling of the HD-Zip transcription factor family in soybean (Glycine max) during dehydration and salt stress. BMC Genomics 15:950. 10.1186/1471-2164-15-950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencke-Malato M., Cabreira C., Wiebke-Strohm B., Bücker-Neto L., Mancini E., Osorio M. B., et al. (2014). Genome-wide annotation of the soybean WRKY family and functional characterization of genes involved in response to Phakopsora pachyrhizi infection. BMC Plant Biol. 14:236. 10.1186/s12870-014-0236-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300. [Google Scholar]
- Cannon S. B., Mitra A., Baumgarten A., Young N. D., May G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. 10.1186/1471-2229-4-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Puryear J., Cairney J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11, 113–116. 10.1007/BF02670468 [DOI] [Google Scholar]
- Chen L., Song Y., Li S., Zhang L., Zou C., Yu D. (2012). The role of WRKY transcription factors in plant abiotic stresses. BBA-Gene Regul. Mech. 1819, 120–128. 10.1016/j.bbagrm.2011.09.002 [DOI] [PubMed] [Google Scholar]
- Chi Y., Yang Y., Zhou Y., Zhou J., Fan B., Yu J. Q., et al. (2013). Protein-protein interactions in the regulation if WRKY transcription factors. Mol. Plant 6, 287–300. 10.1093/mp/sst026 [DOI] [PubMed] [Google Scholar]
- Conant G. C., Wolfe K. H. (2008). Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9, 938–950. 10.1038/nrg2482 [DOI] [PubMed] [Google Scholar]
- Ding M., Chen J., Jiang Y., Lin L., Cao Y., Wang M., et al. (2015). Genome-wide investigation and transcriptome analysis of the WRKY gene family in Gossypium. Mol. Genet. Genomics 290, 151–171. 10.1007/s00438-014-0904-7 [DOI] [PubMed] [Google Scholar]
- Ding Z. J., Yan J. Y., Li G. X., Wu Z. C., Zhang S. Q., Zheng S. J. (2014). WRKY41 controls Arabidopsis seed dormancy via direct regulation of ABI3 transcript levels not downstream of ABA. Plant J. 79, 810–823. 10.1111/tpj.12597 [DOI] [PubMed] [Google Scholar]
- Dou L., Zhang X., Pang C., Song M., Wei H., Fan S., et al. (2014). Genome-wide analysis of the WRKY gene family in cotton. Mol. Genet. Genomics 289, 1103–1121. 10.1007/s00438-014-0872-y [DOI] [PubMed] [Google Scholar]
- Eulgem T., Rushton P., Robatzek S., Somssich I. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. 10.1016/S1360-1385(00)01600-9 [DOI] [PubMed] [Google Scholar]
- Eulgem T., Somssich I. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10, 366–371. 10.1016/j.pbi.2007.04.020 [DOI] [PubMed] [Google Scholar]
- Finn R. D., Clements J., Eddy S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. 10.1093/nar/gkr367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W., Karimi M., Wieczorek K., Van de Cappelle E., Wischnitzki E., Grundler F., et al. (2008). A role for AtWRKY23 in feeding site establishment of plant-parasitic nematodes. Plant Physiol. 148, 358–368. 10.1104/pp.108.119131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Dong Q., Shao Y., Jiang H., Zhu S., Cheng B., et al. (2012). Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 31, 1199–1217. 10.1007/s00299-012-1241-0 [DOI] [PubMed] [Google Scholar]
- Higashi K., Ishiga Y., Inagaki Y., Toyoda K., Shiraishi T., Ichinose Y. (2008). Modulation of defense signal transduction by flagellin induced WRKY41 transcription factor in Arabidopsis thaliana. Mol. Genet. Genomics 279, 303–312. 10.1007/s00438-007-0315-0 [DOI] [PubMed] [Google Scholar]
- Huang S., Gao Y., Liu J., Peng X., Niu X., Fei Z., et al. (2012). Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genomics 287, 495–513. 10.1007/s00438-012-0696-6 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Duan Y., Yin J., Ye S., Zhu J., Zhang F., et al. (2014). Genome-wide identification and characterization of the Populus WRKY transcription factor family and analysis of their expression in response to biotic and abiotic stresses. J. Exp. Bot. 65, 6629–6644. 10.1093/jxb/eru381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. J., Liang G., Yu D. Q. (2012). Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol. Plant 5, 1375–1388. 10.1093/mp/sss080 [DOI] [PubMed] [Google Scholar]
- Johnson C., Kolevski B., Smyth D. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14, 1359–1375. 10.1105/tpc.001404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei A., Seki M., Umezawa T., Ishida J., Satou M., Akiyama K., et al. (2005). Analysis of gene expression profiles in Arabidopsis salt overly sensitive mutants sos2-1 and sos3-1. Plant Cell Environ. 28, 1267–1275. 10.1111/j.1365-3040.2005.01363.x [DOI] [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Bio. Evol. 30, 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayum M. A., Jung H. J., Park J. T., Ahmed N. U., Saha G., Yang T. J., et al. (2015). Identification and expression analysis of WRKY family genes under biotic and abiotic stresses in Brassica rapa. Mol. Genet. Genomics 290, 79–95. 10.1007/s00438-014-0898-1 [DOI] [PubMed] [Google Scholar]
- Li H., Gao Y., Xu H., Dai Y., Deng D., Chen J. (2013). ZmWRKY33, a WRKY maize transcription factor conferring enhanced salt stress tolerances in Arabidopsis. Plant Growth Regul. 70, 207–216. 10.1007/s10725-013-9792-9 [DOI] [Google Scholar]
- Libault M., Thibivilliers S., Bilgin D. D., Radwan O., Benitez M., Clough S. J., et al. (2008). Identification of four soybean reference genes for gene expression normalization. Plant Genome 1, 44–54. 10.3835/plantgenome2008.02.0091 [DOI] [Google Scholar]
- Ling J., Jiang W., Zhang Y., Yu H., Mao Z., Gu X., et al. (2011). Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genomics 12:471. 10.1186/1471-2164-12-471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Leib K., Zhao P., Kogel K. H., Langen G. (2014). Phylogenetic analysis of barley WRKY proteins and characterization of HvWRKY1 and-2 as repressors of the pathogen-inducible gene HvGER4c. Mol. Genet. Genomics 289, 1331–1345. 10.1007/s00438-014-0893-6 [DOI] [PubMed] [Google Scholar]
- Liu H., Yang W., Liu D., Han Y., Zhang A., Li S. (2011). Ectopic expression of a grapevine transcription factor VvWRKY11 contributes to osmotic stress tolerance in Arabidopsis. Mol. Biol. Rep. 38, 417–427. 10.1007/s11033-010-0124-0 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene wxpression sata using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Luo M., Dennis E., Berger F., Peacock W., Chaudhury A. (2005). MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 102, 17531–17536. 10.1073/pnas.0508418102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Bai X., Sun X., Zhu D., Liu B. H., Ji W., et al. (2013a). Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling. J. Exp. Bot. 64, 2155–2169. 10.1093/jxb/ert073 [DOI] [PubMed] [Google Scholar]
- Luo X., Sun X., Liu B., Zhu D., Bai X., Cai H., et al. (2013b). Ectopic expression of a WRKY homolog from Glycine soja alters flowering time in Arabidopsis. PLoS ONE 8:e73295. 10.1371/journal.pone.0073295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Laun T., Zimmermann P., Zentgraf U. (2004). Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 55, 853–867. 10.1007/s11103-005-2142-1 [DOI] [PubMed] [Google Scholar]
- Moore R. C., Purugganan M. D. (2005). The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 8, 122–128. 10.1016/j.pbi.2004.12.001 [DOI] [PubMed] [Google Scholar]
- Ohlrogge J., Benning C. (2000). Unraveling plant metabolism by EST analysis. Curr. Opin. Plant Biol. 3, 224–228. 10.1016/S1369-5266(00)00068-6 [DOI] [PubMed] [Google Scholar]
- Ren X. Z., Chen Z. Z., Liu Y., Zhang H. R., Zhang M., Liu Q. A., et al. (2010). ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 63, 417–429. 10.1111/j.1365-313X.2010.04248.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J. L., Heard J., Martin G., Reuber L., Jiang C. Z., Keddie J., et al. (2000). Arabidopsis transcription factors:genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. 10.1126/science.290.5499.2105 [DOI] [PubMed] [Google Scholar]
- Rinerson C. I., Rabara R. C., Tripathi Q. J., Shen P. J., Rushton P. J. (2015). The evolution of WRKY transcription factors. BMC Plant Biol. 15:66. 10.1186/s12870-015-0456-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S., Somssich I. (2002). Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 16, 1139–1149. 10.1101/gad.222702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton D. L., Tripathi P., Rabara R. C., Lin J., Ringler P., Boken A. K., et al. (2012). WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol. J. 10, 2–11. 10.1111/j.1467-7652.2011.00634.x [DOI] [PubMed] [Google Scholar]
- Rushton P., Somssich I., Ringler P., Shen Q. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. 10.1016/j.tplants.2010.02.006 [DOI] [PubMed] [Google Scholar]
- Scarpeci T. E., Zanor M. I., Mueller-Roeber B., Valle E. M. (2013). Overexpression of AtWRKY30 enhances abiotic stress tolerance during early growth stages in Arabidopsis thaliana. Plant Mol. Biol. 83, 265–277. 10.1007/s11103-013-0090-8 [DOI] [PubMed] [Google Scholar]
- Schmutz J., Cannon S. B., Schlueter J., Ma J., Mitros T., Nelson W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. 10.1038/nature08670 [DOI] [PubMed] [Google Scholar]
- Severin A. J., Woody J. L., Bolon Y. T., Joseph B., Diers B. W., Farmer A. D., et al. (2010). RNA-Seq atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol. 10:160. 10.1186/1471-2229-10-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Yan L., Liu Z. Q., Cao Z., Mei C., Xin Q., et al. (2010). The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to telieve ABA-responsive genes of inhibition. Plant Cell 22, 1909–1935. 10.1105/tpc.110.073874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbe M., Qu N., Galis I., Baldwin I. (2008). Induced plant defenses in the natural environment: Nicotiana attenuata WRKY3 and WRKY6 coordinate responses to herbivory. Plant Cell 20, 1984–2000. 10.1105/tpc.108.058594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Nan Z. (2014). Genome-wide indentification and analysis of WRKY transcription factors in Medicago truncatula (In Chinese). Hereditas (Beijing) 36, 152–168. 10.3724/SP.J.1005.2014.00152 [DOI] [PubMed] [Google Scholar]
- Song H., Wang P., Ma D. C., Xia H., Zhao C. Z., Zhang Y., et al. (2015). Analysis of codon usage bias of WRKY transcription factors in Medicago truncatula (In Chinese). J. Agric. Biotechnol. 23, 203–212. 10.3969/j.issn.1674-7968.2015.02.007 [DOI] [Google Scholar]
- Song H., Wang P., Nan Z., Wang X. (2014). The WRKY transcription factor genes in Lotus japonicus. Int. J. Genomics 2014:420128. 10.1155/2014/420128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Jing S. J., Yu D. Q. (2009). Overexpression of the stress-induced OsWRKY08 improves osmotic stress tolerance in Arabidopsis. Chin. Sci. Bull. 54, 4671–4678. 10.1007/s11434-009-0710-5 [DOI] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Bio. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Wang F., Wang Z., Huang Z., Xiong A., Hou X. (2013). Characterization and co-expression analysis of WRKY orthologs involved in responses to multiple abiotic stresses in Pak-choi (Brassica campestris ssp. chinensis). BMC Plant Biol 13:188. 10.1186/1471-2229-13-188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L., Cai H., Zhai H., Luo X., Wang Z., Cui L., et al. (2014). Overexpression of Glycine soja WRKY20 enhances both drought and salt tolerance in transgenic alfalfa (Medicago sativa L.). Plant Cell Tiss. Org. 115, 77–86. 10.1007/s11240-014-0463-y [DOI] [Google Scholar]
- Tripathi P., Rabara R., Langum T., Boken A., Rushton D., Boomsma D., et al. (2012). The WRKY transcription factor family in Brachypodium distachyon. BMC Genomics 13:270. 10.1186/1471-2164-13-270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Verk M. C., Pappaioannou D., Neeleman L. (2008). A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 146, 1983–1995. 10.1104/pp.107.112789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Deng P., Chen L., Wang X., Ma H., Hu W., et al. (2013). A wheat WRKY transcription factor TaWRKY10 confers tolerance to multiple abiotic stresses in transgenic tobacco. PLoS ONE 8:e65120. 10.1371/journal.pone.0065120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhu W., Fang L., Sun X., Su L., Liang Z., et al. (2014). Genome-wide identification of WRKY family genes and their response to cold stress in Vitis vinifera. BMC Plant Biol. 14:103. 10.1186/1471-2229-4-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Shi X., Hao B., Ge S., Luo J. (2005). Duplication and DNA segmental loss in the rice genome: implications for diploidization. New Phytol. 165, 937–946. 10.1111/j.1469-8137.2004.01293.x [DOI] [PubMed] [Google Scholar]
- Wei K. F., Chen J., Chen Y. F., Wu L. J., Xie D. X. (2012). Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 19, 153–164. 10.1093/dnares/dsr048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen F., Zhu H., Li P., Jiang M., Mao W., Ong C., et al. (2014). Genome-Wide evolutionary characterization and expression analyses of WRKY family genes in Brachypodium distachyon. DNA Res. 21, 327–339. 10.1093/dnares/dst060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Guo Z., Wang H., Li J. (2005). The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 12, 9–26. 10.1093/dnares/12.1.9 [DOI] [PubMed] [Google Scholar]
- Wu X. L., Shiroto Y., Kishitani S., Ito Y., Toriyama K. (2009). Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 28, 21–30. 10.1007/s00299-008-0614-x [DOI] [PubMed] [Google Scholar]
- Yan L., Liu Z. Q., Xu Y. H., Lu K., Wang X. F., Zhang D. P. (2013). Auto- and cross-repression of three Arabidopsis WRKY transcription factors WRKY18, WRKY40, and WRKY60 negatively involved in ABA signaling. J. Plant Growth Regul. 3, 399–416. 10.1007/s00344-012-9310-8 [DOI] [Google Scholar]
- Yang X., Deng C., Zhang Y., Cheng Y., Huo Q., Xue L. (2015). The WRKY transcription factor genes in eggplant (Solanum melongena L.) and turkey berry (Solanum torvum Sw.). Int. J. Mol. Sci. 16, 7608–7626. 10.3390/ijms16047608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Q. Y., Xia E. H., Liu F. H., Gao L. Z. (2015). Genome-wide identification and comparative expression analysis reveal a rapid expansion and functional divergence of duplicated genes in the WRKY gene family of cabbage, Brassica oleracea var. capitata. Gene 557, 35–42. 10.1016/j.gene.2014.12.005 [DOI] [PubMed] [Google Scholar]
- Yin G., Xu H., Xiao S., Qin Y., Li Y., Yan Y., et al. (2013). The large soybean (Glycine max) WRKY TF family expanded by segmental duplication events and subsequent divergent selection among subgroups. BMC Plant Biol. 13:148. 10.1186/1471-2229-13-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R., Zhang Z., Park M., Thomas S., Endo A., Murase K., et al. (2007). Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19, 3037–3057. 10.1105/tpc.107.054999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Z. (2003). Evolution by gene duplication: an update. Trends Ecol. Evol. 18, 292–298. 10.1016/S0169-5347(03)00033-8 [DOI] [Google Scholar]
- Zhang L., Gu L., Ringler P., Smith S., Rushton P. J., Shen Q. (2015). Three WRKY transcription factors additively repress abscisic acid and gibberellin signaling in aleurone cells. Plant Sci. 236, 214–222. 10.1016/j.plantsci.2015.04.014 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wang L. (2005). The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 5:1. 10.1186/1471-2148-5-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Xie Z., Zou X., Casaretto J., Ho T., Shen Q. (2004). A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol. 134, 1500–1513. 10.1104/pp.103.034967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Tian A., Zou H., Xie Z., Lei G., Huang J., et al. (2008). Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stress in transgenic Arabidopsis plants. Plant Biotechnol. J. 6, 486–503. 10.1111/j.1467-7652.2008.00336.x [DOI] [PubMed] [Google Scholar]
- Zou C. S., Jiang W. B., Yu D. Q. (2010). Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in Arabidopsis. J. Exp. Bot. 61, 3901–3914. 10.1093/jxb/erq204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Neuman D., Shen Q. (2008). Interactions of two transcriptional repressors and two transcriptional activators in modulating gibberellin signaling in aleurone cells. Plant Physiol. 148, 176–186. 10.1104/pp.108.123653 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
qRT-PCR primers in this study.
Annotation of GmWRKY genes. The names GmWRKY1-64 and GmWRKY65-182 genes are given according to Zhou et al. (2008) and Bencke-Malato et al. (2014); GmWRKY183-186 genes are given according to the chromosome order in this study.
Digital expression analysis of GmWRKY genes.
GmWRKY genes differentially expressed under dehydration stress at 1, 6, and 12 h.
GmWRKY genes differentially expressed under salt stress at 1, 6, and 12 h.
Optimal codons analysis in GmWRKY genes. Number of codons in high bias dataset 2265; Number of codons in low bias dataset 3255; High bias was assigned to the dataset with the lower average NEC (effective number of codons); RSCU: relative synonymous codon usage; Codon UUC (Phe) chi value was 37.913; Codon UCC (Ser) chi value was 112.912; Codon UAC (Tyr) chi value was 19.766; Codon UGC (Cys) chi value was 5.112; Codon UCG (Ser) chi value was 22.781; Codon CUC (Leu) chi value was 83.801; Codon CCC (Pro) chi value was 7.139; Codon CAC (His) chi value was 60.096; Codon CGC (Arg) chi value was 17.807; Codon CCG (Pro) chi value was 61.074; Codon CGG (Arg) chi value was 13.621; Codon AUC (Ile) chi value was 9.526; Codon ACC (Thr) chi value was 28.927; Codon AAC (Asn) chi value was 43.255; Codon ACG (Thr) chi value was 23.339; Codon GUC (Val) chi value was 10.427; Codon GCC (Ala) chi value was 41.279; Codon GAC (Asp) chi value was 38.179; Codon GGC (Gly) chi value was 27.092; Codon GUG (Val) chi value was 19.760; Codon GCG (Ala) chi value was 39.804; Codon GAG (Glu) chi value was 13.304; Codon GGG (Gly) chi value was 10.177.
cis-element in differentially expressed genes of dehydration.
cis-element in differentially expressed genes of salt.
Phylogenetic tree of AtWRKY and GmWRKY domain. The phylogenetic tree was constructed using MAGE 6.0 by the Neighbor-Joining (NJ) method with 1000 bootstrap replicates.