Abstract
Although genetically modified (GM) plants have improved commercially important traits, such as biomass and biofuel production, digestibility, bioremediation, ornamental value, and tolerance to biotic and abiotic stresses, there remain economic, political, or social concerns over potential ecological effects of transgene flow from GM plants. The current solution for preventing transgene flow from GM plants is genetically engineering sterility; however, approaches to generating both male and female sterility are limited. In addition, existing strategies for creating sterility lead to loss or modifications of entire flowers or floral organs. Here, we demonstrate that instead of the 1.5-kb promoter, the entire SOLO DANCERS (SDS) gene is required for its meiocyte-specific expression. We then developed an efficient method to specifically ablate microspore and megaspore mother cells using the SDS and BARNASE fusion gene, which resulted in complete sterility in both male and female reproductive organs in Arabidopsis (Arabidopsis thaliana) and tobacco (Nicotiana tabacum), but did not affect plant growth or development, including the formation of all flower organs. Therefore, our research provides a general and effective tool to prevent transgene flow in GM plants.
Keywords: completely both male and female sterile plants, flower structure, genetic ablation, gene flow, microspore and megaspore mother cells, SOLO DANCERS
Introduction
Since genetically modified (GM) plants were produced in 1983 (Caplan et al., 1983; Murai et al., 1983), the number of GM plants has been rapidly increasing yearly (Husken et al., 2010). GM trees, turf grasses, biofuel and forage crops, and ornamentals have improved commercially important traits, including biomass and biofuel production, digestibility, bioremediation, ornamental value, and tolerance to biotic and abiotic stresses (Wang and Ge, 2006; Groover, 2007; Harfouche et al., 2011; Xu et al., 2011; Wang and Brummer, 2012; Wilkerson et al., 2014); however, the approval for commercialization of GM plants is subject to complicated and stringent government regulations due to economic, political, or social concerns over potential ecological effects of transgene flow and floral-modified plantations (Goeschl and Swanson, 2003; Hills et al., 2007; Van Acker et al., 2007; Strauss et al., 2009; Lombardo, 2014).
Transgene flow from GM plants to non-GM plants and wild populations is mainly mediated by dispersal of pollen and seeds. Early studies found that the pollen-mediated gene flew from GM Roundup Ready creeping bentgrass (Agrostis stolonifera) occurred within 2 to 21 km (Watrud et al., 2004). The non-GM rabbitfoot grass (Polypogon monspeliensis) could pollinate the GM creeping bentgrass to produce transgenic intergeneric hybrid offspring, suggesting that the transgene escape is also mediated by the female part of GM plants (Zapiola and Mallory-Smith, 2012). Long distance pollen-mediated gene flow occurred between weed beets (Beta vulgaris) as far as 9.6 km and the resulting interfield gene flow was unavoidable (Fenart et al., 2007). Pollen migration from poplars (Populus trichocarpa) often went beyond 10 km (Slavov et al., 2009; DiFazio et al., 2012), indicating that similar issues happened in GM trees. Moreover, gene flow from GM crops to native populations was detected in maize (Zea mays), soybean (Glycine max), wheat (Triticum aestivum), and canola (Brassica napus; Pineyro-Nelson et al., 2009; Liu et al., 2010b; Rieben et al., 2011; Wang and Li, 2012). To overcome regulatory hurdles to field research and, ultimately, commercial uses of GM plants, a practical solution is to create sterile plants by ablating floral organs/tissues using toxic genes under control of specific promoters or by altering flowering time and floral organs via manipulating genes critical for flower development.
Strategies on making male sterility have been extensively and successfully employed to prevent the pollen-mediated transgene flow. In the male reproduction organ anther, tapetum is a layer of nutritive cells, which is required for pollen development. Therefore, genetic ablation of tapetal cells by tapetum-specific promoters driven toxic genes, such as ribonuclease BARNASE and diphtheria toxin fragment A (DTA) genes, is commonly used to create male sterility in various plants. The widely used tobacco tapetum promoter TA29 was first employed to drive BARNASE to create male sterile tobacco and oilseed rape (Brassica napus) plants (Mariani et al., 1990). TA29::DTA tobacco transgenic plants are also male sterile (Koltunow et al., 1990). Since then, various male sterile plants were achieved using other tapetum or anther-specific promoters, including A9, A6, E1, T72, PS1, and PsEND1 in Arabidopsis, rapeseed (Brassica napus), rice (Oryza sativa), and pea (Pisum sativum) plants (Paul et al., 1992; Hird et al., 1993; Zhan et al., 1996; DeBlock et al., 1997; Roque et al., 2007). This strategy was also applied to perennial grasses and trees. The TAP::BARNASE creeping bentgrass is completely pollen sterile (Luo et al., 2005). PrMC2, a pine male cone-specific gene, was successfully used to generate male sterile pine (Pinus radiata) and Eucalyptus (sp.) plants by driving a modified BARNASE gene (Zhang et al., 2012). It was recently reported that the TA29::BARNASE transgenic poplar constantly showed robust male sterility during a 4-year field trial (Elorriaga et al., 2014). Attempts were also made to abolish male and female fertility together. In Arabidopsis, BARNASE driven by the second intron of AGAMOUS resulted in ablation of stamens and carpels (Liu and Liu, 2008). Male and female sterile tobacco plants were generated by expressing BARNASE under control of both the tapetum promoter p108 and the transmitting tract promoter sp41 (Gardner et al., 2009).
In addition, manipulating genes regulating flowering time, floral meristem identify, floral organ identity, and floral organ establishment is used to abolish plant fertility. Silencing the tobacco LEAFY genes NFL1 and NFL2 resulted in plants without flowers (An et al., 2011). Tomato (Lycopersicon lycopersicum) AGAMOUS RNAi lines showed “fruit-in-fruit” phenotype, but did not produce seeds (Pan et al., 2010). Down-regulation of APETALA3 genes OsMADS16 and MtNMH7 caused stamen to carpel transformation and male sterility in rice and Medicago truncatula, respectively (Xiao et al., 2003; Roque et al., 2013). Expression of TFL1, a strong floral repressing gene, led to the non-flowering phenotype in red fescue (Festuca rubra; Jensen et al., 2004). Moreover, overexpression of miR156 inhibited flowering in switchgrass (Panicum virgatum; Fu et al., 2012). Besides generating sterile plants, plastid transformation is also an excellent approach to prevent pollen-mediated transgene flow, since plastids, including chloroplasts, are maternally inherited in most plants (Ruf et al., 2007; Wani et al., 2010).
Although male sterility has been successfully achieved via different approaches in various plant species, it cannot completely prevent transgene flow. Seed development in male sterile GM plants can be rescued by the long-distance transfer of pollen from non-GM plants. The same is also true for female sterile GM plants which disperse pollen to non-GM or male sterile GM plants. Thus, completely abolishing both male and female fertility is the only fail-safe way to prevent transgene flow (Stewart, 2007). So far, approaches to generating complete both male and female sterility are limited. Moreover, existing strategies for creating male and/or female sterility lead to loss or modifications of entire flowers or floral organs (Xiao et al., 2003; Roque et al., 2007; Pan et al., 2010; An et al., 2011), which may cause potential ecological effects on biodiversity of species associated with flowers, such as insects. In economically interesting species, for example ornamentals, altered flowers may also be undesirable. Furthermore, since the remaining toxicity of BARNASE or DTA in non-target organs due to the non-specific basal activities of employed promoters often inhibits plant survival and growth (Lannanpaa et al., 2005; Wei et al., 2007), it is difficult to obtain usable sterile plants that have normal biomass and yield. Therefore, it is imperative to generate sterility in both male and female reproductive organs without affecting plant growth or modifying flower structure.
In Arabidopsis, the SOLO DANCERS (SDS) gene, which encodes a meiosis-specific cyclin, is required for homolog interaction during meiotic prophase I in Arabidopsis (Azumi et al., 2002). With normal growth and development, the sds mutant is both male and female sterile. RNA in situ hybridization analysis showed that SDS transcripts were specifically present in microspore mother cells (male meiocytes) in anthers and megaspore mother cells (female meiocytes) in ovules (Azumi et al., 2002). Here, we report our new approach to create complete both male and female sterility in Arabidopsis and tobacco by specifically ablating microspore and megaspore mother cells using the SDS and BARNASE fusion gene. Our research provides a general and effective tool to prevent transgene flow in GM plants.
Materials and Methods
Plant Materials and Growth Condition
Arabidopsis thaliana Landsberg erecta (Ler) and tobacco (Nicotiana tabacum Petit Havana SR1) were used in this study. Plants were grown in Metro-Mix 360 soil (Sun-Gro Horticulture, Agawam, MA, USA) in a growth chamber under a 16-h light/8-h dark photoperiod regime at 22°C and 50% of humidity.
Generation of Constructs and Transgenic Plants
PCR reactions (see all primers in Supplementary Table S1) were performed using Phusion High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA, USA). The 1.5-kb promoter of the SDS gene (upstream of the SDS coding region and the 3′ non-coding region of the SDS adjacent gene) was amplified and cloned into the pENTR/D-TOPO vector (Invitrogen, Grand Island, NY, USA) to generate pENTR-SDS. The SDS genomic fragment from the beginning of the 1.5-kb promoter region to the last exon was introduced into the pENTR/D-TOPO vector to generate pENTR-SDS::SDS. The BARSTAR gene amplified from the pABGCZ vector (Zhang et al., 2012) was introduced to the pEarleyGate303 vector at the Nsi site to generate pEarleyGate303-BARSTAR. An XhoI site was introduced between BglII and XbaI sites right after attR2 to generate pEarleyGate303-BARSTAR(XhoI). The BARNASE fragment amplified from pABGCZ was cloned into pEarleyGate303-BARSTAR(XhoI) using the XhoI and XbaI sites to generate pEarleyGate303-BARSTAR-BARNASE. Using the Gateway LR recombinase II enzyme mix (Invitrogen, Grand Island, NY, USA), SDS::GUS, SDS::BARNASE, SDS::SDS-GFP, and SDS::SDS-BARNASE binary vectors were generated between pENTR-SDS and pGBW3-GUS, pENTR-SDS and pEarleyGate303-BARSTAR-BARNASE, pENTR-SDS::SDS and pGBW4-GFP, as well as pENTR-SDS::SDS and pEarleyGate303-BARSTAR-BARNASE, respectively.
The floral dip method was used to generate Arabidopsis transgenic plants (Clough and Bent, 1998). Transformants of SDS::GUS and SDS::SDS-GFP were screened on 50 μg/mL of kanamycin and 25 μg/mL of hygromycin. Transformants of SDS::BARNASE and SDS::SDS-BARNASE were screened on 1% of Basta (PlantMedia, Lubbock, TX, USA).
Tobacco transformation was performed as described previously (Curtis et al., 1995). Briefly, leaf disks were inoculated with the Agrobacterium strain GV3101 containing the SDS::SDS-BARNASE binary vector and cultured for 1 day in the dark, followed by 2 days under light. Then, leaf disks were screened on shoot and root selection medium containing 4% of Basta. The regenerated seedlings were transferred into soil and sprayed with 4% of Basta solution one week later. The surviving plants were used for further analyses.
Pollen Staining and Anther Semi-Thin Sections
To examine pollen viability in Arabidopsis plants, Alexander pollen staining was carried out as described previously (Zhao et al., 2002). Briefly, main inflorescences were collected when 1–4 flower(s) were opened. Inflorescences were fixed for 24 h in the fixative containing methanol, 60 mL; chloroform, 30 mL; distilled water, 20 mL; picric acid, 1 g; and HgCl2, 1 g. After transferring through 70, 50, and 30% ethanol series (30 min in each change), inflorescences were finally incubated with water. Inflorescences were them transferred into the staining solution (ethanol 95%, 10 ml; malachite green, 10 mg; acid fuchsin, 50 mg; orange G, 5 mg; phenol, 5 g; glacial acetic acid, 2 ml; glycerol, 25 ml; and distilled water 50 ml) and kept at 50°C for 48 h. Individual anthers were dissected out from flowers and then mounted on the glass slides together with the staining solution for observation. Mature anthers from tobacco plants were collected and analyzed using the same method. Pollen grains were released from anthers before imaging.
Semi-thin sectioning was performed as described in our previous studies (Zhao et al., 2002; Jia et al., 2008). Briefly, dissected floral buds were fixed in 2.5% (vol/vol) glutaraldehyde in 0.1 M HEPES (N-2-Hydroxyethyl piperazine-N_-2-ethanesulfonic acid) buffer (pH 7.2) and 0.02% TritonX-100 overnight at room temperature. Samples were washed three times for 15 min each in 0.1 M HEPES buffer with 0.02% Triton X-100 and then fixed in 1% OsO4 overnight at room temperature. Samples were then dehydrated in a graded acetone series (10% increments) for 60 min each. Infiltration started with 20% Spurr’s resin and then 40, 60, and 80% Spurr’s resin every 3 h. Samples were transferred to 100% Spurr’s resin three times for 24 hours each. Samples were finally embedded in 100% Spurr’s resin and polymerized at 60°C overnight. Semi-thin (0.5 μm) sections were made using an Ultracut E ultramicrotome (Reichert–Jung) and were stained with 0.25% Toluidine Blue O.
GUS Staining Assay
Histochemical GUS staining assay was performed as previously described (Liu et al., 2010a).
Briefly, tissues were collected and fixed for 1 h in 90% acetone at –20°C. After washing tissues in washing buffer [0.1 M phosphate (pH 7.0), 10 mM EDTA, and 2 mM K3Fe(CN)6] twice for 5 min under the vacuum, the drained tissues were transferred into the GUS staining buffer [0.1 M phosphate (pH 7.0), 10 mM EDTA, 1 mM K3Fe(CN)6, 1 mM K4Fe(CN)6⋅3H2O, and 1 mg/ml X-GLUC)] and incubated overnight at 37°C. GUS-stained tissues were then fixed in a 3:1 mixture of ethanol and acetic acid. Tissues were mounted onto the glass slides for observation.
Real-Time qRT-PCR
Inflorescences of wild-type (WT) and SDS::SDS-BARNASE independent Arabidopsis transgenic plants were collected for RNA isolation using the RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA). After determining the RNA quantification by the NanoDrop 2000c (Thermo Scientific, Bannockburn, IL, USA), RNA reverse transcription was conducted using the QuantiTect Reverse Transcription Kit (Qiagen, Valencia, CA, USA). Real-time PCR (DNA Engine Opticon 2 system, Hercules, CA, USA) and data analysis were performed as previously described (Liu et al., 2010a) to evaluate expressions of A9, ATA7, DMC1, and SWI1 (Supplementary Table S1). The ACTIN2 gene was used as an internal control. Three independent biological repeats were carried out.
Microscopy
Pollen staining samples, GUS staining and semi-thin sections were observed and imaged with Olympus SZX7 and BX51 microscopes (Olympus, Center Valley, PA, USA), respectively. Images were obtained with an Olympus DP 70 digital camera (Olympus, Center Valley, PA, USA). For confocal microscopy analysis, anthers and ovules were dissected and mounted in water. The GFP signal was observed with a Leica TCS SP2 laser scanning confocal microscope (Leica, Buffalo Grove, IL, USA) using a 63×/1.4 water immersion objective lens. The 488-nm laser line was used to excite GFP and it also induced chlorophyll autofluorescence. The PMT gain settings was held at 650. GFP and chlorophyll autofluorescence were detected at 505–530 nm and 644–719 nm, respectively.
Results
BARNASE Driven by the 1.5-kb Promoter of the SDS Gene Caused Various Defects in Growth and Reproduction
To create completely both male and female sterile plants without altering flower structure, we first generated the SDS::BARNASE construct using the 1.5-kb promoter of the SDS gene and a modified BARNASE (Zhang et al., 2012) to genetically ablate microspore and megaspore mother cells in Arabidopsis (Figure 1A). Among 66 examined SDS::BARNASE transgenic plants, none of them showed the specific phenotype in sterility. Instead, compared with the wild type (Figure 2A), SDS::BARNASE young plants were defective in vegetative growth, indicated by abnormal shape and numbers of rosette leaves (Figures 2B,C). Different from the WT adult plant (Figure 2D), SDS::BARNASE adult plants also exhibited various abnormal phenotypes, such as dwarf and fertile (Figure 2E), dwarf and sterile (Figure 2F), and even no inflorescence (Figure 2G). The height of mature SDS::BARNASE plants was significantly reduced (Figure 2H). Moreover, SDS::BARNASE plants produced significantly fewer rosette leaves than that of wild type (Figure 2I). Various defects of SDS::BARNASE plants in growth and development suggest that the 1.5-kb promoter of the SDS gene failed to drive the specific expression of BARNASE in microspore and megaspore mother cells.
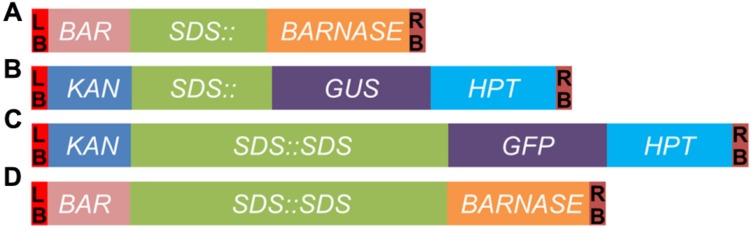
FIGURE 1.
Schematic diagrams of constructs generated in this study. (A) SDS::BARNASE. (B) SDS::GUS. (C) SDS::SDS-GFP. (D) SDS::SDS-BARNASE. LB and RB, the T-DNA left and right border, respectively; BAR, the gene conferring resistance to the herbicide Basta; SDS::, the 1.5-kb promoter of the SDS gene; BARNASE, the bacterial ribonuclease; KAN, the kanamycin resistance gene; GUS, the gene encoding β-glucuronidase; GFP, the gene encoding green fluorescent protein; HPT, the hygromycin phosphotransferase gene; and SDS::SDS, the SDS genomic fragment containing a 1.5-kb promoter followed by a DNA fragment consisting of seven exons and six introns.
FIGURE 2.
SDS::BARNASE Arabidopsis plants were abnormal in growth and development. (A–C) Compared to wild type (A), three-week old SDS::BARNASE (B,C) plants produced less rosette leaves with irregular shape. Bars = 0.5 cm. (D–G) Six-week old wild-type (WT, D) and SDS::BARNASE plants showing fertile but dwarf (E), dwarf and sterile (F), and no inflorescence (G) phenotypes. Bars = 1 cm. (H) Six-week old SDS::BARNASE plants were significantly shorter than the wild type. (I) The rosette leaf number of SDS::BARNASE adult plants was significantly reduced. “n” indicates the number of examined plants. Stars indicate significant difference (P < 0.01).
The 1.5 kb Upstream Region of the SDS Gene did not Confer its Meiocyte-Specific Expression
Genetic ablation relies on the specificity of employed promoters. To examine why BARNASE under the control of the 1.5-kb SDS promoter did not achieve specific ablation effects on microspore and megaspore mother cells, we generated SDS::GUS plants to test the transcriptional activity of the 1.5-kb promoter (Figure 1B). Among 25 examined SDS::GUS transgenic plants, GUS signals were detected in cotyledons, true leaves, and shoot apical meristem of young seedlings (Figure 3A), as well as in carpels and stigmas of young buds (Figures 3B–D). Thus, our results suggest that the 1.5-kb promoter of the SDS gene was not sufficient for conferring its meiocyte-specific expression, which resulted in abnormal plant growth and development when it drove the expression of BARNASE.
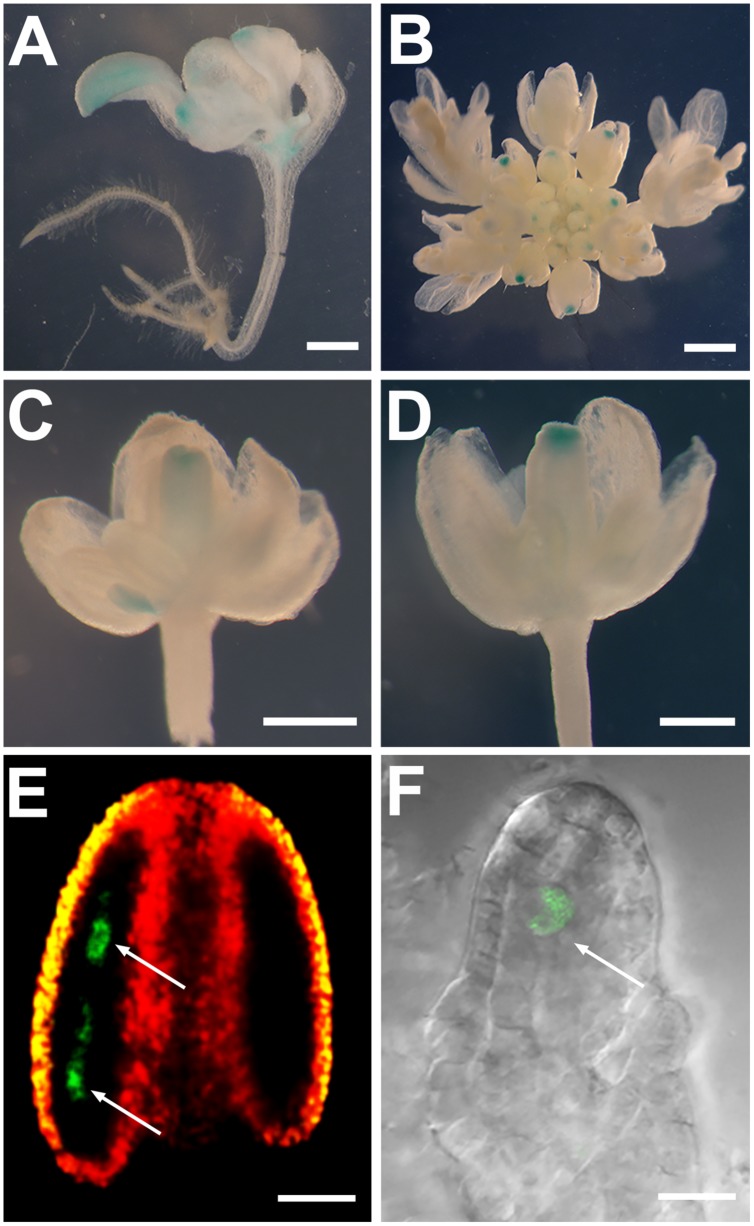
FIGURE 3.
The entire SDS gene but not the SDS 1.5-kb promoter confers the SDS meiocyte-specific expression. (A–D) GUS staining of SDS::GUS plants showing GUS signals in cotyledons, true leaves, and shoot apical meristem of a young seedling (A), as well as in carpels and stigmas of young buds (B–D). (E) A confocal image from an SDS::SDS-GFP stage-5 anther showing the GFP signal (green color) only in microspore mother cells (arrows). Red and yellow colors showing merged autofluorescences. (F) A confocal image from an SDS::SDS-GFP stage 2-IV ovule showing the GFP signal only in the megaspore mother cell (arrow). Bars = 0.1 cm (A,B), 0.5 mm (C,D), 50 μm (E), and 10 μm (F).
The Entire SDS Gene Led to the Meiocyte-Specific Expression of the SDS Protein
The possible existence of regulatory elements in SDS introns may contribute to the SDS meiocyte-specific expression. To test, how to achieve the specific expression of SDS in microspore and megaspore mother cells, we generated SDS::SDS-GFP constructs by fusing the SDS genomic fragment, containing the 1.5-kb promoter, seven exons and six introns, with the GFP gene (Figure 1C). In examined 18 SDS::SDS-GFP transgenic plants, we did not detect the GFP signal during the seedling stage and later in the vegetative growth stage. We, however, observed GFP signals only in microspore mother cells in anthers (Figure 3E) and megaspore mother cell in ovule during the reproductive stage (Figure 3F). Therefore, our results indicate that the entire SDS gene led to the meiocyte-specific expression of the SDS protein.
SDS::SDS-BARNASE Caused Complete Both Male and Female Sterility but did not Affect Growth or Development in Arabidopsis
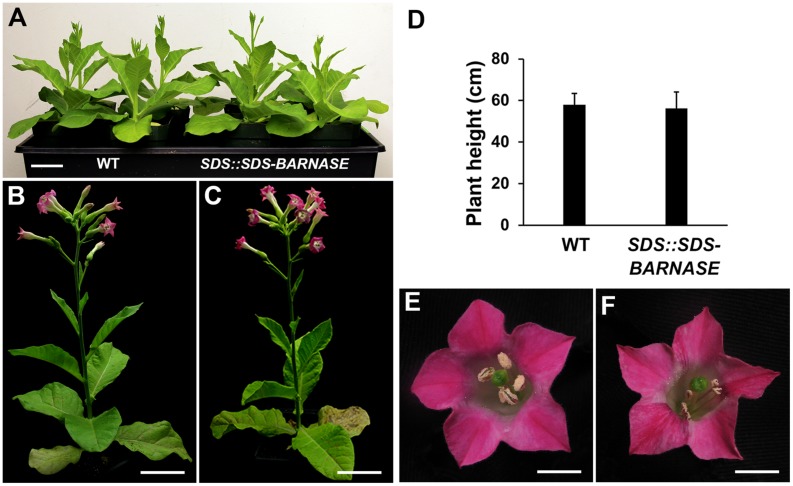
To generate complete both male and female sterility by specifically ablating microspore and megaspore mother cells, we made the SDS::SDS-BARNASE construct by fusing the SDS entire gene with the BARNASE gene (Figure 1D). We performed three transformations, resulting in 97, 80, and 126 SDS::SDS-BARNASE transgenic plants, respectively. All independent transgenic plants were sterile. We first evaluated the effects of SDS::SDS-BARNASE on growth and development. SDS::SDS-BARNASE transgenic plants produced rosette leaves with the same number, size, and shape as that of WT plants (Figures 4A,B). No morphological changes were observed in SDS::SDS-BARNASE inflorescences and flowers (Figures 4C,D). Moreover, mature SDS::SDS-BARNASE plants had a similar height to the wild type (Figures 4E–G). The flowering time of SDS::SDS-BARNASE plants was not affected either, because the same rosette leaf numbers as the wild type were produced when flowering (Figure 4H).
FIGURE 4.
SDS::SDS-BARNASE Arabidopsis plants showed normal growth and development. (A,B) Three-week old WT (A) and SDS::SDS-BARNASE (B) plants. Bars = 0.5 cm. (C,D) Five-week old WT (C) and SDS::SDS-BARNASE (D) inflorescences. Bars = 0.5 cm. (E,F) Six-week old WT (E) and SDS::SDS-BARNASE (F) plants. Bars = 1 cm. (G) No difference in average height between six-week old WT and SDS::SDS-BARNASE plants. (H) Similar rosette leaf numbers indicating no difference in flowering time between WT and SDS::SDS-BARNASE plants. “n” in (G,H) indicates the number of examined plants.
To further investigate sterility of SDS::SDS-BARNASE transgenic plants, we analyzed both male and female fertilities. Compared with the wild type (Figures 5A,H), SDS::SDS-BARNASE plants produced short siliques (Figures 5B,I). Except short filaments, SDS::SDS-BARNASE plants formed flowers that were the same as the wild type, indicated by four sepals, four petals, six stamens, and two carpels (Figures 5D,E). In the WT flower, pollen grains were released from anthers that reached the stigma (Figure 5D), whereas in the SDS::SDS-BARNASE flower, no pollen grains were observed on the anther surface and anthers did not reach the stigma (Figure 5E). Furthermore, different from the WT anther (Figure 5F), the SDS::SDS-BARNASE anther did not produce pollen grains (Figure 5G), indicating that SDS::SDS-BARNASE plants were male sterile. Because pollination using the WT pollen did not rescue the fertility (Figures 5C,J), SDS::SDS-BARNASE plants were female sterile too. Thus, using SDS::SDS-BARNASE, we efficiently created completely both male and female sterile Arabidopsis plants that had normal vegetative and reproductive growth and development, including the formation of all flower organs.
FIGURE 5.
SDS::SDS-BARNASE Arabidopsis plants were completely both male and female sterile. (A–C) Primary branches showing normal siliques in wild type (A) and short siliques indicating no developing seeds in SDS::SDS-BARNASE plants without (B) and with (C) pollination. Bars = 1 cm. (D,E) Side view of mature flowers (One sepal was removed, respectively) showing the SDS::SDS-BARNASE flower (E) is similar to the wild type (D) except short filaments. Pollen grains released from WT anthers (D, inset), while no pollen grains from SDS::SDS-BARNASE anthers (E, inset). Bars = 0.5 mm. (F,G) Pollen staining showing the WT anther full of viable pollen grains (F), but no pollen grains from the SDS::SDS-BARNASE anther (G). Bars = 30 μm. (H–J) Dissected individual siliques from primary inflorescences (positions 1–9) were long in wild type (H), but short in SDS::SDS-BARNASE plants (I, without pollination; J, pollinated with WT pollen). Bars = 1 cm.
SDS::SDS-BARNASE Inhibited Both Male and Female Gamete Formation
To further understand ablation effects on microspore and megaspore mother cells, we did semi-thin sectioning of anthers and whole-mount squashes of ovules. At stage 5 (Sanders et al., 1999; Zhao et al., 2002), when compared with the WT anther (Figure 6A), the SDS::SDS-BARNASE anther showed vacuolated microsporocytes (microspore mother cells) and tapetal cells (Figure 6D), indicating the degeneration of both cells. At stage 7 in the WT anther, successful male meiosis resulted in the formation of tetrads (Figure 6B), whereas in the SDS::SDS-BARNASE anther, tetrads, and tapetal cells were collapsed (Figure 6E). At stage 9, the WT anther contains developing pollen grains (Figure 6C), but the SDS::SDS-BARNASE anther lacked developing microspores (Figure 6F).
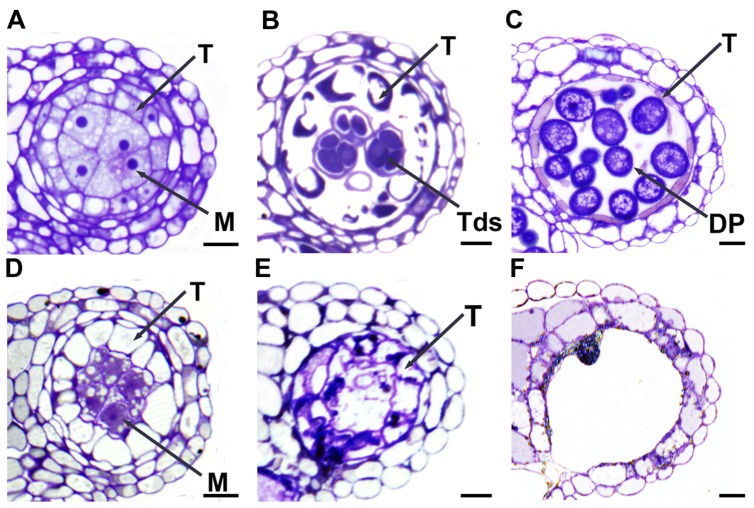
FIGURE 6.
Formation of male gametes was arrested in SDS::SDS-BARNASE Arabidopsis plants. (A–C) WT anthers showing microsporocytes (microspore mother cells) and surrounding tapetal cells at stage 5 (A), tetrads and tapetal cells at stage 7 (B), and developing pollen grains at stage 9 (C). (D–F) SDS::SDS-BARNASE anthers showing degenerating microsporocytes and precociously vacuolated tapetal cells at stage 5 (D), dead microsporocytes and tapetal cells at stage 7 (E), and a nearly empty anther lobe at stage 9 (only one dead pollen, F). M, microsporocytes (microspore mother cells); DP, developing pollen; T, tapetal cell; and Tds, tetrads.
In embryo sacs of WT ovules, two nuclei at stage FG3 (Pagnussat et al., 2009) (Figure 7A) and four nuclei at stage FG4 (Figure 7B) were observed; however, in SDS::SDS-BARNASE embryo sacs, only a single nucleus was produced (Figures 7D,E). At stage FG6, the WT embryo sac showed the central cell, the egg cell, and synergid cells (Figure 7C), but the SDS::SDS-BARNASE embryo sac is empty (Figure 7F).
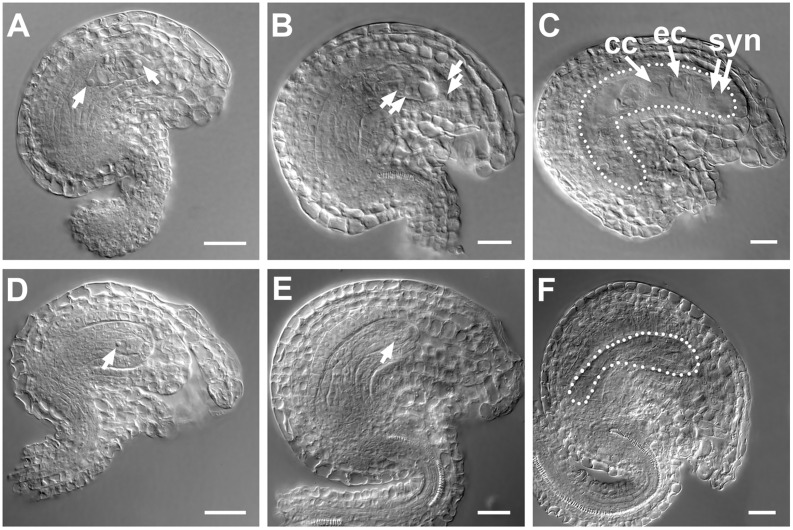
FIGURE 7.
Formation of female gamete was arrested in SDS::SDS-BARNASE Arabidopsis plants. (A–C) WT ovules showing two separated nuclei (arrows) at the FG3 stage (A), four nuclei (arrows) at the FG4 stage (B), and the central cell, the egg cell, and synergid cells in a mature embryo sac (white dots outlined) at the FG6 stage (C). (D–F) SDS::SDS-BARNASE ovules showing one small nucleus (arrow) at both FG3 (D) and FG4 (E) stages and a small empty embryo sac (white dots outlined) at the FG6 stage (F). Bars = 10 μm. cc, central cell; ec, egg cell; and syn, synergid cells.
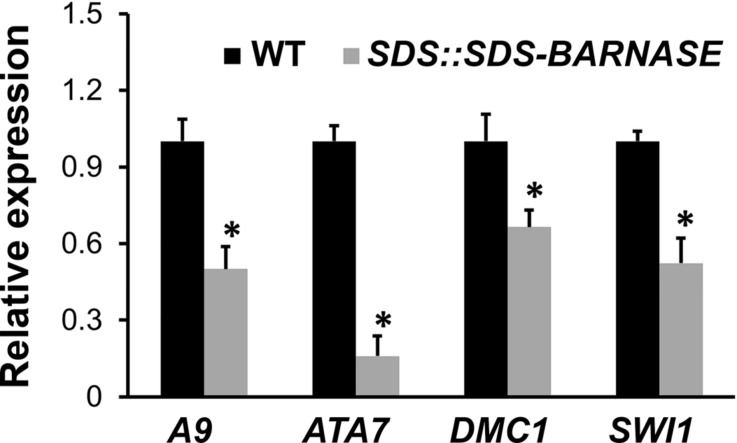
Furthermore, our results showed that expressions of tapetal cell marker genes A9 and ATA7 as well as microspore and megaspore mother cell marker genes DMC1 and SWI1 were significantly decreased in SDS::SDS-BARNASE buds in comparison to the wild type (Figure 8). In summary, the specific expression of the SDS-BARNASE toxic fusion protein in microspore and megaspore mother cells efficiently impaired the production of both male and female gametes, which led to absolute both male and female sterility, but did not affect flower organ formation or plant growth and development.
FIGURE 8.
Expressions of tapetal cell as well as microspore and megaspore mother cell marker genes. Real-time qRT-PCR showing decreased expressions of tapetal cell marker genes A9 and ATA7 as well as microspore and megaspore mother cell marker genes DMC1 and SWI1. Stars indicate significant difference (P < 0.01).
SDS::SDS-BARNASE Caused Complete Both Male and Female Sterility but did not Affect Growth or Development in Tobacco
To test whether SDS::SDS-BARNASE can provide a general tool to create both male and female sterile plants, we transformed it into tobacco and generated SDS::SDS-BARNASE tobacco transgenic plants by tissue culture. Among 14 examined SDS::SDS-BARNASE tobacco transgenic lines, leaf shape and size (Figures 9A–C), as well as the plant height (Figures 9B–D) were the same as that of WT plants. In addition, the SDS::SDS-BARNASE tobacco flower had the same size, color, and structure as that of wild type (Figures 9E,F). Therefore, SDS::SDS-BARNASE did not affect growth or development in tobacco plants.
FIGURE 9.
SDS::SDS-BARNASE tobacco plants showed normal growth and development. (A) Forty-day old tobacco WT and SDS::SDS-BARNASE plants. Bar = 5 cm. (B,C) Sixty-day old WT (B) and SDS::SDS-BARNASE (C) plants. Bars = 10 cm. (D) No difference in average height between WT and SDS::SDS-BARNASE adult plants. (E,F) Flower size, color, and structure remained the same in WT and SDS::SDS-BARNASE plants. Bars = 1 cm.
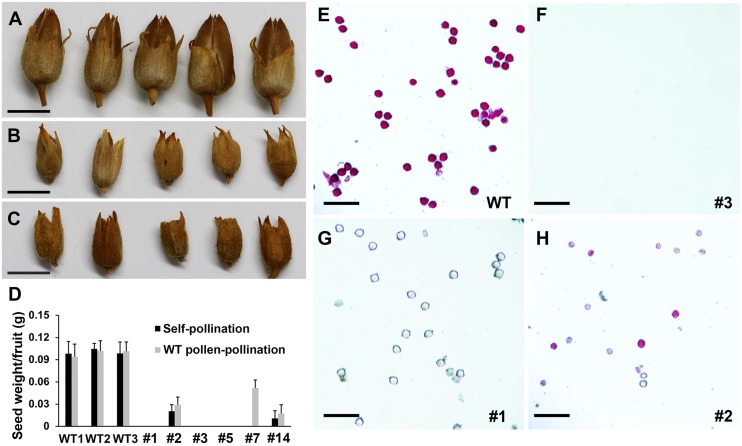
Ten examined SDS::SDS-BARNASE tobacco transgenic lines were completely sterile. WT tobacco plants produced large fruits and per fruit averagely contained 0.11 g of seeds (Figures 10A,D). Conversely, SDS::SDS-BARNASE plants produced small fruits and no seeds were found when self-pollenated (Figures 10B,D, e.g., plants #1, 3, 5, and 7). Further pollen viability analysis showed that WT tobacco anthers produced viable pollen, indicated by red color (Figure 10E), whereas anthers from sterile tobacco plants either lacked pollen grains (Figure 10F) or formed dead pollen grains (Figure 10G). The four non-absolutely sterile lines produced a few seeds (Figure 10D, e.g., plants #2, and 14) and only some functional pollen grains were found in anthers of those lines (Figure 10H, e.g., plant #2). Our results suggest that SDS::SDS-BARNASE impaired male fertility in tobacco.
FIGURE 10.
SDS::SDS-BARNASE tobacco plants were completely both male and female sterile. (A–C) Large fruits from the WT plant (A) and small fruits from SDS::SDS-BARNASE plants without (B) and with (C) manual pollination with WT pollen grains. Bars = 1 cm. (D) The weight of seeds per self-pollinated and manually pollinated fruit (n = 5), respectively. Numbers indicate examined independent transgenic lines. (E) WT viable pollen grains in red color. (F–H) no (F), all dead (G) and a few viable (H) pollen grains in SDS::SDS-BARNASE plants. Numbers indicate examined independent transgenic lines. Bars = 100 μm.
We then examined the female fertility in sterile tobacco transgenic plants. The fertility of manually male-sterilized WT flowers could be rescued by cross-pollination with WT pollen (Figure 10D), but following cross-pollination with WT pollen, the fruit size of SDS::SDS-BARNASE sterile tobacco plants did not change (Figure 10C) and no seeds were produced (Figure 10D, e.g., plants #1, 3, and 5). Thus, SDS::SDS-BARNASE tobacco transgenic plants were also female sterile. Manual pollination partially rescued the fertility of line #7, indicating that the line #7 is a completely male but partially female sterile plant, while lines #2 and 14 were nearly male and female sterile plants (Figure 10D). Collectively, a majority of SDS::SDS-BARNASE tobacco transgenic plants were completely male and female sterile, suggesting that SDS::SDS-BARNASE is functionally conserved, which can be used to create both male and female sterility in general.
Discussion
SOLO DANCERS, a unique type of (SDS-type) meiosis-specific cyclin, is conserved in flowering plants (Wang et al., 2004; Zhang et al., 2014; Wu et al., 2015). In our studies, the 1.5-kb SDS promoter did not achieve the specific expression of SDS in either microspore or megaspore mother cells. Conversely, the entire SDS gene genomic fragment did. The intron-dependent spatial expression has been revealed in different genes from various species, including SUS3 and SUS4 in potato (Fu et al., 1995a,b), OsTubA1 in rice (Jeon et al., 2000; Giani et al., 2009), PhADF1 in petunia (Jeong et al., 2007), as well as AGAMOUS, ACT1 and PRF in Arabidopsis (An et al., 1996; Sieburth and Meyerowitz, 1997; Jeong et al., 2006). Therefore, it is possible that regulatory motifs in SDS introns contribute to its specific spatial and temporal expression. Future studies should be focused on dissecting the functions of unknown regulatory motifs and then making a synthetic promoter that confers the strong and specific expression of SDS in microspore and megaspore mother cells. We found a few non-completely sterile tobacco transgenic plants, suggesting that the Arabidopsis SDS gene did not work efficiently in tobacco. In order to achieve accurate and efficient ablation effects, it would be more practical to use SDS orthologous genes of target species or the synthetic promoter to drive BARNASE.
The existing methods for creating male sterile only GM plants are not able to completely prevent transgene flow, because pollen from non-GM plants can rescue seed development. In addition, current sterilization technologies either suppress the production of entire flower or some floral organs, which may cause potential ecological effects besides transgene flow. Furthermore, BARNASE and DTA are very toxic. Many “specific” promoters still have basal activities in other organs; therefore, the low expression of BARNASE/DTA in non-target organs often reduces plant survival rate and inhibits plant growth. Microspore and megaspore mother cells are two small groups of male and female reproductive cells, which are differentiated after all floral organs are established. Ablating microspore and megaspore mother cells only leads to elimination of male and female gametes, but it does not affect other somatic cell differentiation and flower development. In this study, we specifically ablated microspore and megaspore mother cells using the SDS and BARNASE fusion protein. Thus, our research developed an efficient strategy to successfully create completely both male and female sterile plants; however, the plant growth and development, including the formation of all flower organs, were not affected.
Genetically modified crops have been widely cultivated in many countries due to their improved agronomic traits; however, the adoption of GM trees (e.g., poplar, eucalypts, and pines) and perennial grasses (e.g., miscanthus and switchgrass) is limited, because those plants are long-lived, weakly domesticated, and important to ecosystems. Various studies has been done to increase cold tolerance and biomass, modify lignin and cellulose biosynthesis, or alter growth and flowering of Eucalyptus (Girijashankar, 2011; Hinchee et al., 2011; Klocko et al., 2015), aspen (Etchells et al., 2015), poplar (Wilkerson et al., 2014), and switchgrass (Fu et al., 2011; Shen et al., 2013; Baxter et al., 2014; Poovaiah et al., 2015). Our research developed a general and effective approach to create completely both male and female sterile plants by specifically ablating microspore and megaspore mother cells, which provides a solution for overcoming regulatory hurdles to field research and ultimately commercial uses of GM plants.
Besides the transgene containment, our method can be applied for modifying invasive and ornamental plants. Male and female sterilized invasive plants generated by our method can be planted for multiple purposes, such as forestry and horticulture. The main valuable trait for many ornamental trees, such as cherries and plums, is the beauty of flowers. Fruits often make the garden messy. Moreover, nutrient competition from fruit setting affects flower organ differentiation, and consequently reduces flower numbers in the coming year. Our new method serves as an excellent tool to engineer ornamental trees that still produce attractive intact flowers without fruit setting, which, therefore, maintains their ornamental value.
Author Contributions
DZ conceived and designed the experiments. JH, AS, and TZ performed experiments. All authors analyzed data. DZ and JH wrote the article.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. Dazhong Zhao has filed a patent application for the sterilization technique reported in this article.
Acknowledgments
We thank H.A. Owen for critical reading of this manuscript, C. Zhang for the pABGCZ vector, T. Nakagawa for Gateway binary vectors, C. Pikaard for the pEARLEY303 vector, and J. Gonnering and Tom Schuck for plant care.
Footnotes
Funding. This work was supported in part by the National Science Foundation (NSF IOS-0721192 and IOS-1322796), the Research Growth Initiative (RGI) at the University of Wisconsin-Milwaukee, and the UW-Madison/UW-Milwaukee Intercampus Research Incentive Grants Program to DZ. DZ also gratefully acknowledges the support of the Shaw Scientist Award from the Greater Milwaukee Foundation.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00030
References
- An X. M., Wang D. M., Wang Z. L., Li B., Bo W. H., Cao G. L., et al. (2011). Isolation of a LEAFY homolog from Populus tomentosa: expression of PtLFY in P. tomentosa floral buds and PtLFY-IR-mediated gene silencing in tobacco (Nicotiana tabacum). Plant Cell Rep. 30 89–100. 10.1007/s00299-010-0947-0 [DOI] [PubMed] [Google Scholar]
- An Y. Q., Huang S., McDowell J. M., McKinney E. C., Meagher R. B. (1996). Conserved expression of the Arabidopsis ACT1 and ACT 3 actin subclass in organ primordia and mature pollen. Plant Cell 8 15–30. 10.1105/tpc.8.1.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azumi Y., Liu D., Zhao D., Li W., Wang G., Hu Y., et al. (2002). Homolog interaction during meiotic prophase I in Arabidopsis requires the solo dancers gene encoding a novel cyclin-like protein. EMBO J. 21 3081–3095. 10.1093/emboj/cdf285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter H. L., Mazarei M., Labbe N., Kline L. M., Cheng Q. K., Windham M. T., et al. (2014). Two-year field analysis of reduced recalcitrance transgenic switchgrass. Plant Biotechnol. J. 12 914–924. 10.1111/Pbi.12195 [DOI] [PubMed] [Google Scholar]
- Caplan A., Herrera-Estrella L., Inze D., Van Haute E., Van Montagu M., Schell J., et al. (1983). Introduction of genetic material into plant cells. Science 222 815–821. 10.1126/science.222.4625.815 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Curtis I. S., Davey M. R., Power J. B. (1995). Leaf disk transformation. Methods Mol. Biol. 44 59–70. 10.1385/0-89603-302-3:59 [DOI] [PubMed] [Google Scholar]
- DeBlock M., Debrouwer D., Moens T. (1997). The development of a nuclear male sterility system in wheat. Expression of the barnase gene under the control of tapetum specific promoters. Theor. Appl. Genet. 95 125–131. 10.1016/j.gene.2014.01.001 [DOI] [Google Scholar]
- DiFazio S. P., Leonardi S., Slavov G. T., Garman S. L., Adams W. T., Strauss S. H. (2012). Gene flow and simulation of transgene dispersal from hybrid poplar plantations. New Phytol. 193 903–915. 10.1111/j.1469-8137.2011.04012.x [DOI] [PubMed] [Google Scholar]
- Elorriaga E., Meilan R., Ma C., Skinner J. S., Etherington E., Brunner A., et al. (2014). A tapetal ablation transgene induces stable male sterility and slows field growth in Populus. Tree Genet. Genomes. 10 1583–1593. 10.1007/s11295-014-0781-6 [DOI] [Google Scholar]
- Etchells J. P., Mishra L. S., Kumar M., Campbell L., Turner S. R. (2015). Wood formation in trees is increased by manipulating PXY-regulated cell division. Curr. Biol. 25 1050–1055. 10.1016/j.cub.2015.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenart S., Austerlitz F., Cuguen J., Arnaud J. F. (2007). Long distance pollen-mediated gene flow at a landscape level: the weed beet as a case study. Mol. Ecol. 16 3801–3813. 10.1111/j.1365-294X.2007.03448.x [DOI] [PubMed] [Google Scholar]
- Fu C., Mielenz J. R., Xiao X., Ge Y., Hamilton C. Y., Rodriguez M., et al. (2011). Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc. Natl. Acad. Sci. U.S.A. 108 3803–3808. 10.1073/pnas.1100310108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C. X., Sunkar R., Zhou C. E., Shen H., Zhang J. Y., Matts J., et al. (2012). Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnol. J. 10 443–452. 10.1111/j.1467-7652.2011.00677.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Kim S. Y., Park W. D. (1995a). High-level tuber expression and sucrose inducibility of a potato Sus4 sucrose synthase gene require 5′ and 3′ flanking sequences and the leader intron. Plant Cell 7 1387–1394. 10.2307/3870129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H. Y., Kim S. Y., Park W. D. (1995b). A potato Sus3 sucrose synthase gene contains a context-dependent 3′ element and a leader intron with both positive and negative tissue-specific effects. Plant Cell 7 1395–1403. 10.1105/tpc.7.9.1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner N., Felsheim R., Smith A. G. (2009). Production of male- and female-sterile plants through reproductive tissue ablation. J. Plant Physiol. 166 871–881. 10.1016/j.jplph.2008.10.002 [DOI] [PubMed] [Google Scholar]
- Giani S., Altana A., Campanoni P., Morello L., Breviario D. (2009). In trangenic rice, α- and β-tubulin regulatory sequences control GUS amount and distribution through intron mediated enhancement and intron dependent spatial expression. Transgenic Res. 18 151–162. 10.1007/s11248-008-9202-7 [DOI] [PubMed] [Google Scholar]
- Girijashankar V. (2011). Genetic transformation of Eucalyptus. Physiol. Mol. Biol. Plants 17 9–23. 10.1007/s12298-010-0048-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeschl T., Swanson T. (2003). The development impact of genetic use restriction technologies: a forecast based on the hybrid crop experience. Environ. Dev. Econ. 8 149–165. 10.1017/S1355770x03000081 [DOI] [Google Scholar]
- Groover A. T. (2007). Will genomics guide a greener forest biotech? Trends Plant Sci. 12 234–238. 10.1016/j.tplants.2007.04.005 [DOI] [PubMed] [Google Scholar]
- Harfouche A., Meilan R., Altman A. (2011). Tree genetic engineering and applications to sustainable forestry and biomass production. Trends Biotech. 29 9–17. 10.1016/j.tibtech.2010.09.003 [DOI] [PubMed] [Google Scholar]
- Hills M. J., Hall L., Arnison P. G., Good A. G. (2007). Genetic use restriction technologies (GURTs): strategies to impede transgene movement. Trends Plant Sci. 12 177–183. 10.1016/j.tplants.2007.02.002 [DOI] [PubMed] [Google Scholar]
- Hinchee M., Zhang C., Chang S., Cunningham M., Hammond W., Nehra N. (2011). Biotech Eucalyptus can sustainably address society’s need for wood: the example of freeze tolerant Eucalyptus in the southeastern U.S. BMC Proc. 5(Suppl. 7):I24 10.1186/1753-6561-5-S7-I24 [DOI] [Google Scholar]
- Hird D. L., Worrall D., Hodge R., Smartt S., Paul W., Scott R. (1993). The anther-specific protein encoded by the Brassica napus and Arabidopsis thaliana A6 gene displays similarity to β-1,3-glucanases. Plant J. 4 1023–1033. 10.1046/j.1365-313X.1993.04061023.x [DOI] [PubMed] [Google Scholar]
- Husken A., Prescher S., Schiemann J. (2010). Evaluating biological containment strategies for pollen-mediated gene flow. Environ. Biosafety Res. 9 67–73. 10.1051/ebr/2010009 [DOI] [PubMed] [Google Scholar]
- Jensen C. S., Salchert K., Gao C. X., Andersen C., Didion T., Nielsen K. K. (2004). Floral inhibition in red fescue (Festuca rubra L.) through expression of a heterologous flowering repressor from Lolium. Mol. Breed. 13 37–48. 10.1023/B:MOLB.0000012327.47625.23 [DOI] [Google Scholar]
- Jeon J. S., Lee S., Jung K. H., Jun S. H., Kim C., An G. (2000). Tissue-preferential expression of a rice α-tubulin gene, OsTubA1, mediated by the first intron. Plant Physiol. 123 1005–1014. 10.1104/pp.123.3.1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y. M., Mun J. H., Kim H., Lee S. Y., Kim S. G. (2007). An upstream region in the first intron of petunia actin-depolymerizing factor 1 affects tissue-specific expression in transgenic Arabidopsis (Arabidopsis thaliana). Plant J. 50 230–239. 10.1111/j.1365-313X.2007.03053.x [DOI] [PubMed] [Google Scholar]
- Jeong Y. M., Mun J. H., Lee I., Woo J. C., Hong C. B., Kim S. G. (2006). Distinct roles of the first introns on the expression of Arabidopsis profilin gene family members. Plant Physiol. 140 196–209. 10.1104/pp.105.071316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G., Liu X., Owen H. A., Zhao D. (2008). Signaling of cell fate determination by the TPD1 small protein and EMS1 receptor kinase. Proc. Natl Acad. Sci. U.S.A. 105 2220–2225. 10.1073/pnas.0708795105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klocko A. L., Ma C., Robertson S., Esfandiari E., Nilsson O., Strauss S. H. (2015). FT overexpression induces precocious flowering and normal reproductive development in Eucalyptus. Plant Biotechnol. J. 10.1111/pbi.12431 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow A. M., Truettner J., Cox K. H., Wallroth M., Goldberg R. B. (1990). Different temporal and spatial gene expression patterns occur during anther development. Plant Cell 2 1201–1224. 10.2307/3869340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lannanpaa M., Hassinen M., Ranki A., Holtta-Vuori M., Lemmetyinen J., Keinonen K., et al. (2005). Prevention of flower development in birch and other plants using a BpFULL1::BARNASE construct. Plant Cell Rep. 24 69–78. 10.1007/s00299-004-0903-y [DOI] [PubMed] [Google Scholar]
- Liu X., Huang J., Wang Y., Khanna K., Xie Z., Owen H. A., et al. (2010a). The role of floral organs in carpels, an Arabidopsis loss-of-function mutation in MicroRNA160a, in organogenesis and the mechanism regulating its expression. Plant J. 62 416–428. 10.1111/j.1365-313X.2010.04164.x [DOI] [PubMed] [Google Scholar]
- Liu Y. B., Wei W., Ma K. P., Darmency H. (2010b). Backcrosses to Brassica napus of hybrids between B. juncea and B. napus as a source of herbicide-resistant volunteer-like feral populations. Plant Sci. 179 459–465. 10.1016/j.plantsci.2010.07.005 [DOI] [PubMed] [Google Scholar]
- Liu Z., Liu Z. (2008). The second intron of AGAMOUS drives carpel- and stamen-specific expression sufficient to induce complete sterility in Arabidopsis. Plant Cell Rep. 27 855–863. 10.1007/s00299-008-0511-3 [DOI] [PubMed] [Google Scholar]
- Lombardo L. (2014). Genetic use restriction technologies: a review. Plant Biotechnol. J. 12 995–1005. 10.1111/Pbi.12242 [DOI] [PubMed] [Google Scholar]
- Luo H., Kausch A. P., Hu Q., Nelson K., Wipff J. K., Fricker C. C. R., et al. (2005). Controlling transgene escape in GM creeping Bentgrass. Mol. Breed. 16 185–188. 10.1007/s11032-005-4784-8 [DOI] [Google Scholar]
- Mariani C., Debeuckeleer M., Truettner J., Leemans J., Goldberg R. B. (1990). Induction of male-sterility in plants by a chimaeric ribonuclease gene. Nature 347 737–741. 10.1038/347737a0 [DOI] [Google Scholar]
- Murai N., Kemp J. D., Sutton D. W., Murray M. G., Slightom J. L., Merlo D. J., et al. (1983). Phaseolin gene from bean is expressed after transfer to sunflower via tumor-inducing plasmid vectors. Science 222 476–482. 10.1038/347737a0 [DOI] [PubMed] [Google Scholar]
- Pagnussat G. C., Alandete-Saez M., Bowman J. L., Sundaresan V. (2009). Auxin-dependent patterning and gamete specification in the Arabidopsis female gametophyte. Science 324 1684–1689. 10.1126/science.1167324 [DOI] [PubMed] [Google Scholar]
- Pan I. L., McQuinn R., Giovannoni J. J., Irish V. F. (2010). Functional diversification of AGAMOUS lineage genes in regulating tomato flower and fruit development. J. Exp. Bot. 61 1795–1806. 10.1093/jxb/erq046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul W., Hodge R., Smartt S., Draper J., Scott R. (1992). The isolation and characterisation of the tapetum-specific Arabidopsis thaliana A9 gene. Plant Mol. Biol. 19 611–622. 10.1007/BF00026787 [DOI] [PubMed] [Google Scholar]
- Pineyro-Nelson A., Van Heerwaarden J., Perales H. R., Serratos-Hernandez J. A., Rangel A., Hufford M. B., et al. (2009). Transgenes in Mexican maize: molecular evidence and methodological considerations for GMO detection in landrace populations. Mol. Ecol. 18 750–761. 10.1111/j.1365-294X.2008.03993.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poovaiah C. R., Mazarei M., Decker S. R., Turner G. B., Sykes R. W., Davis M. F., et al. (2015). Transgenic switchgrass (Panicum virgatum L.) biomass is increased by overexpression of switchgrass sucrose synthase (PvSUS1). Biotechnol. J. 10 552–563. 10.1002/biot.201400499 [DOI] [PubMed] [Google Scholar]
- Rieben S., Kalinina O., Schmid B., Zeller S. L. (2011). Gene flow in genetically modified wheat. PLoS ONE 6:e29730 10.1371/journal.pone.0029730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque E., Gómez M. D., Ellul P., Wallbraun M., Madueño F., Beltran J. P., et al. (2007). The PsEND1 promoter: a novel tool to produce genetically engineered male-sterile plants by early anther ablation. Plant Cell Rep. 26 313–325. 10.1007/s00299-006-0237-z [DOI] [PubMed] [Google Scholar]
- Roque E., Serwatowska J., Cruz Rochina M., Wen J., Mysore K. S., Yenush L., et al. (2013). Functional specialization of duplicated AP3-like genes in Medicago truncatula. Plant J. 73 663–675. 10.1007/s00299-006-0237-z [DOI] [PubMed] [Google Scholar]
- Ruf S., Karcher D., Bock R. (2007). Determining the transgene containment level provided by chloroplast transformation. Proc. Natl. Acad. Sci. U.S.A. 104 6998–7002. 10.1073/pnas.0700008104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders P. M., Bui A. Q., Weterings K., McIntire K. N., Hsu Y. C., Lee P. Y., et al. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11 297–322. 10.1007/s004970050158 [DOI] [Google Scholar]
- Shen H., Poovaiah C. R., Ziebell A., Tschaplinski T. J., Pattathil S., Gjersing E., et al. (2013). Enhanced characteristics of genetically modified switchgrass (Panicum virgatum L.) for high biofuel production. Biotechnol. Biofuels 6:71 10.1186/1754-6834-6-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth L. E., Meyerowitz E. M. (1997). Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9 355–365. 10.1105/tpc.9.3.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavov G. T., Leonardi S., Burczyk J., Adams W. T., Strauss S. H., Difazio S. P. (2009). Extensive pollen flow in two ecologically contrasting populations of Populus trichocarpa. Mol. Ecol. 18 357–373. 10.1111/j.1365-294X.2008.04016.x [DOI] [PubMed] [Google Scholar]
- Stewart C. N., Jr. (2007). Biofuels and biocontainment. Nat. Biotechnol. 25 283–284. 10.1038/nbt0307-283 [DOI] [PubMed] [Google Scholar]
- Strauss S. H., Tan H. M., Boerjan W., Sedjo R. (2009). Strangled at birth? Forest biotech and the convention on biological diversity. Nat. Biotechnol. 27 519–527. 10.1038/Nbt0609-519 [DOI] [PubMed] [Google Scholar]
- Van Acker R. C., Szumgalski A. R., Friesen L. F. (2007). The potential benefits, risks and costs of genetic use restriction technologies. Can. J. Plant Sci. 87 753–762. 10.4141/CJPS06033 [DOI] [Google Scholar]
- Wang G., Kong H., Sun Y., Zhang X., Zhang W., Altman N., et al. (2004). Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol. 135 1084–1099. 10.1104/pp.104.040436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. J., Li X. H. (2012). Genetic characterization and gene flow in different geographical-distance neighbouring natural populations of wild soybean (Glycine soja Sieb. & Zucc.) and implications for protection from GM soybeans. Euphytica 186 817–830. 10.1007/s10681-012-0621-4 [DOI] [Google Scholar]
- Wang Z. Y., Brummer E. C. (2012). Is genetic engineering ever going to take off in forage, turf and bioenergy crop breeding? Ann. Bot. 110 1317–1325. 10.1093/Aob/Mcs027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. Y., Ge Y. X. (2006). Invited review: recent advances in genetic transformation of forage and turf grasses. In Vitro Cell. Dev. Biol. Plant 42 1–18. 10.1079/Ivp2005726 [DOI] [Google Scholar]
- Wani S. H., Haider N., Kumar H., Singh N. B. (2010). Plant plastid engineering. Curr. Genomics 11 500–512. 10.2174/138920210793175912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrud L. S., Lee E. H., Fairbrother A., Burdick C., Reichman J. R., Bollman M., et al. (2004). Evidence for landscape-level, pollen-mediated gene flow from genetically modified creeping bentgrass with CP4 EPSPS as a marker. Proc. Natl. Acad. Sci. U.S.A. 101 14533–14538. 10.1073/pnas.0405154101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Meilan R., Brunner A. M., Skinner J. S., Ma C. P., Gandhi H. T., et al. (2007). Field trial detects incomplete barstar attenuation of vegetative cytotoxicity in Populus trees containing a poplar LEAFY promoter::barnase sterility transgene. Mol. Breed. 19 69–85. 10.1007/s11032-006-9045-y [DOI] [Google Scholar]
- Wilkerson C. G., Mansfield S. D., Lu F., Withers S., Park J. Y., Karlen S. D., et al. (2014). Monolignol ferulate transferase introduces chemically labile linkages into the lignin backbone. Science 344 90–93. 10.1126/science.1250161 [DOI] [PubMed] [Google Scholar]
- Wu Z., Ji J. H., Tang D., Wang H. J., Shen Y., Shi W. Q., et al. (2015). OsSDS is essential for DSB formation in rice meiosis. Front. Plant Sci. 6:21 10.3389/Fpls.2015.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H., Wang Y., Liu D., Wang W., Li X., Zhao X., et al. (2003). Functional analysis of the rice AP3 homologue OsMADS16 by RNA interference. Plant Mol. Biol. 52 957–966. 10.1023/A:1025401611354 [DOI] [PubMed] [Google Scholar]
- Xu J. L., Wang Z. Y., Cheng J. J. (2011). Bermuda grass as feedstock for biofuel production: a review. Bioresour. Technol. 102 7613–7620. 10.1016/j.biortech.2011.05.070 [DOI] [PubMed] [Google Scholar]
- Zapiola M. L., Mallory-Smith C. A. (2012). Crossing the divide: gene flow produces intergeneric hybrid in feral transgenic creeping bentgrass population. Mol. Ecol. 21 4672–4680. 10.1111/j.1365-294X.2012.05627.x [DOI] [PubMed] [Google Scholar]
- Zhan X. Y., Wu H. M., Cheung A. Y. (1996). Nuclear male sterility induced by pollen-specific expression of a ribonuclease. Sex. Plant Reprod. 9 35–43. 10.1007/BF00230364 [DOI] [Google Scholar]
- Zhang C., Norris-Caneda K. H., Rottmann W. H., Gulledge J. E., Chang S., Kwan B. Y., et al. (2012). Control of pollen-mediated gene flow in transgenic trees. Plant Physiol. 159 1319–1334. 10.1104/pp.112.197228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Wang X., Lu Y., Cai X., Ye Z., Zhang J. (2014). Genome-wide analysis of the cyclin gene family in tomato. Int. J. Mol. Sci. 15 120–140. 10.3390/ijms15010120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D. Z., Wang G. F., Speal B., Ma H. (2002). The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 16 2021–2031. 10.1101/gad.997902 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.