Abstract
In order to evaluate potential therapeutic targets for treatment of amyloidoses such as Alzheimer’s disease (AD), it is essential to determine the structures of toxic amyloid oligomers. However, for the amyloid β-protein peptide (Aβ), thought to be the seminal neuropathogenetic agent in AD, its fast aggregation kinetics and the rapid equilibrium dynamics among oligomers of different size pose significant experimental challenges. Here we use ion-mobility mass spectrometry, in combination with electron microscopy, atomic force microscopy, and computational modeling, to test the hypothesis that Aβ peptides can form oligomeric structures resembling cylindrins and β-barrels. These structures are hypothesized to cause neuronal injury and death through perturbation of plasma membrane integrity. We show that hexamers of C-terminal Aβ fragments, including Aβ(24-34), Aβ(25-35) and Aβ(26-36), have collision cross-sections similar to those of cylindrins. We also show that linking two identical fragments head-to-tail using di-glycine increases the proportion of cylindrin-sized oligomers. In addition, we find that larger oligomers of these fragments may adopt β-barrel structures and that β-barrels can be formed by folding an out-of-register β-sheet, a common type of structure found in amyloid proteins.
INTRODUCTION
Structure-neurotoxicity relationships of Aβ oligomers have been the subject of intense research efforts. Some Aβ oligomers have been found to be precursors of the classical 10 nm-diameter amyloid fibrils, while others form independently of fibril formation. Although fibril formation is a defining pathological feature of many devastating diseases including Alzheimer’s, Parkinson’s and type II diabetes,1–5 multiple lines of evidence indicate that oligomers rather than fibrils are likely to be the most important toxic agents.6–9 Of note, a variety of amyloid proteins and peptides with different primary structures form oligomers with similar quaternary structures.8,10 These structures are more stable than their monomeric and smaller oligomeric precursors, but less stable than their ultimate fibrillar products.11,12 From a structure-function perspective, toxic oligomers would be predicted to have relatively well-organized structures that interact with cellular membranes, receptors, or other proteins. Recently, a novel class of oligomer structure, the cylindrin, was defined.13 Cylindrins contain six single β-strands arrayed near-vertically around a central axis, thus forming a cylinder. Computational studies predict that larger cylindrins are possible, but evidence for these remains lacking.14
To date, the most detailed structural findings have come from studies of amyloid oligomers in stable, homogeneous populations.13,15–17 X-ray crystallographic studies were the first to determine the three-dimensional structures of oligomers and fibrils,18–22 including those of model peptides from αB-crystallin13 and prion fragments.20 These two peptides were found to form cylindrins. The success of those studies was highly dependent on the availability of homogeneous, stable oligomers. However, for many biologically-relevant amyloid proteins, it remains quite challenging to perform the same kind of experiment. Reasons include: (a) the extremely high aggregation propensity of many of these proteins, which produces polydisperse aggregates;13 and (b) the existence of multiple conformational states for oligomers of identical molecular weight.23 In addition, x-ray crystallographic analyses often yield data only on a dominant conformational state, thus a complete definition of the oligomer conformational space is not possible.
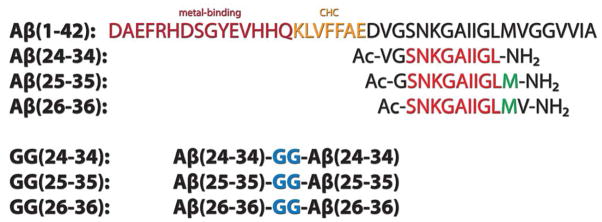
Many of these experimental difficulties can be overcome by mass spectrometry (MS). The maturation of MS in recent years and has led to significant advancements in studies of protein structure-function relationships, especially in the area of protein assembly and aggregation.23–29 Native ion-mobility mass spectrometry (IM-MS) offers an additional dimension of measurement, in that it allows a variety of oligomers to be separated by both their mass to charge ratios (m/z) as in basic MS, and by their sizes and shapes. With IM-MS, the overall structure of a specific oligomer can be captured through means of collision cross-section measurement, which can then be directly compared with structures obtained using other experimental techniques or theoretical calculation.24,25,30–32 Here, we have applied IM-MS, in combination with transmission electron microscopy, atomic force microscopy and computational modeling, to investigate possible cylindrin formation by fragments of Aβ. We examined three overlapping fragments: Aβ(24-34), Aβ(25-35), and Aβ(26-36). The Aβ(25-35) fragment is known to exist in the brain and is cytotoxic.33,34 The other two fragments were predicted to be compatible with the cylindrin structure. In addition, we examined tandem-repeat versions of each fragment, in which two copies of the same fragment were connected head-to-tail by a di-glycine (GG) linker. This linking strategy was used successfully with cylindrin-forming fragments of αB-crystallin.13 The tandem-repeat peptides of Aβ are annotated as GG(24-34), GG(25-35) and GG(26-36). Scheme 1 shows the sequences of full length Aβ(1-42) and of the three single and tandem-repeat peptide fragments used in this study.
Scheme 1.
Primary structures of Aβ(1-42), Aβ(24-34), Aβ(25-35), Aβ(26-36) and their tandem repeats. The postulated metal-binding region and the central hydrophobic core are annotated. The sequence common to all three peptides is colored red. Methionine in the peptide fragments is colored green.
MATERIALS AND METHODS
All peptides were synthesized by FMOC (N-(9-fluorenyl)methoxycarbonyl) chemistry with acetylated N-termini and amidated C-termini. Dried peptides were dissolved in water or in 20 mM ammonium acetate or sodium phosphate buffer, pH 7.0, to the final concentration of 50–100 μM. The samples were incubated at room temperature for 24 hours to one week.
Prediction of cylindrin-compatible Aβ fragments
Using ROSETTADESIGN (www.rosettacommons.org), the sequence of Aβ was threaded onto the backbone structure of the hexameric αB-crystallin cylindrin (PDB ID 3SGO). After side-chain repacking, the energy of each 11-aa stretch of Aβ (in the cylindrin conformation) was calculated. C-terminal fragments Aβ(24-34), Aβ(28-38) and Aβ(32-42) scored well, that is, they had energies that were lower than that of the native cylindrin sequence (Table 1). Each of these fragments contains a glycine at position 6, which allows space for packing side chains of the adjacent internal site, position 4. Further manual predictions of cylindrin-compatible fragments were made based on having a pattern of internal glycines adjacent to aliphatic residues. Specifically, sequences containing an aliphatic residue at position 6, with glycines at positions 4 and 8, were predicted to have favorable internal packing. Sequences matching this pattern include Aβ(26-36) and Aβ(30-40). We note that since these two sequences were chosen manually, their Rosetta Energy scores were not available. In the present work, we chose to study the predicted cylindrin-compatible Aβ(24-34) and Aβ(26-36) fragments, the closely related Aβ(25-35) fragment, and the GG-linked tandem-repeats of these three fragments.
Table 1.
Sequences and ROSETTADESIGN energies of 11-residue cylindrin-compatible fragments.
| Protein Fragment | Sequence | Rosetta Energy Units |
|---|---|---|
| α B crystallin cylindrin | KVKVLGDVIEV | −166.00 |
| Aβ(24-34) | VGSNKGAIIGL | −199.00 |
| Aβ(26-36) | SNKGAIIGLMV | n/a |
| Aβ(28-38) | KGAIIGLMVGG | −217.88 |
| Aβ(30-40) | AIIGLMVGGVV | n/a |
| Aβ(32-42) | IGLMVGGVVIA | −205.81 |
Ion-mobility mass spectrometry
In IM-MS, species at a particular m/z (m = mass, z = charge) with either different conformations or different n/z (n = oligomer number, or number of monomer subunits) can be separated by measuring arrival time distributions (ATDs). In these experiments, ions are generated from solution by nano-electrospray ionization (n-ESI), captured by an ion funnel and then pulsed, via a ‘drift voltage’, into a drift cell filled with helium gas. Species with larger charge are ‘pushed’ harder by the drift voltage and travel faster than species with smaller charge. In contrast, species having the same charge state but a larger shape will collide with helium atoms more frequently, and be slowed to a greater degree, than species with smaller shape. Upon exiting the drift cell, the species of interest are selected by a mass analyzer and passed on to the detector. The ATDs of these species are measured with the pulse occurring at time t = 0 and the arrival at the detector occurring at time tA. By measuring ATDs at different pressure-to-drift-voltage ratios (P/V), the mobility K0 can be measured,35 and the cross section σ can be calculated (see Eq. 1).36 These cross-section values are independent of instrumental parameters and can be compared with cross sections generated from theoretical structures.
| (Eq. 1) |
Here, N is the buffer gas number density, μ is the reduced mass of the collision system (ion + helium), kB is Boltzmann’s factor and T is the drift cell temperature. The flux of ions exiting the drift tube can be calculated. It is assumed that the ion packet takes the form of a periodic delta function and the flux is given by Eq. 2
| (Eq. 2) |
where z is the distance the ions travel, r0is the radius of the intial ion packet, a is the area of the exit aperture, DL and DT are the longitudinal and transverse diffusion coefficients, s is the initial ion density and α is the loss of ions due to the reactions in the drift tube.35 The line shape generated from Eq. 2 would correspond to that of an ion packet composed of a single ion conformation. Experimental line shapes broader than this limit indicate more than one conformation is generating the experimental peak.
Here we use two different IM-MS instruments with somewhat different capabilities, for reasons described below:
Instrument I. This lab-built instrument37 consists of an n-ESI source, an ion funnel, a 200-cm-long drift cell, and a quadrupole mass filter. The long drift cell allows for good separation of oligomers of different size.
Instrument II. This instrument is similar to instrument I, except with a shorter, 5.0-cm-long drift cell.38 The injection voltages on this instrument can be manually controlled, so it is possible to perform injection energy studies on this instrument. In brief, by gradually increasing the injection energy applied to the ions, larger, less-stable oligomers can be broken apart into smaller, more-stable oligomers.39 This method is useful in determining the oligomer-charge ratios (n/z) of features in the ATDs that contain multiple peaks. In addition, this instrument can detect oligomers with lower charge states than instrument I, which is useful for a more-detailed investigation of large oligomers and their conformations.
Transmission Electron Microscopy (TEM), Atomic Force Microscopy (AFM), Gel-Filtration/Size Exclusion Chromatography, Dot Blot Assay, Thioflavin-T (ThT) Assays, Circular Dichroism (CD) Spectroscopy and Molecular Dynamics (MD) Simulations
Details of experimental procedures for obtaining TEM and AFM images, gel filtration/size exclusion chromatography, dot blot assays, ThT assays, CD spectra and parameters for standard and replica-exchange MD simulations, can be found in Supporting Information section S1.
RESULTS AND DISCUSSION
Imaging data indicate different aggregation characteristics for similar Aβ fragments
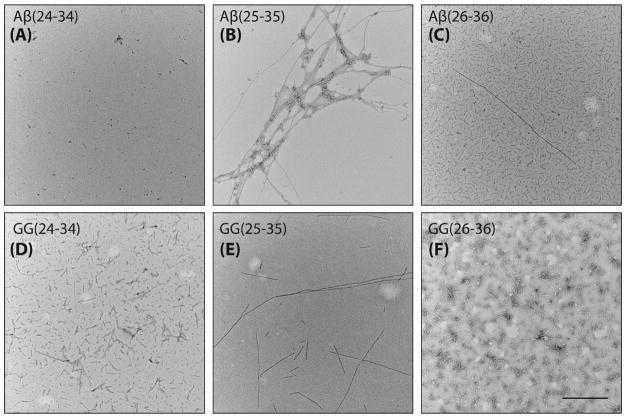
We examined the aggregation characteristics of the six peptides by AFM and TEM (see Figure 1 and Supporting Information Figures S1–S3). All peptides formed fibrils after a 24-hour incubation except Aβ(24-34), which did not form fibrils observable by microscopy even after one week (Figure 1A). The weaker aggregation propensity of Aβ(24-34) is consistent with previous studies by Pike et al. and Hou et al. showing that methionine M35 is crucial for fibril formation.24,33,40 Circular dichroism studies indicate that the single-repeat Aβ(24-34) and Aβ(25-35) remained intrinsically disordered over time whereas Aβ(26-36) might show a presence of mixed α/β structure, or β-sheet structure with an higher than typical minimum at ~220 nm (see Supporting Information Figure S4, top panels). On the other hand, the tandem-repeat peptides possessed mixed α/β elements (see Figure S4, bottom panels), suggesting that the tandem-repeat peptides become structured more readily than the single-repeat peptides.
Figure 1.
Representative TEM images of peptides incubated at 150 μM in water for one week. The scale bar is 100 nm.
We observed a variety of aggregate morphologies for GG(26-36) (Figure 1F), including a mix of elongated twisted fibrils, short fibrils, non-fibrillar aggregates, and ring-like structures. A comparison to microtubule morphology reveals that some of the short fibrils found in GG(26-36) may be similar in shape (see Supporting Information Figure S2, blue arrows). Similar results were observed from the set of AFM images (see Supporting Information Figure S3) with Aβ(25-35) forming the most fibrils out of the two single-repeat peptides while all three tandem repeat peptides show abundant fibrils and non-amyloid aggregates.
The overall microscopy data suggest that these peptides have different aggregation characteristics in which Aβ(24-34) does not appear to form fibrils and Aβ(26-36) forms less regular fibrils. The effect of the GG linker also varies. Our Thioflavin-T assays indicate that both Aβ(24-34) and Aβ(26-36) have weaker aggregation propensities than Aβ(25-35) (see Supporting Information Figure S5A). On the other hand, the same experiments reveal that GG(26-36) is the most aggregation-prone tandem-repeat out of the three, followed by GG(25-35), and GG(24-34) remains the weakest (see Figure S5B).
The Aβ fragments and tandem GG repeats form stoichiometric oligomers with similar cross sections
We next turned to IM-MS to investigate oligomer structure. All mass spectral data reported here are of the six peptides in water. Ammonium acetate buffer (an ESI-friendly solvent) yielded similar charge state distributions and aggregate morphologies (see Supporting Information Figures S6–S7 for TEM and AFM images obtained in buffered conditions), but the signals for oligomer peaks were less intense and the arrival time distributions (ATDs) were less resolved than in water.
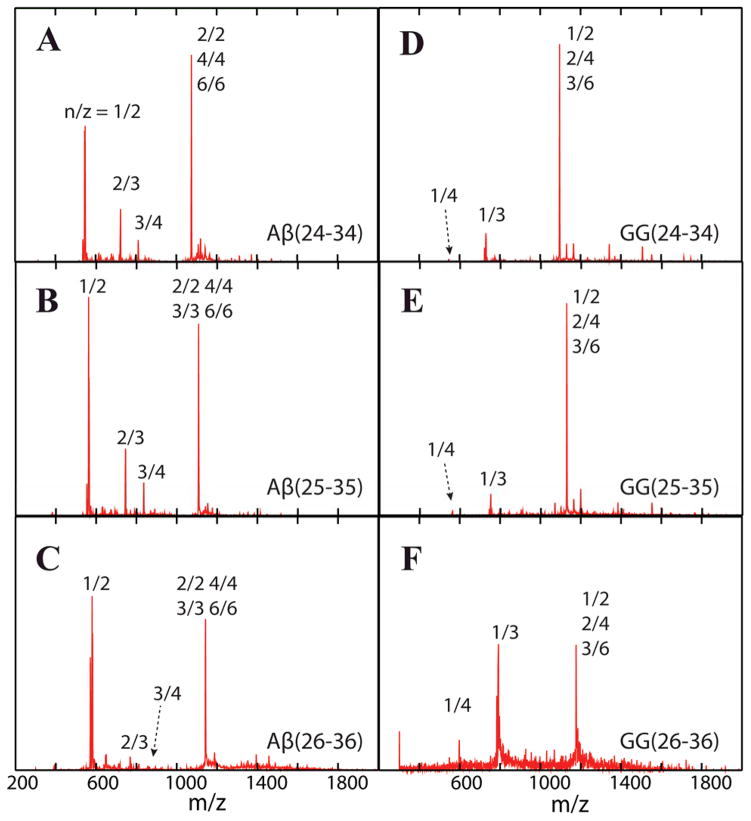
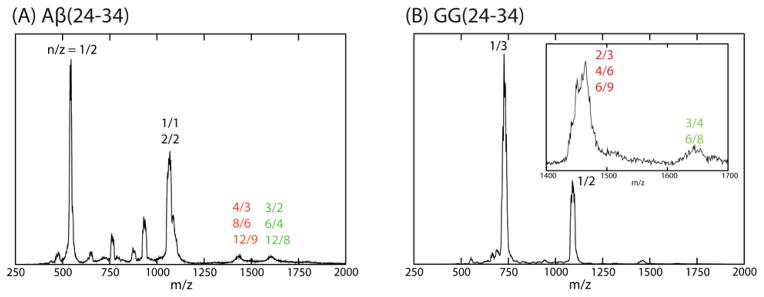
Figure 2 shows the mass spectra of the six peptides obtained from the high-resolution instrument I. For the single repeat peptides, panels A, B and C, there are two dominant peaks. At lowest m/z are peaks designated n/z = 1/2. The ATDs for these peaks show a single feature (Figure S8), which can be assigned as the doubly charged monomers. Small features labeled n/z = 2/3 and 3/4 are next highest in m/z. The ATDs (Fig S8) indicate the n/z = 2/3 peaks are exclusively the triply charged dimers and the n/z = 3/4 peaks dominantly the tetra-charged trimers. The final noted peaks in the mass spectra, in the absence of ATD information, would be assigned as n/z = 1/1. However, analysis of the ATDs given in Figure 3 indicate that there actually is no singly charged monomer in any of these nominal n/z = 1 peaks but rather only multiply charged oligomers (2/2, 3/3 etc.). Similar analysis leads to the assignments of the various peaks in the GG tandem repeat mass spectra shown in panels D, E and F.
Figure 2.
n-ESI-quadrupole mass spectra of Aβ(24-34), Aβ(25-35), Aβ(26-36) and their GG tandem repeats. Each mass spectral peak is annotated with an n/z ratio where n is the oligomer number and z is the charge. When multiple designations occur, they come from analysis of the ATD of that peak. The peptide concentration is 100 μM.
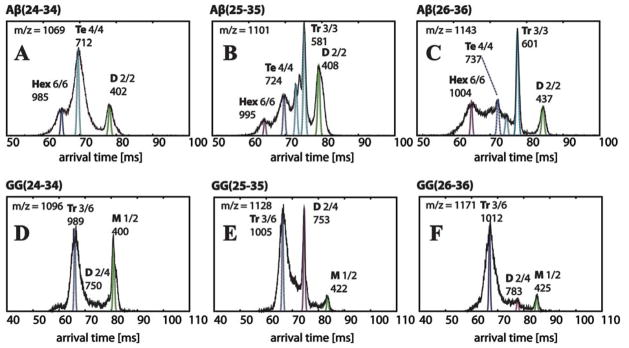
Figure 3.
Representative arrival time distributions (ATDs) of the natural charge state (one charge per monomer) peaks of Aβ(24-34), Aβ(25-35), Aβ(26-36) and their GG versions obtained from instrument I. Each feature is labeled with oligomer size (M = monomer, D = dimer, Tr = trimer, Te = tetramer, Hex = hexamer), n/z ratio and experimental cross section σ in Å2. The peptide concentration is 100 μM. The narrow dashed lines are the peak shapes predicted for a single conformer of the cross sections given in the Figure. The ATD features are broader than the predicted shape for a single conformer, suggesting there are multiple families of structures with similar cross sections. The cross sections listed above the peaks and in Table 2 correspond to these dotted line peaks.
We arrived at these assignments using several considerations. Of most importance are the values of the cross sections of the ATD features at longest times.
The assignment of features in the Aβ(25-35) ATD (n/z = 1/1, 1101 m/z; Figure 3B) is representative of the process we followed for each sample. The feature with the longest arrival time must be either a monomer or a small oligomer. If assigned as a monomer, the arrival time indicates a cross section of 204 Å2, which is significantly too small. Our previous work23,41 shows that the smallest Aβ(25-35) monomer should have a cross section of about 250 Å2. In this work, IM-MS experiments (Figure S8) and T-REMD simulations (Figure S9) show the monomer has a cross section in the 260 to 280 Å2 range. Hence, we assigned this feature as a dimer.
Assignment of the remaining features in the ATD took advantage of the fact that for oligomers of the same n/z the oligomer with the highest n travels fastest through the cell because the charge increases linearly with n but the cross section more slowly with n.39 For example, a dimer with two charges will travel through the cell faster than a monomer with one charge since the summed cross sections of two monomers is always greater than the angle averaged cross section of the corresponding dimer. Hence, we assigned the next feature as a trimer, since the cross section would be too small for a dimer. The two partially resolved features at immediately shorter times are also trimers. The next feature, near 70 ms, is therefore the tetramer. We assigned the feature at the shortest time as a hexamer (n/z = +6/6), rather than a pentamer (n/z = +5/5), based on the trend in the spacing between each pair of features. As we move from dimer to trimer to tetramer, the spacing between features decreases, since adding a monomer adds a proportionately smaller volume as oligomers get larger. However, the spacing increases between the tetramer peak and the shortest time peak, indicating an oligomer larger than a pentamer.
Using the same analysis, we assigned all of the features in the ATDs of Aβ(24-34) and Aβ(26-36) (Figure 3, panels A and C). In the Aβ(24-34) ATD (n/z = 1/1, 1069 m/z, Figure 3A), the trimer feature near 75 ms is missing. We note that the cross sections of dimer, trimer, tetramer, and hexamer species are similar among all three Aβ fragments, as are the spacings between their features in the ATDs (Table 2, Figure 3A–C).
Table 2.
Experimental cross sections (σ, Å2) of single-repeat Aβ and GG tandem-repeat monomers and oligomers.
| Single-repeat | Dimer | Tetramer | Hexamer | Tandem-repeat | Monomer | Dimer | Trimer |
|---|---|---|---|---|---|---|---|
| Aβ(24-34) | 402 | 712 | 985 | GG(24-34) | 400 | 750 | 989 |
| Aβ(25-35) | 408 | 724 | 995 | GG(25-35) | 422 | 753 | 1005 |
| Aβ(26-36) | 437 | 737 | 1004 | GG(26-36) | 425 | 782 | 1012 |
The ATDs of the tandem-repeat GG peptides (Figure 3D–F) were less challenging to assign. These ATDs of species with nominal n/z = 1/2 have oligomers whose number of charges per Aβ repeat are identical to the species of nominal n/z = 1/1 of the single-repeat Aβ. We assigned the features near 80 ms to be monomers whose cross sections are comparable to the dimers of single-repeat Aβ peptides. The middle features are thus dimers, whose cross sections correlate with those of the single-repeat tetramers (see Table 2). The remaining features are assigned as trimers, having cross sections comparable to single-repeat hexamers. Taken together, these cross-section data suggest that the GG linkers do not significantly affect the quaternary structures of the oligomers. In summary, ion-mobility experiments by instrument I suggest that all of the three Aβ fragments can form hexamers, whereas GG tandem-repeats populate trimers.
Of note, size exclusion chromatography (SEC) reveals that GG(24-34) forms a trimer (see Supporting Information Figure S10). Furthermore, this trimer is recognized by the oligomer-specific antibody, A11, while fibrils of the same segment are not recognized by A11. GG(25-35) and GG(26-36) oligomers could not be resolved with SEC, most likely because GG(25-35) and GG(26-36) are significantly less soluble than GG(24-34). Therefore, we did not pursue these methods to characterize their oligomeric forms. However, IM-MS data on both these peptides strongly support the notion that both segments are capable of forming trimers of tandem-repeat Aβ fragments.
The cross sections of Aβ-fragment hexamers and tandem-repeat GG trimers are in good agreement with cylindrin model structures
In order to determine whether or not the observed single-repeat Aβ hexamers and GG tandem-repeat trimers could be cylindrins, we made cylindrin models of all six peptides and calculated their cross sections. Beginning with the x-ray crystal structures of the αB-crystallin cylindrin hexamer and tandem GG trimer (PDB ID 3SGO and 3SGR),13 we substituted the side chains to match each of the six Aβ constructs using Swiss-PDB (http://www.expasy.org/spdbv/).42,43 This modeling was followed by MD relaxation using the GROMACS package,44,45 to allow the side chains to structurally equilibrate. The final model structures are shown in Figure 4. Because it is challenging to calculate accurate cross sections for such complex structures, we used two methods: the trajectory (TJ) method available from the Mobcal package46,47 and the projected superposition approximation (PSA) method48,49. The calculated cross sections agree reasonably well with each other and with the experimental cross sections (Table 3), especially given the approximate nature of the theoretical structures. We note that the experimental cross sections of the single-repeat Aβ hexamers and GG tandem-repeat trimers are smaller than the β-sheet-like hexamers (σ > 1098 Å2) that were previously observed for the uncapped Aβ(25-35) peptide.23
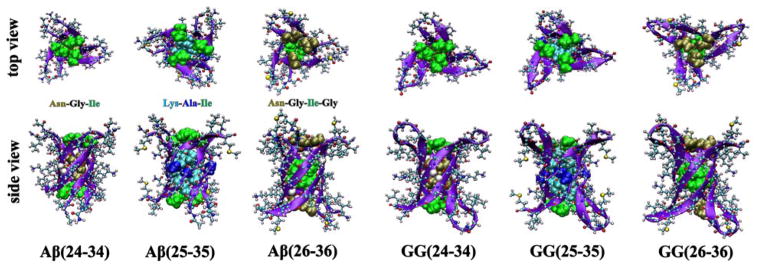
Figure 4.
Cylindrin models of single-repeat Aβ hexamers and tandem-repeat GG trimers. Each peptide chain is shown as a violet β-strand in CPK representation. The side chains inside the cylindrin cavities are shown in space filling representation.
Table 3.
Experimental and theoretical cylindrin cross sections (σ, Å2) of the hexamers of Aβ(24-34), Aβ(25-35) and Aβ(26-36) and the trimers of GG(24-34), GG(25-35) and GG(26-36). The cross section data are from instrument I. The theoretical cross sections were calculated using the trajectory (TJ)46,47 and the projected superposition approximation (PSA)48,49 methods.
| Peptide | Aβ(24-34) | Aβ(25-35) | Aβ(26-36) | GG(24-34) | GG(25-35) | GG(26-36) |
|---|---|---|---|---|---|---|
| σEXP (Å2) | 985 | 995 | 1004 | 989 | 1005 | 1012 |
| σTJ (Å2) | 1038 | 1041 | 1074 | 1058 | 1067 | 1101 |
| σPSA (Å2) | 901 | 949 | 942 | 938 | 974 | 965 |
It is important to determine whether the Aβ and GG cylindrin structures uniquely explain the experimental data. Other possible structures are β-sheets and steric zippers (i.e., the multi-layer β-sheet structures) which are frequently encountered in the aggregation mechanism of short peptides. We constructed both parallel and anti-parallel β-sheets of Aβ(25-35) and calculated their cross sections (obtained by averaging the TJ46,47 and PSA48,49 cross sections). We found both parallel (σav = 1059 Å2) and anti-parallel (σav = 1116 Å2) β-sheets are significantly larger than experiment (σ EXP = 995 Å2) indicating that the cylindrin is a more realistic structure.
The steric zipper, is stabilized through backbone hydrogen bonds between peptide strands within the same β-sheet layer, and side chain interactions within the dry interfaces between the two mating sheets.19 Eisenberg and co-workers, using x-ray crystallography, obtained a steric zipper model of Aβ(27-32) and quasi-ordered diffraction of Aβ(22-35) microcrystals, and used them to construct an ideal Aβ(25-35) steric zipper (see Supporting Information Figure S11).50 This structure indicates that the peptide strands are parallel to each other within one sheet and anti-parallel between two face-to-face mating sheets. The anti-parallel interactions between the two face-to-face mating sheets are important factors contributing to Aβ(22-35) steric zipper stability. Hence, we constructed a steric zipper of Aβ(25-35) using the same atom coordinates of the steric zipper of Aβ(22-35), minimized the structure in vacuum and computed its theoretical cross section. The theoretical cross section of this model (σav = 978 Å2) is smaller than both the theoretical cross section of the cylindrin model and experimental cross section (σav = σEXP = 995 Å2).
The excellent agreement between the experimental cross sections of the single repeat Aβ hexamers and the tandem repeat GG trimers (Table 2) supports the conclusion that they adopt the same type of structure. However, for the GG tandem repeats to form steric zippers, the GG repeat would need to adopt a β-arch structure (i.e., a strand-turn-strand motif in which the two β-strands interacting via their side chains, but not hydrogen bonds) instead of a β-hairpin conformation. This is very unlikely due to the short length of the GG linker. Finally, the experimental cross section of GG(25-35) trimer (σEXP = 1005 Å2) is significantly smaller than theoretical cross sections of the corresponding β-hairpin stacking models (σav = 1075 and 1100 Å2, Figure S11). Hence, we conclude that the cylindrins (Figure 4) are the best models to explain the experimental cross section data.
The models shown in Figure 4 highlight some differences among the peptides. The cylindrins of Aβ(25-35) and GG(25-35) have less β-strand content than the other four models due to the lysine residues being positioned inside the cavity. In Aβ(24-34), Aβ(26-36), GG(24-34) and GG(26-36), Asn and Ile are the only two amino acids of each chain that participate in forming the cylindrin core. As a result, the cylindrin core of these four peptides are relatively hydrophobic whereas those of Aβ(25-35) and GG(25-35) are amphipathic. Previous toxicity assays on cylindrin models with dry and hydrophobic cores show that membrane disruption is not responsible for their toxicity.13 However, we hypothesize that when inserting into a cell membrane, the amphipathic core of Aβ(25-35) cylindrin may create an ion transport channel leading to membrane leakage. A similar mechanism has been proposed for phenylalanine oligomers in Phenylketonuria disease.51
Injection energy studies reveal octamers and dodecamers
To verify the existence of single-repeat Aβ hexamers and GG tandem-repeat trimers, we collected IM-MS data on instrument II. Instrument II is better at generating low-charge-state oligomers than instrument I, and thus larger oligomers can often be detected. Figure 5 shows the n-ESI mass spectra of Aβ(24-34) and GG(24-34) in water obtained from instrument II. The mass spectra of the other peptides are shown in Supporting Information Figure S13.
Figure 5.
n-ESI-quadrupole mass spectra of (A) Aβ(24-34) and (B) GG(24-34). Each mass spectral peak is annotated with n/z ratio where n is oligomer number and z is charge. When multiple designations occur, they come from analysis of the ATD of that peak. The peptide concentration is 50 μM.
One major difference between the mass spectra in Figures 2 and 5 (and Figure S13) is the presence of low charge state species (z < n) whose ATDs can be recorded using instrument II. Due to the difference in construction, the same oligomers present in solution can have different charge states when sprayed from instruments I and II (i.e., large ions generated from instrument II tend to have lower charge states than the same ions generated by instrument I). The mass spectrum of Aβ(24-34) (Figure 5A) shows the presence of n/z = 3/2 (1604 m/z) and 4/3 (1427 m/z) while that of its GG version shows n/z = 2/3 (1644 m/z) and 3/4 (1462 m/z). It is important to note the species of Aβ(24-34) oligomers having n/z = 3/2 have approximately 0.67 charge per Aβ(24-34) repeat, which is approximately the same as the species of GG(24-34) having n/z = 3/4. Similarly, the n/z = 4/3 species of Aβ(24-34) contains 0.75 charge per Aβ(24-34) repeat, similar to the n/z = 2/3 species of GG(24-34). The ATDs are given in Figure 6. The overall ATDs of the species with the same charge per Aβ(24-34) are similar, indicating similar oligomer formation in both cases, consistent with the data obtained from instrument I.
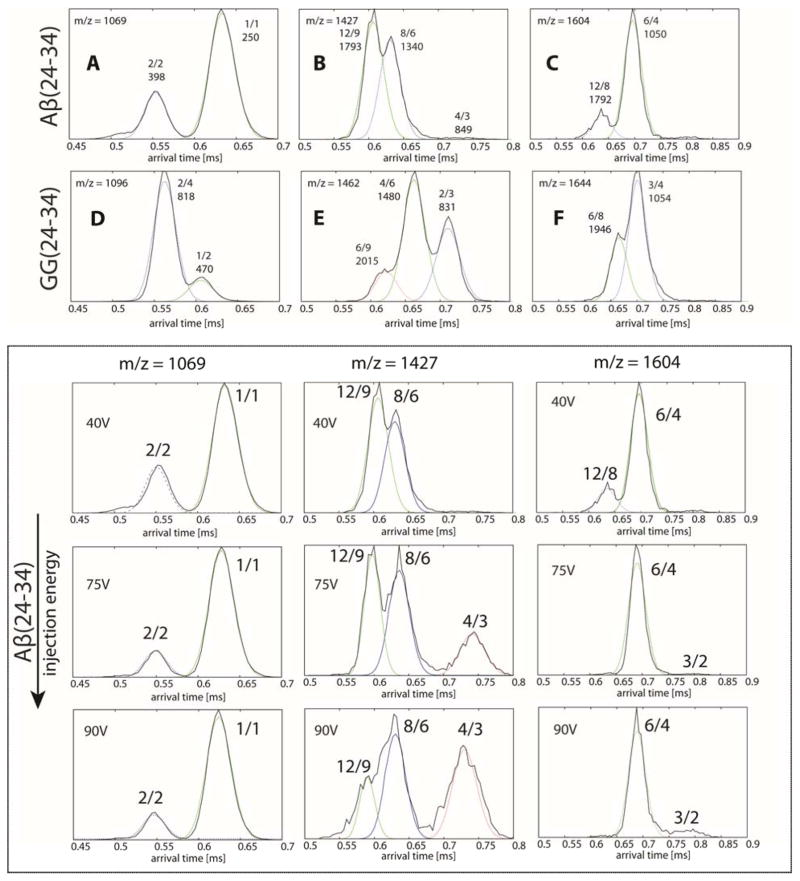
Figure 6.
(Top panel) Representative arrival time distributions (ATDs) of the n/z = 1/1, 4/3 and 3/2 peaks of Aβ(24-34), and 1/2, 2/3 and 3/4 of its GG tandem version. The features in the ATDs of the low charge state peaks are assigned based on injection energy studies. Each feature is annotated with n/z ratio and experimental cross section σ in Å2. The narrow dashed lines are the peak shapes predicted for a single conformer of the cross sections given in the Figure. The peptide concentration is 50 μM. (Bottom panel, in box) Representative ATDs illustrating the injection energy studies for Aβ(24-34).
A second difference between the data obtained from the two instruments is the ATDs of n/z = 1/1 of Aβ(24-34) (Figure 6A) and n/z = 1/2 of GG(24-34) (Figure 6D). The largest oligomers detected from these ATDs are only Aβ(24-34) and GG(24-34) dimers. Larger oligomers are not detected at these charge states. However, a hexamer and a dodecamer of Aβ(24-34) are observed at 1604 m/z (Figure 6C). Similarly, an octamer and another dodecamer are observed at 1427 m/z. These large oligomers (i.e., the shorter time features in the ATDs) dissociate into trimer and tetramer, respectively, at high injection voltages (see bottom panels of Figure 6).
Of note, a simple calculation using the tetramer (n/z = 4/3, σ = 849 Å2) predicts the cross sections of octamer and dodecamer to be σpredict (n/z = 8/6) = 849 × 22/3 = 1347 Å2 and σpredict (n/z = 12/9) = 849 × 32/3 = 1766 Å2 in very good agreement with experiment suggesting the larger oligomers are multimers of the tetramer. On the other hand, a similar procedure for the n/z = 12/8 dodecamer cross section starting from the n/z = 6/4 hexamer yields a smaller cross section than the experiment (1667 vs. 1792 Å2), indicating the larger dodecamer does not have a cylindrin structure and possibly has a steric zipper or other β-sheet type structure.
Similar results are obtained for GG(24-34) where oligomers as large as hexamers (i.e., stoichiometrically equivalent to an Aβ(24-34) dodecamer) are found in the ATDs (see also Supporting Information Figure S14). The cross sections of the Aβ(24-34) hexamer (σexp = 1050 Å2) and octamer (σexp = 1340 Å2) are very similar to the cross sections of the GG(24-34) trimer (σexp = 1054 Å2) and tetramer (σexp = 1343 Å2). The cross sections measured on instrument II for the hexamers are somewhat larger (+5%, see Table S1) than those measured on the more accurate instrument I. Nevertheless, the charge state distributions in the mass spectra and injection studies unambiguously support the presence of hexamer, octamer and dodecamer of Aβ(24-34) as well as trimer, tetramer and hexamer of GG(24-34). The large oligomers are also found in Aβ(25-35), Aβ(26-36) and their tandem GG versions (see Supporting Information Figures S15–16).
A minor, but interesting difference between the data obtained from the two instruments is that the ATD peaks in instrument II appear to better fit the expected single species line width than in instrument I. Since instrument II provides less gentle conditions than instrument I, several metastable structures could be annealed into fewer families of structures during the ion-trapping and injection process. These less gentle conditions may also account for the absence of higher order oligomers in the n/z = 1/1 peaks of the single-repeat peptides and in the n/z = 1/2 peaks of the GG tandem repeats
Cylindrical octamer of amyloid peptides can be formed from anti-parallel β-sheet constructed with a high shear number
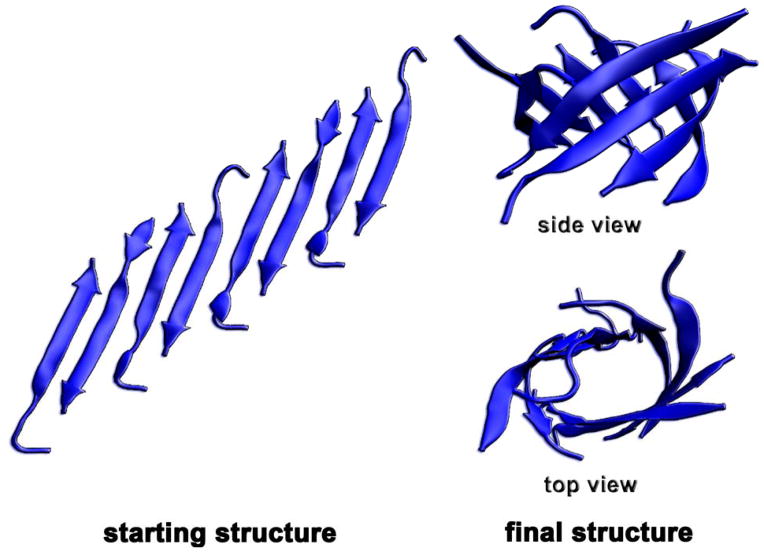
Low charge state species observed in instrument II unambiguously support the presence of octamers of the single-repeat Aβ fragments (Figure 6). In order to elucidate the structure of octamers, a standard MD simulation was performed starting with a pre-built out-of-register β-sheet. The simulation started with an out-of-register (triclinic) antiparallel Aβ(25-35) β-sheet (octamer) (see Figure 7 for the starting structure). Of the three single repeat peptides, Aβ(25-35) is more biologically active than either Aβ(24-34) or Aβ(26-36).33 Further, Aβ(25-35) shows similar fibril morphology to the full-length protein for both single- and tandem-repeat versions.23,33,34 This peptide also forms the highest populations of octamers and GG tandem-repeat tetramers. Of note, the simulation demonstrates that the cylindrins/β-barrels can be formed from out-of-register β-sheets. Some snapshots obtained from the simulation are shown in Figure 7.
Figure 7.
Initial and final structures of Aβ(25-35) octamers obtained from standard explicit solvent MD simulation.
There is abundant evidence suggesting that in-register β-sheets are the architecture of the cores of amyloid fibrils.52 Thus the formation of such a β-sheet should favor the formation of amyloid fibrils, rather than a cylindrin, although a cylindrin may have a lower free energy than a β-sheet in general according to Laganowsky et al.13 They also show that unrolling of a cylindrin hexamer yields an antiparallel β-sheet with the shear number S = 6 (i.e., a measure of the stagger of the strands within the sheet)53 and the mean slope of strands to the central axis of the barrel of 35°. However, we show here by MD simulation that a triclinic antiparallel β-sheet with higher shear number can fold into a β-barrel which resembles a large cylindrin. A cylindrin can be considered as a specific type of β-barrel that exists for small oligomers. These cylindrins and β-barrels can become toxic agents mainly by interacting with cell membranes as proposed by other research groups.54–56 The cross section obtained from the TJ method is 1355 Å2 and that from the PSA method is 1205 Å2, which is very similar to Aβ(25-35) octamer (σexp = 1320 Å2) and GG(25-35) tetramer (σexp = 1426 Å2) obtained from instrument II.
SUMMARY AND CONCLUSIONS
A central question in the assembly of amyloid systems is whether or not there exists a common oligomeric structure, or family of structures, responsible for disease initiation in these systems. Evidence now strongly supports the fact that oligomers are the dominant toxic agents in Alzheimer’s disease, type 2 diabetes and other amyloid diseases. This is a very difficult question to address and successfully answer since oligomers in amyloid systems exist in a dynamic and evolving environment that resists study by standard structural methods. Two systems have, however, been shown recently to allow crystal growth and subsequent x-ray analysis of peptide fragments: The αB-crystallin and human prion protein amyloid systems.13,20 In both cases cylindrical, hexameric, β-strand structures were observed and, in the case of αB-crystallin, named a cylindrin.13 However, due to the heterogeneous and dynamic nature of most amyloid systems in solution it is very difficult to apply these methods broadly to investigate whether cylindrin type structures are common or only occur in select systems. Here we have chosen to apply ion mobility based mass spectrometry, high level molecular dynamics simulations and a variety of supporting techniques to this difficult but important problem. IM-MS has been shown to successfully obtain both oligomer distributions and structures in a number of amyloid systems23–25 and hence is an ideal technique to apply to this problem.
In this paper we have chosen to study three peptide fragments of the amyloid β-protein Aβ42 responsible for Alzheimer’s disease: Aβ(25-35) and its two nearest neighbors Aβ(24-34) and Aβ(26-36). Aβ(25-35) was chosen as it is known to both exist in the brain and to be cyto-toxic while Aβ(24-34) and Aβ(26-36) fulfill known sequence requirements for possible cylindrin formation.13 We also studied the GG tandem repeats of all three peptides in order to be consistent with the earlier study of the αB-crystallin fragment.13 The IM-MS data reveal the existence of hexamers in the aggregation cascades of all single-stranded Aβ fragments used in this study. The GG linker connecting two Aβ fragments head-to-tail stabilizes the GG tandem-repeat trimer, which is the stoichiometric equivalent of a single-repeat Aβ hexamer. Some important conclusions can be drawn from these data:
The experimental cross sections of the Aβ fragment hexamers and the GG tandem-repeat trimers are in good agreement with each other and with the cross sections of cylindrin model structures constructed from the experimental x-ray crystal structure of αB-crystallin peptide. This result suggests that cylindrin formation may be a common event in amyloid systems although further research is needed to verify this suggestion.
The Aβ-fragment octamers and corresponding GG tandem-repeat tetramers are also observed. The majority of these structures have cross sections similar to a β-barrel obtained from the folding of a triclinic anti-parallel β-sheet with a high shear number. Hence there may be families of β-barrel structures found in amyloid oligomers of which the cylindrin is the smallest one.
The formation of these cylindrin and β-barrel structures requires a specific kind of β-sheet. Due to a relatively low population in vitro, it is difficult for conventional techniques to isolate and characterize these oligomers. IM-MS provides a new approach to search for cylindrin and barrel-like oligomer structures that may well be important in initiating disease in amyloid systems.
Finally, the results presented here provide important new evidence for structures that may be involved in amyloid disease initiation. However, more research is needed to determine how wide spread cylindrin/β-barrel structures are, whether these structures are always toxic and if they are toxic what is the mechanism involved.
Supplementary Material
Acknowledgments
The authors thank Dr. Nicholas Economou and Prof. Steven Buratto at UCSB for the microscopy data and Ms. Margaret Condron at UCLA for synthesizing the peptides. We gratefully acknowledge support from the National Science Foundation grants CHE-1301032 (M.T.B.) and MCB-1158577 (J.-E.S.) for personnel support, the Air Force Office of Scientific Research grant FA9550-11-0113 (M.T.B.) for instrumental support, the National Institutes of Health grants 1RO1AG047116-01 (M.T.B.), NS038328 (D.B.T.) and AG041295 (D.B.T.) for material support and partial personal support, and a Special Santa Barbara Cottage Hospital-UCSB Research Award (N.E.L.). We acknowledge support from the Center for Scientific Computing from the CNSI and MRL: an NSF MRSEC (DMR-1121053) and NSF CNS-0960316. We acknowledge the use of the NRI-MCDB Microscopy Facility at UC, Santa Barbara.
Footnotes
The authors declare no competing financial interest.
Supporting Information. Additional AFM and TEM images of the incubated peptides, Circular dichroism and Thioflavin-T time courses, ATDs of other spectral peaks obtained using the high-resolution ion-mobility mass spectrometer, additional ATDs showing the injection studies, and a movie showing the folding of a triclinic β-sheet into a β-barrel. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hardy J, Selkoe DJ. Science. 2002;297:353. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 2.Chiti F, Dobson CM. Annu Rev Biochem. 2006;75:333. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 3.Chiti F, Dobson CM. Nat Chem Biol. 2008;5:15. doi: 10.1038/nchembio.131. [DOI] [PubMed] [Google Scholar]
- 4.Eisenberg D, Nelson R, Sawaya MR, Balbirnie M, Sambashivan S, Ivanova MI, Madsen AO, Riekel C. Acc Chem Res. 2006;39:568. doi: 10.1021/ar0500618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy RM. Ann Rev Biomed Eng. 2002;4:155. doi: 10.1146/annurev.bioeng.4.092801.094202. [DOI] [PubMed] [Google Scholar]
- 6.Caughey B, Lansbury PT. Ann Rev Neurosci. 2003;26:267. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 7.Xue WF, Hellewell AL, Gosal WS, Homans SW, Hewitt EW, Radford SE. J Biol Chem. 2009;284:34272. doi: 10.1074/jbc.M109.049809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Science. 2003;300:486. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 9.Glabe CG. J Biol Chem. 2008;283:29639. doi: 10.1074/jbc.R800016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishnan R, Goodman JL, Mukhopadhyay S, Pacheco CD, Lemke EA, Deniz AA, Lindquist S. Proc Natl Acad Sci U S A. 2012;109:11172. doi: 10.1073/pnas.1209527109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kodali R, Wetzel R. Curr Opin Struc Biol. 2007;17:48. doi: 10.1016/j.sbi.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 12.Do TD, LaPointe NE, Sangwan S, Teplow DB, Feinstein SC, Sawaya MR, Eisenberg DS, Bowers MT. J Phys Chem B. 2014;118:7247. doi: 10.1021/jp502473s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laganowsky A, Liu C, Sawaya MR, Whitelegge JP, Park J, Zhao ML, Pensalfini A, Soriaga AB, Landau M, Teng PK, Cascio D, Glabe C, Eisenberg D. Science. 2012;335:1228. doi: 10.1126/science.1213151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Simone A, Derreumaux P. J Chem Phys. 2010;132:165103. doi: 10.1063/1.3385470. [DOI] [PubMed] [Google Scholar]
- 15.Liu C, Zhao M, Jiang L, Cheng PN, Park J, Sawaya MR, Pensalfini A, Gou D, Berk AJ, Glabe CG, Nowick J, Eisenberg D. Proc Natl Acad Sci U S A. 2012;109:20913. doi: 10.1073/pnas.1218792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu L, Liu C, Stroud JC, Ngo S, Jiang L, Guo Z. J Biol Chem. 2014;289:27300. doi: 10.1074/jbc.M114.569004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgado I, Wieligmann K, Bereza M, Ronicke R, Meinhardt K, Annamalai K, Baumann M, Wacker J, Hortschansky P, Malesevic M, Parthier C, Mawrin C, Schiene-Fischer C, Reymann KG, Stubbs MT, Balbach J, Gorlach M, Horn U, Fandrich M. Proc Natl Acad Sci U S A. 2012;109:12503. doi: 10.1073/pnas.1206433109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Nature. 2005;435:773. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJW, MacFarlane HT, Madsen AØ, Riekel C, Eisenberg D. Nature. 2007;447:453. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 20.Apostol MI, Perry K, Surewicz WK. J Am Chem Soc. 2013;135:10202. doi: 10.1021/ja403001q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pham JD, Chim N, Goulding CW, Nowick JS. J Am Chem Soc. 2013;135:12460. doi: 10.1021/ja4068854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spencer RK, Li H, Nowick JS. J Am Chem Soc. 2014;136:5595. doi: 10.1021/ja5017409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bleiholder C, Do TD, Wu C, Economou NJ, Bernstein SS, Buratto SK, Shea JE, Bowers MT. J Am Chem Soc. 2013;135:16926. doi: 10.1021/ja406197f. [DOI] [PubMed] [Google Scholar]
- 24.Bernstein SL, Dupuis NF, Lazo ND, Wyttenbach T, Condron MM, Bitan G, Teplow DB, Shea JE, Ruotolo BT, Robinson CV, Bowers MT. Nat Chem. 2009;1:326. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bleiholder C, Dupuis NF, Wyttenbach T, Bowers MT. Nat Chem. 2011;3:172. doi: 10.1038/nchem.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uetrecht C, Versluis C, Watts NR, Roos WH, Wuite GJL, Wingfield PT, Steven AC, Heck AJR. Proc Natl Acad Sci U S A. 2008;105:9216. doi: 10.1073/pnas.0800406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pierson EE, Keifer DZ, Selzer L, Lee LS, Contino NC, Wang JC, Zlotnick A, Jarrold MF. J Am Chem Soc. 2014;136:3536. doi: 10.1021/ja411460w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laganowsky A, Reading E, Allison TM, Ulmschneider MB, Degiacomi MT, Baldwin AJ, Robinson CV. Nature. 2014;510:172. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benesch JLP, Ruotolo BT. Curr Opin Struc Biol. 2011;21:641. doi: 10.1016/j.sbi.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Susa AC, Wu C, Bernstein SL, Dupuis NF, Wang H, Raleigh DP, Shea JE, Bowers MT. J Am Chem Soc. 2014;136:12912. doi: 10.1021/ja504031d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi LQ, Holliday AE, Shi HL, Zhu FF, Ewing MA, Russell DH, Clemmer DE. J Am Chem Soc. 2014;136:12702. doi: 10.1021/ja505899g. [DOI] [PubMed] [Google Scholar]
- 32.Wyttenbach T, Pierson NA, Clemmer DE, Bowers MT. Ann Rev Phys Chem. 2014;65:175. doi: 10.1146/annurev-physchem-040513-103644. [DOI] [PubMed] [Google Scholar]
- 33.Pike CJ, Walencewiczwasserman AJ, Kosmoski J, Cribbs DH, Glabe CG, Cotman CW. J Neurochem. 1995;64:253. doi: 10.1046/j.1471-4159.1995.64010253.x. [DOI] [PubMed] [Google Scholar]
- 34.Kubo T, Nishimura S, Kumagae Y, Kaneko I. J Neurosci Res. 2002;70:474. doi: 10.1002/jnr.10391. [DOI] [PubMed] [Google Scholar]
- 35.Gidden J, Ferzoco A, Baker ES, Bowers MT. J Am Chem Soc. 2004;126:15132. doi: 10.1021/ja046433+. [DOI] [PubMed] [Google Scholar]
- 36.Mason EA. Transport Properties of Ions in Gases. 99. John Wiley & Sons; 1988. [Google Scholar]
- 37.Kemper PR, Dupuis NF, Bowers MT. Int J Mass Spectrom. 2009;287:46. [Google Scholar]
- 38.Wyttenbach T, Kemper PR, Bowers MT. Int J Mass Spectrom. 2001;212:13. [Google Scholar]
- 39.Bernstein SL, Wyttenbach T, Baumketner A, Shea JE, Bitan G, Teplow DB, Bowers MT. J Am Chem Soc. 2005;127:2075. doi: 10.1021/ja044531p. [DOI] [PubMed] [Google Scholar]
- 40.Hou LM, Kang I, Marchant RE, Zagorski MG. J Biol Chem. 2002;277:40173. doi: 10.1074/jbc.C200338200. [DOI] [PubMed] [Google Scholar]
- 41.Larini L, Shea JE. Biophys J. 2012;103:576. doi: 10.1016/j.bpj.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guex N, Peitsch MC. Electrophoresis. 1997;18:2714. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 43.Johansson MU, Zoete V, Michielin O, Guex N. Bmc Bioinformatics. 2012;13 doi: 10.1186/1471-2105-13-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hess B, Kutzner C, Spoel Dvd, Lindahl E. J Chem Theory Comput. 2008;4:435. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 45.Spoel DVD, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. J Comp Chem. 2005;26:1701. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 46.Shvartsburg AA, Jarrold MF. Chem Phys Lett. 1996;261:86. [Google Scholar]
- 47.Mesleh MF, Hunter JM, Shvartsburg AA, Schatz GC, Jarrold MF. J Phys Chem A. 1996;100:16082. [Google Scholar]
- 48.Bleiholder C, Wyttenbach T, Bowers MT. Int J Mass Spectrom. 2011;308:1. [Google Scholar]
- 49.Bleiholder C, Contreras S, Do TD, Bowers MT. Int J Mass Spectrom. 2013;345:89. [Google Scholar]
- 50.Colletier J-P, Laganowsky A, Landau M, Zhao M, Soriaga AB, Goldschmidt L, Flot D, Cascio D, Sawaya MR, Eisenberg D. Proc Natl Acad Sci USA. 2011:16938. doi: 10.1073/pnas.1112600108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Do TD, Kincannon WM, Bowers MT. J Am Chem Soc. 2015;137:10080. doi: 10.1021/jacs.5b05482. [DOI] [PubMed] [Google Scholar]
- 52.Groveman BR, Dolan MA, Taubner LM, Kraus A, Wickner RB, Caughey B. J Biol Chem. 2014;289:24129. doi: 10.1074/jbc.M114.578344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murzin AG, Lesk AM, Chothia C. J Mol Biol. 1994;236:1369. doi: 10.1016/0022-2836(94)90064-7. [DOI] [PubMed] [Google Scholar]
- 54.Jang H, Arce FT, Ramachandran S, Capone R, Lal R, Nussinov R. J Mol Biol. 2010;404:917. doi: 10.1016/j.jmb.2010.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang Z, Luo Y, Zhang Y, Wei G. J Phys Chem B. 2011;115:1165. doi: 10.1021/jp107558e. [DOI] [PubMed] [Google Scholar]
- 56.Jang HB, Zheng J, Lal R, Nussinov R. Trends Biochem Sci. 2008;33:91. doi: 10.1016/j.tibs.2007.10.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.