FleN from P. aeruginosa has been cloned, purified and crystallized.
Keywords: FleN, transcription, flagella, Pseudomonas
Abstract
The assembly of bacterial flagella requires the coordinated expression of a large number of genes in a hierarchical manner. These genes code for structural components of flagella, regulatory components and components that are required for chemotaxis. Stringent spatial and numerical control of flagella biosynthesis is essential for promoting motility and pathogenesis in bacteria. These genes are regulated at the level of transcription. FleN, a P-loop-containing ATPase, plays an important role in maintaining flagellar number in Pseudomonas aeruginosa. FleN exhibits anti-activator activity against FleQ, the global transcriptional regulator of flagellar genes. In order to gain insights into the regulatory mechanism of flagella synthesis, full-length FleN was crystallized in complex with the nonhydrolyzable ATP analogue β,γ-imidoadenosine 5′-triphosphate (AMPPNP) in space group C2221, with unit-cell parameters a = 49.1, b = 206.9, c = 103.3 Å. The Matthews coefficient is 2.19 Å3 Da−1 assuming the presence of two molecules in the asymmetric unit, and the corresponding solvent content is 43.7%. X-ray diffraction data were collected to a minimum Bragg spacing of 2.21 Å and crystals of FleN–AMPPNP were prepared with selenomethionine-labelled FleN for ab initio phasing.
1. Introduction
Pseudomonas aeruginosa has a single polar flagellum which is involved in the pathogenesis of infection and biofilm formation (Gellatly & Hancock, 2013 ▸; O’Toole & Kolter, 1998 ▸). Flagellar number and chemotactic motility in Pseudomonas are regulated by the protein FleN (Dasgupta et al., 2000 ▸). Deletion of the fleN gene results in upregulation of flagellar genes that code for structural and regulatory proteins, leading to the multi-flagellated phenotype, which shows motility defects (Dasgupta et al., 2000 ▸). These genes are known to be activated by FleQ, a multidomain σ54-dependent activator of flagellar genes in Pseudomonas (Arora et al., 1997 ▸; Dasgupta et al., 2003 ▸; Hickman & Harwood, 2008 ▸). Thus, FleN exhibits an anti-activator effect on FleQ-dependent transcription and this activity is important in maintaining the correct number of flagella. Homologues of FleN are present in several bacterial species, including Helicobacter pylori, Bacillus subtilis, Pseudomonas putida, Vibrio cholerae and Aquifex aeolicus (Dasgupta & Ramphal, 2001 ▸). FleN is a 280 amino-acid protein that belongs to the P-loop NTPase family. FleN has a nucleotide-binding motif that deviates from the classical Walker A motif (Dasgupta & Ramphal, 2001 ▸; Koonin, 1993 ▸). FleN does not have a DNA-binding sequence and its lack of DNA-binding ability has been confirmed by gel-retardation assays (Dasgupta et al., 2000 ▸; Baraquet et al., 2012 ▸). FleN has been shown to exert its effect through direct protein–protein interaction with FleQ in the presence as well as in the absence of DNA (Hickman & Harwood, 2008 ▸; Dasgupta & Ramphal, 2001 ▸; Baraquet et al., 2012 ▸; Baraquet & Harwood, 2013 ▸). The exact mechanism of downregulation of FleQ by FleN is not yet known. In order to understand the regulatory mechanism utilized by FleN to control flagella synthesis, we have cloned, expressed and purified FleN and have obtained crystals of the FleN–AMPPNP complex.
2. Materials and methods
2.1. Macromolecule production
2.1.1. Cloning of FleN
The FleN coding sequence (840 bp) was amplified by PCR from genomic DNA of P. aeruginosa (ATCC) and cloned into the pGEX-6P1 (GE Healthcare) expression vector between BamHI and NotI sites. The construct was verified by sequencing. Expression of protein using this vector (pGEX6P1_FleN) gives rise to GST-FleN fusion protein. The PCR amplification was performed using the following forward and reverse primers: 5′-CGTATGGATCCATGAAGCAGATGGGTAGCATGC-3′ and 5′-GCATGCGGCCGCTCATACGGCCGAACCTGTCGC-3′, respectively.
2.1.2. Overexpression and purification
The fusion protein was expressed in Escherichia coli C41(DE3) cells induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 18°C. The cells were harvested by centrifugation at 9000g for 30 min at 4°C and the cell pellet was resuspended in lysis buffer (25 mM HEPES pH 7.0, 500 mM NaCl, 5% glycerol, 0.5 mM EDTA, 2 mM DTT, 0.01% IGEPAL). PMSF was added to the cells at a concentration of 1 mM and the cells were lysed by sonication. After sonication on ice, a clear supernatant was obtained by centrifugation at 18 000 rev min−1 for 45 min at 4°C. The protein was subjected to affinity chromatography utilizing a 5 ml GST Sepharose column (GE Healthcare) pre-equilibrated with buffer A (25 mM HEPES pH 7.0, 300 mM NaCl, 10% glycerol, 2 mM DTT, 0.1 mM EDTA). The clear supernatant of the bacterial lysate was applied onto this column and then washed with 45 ml buffer A at 4°C. This was followed by washing the column with 45 ml buffer B (25 mM HEPES pH 7.0, 1000 mM NaCl, 10% glycerol, 2 mM DTT, 0.1 mM EDTA) to remove any nonspecifically bound proteins. The fusion protein was then eluted with 15 mM glutathione in buffer B. To cleave the GST tag, this protein was incubated with PreScission protease overnight at 4°C. FleN was then purified from the resulting mixture by gel-filtration chromatography using a Superdex 200 column (GE Healthcare) pre-equilibrated with buffer C (20 mM HEPES pH 7.0, 500 mM NaCl, 1 mM DTT). A 1 ml GST Sepharose column was connected in series to trap the cleaved GST tag and the GST-tagged PreScission protease. The purity of the protein was assessed by SDS–PAGE. The purified protein was concentrated using Vivaspin 6 (Sartorius) centrifugal concentrators with a molecular-weight cutoff of 10 kDa.
2.1.3. Preparation of selenomethionine-labelled FleN
To prepare selenomethionine-labelled FleN (Se-FleN), the protein was expressed in the B834 strain of E. coli, which is auxotrophic for methionine, and the cells were grown using a SelenoMet Medium kit (Molecular Dimensions). Fresh competent E. coli B834 cells were transformed with the pGEX6P1_FleN plasmid and the cells were plated onto LB agar containing ampicillin (100 µg ml−1). A single colony was used to inoculate a starter culture (20 ml) that was grown overnight at 37°C. 2 l of reconstituted SelenoMet Medium (Molecular Dimensions) were supplemented with 50 mg l−1 selenomethionine (Sigma–Aldrich) and this medium was used to wash the pelleted cells from the starter culture three times before inoculation. When the OD reached 0.8 (∼6 h), expression of FleN was induced with 0.5 mM IPTG and the flasks were incubated overnight at 18°C. Se-FleN was purified using a protocol identical to that for native protein except that all of the buffers were supplemented with 5 mM DTT.
2.2. Crystallization
Crystallization trials were carried out in 96-well plates (TTP Labtech) with a Mosquito robot using the hanging-drop vapour-diffusion method. FleN and AMPPNP (Sigma–Aldrich) were mixed in a 1:2 molar ratio. Equal volumes of protein–AMPPNP complex and reservoir solutions were mixed and 600 nl drops were equilibrated against 100 µl of the same precipitant solution in the reservoir. A number of crystallization conditions were tested using commercially available kits from Emerald Bio (Wizard I and Wizard II). Positive hits were further optimized by vapour diffusion using 24-well crystallization trays (Molecular Dimensions).
2.3. Data collection and processing
Mounted CryoLoops (Hampton Research) were used to separate and pick up the crystals. For cooling, crystals were treated with different concentrations of cryoprotectants such as glycerol and glucose for varying periods. The quality of diffraction was tested using a MAR Research GeniX X-ray generator with X-ray optics and the frames were recorded on a MAR345 imaging plate (Regional Centre for Biotechnology). The frames collected were indexed using iMosflm (Leslie, 1992 ▸). X-ray data were collected from a single crystal. The data were scaled using SCALA (Evans, 2006 ▸).
3. Results and discussion
3.1. Expression and purification
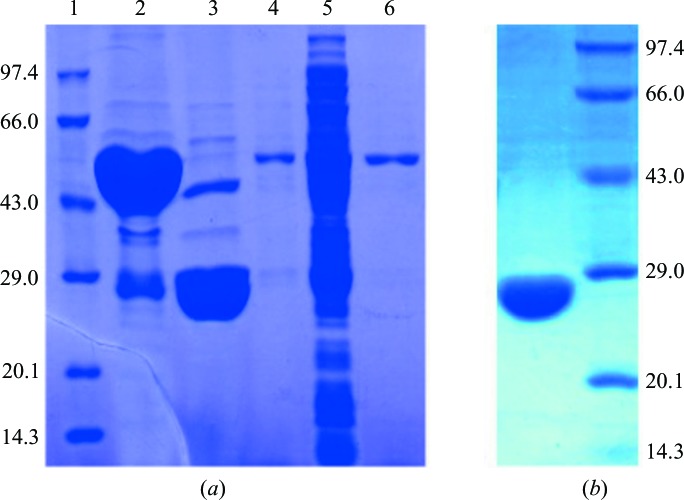
Preliminary expression studies showed that FleN is optimally expressed in E. coli C41(DE3) cells, with a significant amount of soluble protein. The heterologously expressed protein was then purified by affinity and size-exclusion chromatography (Figs. 1 ▸ a and 1 ▸ b). The sample was 99% pure as estimated by SDS–PAGE. The final yield of FleN from 5 l of culture was 6.8 mg and the protein was concentrated to a final concentration of 0.7 mM. Macromolecule-production information is summarized in Table 1 ▸.
Figure 1.
(a) Coomassie-stained SDS–PAGE analysis of FleN purified by GST column chromatography. Lane 1, marker; lane 2, target protein eluted from the GST column; lane 3, FleN after digestion of the GST tag; lane 4, wash with buffer A; lane 5, flowthrough after the supernatant had been loaded onto the column; lane 6, wash with buffer B. The molecular weights of the marker are shown in kDa on the left and the molecular weight of FleN is 30 kDa. (b) Coomassie-stained SDS–PAGE analysis of FleN after elution from the gel-filtration column. Lane 1, pure FleN; lane 2, marker.
Table 1. Macromolecule-production information.
| Source organism | P. aeruginosa POA1 |
| DNA source | P. aeruginosa POA1 |
| Forward primer† | 5′-CGTATGGATCCATGAAGCAGATGGGTAGCATGC-3′ |
| Reverse primer† | 5′-GCATGCGGCCGCTCATACGGCCGAACCTGTCGC-3′ |
| Cloning vector | pGEX-6P1 |
| Expression vector | pGEX-6P1 |
| Expression host | E. coli C41(DE3) |
| Complete amino-acid sequence of the construct produced‡ | GPLGSMKQMGSMHPVQVIAVTGGKGGVGKTNVSVNLALALADLGRRVMLLDADLGLANVDVLLGLTPKRTLADVIEGRCELRDVLLLGPGGVRIVPAASGTQSMVHLSPMQHAGLIQAFSDISDNLDVLVVDTAAGIGDSVVSFVRAAQEVLLVVCDEPTSITDAYALIKLLNRDHGMTRFRVLANMAHSPQEGRNLFAKLTKVTDRFLDVALQYVGVIPYDESVRKAVQKQRAVYEAFPRSKASLAFKAVAQKVDSWPLPANPRGHLEFFVERLVQHPATGSAV |
Restriction sites are underlined.
Amino acids from the vector are underlined.
3.2. Crystallization
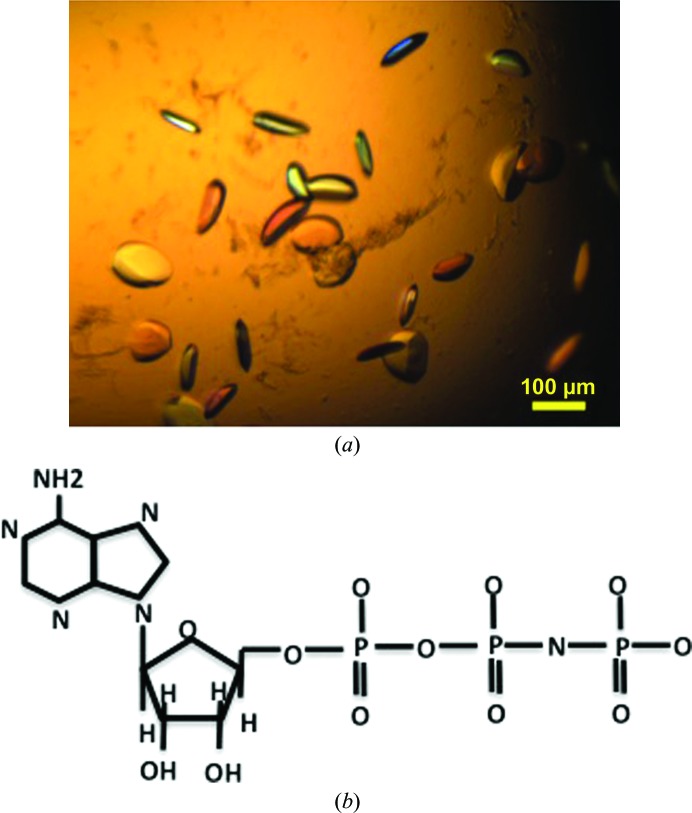
Using the hanging-drop method, crystals of the FleN–AMPPNP complex (Figs. 2 ▸ a and 2 ▸ b) were obtained at 277 K in condition No. 13 (1.26 M ammonium sulfate, 0.1 M sodium cacodylate pH 6.5) of the Wizard I screen. The concentration of NaCl and the pH of the buffer were varied to optimize the size of the crystals. The crystal grew to average dimensions of 0.11 × 0.06 × 0.02 mm. Crystallization information is summarized in Table 2 ▸.
Figure 2.
(a) Crystals of the FleN–AMPPNP complex. (b) Chemical structure of AMPPNP.
Table 2. Crystallization.
| Method | Hanging-drop vapour diffusion |
| Plate type | 24-well |
| Temperature (K) | 277 |
| Composition of reservoir solution | 1.26 M ammonium sulfate, 0.1 M sodium cacodylate pH 6.5 |
| Volume and ratio of drop | 800 nl, 1:1 |
| Volume of reservoir (µl) | 500 |
3.3. Data collection
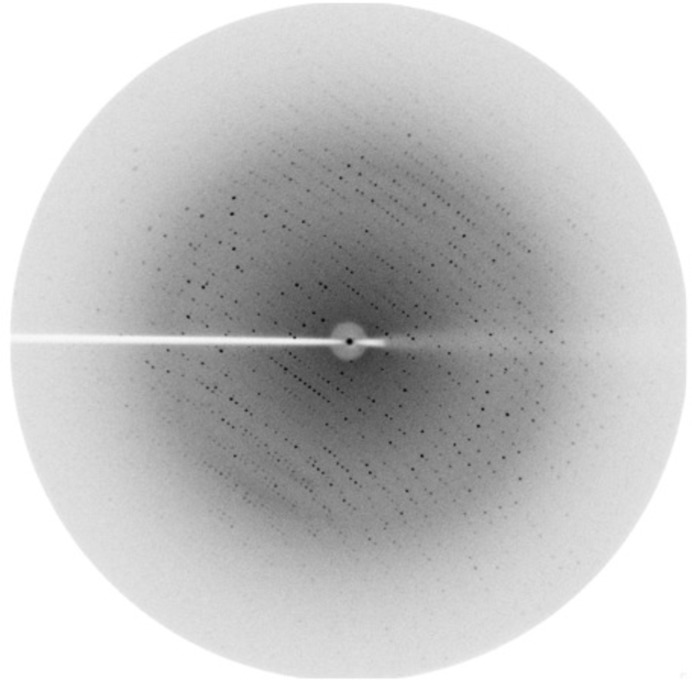
The optimized cryoprotection strategy involved soaking the crystal sequentially for 60 s in reservoir solution supplemented with 5, 10, 15 and 20% glycerol. The crystals diffracted to a resolution of 2.21 Å. The crystals belonged to the orthorhombic space group C2221, with unit-cell parameters a = 49.1, b = 206.9, c = 103.3 Å. A complete X-ray diffraction data set with a multiplicity of about 4.6 was collected with a minimum Bragg spacing of 2.21 Å. A representative frame recorded during this data collection is shown in Fig. 3 ▸. The volume of the asymmetric unit allows the presence of a dimer, giving a Matthews volume (V M) of 2.19 Å3 Da−1 and a solvent content of 43.7% (Matthews, 1968 ▸). The data-collection statistics are reported in Table 3 ▸.
Figure 3.
Diffraction image of an FleN–AMPPNP crystal.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Wavelength (Å) | 1.54 |
| Temperature (K) | 100 |
| Detector | MAR345 |
| Crystal-to-detector distance (mm) | 200 |
| Rotation range per image (°) | 1 |
| Total rotation range (°) | 130 |
| Exposure time per image (s) | 600 |
| Space group | C2221 |
| a, b, c (Å) | 49.1, 206.9, 103.3 |
| α, β, γ (°) | 90.00, 90.00, 90.00 |
| Resolution range (Å) | 51.71–2.21 (2.33–2.21) |
| Total No. of reflections | 119621 |
| No. of unique reflections | 26128 |
| Completeness (%) | 97.4 (83.3) |
| Multiplicity | 4.6 (4.4) |
| 〈I/σ(I)〉 | 8.5 (2.9) |
| R meas (%) | 17.5 (57.0) |
| Overall B factor from Wilson plot (Å2) | 12.2 |
3.4. Preparation and crystallization of selenomethionine-labelled protein
Selenomethionine-labelled FleN (Se-FleN) could be purified using the same protocol as used for the native protein. The yield of Se-FleN from 5 l of culture was 2 mg and the protein was concentrated to a final concentration of 0.5 mM. Crystals of Se-FleN were obtained under identical conditions as for the native protein and will be used for ab initio phasing.
Acknowledgments
DJ is the recipient of an Innovative Young Biotechnologist Award from Department of Biotechnology, Government of India. Chanchal acknowledges the fellowship from the Department of Biotechnology.
References
- Arora, S. K., Ritchings, B. W., Almira, E. C., Lory, S. & Ramphal, R. (1997). J. Bacteriol. 179, 5574–5581. [DOI] [PMC free article] [PubMed]
- Baraquet, C. & Harwood, C. S. (2013). Proc. Natl Acad. Sci. USA, 110, 18478–18483. [DOI] [PMC free article] [PubMed]
- Baraquet, C., Murakami, K., Parsek, M. R. & Harwood, C. S. (2012). Nucleic Acids Res. 40, 7207–7218. [DOI] [PMC free article] [PubMed]
- Dasgupta, N., Arora, S. K. & Ramphal, R. (2000). J. Bacteriol. 182, 357–364. [DOI] [PMC free article] [PubMed]
- Dasgupta, N. & Ramphal, R. (2001). J. Bacteriol. 183, 6636–6644. [DOI] [PMC free article] [PubMed]
- Dasgupta, N., Wolfgang, M. C., Goodman, A. L., Arora, S. K., Jyot, J., Lory, S. & Ramphal, R. (2003). Mol. Microbiol. 50, 809–824. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Gellatly, S. L. & Hancock, R. E. W. (2013). Pathog. Dis. 67, 159–173. [DOI] [PubMed]
- Hickman, J. W. & Harwood, C. S. (2008). Mol. Microbiol. 69, 376–389. [DOI] [PMC free article] [PubMed]
- Koonin, E. V. (1993). J. Mol. Biol. 229, 1165–1174. [DOI] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr. 26.
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- O’Toole, G. A. & Kolter, R. (1998). Mol. Microbiol. 30, 295–304. [DOI] [PubMed]