The crystal structures of two conformations of M. tuberculosis EccB1 (states III and IV) are reported and are compared with two previously reported conformations (states I and II) in order to better understand the structure–function relationship of EccB1.
Keywords: Rv3869, type VII secretion system, Mycobacterium tuberculosis, ESX-1
Abstract
The protein EccB1, a core component of the type VII secretion system (T7SS) of Mycobacterium tuberculosis, has been identified as an ATPase and is essential for the secretion of virulence factors by the ESX-1 system. In a previous study, EccB1 structures were determined in two different conformations. Here, two new conformations are identified and described. These four conformations present snapshots of the swinging movement of the membrane-distal domain A2. The movement of this domain involves conformational changes in two flexible loops (loop A, residues 243–264, and loop B, residues 324–341) which are rich in proline and glycine residues and connect domain A2 to domains C1 and B2. It is proposed that the movement of this domain is related to the ATPase activity of EccB1 and its homologues, as well as to the substrate transport of ESX secretion systems.
1. Introduction
Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis, a disease that led to 1.5 million deaths in the year 2014 alone (World Health Organization, 2015 ▸), contains five paralogous ESX secretion systems, also known as type VII secretion systems (T7SS; Gey van Pittius et al., 2006 ▸), that are designated ESX-1 through ESX-5. These secretion systems recruit secreted virulence proteins to break down the host immune system during infection (Abdallah et al., 2007 ▸). Understanding the architecture and secretory mechanisms of the T7SS will be key to understanding mycobacterial pathogenesis, and should help to facilitate the identification of novel drug targets and vaccine candidates.
EccB1 (Rv3869) from Mtb H37Rv is a core component of the ESX-1 secretion system (Das et al., 2011 ▸) and has previously been shown to be essential for substrate transport and bacterial virulence (Brodin et al., 2006 ▸); secretion of ESAT-6/CFP-10 and bacterial virulence are attenuated on knocking out the Rv3869 gene. We have previously shown that EccB1 is an ATPase that requires divalent ions: Ca2+ or Mg2+ (Zhang et al., 2015 ▸).
Here, in order to further understand the structural basis of this ATPase activity and its role in the transportation of substrates by T7SS, we describe the structures of two additional conformations of EccB1. Comparing these with two previously reported conformations (Zhang et al., 2015 ▸), we found that the membrane-distal domain A2 can swing. Sequence alignment of EccB1 homologues from the ESX-1 to ESX-5 systems indicates that two loops are rich in proline and glycine residues. Based on structure superposition, we propose that structural changes in these two loop regions are the basis for the swinging of domain A2. Our results suggest that membrane-distal domain A2 is flexible and that the oscillation of domain A2 is involved in the ATPase activity and substrate-transport mechanism of EccB1.
2. Materials and methods
2.1. Macromolecule production
The gene segment expressing the periplasmic region (residues 72–480) of Rv3869 was PCR-amplified from genomic DNA of M. tuberculosis strain H37Rv and ligated into a modified pET-28a(+) vector (pLW). Its sequence was verified by DNA sequencing. The plasmid was then transformed into Escherichia coli BL21(DE3) cells for protein expression. Large-scale cultures were grown to an OD600 of 0.8 in standard LB medium at 37°C before adding IPTG to a final concentration of 0.4 mM to induce protein expression. Incubation was continued for 16 h at 16°C (Li et al., 2011 ▸).
After harvesting, the bacterial cells were resuspended in ice-cold lysis buffer (20 mM Tris pH 8.0, 500 mM NaCl) and were subjected to high-pressure homogenization followed by centrifugation for 40 min at 4°C and 15 000 rev min−1. The supernatant was loaded onto an Ni2+-affinity resin chromatography gravity column pre-equilibrated with lysis buffer containing 10 mM imidazole. After washing with six column volumes of lysis buffer containing 60 mM imidazole, the Rv3869 protein was eluted with two column volumes of lysis buffer containing 300 mM imidazole. The eluted protein was purified by ion exchange using a HiTrap Q Sepharose FF column (GE Healthcare, New Jersey, USA), eluting with a linear gradient of Tris buffer pH 8.0 containing 1 M NaCl. Fractions containing the pure protein were pooled and concentrated for further purification on a Superdex 200 (GE Healthcare) size-exclusion column pre-equilibrated with buffer consisting of 20 mM Tris pH 7.5, 100 mM NaCl. Protein from the elution peak was quantified using a BCA protein assay kit (Pierce, Rockford, Illinois, USA) and stored at −80°C until required (Zhang et al., 2015 ▸). Macromolecule-production information is summarized in Table 1 ▸.
Table 1. Macromolecule-production information.
| Source organism | M. tuberculosis strain H37Rv |
| DNA source | M. tuberculosis strain H37Rv genome |
| Forward primer | GAAAAAACATATGGGCGGCACCAGCCTGTTCAC |
| Reverse primer | TGGGAATTCTCAGGGGGCTCCGGAGGCT |
| Cloning vector | pET-28a(+) (pLW) |
| Expression vector | pET-28a(+) (pLW) |
| Expression host | E. coli BL21(DE3) |
| Complete amino-acid sequence of the construct produced | MGSSHHHHHHSSGLEVLFQGPHMGTSLFTDRATNQLYVLLSGQLHPVYNLTSARLVLGNPANPATVKSSELSKLPMGQTVGIPGAPYATPVSAGSTSIWTLCDTVARADSTSPVVQTAVIAMPLEIDASIDPLQSHEAVLVSYQGETWIVTTKGRHAIDLTDRALTSSMGIPVTARPTPISEGMFNALPDMGPWQLPPIPAAGAPNSLGLPDDLVIGSVFQIHTDKGPQYYVVLPDGIAQVNATTAAALRATQAHGLVAPPAMVPSLVVRIAERVYPSPLPDEPLKIVSRPQDPALCWSWQRSAGDQSPQSTVLSGRHLPISPSAMNMGIKQIHGTATVYLDGGKFVALQSPDPRYTESMYYIDPQGVRYGVPNAETAKSLGLSSPQNAPWEIVRLLVDGPVLSKDAALLEHDTLPADPSPRKVPAGASGAP |
2.2. Crystallization
Initial crystallization experiments were performed at 16°C using the hanging-drop vapour-diffusion method with kits from Hampton Research (PEG/Ion, Crystal Screen and Crystal Screen 2) and a protein concentration of 4 mg ml−1 in 20 mM Tris pH 7.5, 100 mM NaCl. As in our previous study, the protein crystals of the two new structures were obtained in 20 mM Tris pH 7.5, 100 mM magnesium formate, 15%(w/v) PEG 3350 using 1 µl drops of protein solution mixed with equal volumes of reservoir solution and equilibrated over a reservoir containing 0.45 ml reservoir solution (Zhang et al., 2015 ▸). Crystallization information is summarized in Table 2 ▸.
Table 2. Crystallization.
| Method | Hanging-drop vapour diffusion |
| Plate type | 16-well hanging-drop plate |
| Temperature (K) | 289 |
| Protein concentration (mg ml−1) | 4 |
| Buffer composition of protein solution | 20 mM Tris pH 7.5, 100 mM NaCl |
| Composition of reservoir solution | 20 mM Tris pH 7.5, 100 mM magnesium formate, 15%(w/v) PEG 3350 |
| Volume and ratio of drop | 1.0 µl, 1:1 |
| Volume of reservoir (ml) | 0.45 |
2.3. Data collection and processing and structure solution and refinement
For X-ray data collection, the crystals were soaked for 40 s in reservoir solution supplemented with 10% glycerol and were then flash-cooled to 100 K in a liquid-nitrogen stream. Diffraction data sets were collected with an in-house X-ray facility using a Rigaku R-AXIS IV++ image plate and a Rigaku MICROSTAR copper rotating-anode generator operating at 40 mA and 40 kV. A total of 180 frames were collected with an oscillation step of 1° and an exposure of 5 min per frame. The crystal-to-detector distance was maintained at 200 mm. Diffraction data were processed with MOSFLM (Leslie & Powell, 2007 ▸) and scaled with SCALA from the CCP4 suite (Winn et al., 2011 ▸). Molecular replacement and structural refinement were carried out with the PHENIX software package (Adams et al., 2002 ▸) and manual model rebuilding was performed using Coot (Emsley et al., 2010 ▸). All structural figures were rendered in PyMOL (v.1.2r3pre; Schrödinger). Data collection and structure refinement are summarized in Tables 3 ▸ and 4 ▸.
Table 3. Data collection and processing.
Values in parentheses are for the outermost shell.
| State III | State IV | |
|---|---|---|
| Diffraction source | Rigaku MICROSTAR copper rotating-anode generator | Rigaku MICROSTAR copper rotating-anode generator |
| Wavelength (Å) | 1.5418 | 1.5418 |
| Temperature (K) | 100 | 100 |
| Detector | Rigaku R-AXIS IV++ image plate | Rigaku R-AXIS IV++ image plate |
| Crystal-to-detector distance (mm) | 200 | 200 |
| Rotation range per image (°) | 1 | 1 |
| Total rotation range (°) | 180 | 180 |
| Exposure time per image (s) | 300 | 300 |
| Space group | P21 | P21 |
| a, b, c (Å) | 31.61, 120.21, 61.26 | 30.81, 111.16, 62.74 |
| α, β, γ (°) | 90, 102.92, 90 | 90, 103.07, 90 |
| Mosaicity (°) | 1.06 | 1.84 |
| Resolution range (Å) | 59.71–3.00 (3.16–3.00) | 55.58–2.60 (2.74–2.60) |
| Total No. of reflections | 28708 (3610) | 32753 (4559) |
| No. of unique reflections | 8613 (1168) | 9779 (1328) |
| Completeness† (%) | 96.1 (88.9) | 77.8 (73.2) |
| Multiplicity | 3.3 (3.1) | 3.3 (3.4) |
| 〈I/σ(I)〉 | 10.3 (2.5) | 11.9 (2.5) |
| R r.i.m. | 0.066 (0.532) | 0.061 (0.451) |
| Overall B factor from Wilson plot (Å2) | 85.1 | 60.5 |
The state IV crystal was anisotropic, resulting in the low completeness of the diffraction data. Although we tried many crystals, it was difficult to collect a data set with higher completeness.
Table 4. Structure refinement.
Values in parentheses are for the outer shell.
| State III | State IV | |
|---|---|---|
| Resolution range (Å) | 30.27–3.00 | 41.11–2.60 |
| Completeness (%) | 95.68 | 76.92 |
| σ Cutoff | 0 | 0 |
| No. of reflections, working set | 7719 | 8779 |
| No. of reflections, test set | 860 | 972 |
| Final R cryst | 0.231 | 0.219 |
| Final R free | 0.286 | 0.282 |
| No. of non-H atoms | ||
| Protein | 2852 | 2813 |
| Ion | 1 | 3 |
| Water | — | 46 |
| Total | 2853 | 2862 |
| R.m.s. deviations | ||
| Bonds (Å) | 0.003 | 0.004 |
| Angles (°) | 0.856 | 0.933 |
| Average B factors (Å2) | ||
| Protein | 77.5 | 50.1 |
| Ion | 100.1 | 63.39 |
| Water | — | 36.2 |
| Total | 77.5 | 49.99 |
| Ramachandran plot | ||
| Favoured regions (%) | 88.16 | 93.33 |
| Additionally allowed (%) | 8.95 | 4.80 |
| Outliers (%) | 2.89† | 1.87† |
Most of the residues with main-chain conformation outliers are prolines or glycines; others either form hydrogen bonds to other atoms that affect the main-chain conformation or are located in turns and adjacent to prolines or glycines.
3. Results and discussion
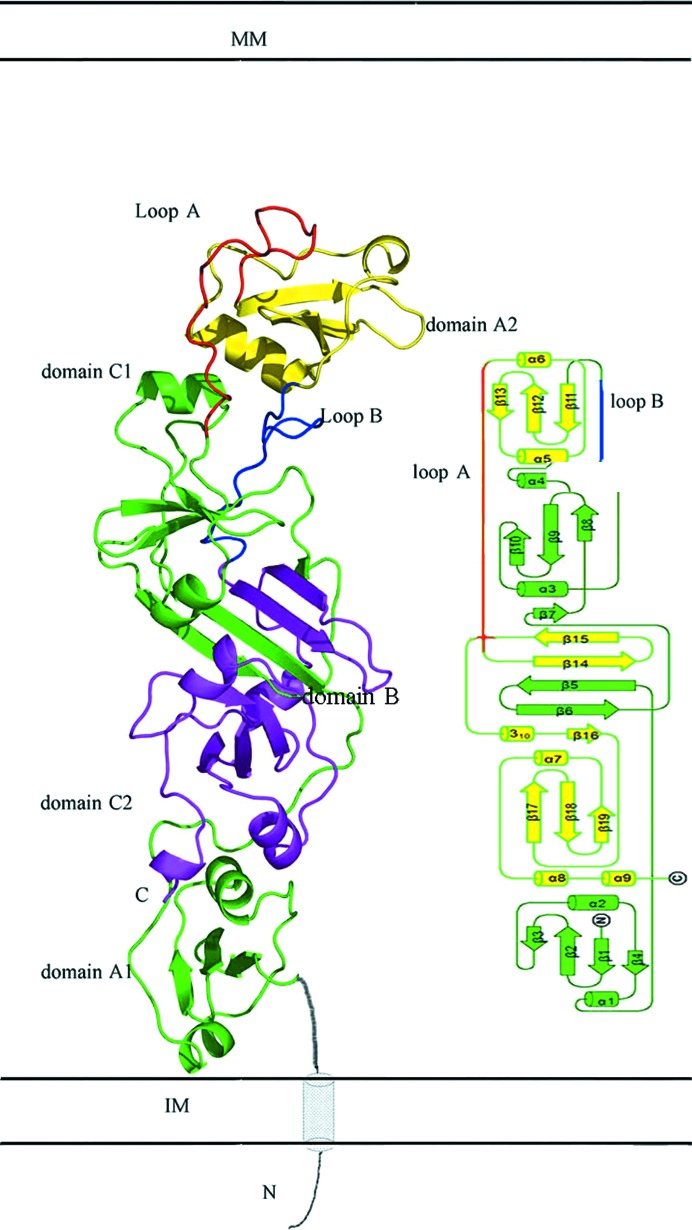
In a previous study, we showed that EccB1 is a periplasmic ATPase and determined its native (referred to here as state I) and selenomethionine-derivative (referred to here as state II) crystal structures. Interestingly, both crystal forms were obtained in the same condition, even though they had different conformations and belonged to different space groups. Here, we obtained two further crystal forms (referred to as states III and IV) using the same condition. These new crystal structures represent conformations of EccB1 that were different from those that we reported previously (PDB entries 3x3m and 3x3n; Zhang et al., 2015 ▸), and were determined at 3.0 and 2.6 Å resolution, respectively, with R factors of 0.231 and 0.219 and R free values of 0.286 and 0.282, respectively. Like the previously reported structures, the overall structures of the two new crystal forms consist of five domains, namely A1 (residues 72–135), B (B1, residues 143–180; B2, residues 342–390), C1 (residues 181–242), A2 (residues 265–323) and C2 (residues 391–451) (Fig. 1 ▸). Domains A1, C1, A2 and C2 all adopt an α/β/α sandwich fold, whereas domain B is a β-sheet. These five domains are arranged in the order A1–B1–C1–A2–B2–C2 according to the amino-acid sequence, but spatially are in the order A1–C2–B1–B2–C1–A2 from the membrane-proximal end to the membrane-distal end and adopt an ‘S’ shape.
Figure 1.
Cartoon and topological presentations of the overall structure of EccB1. EccB1 contains five domains: A1 and C1 are shown in green, A2 is shown in yellow and B1 and B2 are shown in green and purple, respectively. The N-terminus (predicted structure shown in grey) is likely to span the IM and is located in proximity to the C-terminus. In the topological structure, domains A1–B1–C1 are shown in green and A2–B2–C2 are shown in yellow. Loops A and B are shown in red and blue, respectively. MM, mycomembrane. IM, inner membrane.
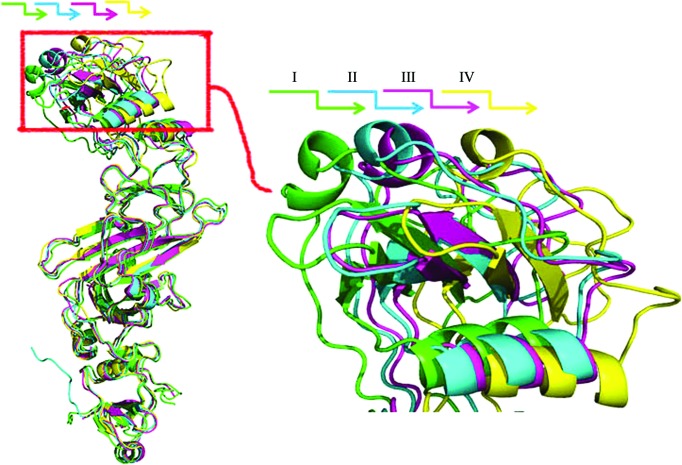
The conformations of states III (PDB entry 5ebc) and IV (PDB entry 5ebd) are similar to the previously reported native (state I; PDB entry 3x3m) and selenomethionine-derivative (state II; PDB entry 3x3n) conformations, and have overall Cα r.m.s.d.s of 1.78 and 2.17 Å from state I, respectively (Supplementary Table S1). Comparing these four conformations in order to understand the differences between them and the structural basis of the conformational changes, we observed that all of the domains are structurally conserved between the four conformations, with an inter-conformation r.m.s.d. of less than 1.0 Å (Supplementary Table S1), and that the assembly of all of the domains except A2 is also structurally conserved among the four conformations, with an r.m.s.d. of less than 1.0 Å (Supplementary Table S1). Superposing the four conformations through the A1, B, C1 and C2 domains revealed that the major difference between the four conformations is that domain A2 has different orientations in different conformations (Fig. 2 ▸). As shown in Fig. 1 ▸, domain A1 is located at the inner membrane end and A2 at the mycomembrane end of EccB1, with the N- and C-termini being located on the left and right sides of domain A1, respectively. We found that domain A2 displays different orientations in the four states, representing snapshots of a swing from left to right which causes domain A2 to move away step by step from domains B and C1. One further minor difference that was observed, a slight translation of α3, the N-terminal helix of domain C1, is likely to be owing to the movement of domain A2. Close inspection of the crystal packing of the different states of EccB1 shows that domain A2 mainly packs against itself in the crystal and has limited interaction with domain B (Supplementary Fig. S1). The domain movement of A2 observed in the different states thus only causes movement of the packing interface of A2 and does not affect crystal packing, explaining how the different conformations of the crystals belong to the same space group and have almost the same unit-cell parameters. In addition, the recently resolved structures of M. tuberculosis EccB1 (PDB entry 4kk7; K. V. Korotkov & T. J. Evans, unpublished work) and M. smegmatis EccB1 (PDB entry 5cyu; TB Structural Genomics Consortium, unpublished work) have similar conformations to those of states I and III, with Cα r.m.s.d.s of 0.72 and 1.38 Å, respectively, suggesting this movement might be common across EccB1 homologues.
Figure 2.
Structure superimposition of the four conformations of EccB1. States I, II, III and IV are shown in green, blue, pink and yellow, respectively.
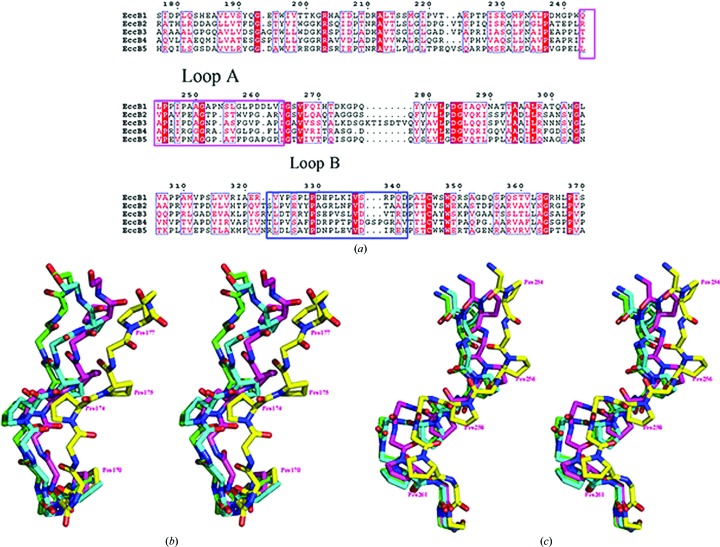
Structure superposition of all four conformations revealed that the movement of domain A2 was based on the swing of two loops, one named loop A (residues 243–264) that connects domains C1 and A2, and the second named loop B (residues 324–341) that connects domains A2 and B2. Except for these loops, significant conformational changes were only observed in the adjacent residues 240–242 and 342–344. The first loop consists of five proline and two glycine residues, and the second loop consists of five proline residues (Fig. 3 ▸). The fact that both loops are rich in proline and/or glycine residues in their amino-acid sequences is likely to be the reason why EccB1 can adopt four different conformations in these loops, resulting in the different orientations of domain A2 (Fig. 3 ▸). Sequence alignments show that these two loops are also rich in proline and/or glycine residues in EccB1 homologues, suggesting that the swinging movement of membrane-distal domain A2 could be a common feature of EccB1 and its homologues.
Figure 3.
(a) Sequence alignment of M. tuberculosis ESX-1–ESX-5 EccB homologues. Alignment of the sequences of EccB homologues from the ESX-1–ESX-5 secretion systems. Loops A (residues 243–264) and B (residues 324–341) are boxed in pink and blue. (b, c) Structure superimposition of loops A (b) and B (c) in the four states. Main-chain atoms and proline residues are shown as sticks. States I, II, III and IV are shown in green, blue, pink and yellow, respectively. N and C atoms are shown in blue and red, respectively.
As reported previously, EccB1 and its homologues are ATPases that are essential for the substrate transportation of ESX-1 systems and share highly conserved sequences, including two conserved motifs involved in ATPase activity. Here, we further propose that two flexible proline-rich and/or glycine-rich loops, loops A and B, that connect domain A2 to domains C1/B2 are a common feature of EccB1 and its homologues and may enable the swinging movement of domain A2 (Fig. 3 ▸). The conservation of these two flexible loops and the swinging of domain A2 are likely to be related to the ATPase activity of EccB1 and its homologues and may be involved in the substrate transport of ESX secretion systems.
Supplementary Material
PDB reference: Rv3869, state III, 5ebc
PDB reference: state IV, 5ebd
Supplementary figure and table.. DOI: 10.1107/S2053230X16000212/nw5034sup1.pdf
Acknowledgments
We would like to acknowledge the help of the staff of the Protein Science Core Facility Center at the Institute of Biophysics, Chinese Academy of Sciences. Financial support for this project was provided by the National Basic Research Program of China (2011CB910300, 2012CB518700 and 2013CB911500), the Key Project Specialized for Infectious Diseases of the Chinese Ministry of Health (2013ZX10003006 and 2012ZX10003002) and the National Natural Science Foundation of China (31170132 and 31270792).
References
- Abdallah, A. M., Gey van Pittius, N. C., DiGiuseppe Champion, P. A., Cox, J., Luirink, J., Vandenbroucke-Grauls, C. M. J. E., Appelmelk, B. J. & Bitter, W. (2007). Nature Rev. Microbiol. 5, 883–891. [DOI] [PubMed]
- Adams, P. D., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K. & Terwilliger, T. C. (2002). Acta Cryst. D58, 1948–1954. [DOI] [PubMed]
- Brodin, P., Majlessi, L., Marsollier, L., de Jonge, M. I., Bottai, D., Demangel, C., Hinds, J., Neyrolles, O., Butcher, P. D., Leclerc, C., Cole, S. T. & Brosch, R. (2006). Infect. Immun. 74, 88–98. [DOI] [PMC free article] [PubMed]
- Das, C., Ghosh, T. S. & Mande, S. S. (2011). PLoS One, 6, e27980. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Gey van Pittius, N. C., Sampson, S. L., Lee, H., Kim, Y., van Helden, P. D. & Warren, R. M. (2006). BMC Evol. Biol. 6, 95. [DOI] [PMC free article] [PubMed]
- Leslie, A. G. W. & Powell, H. R. (2007). Evolving Methods for Macromolecular Crystallography, edited by R. J. Read & J. L. Sussman, pp. 41–51. Dordrecht: Springer.
- Li, H., Zhang, X., Bi, L., He, J. & Jiang, T. (2011). PLoS One, 6, e26743. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- World Health Organization (2015). Global Tuberculosis Report 2015. Geneva: World Health Organization. http://www.who.int/tb/publications/global_report/en/.
- Zhang, X.-L., Li, D.-F., Fleming, J., Wang, L.-W., Zhou, Y., Wang, D.-C., Zhang, X.-E. & Bi, L.-J. (2015). FASEB J. 29, 4804–4814. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Rv3869, state III, 5ebc
PDB reference: state IV, 5ebd
Supplementary figure and table.. DOI: 10.1107/S2053230X16000212/nw5034sup1.pdf